Abstract
To understand movement of amoeboid cells we have developed an information tool that automatically detects protrusions of moving cells. The algorithm uses digitized cell recordings at a speed of ∼1 image per second that are analyzed in three steps. In the first part, the outline of a cell is defined as a polygon of ∼150 nodes, using the previously published Quimp2 program. By comparing the position of the nodes in place and time, each node contains information on position, local curvature and speed of movement. The second part uses rules for curvature and movement to define the position and time of start and end of a growing pseudopod. This part of the algorithm produces quantitative data on size, surface area, lifetime, frequency and direction of pseudopod extension. The third part of the algorithm assigns qualitative properties to each pseudopod. It decides on the origin of a pseudopod as splitting of an existing pseudopod or as extension de novo. It also decides on the fate of each pseudopod as merged with the cell body or retracted. Here we describe the pseudopod tool and present the first data based on the analysis of ∼1,000 pseudopodia extended by Dictyostelium cells in the absence of external cues.
Keywords: pseudopod, locomotion, computer-aided, dictyostelium, chemotaxis, cell movement, protrusion
Introduction
Amoeboid cells move by extending protrusions, called pseudopodia.1–3 The speed and trajectory of cells are determined primarily by the frequency, size and direction of pseudopod extensions. Cells may modulate the extension of pseudopodia depending on the presence of internal or external cues, which allows them to move to a desired place with food or bacterial infections, or develop structures during morphogenesis. Although pseudopodia are the basic elements of cell movement, little is known about why a cell extends a new pseudopod at a specific time or place. Pseudopodia are not well-defined uniform structures, but exhibit a large variation in spatial/temporal dimensions, which make them a difficult object to study quantitatively. Nevertheless, for a fundamental insight of cell movement and chemotaxis it is important to combine the detailed information on the localization of proteins with the decisions when and where to extend a pseudopod. The tools to quantify motility of protrusions from time-lapse movies are in constant development.4–6 Most methods are based on the detection of the difference between subsequent images yielding new extending areas or volumes.4,6,7 Although these methods may correctly identify pseudopodia, each pseudopod has complex topological properties from which it is difficult to derive general statements on precisely pseudopod dynamics. We have started to quantitatively describe pseudopod extensions in Dictyostelium, first manually by recording the position and time of the tip of the pseudopod at the start and end of its growing period. The novelty of the method is that a pseudopod is defined as a simple vector with length, timing and direction that can be used for large scale statistical analysis to investigate the ordered extension of pseudopodia.8 After analyzing a few thousand pseudopodia manually, we learned how to design formal rules for pseudopod extension that can be implemented in a computer algorithm, Quimp3.
The pseudopod tracking algorithm is based on the active contour analysis program Quimp2.9,10 This program identifies the outline of a cell as a polygon of bar-coded nodes. The position of the nodes relative to adjacent nodes provides information on local curvature, whereas the position of a node in time yields information on speed and area change of that node. The pseudopod macro is an algorithm to identify protrusions of the cell, based on an area of convex curvature of the contour and extension speed of this curved area. The output of the macro contains quantitative information on pseudopodia, such as position and time of start and end of growth period, surface area and speed. The output also contains qualitative data, such as whether the pseudopod is retracted or fuses with the cell body, and whether it arises de novo or from a parental pseudopod by splitting. For fluorescent movies, the fluorescent intensities of the pseudopodia at or below the membrane can also be quantitated and correlated with extension or retraction of pseudopodia. Here we describe the algorithm and compare the manual and fully automated pseudopod detection tool. Finally we provide the first quantitative data on the stochastic variation in pseudopod size, growth period, interval and direction.
Results and Discussion
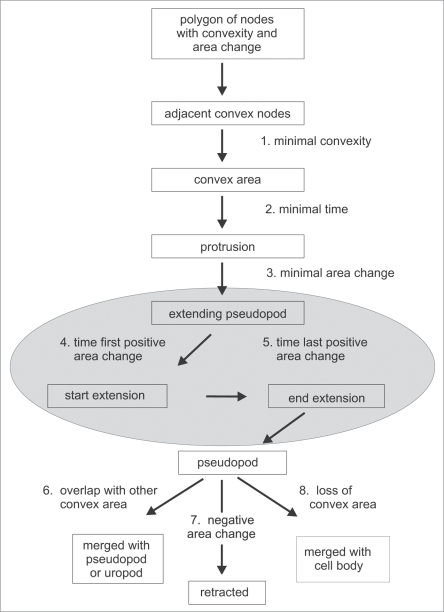
A protrusion of the membrane contains a convex region (a number of consecutive convex nodes) at the tip and often two concave regions at the base where the protrusion meets the cell body. This feature was exploited to build an automated detection algorithm for pseudopodia and the uropod (Fig. 1). By including the extending or retracting displacement data of the nodes that belong to a certain convex region, the protrusions are detected as extending or retracting pseudopodia. The space coordinates of the tip of the pseudopod is identified by the position of the center node of the convex region; the time coordinates of start and end of the extension is given by a user-defined minimal setting of the area change of the convex region. The path of a moving cell can then be described as a series of pseudopod vectors.
Figure 1.
Decision tree for identification of extending pseudopodia (gray area). The algorithm searches for adjacent convex nodes. Using criteria for convexity, life time and area change, extending pseudopodia are identified. The algorithm searches in the frames of the movie where the area change started and ends, and exports the x,y,t coordinates of the tip of the pseudopod at start and end. After growth stops, the protrusion is still assigned as a pseudopod. A pseudopod disappears because it may merge with other convex areas such as pseudopod or uropod, merge with the cell body, or is actively retracted.
Movement as the basis of pseudopod tracking.
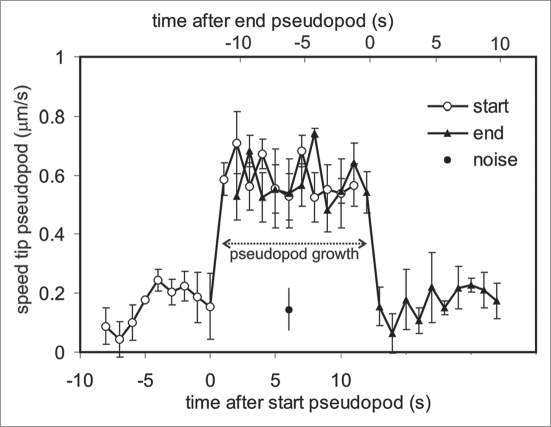
We determined the position of the tip of 20 pseudopodia during ∼30 seconds and calculated the speed (Fig. 3). The start of a pseudopod is defined here as the first frame of at least three frames in which the speed surpasses the user-defined threshold of 0.4 μm/s, and a pseudopod ends when the speed declines below this value. The results show that the tip of an active pseudopod moves at a high speed of 0.59 +/− 0.08 μm/s (means and SD). Before and after the growing period the tip node moves at a much lower speed of 0.16 +/− 0.06 μm/s. We determined the instrument noise by tracking small particles that are nearly stationary during 6 min, and obtained a speed of 0.14 +/− 0.07 µm/s, slightly smaller than the speed of the tip node before or after growth. Importantly, at the start of the growth period, the speed of the tip node increases within one frame (1 s) between the basal and maximal level; at the end of the growth period, a similar sharp decline of the pseudopod speed is observed. In addition, the speed of the tip of the pseudopod is approximately constant during the entire growth period of the pseudopod. Due to these large and sudden changes in speed, the start and end of pseudopod growth are relatively easy to determine by eye or by computer algorithms.
Figure 3.
Speed of pseudopod movement. The speed of the center convex node was determined before, during and after growth of 20 pseudopodia. For open symbols the data were aligned (at t = 1) at the first frame that the speed increased above 0.4 μm/s, whereas for the closed triangles the data were aligned (at t = 1) at the first frame that the speed decreased below this value. The two curves were placed 12 sec apart, because that is the average growth period of Dictyostelium pseudopodia. Due to equipment vibrations, a point fixed in space will have an apparent speed. This noise was determined for two “stationary” particles (2 × 2 pixels and 3 × 3 pixels) that remained at nearly the same place during 6 minutes (closed circle; 0.14 +/− 0.07 μm/s). The results show that the tip of a growing pseudopod moves at a high speed of 0.59 +/− 0.08 μm/s (means and SD) and moves at a much lower speed of 0.16 +/− 0.06 μm/s before and after the growing period. Furthermore, the switch between slow and fast movement occurs within one frame (1 s). Due to these large and sudden changes in speed, the start and end of pseudopod growth are relatively easy to determine by eye or by computer algorithms.
Validation.
To validate the method, pseudopod extension of six cells during 15 minutes was analyzed independently by the manual and by the fully automated methods. The results are presented in Table 2, showing a very good correspondence between manual and fully automated pseudopod tracking. More than 90% of the pseudopodia are assigned identical by the two methods as split/de novo, left/right, and lost/maintained. The root mean square (RMS) deviation between the methods for the time and position of the pseudopodia is 1.1 frames and 1.4 pixels, respectively, which correspond to 1.1 s and 0.7 μm. As presented below for 896 pseudopodia, the average growth period is 12.9 +/− 5.9 s, and the pseudopod size is 5.3 +/− 2.2 μm (means and SD). This reveals that the observed SD of pseudopod period or size is ∼5 times larger than the RMS deviation of the two methods, suggesting that the x,y and time coordinates of pseudopodia are defined with sufficient spatial and temporal resolution to obtain detailed quantitative data on timing, size and direction of pseudopodia.
Table 2.
Comparison of manual and automated pseudopod tracking
| Property | % different |
| Assignment pseudopod as de novo or splitting | 5% |
| Assignment parental pseudopod | 7% |
| Assignment split as step or hop | 9% |
| RMS time difference pseudopod | 1.08 s |
| RMS position difference pseudopod | 0.67 µm (1.37 pixels) |
The extension of pseudopodia was determined by the manual and the fully automated method for six cells from two movies (15 minutes) giving a total of about 330 pseudopodia; The percentage of pseudopodia that were assigned differently is indicated; The difference of time and position of the pseudopodia between the two methods is calculated as root mean square (RMS) deviation; The RMS deviation of the manual method with two experienced investigators is smaller (∼0.8 s and ∼0.5 µm); The RMS deviation of the two methods may be compared with the variation within the population; The SD of the pseudopod growth period and pseudopod length is 5.36 sec and 2.24 µm, respectively; This reveals that the difference between the methods is 3–5 times smaller than the variation of the pseudopod population, suggesting that variation due to the method does not contribute strongly to the observed variation of the population.
Properties of splitting and de novo pseudopodia.
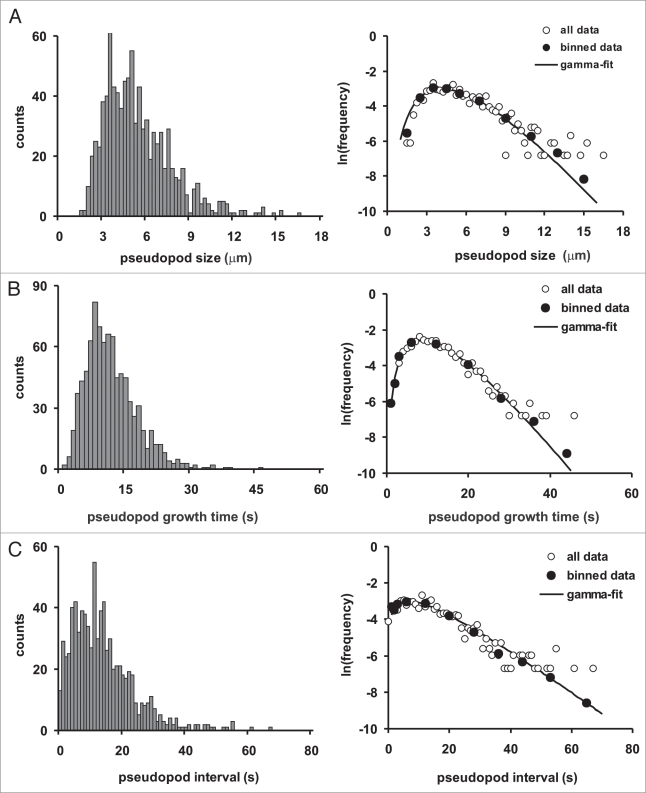
The quantitative data of 896 pseudopodia extended in buffer are summarized in Figure 4 and Table 3. The probability frequency distributions are not symmetric and follow a gamma distribution with exponential tails (Fig. 4). The data were fitted according to a Maximum-likelihood Gamma Distribution,15 yielding estimates for the shape parameter (k) and rate parameter (λ), which were used to estimate the mean (k/λ) and variance (k/λ2). The average size of a pseudopod is 5.3 +/− 2.2 μm (mean and SD). The growth period of a pseudopod is 12.9 +/− 5.9 s. The growth rate of the pseudopodia is 0.46 +/− 0.14 μm/s. We observed a weak but significant correlation between growth period and pseudopod size (R2 = 0.24; n = 896, p < 0.01 data not shown), which is confirmed by the observation that the relative standard deviation of growth rate (30%) is smaller than the relative standard deviation of growth period (45%) or pseudopod size (42%). Thus, pseudopodia grow at a somewhat variable rate, and reach a larger size when they grow for a longer period. The average pseudopod interval, i.e., the time period between the start of two pseudopodia, is 15.6 +/− 10.7 s; with this interval, a cell extends ∼4 pseudopodia per minute. The variation in size, growth time and especially pseudopod interval is very large, demonstrating the heterogeneity in pseudopod extension (Table 3). Finally, we determined the size, growth period and pseudopod interval for 396 split pseudopodia and 94 de novo pseudopodia, and did not observe a difference that is statistically significant (data not shown).
Figure 4.
Probability frequency distributions. The distributions of pseudopod size (A), life time (B) and interval (C) are presented at the left on a linear scale of number of observations, and at the right on a logarithmic scale of frequency. The distributions have exponential tails, and were fitted according to a maximum-likelihood gamma distribution, yielding estimates for the shape parameter k and the rate parameter λ: pseudopod size (A), k = 5.80 +/− 0.27; λ = 1.10 +/− 0.05; pseudopod growth time (B), k = 4.87 +/- 0.26; λ = 0.38.10 +/- 0.02; pseudopod interval (C), k = 1.86 +/− 0.08; λ = 0.128 +/− 0.007. The mean and SD are presented in Table 3.
Table 3.
Pseudopod properties of Dictyostelium cells in buffer
| Property | Mean | SD | n |
| Quantitative properties | |||
| size (µm) | 5.27 | 2.19 | 896 |
| growth period (s) | 12.92 | 5.86 | 896 |
| growth speed (µm/s) | 0.46 | 0.14 | 896 |
| interval (s) | 15.56 | 10.69 | 803 |
| frequency (min−1) | 3.93 | 1.66 | 24 |
| Pseudopod types | |||
| de novo (%) | 14 | 5 | 24 |
| Split (%) | 76 | 5 | 24 |
| - steps (alternating right/left) | 53 | 6 | 24 |
| - hops (consecutive right/right or left/left) | 18 | 5 | 24 |
| - Y-split (simultaneous) | 6 | 3 | 24 |
| undefined (%) | 10 | 3 | 24 |
| Pseudopodia for cell movement | |||
| maintained (%) (other lost) | 55 | 6 | 24 |
Data obtained from 896 pseudopodia extended by 24 cells from 6 movies of starved Dictyostelium cells in buffer; The results show the means and SD with n the number of pseudopodia or the number of cells; The pseudopod interval contains less data because Y-splits and the first pseudopod of a cell in the movie are not included; The pseudopodia are either de novo, a split, or undefined (mostly beginning of movie); A split pseudopod may give rise to two growing extensions (Y-split), or only one extension; this one extension may be to the right or left compared to the parental pseudopod from which it is formed; alternating right/left steps occur ∼3-fold more often than consecutive right/right or left/left hops; Maintained pseudopodia contribute to the translocation of the cell.
Pseudopod extension and cell tracks.
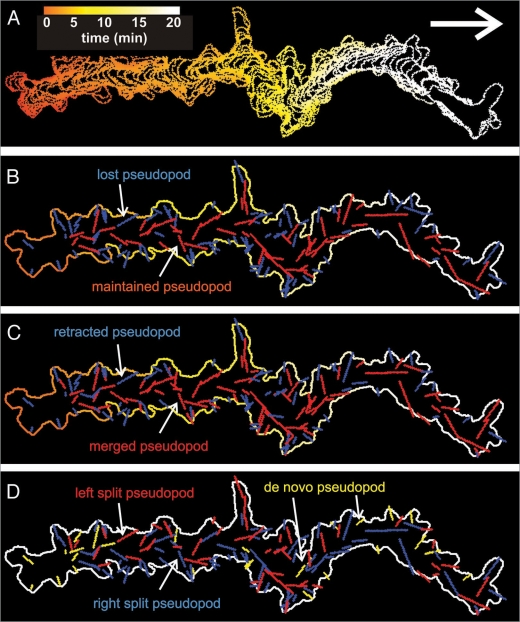
Figure 5 presents the path and pseudopodia extended by a Dictyostelium cell moving for 20 minutes in buffer. The track of the cell is presented in panel A showing that the cell moves for several minutes in approximately the same direction. The 137 pseudopodia that are detected by the algorithm are presented using different color codes. In panel B maintained (red) and lost (blue) pseudopodia are discriminated, showing that the path of the cell is determined predominantly by the maintained pseudopodia. The lost pseudopodia are often retracted within one minute after they appeared and therefore do not contribute to the movement of the cell (panel C). The maintained pseudopodia originate often by splitting of an existing pseudopod, whereas the lost or retracted pseudopodia more often are formed de novo from areas of the cell that did not extend a pseudopod recently. Finally, the track of panel D and the data of Table 3 reveal that split pseudopodia are frequently extended alternating to the right and left, about 3-fold more often than consecutive right/right and left/left, which may explain the relatively straight path of the cell.8
Figure 5.
Tracks of a moving Dictyostelium cell with annotated pseudopodia. A 20-minute phase contrast movie was recorded with wild type Dictyostelium amoeba crawling in buffer. The movie was converted to black and white and analyzed with the pseudopod macro of Quimp3. In (A), the cell track with time-driven coloration is displayed. In (B), the line segments representing all 137 detected pseudopodia are projected. Maintained pseudopodia are colored red, whereas lost pseudopodia are shown in blue. In (C) the fate of the pseudopodia is colored red for retracted and blue for non-retracted pseudopodia (see Table 1 fate of pseudopod with negative (red) and positive (blue) sign). In (D) the de novo pseudopodia are indicated in yellow, the split pseudopodia to the right relative to the parent are colored blue and split pseudopodia to the left are red.
Methods
Recording of movies.
Wild-type Dictyostelium discoideum AX3 amoebae were starved in the well of a 6-well Nunc plate at a density of about 106 cells/cm2 under a layer of 1 ml of 10 mM phosphate buffer (pH 6.5). At the onset of aggregation, when cells make the first cell-cell contacts in streams (after about 5 hours), cells were harvested and seeded at low density of about 5 × 104 cells/cm2 on a glass cover slip in phosphate buffer supplemented with 2.5 mM caffeine to block autonomous cAMP signaling. The cells were recorded at 1 second time intervals on a standard inverted microscope using a 20x lens with a JVC TK-C1381 camera and Virtual dub. The speed of 1 frame/s is the optimum between computing time and temporal/spatial resolution of the pseudopodia. Frame rates below 0.3 frames/s are insufficient to identify start and end of the growing period of pseudopodia; as a rule of thumb we advise a frame rate of ∼1/10th the average growing period of the pseudopod (12 seconds in wild type cells).
Manual pseudopod tracking.
Images were analyzed using ImageJ (http://rsb.info.nih.gov/ij/) with a custom made macro that provides a semi-automatic method to characterize pseudopodia. The investigator identifies the start and final position of a pseudopod growth. The macro exports the frame number and x,y-coordinates of these positions, and prints a hard-copy arrow on the relevant frames of the movie. In the result file the investigator can annotate each pseudopod as split versus de novo, maintained versus retracted, and extended to the right or left relative to the previous pseudopod.
Fully automatic pseudopod tracking.
The automated pseudopod tracking algorithm is a macro for the open source program ImageJ (http://rsb.info.nih.gov/ij/) and is written as an extension of the Quimp2 program.11 The package can be downloaded as Quimp3 from the site that also contains the previous versions of Quimp: http://go.warwick.ac.uk/bretschneider/quimp. A detailed description of Quimp3 is presented in the help file of the package.
The phase contrast movie was converted to a black and white movie using the “phase contrast to BW” macro11 that is also included in the Quimp3 package. This macro converts the darker grey area of the cell into black, while the lighter grey area outside the cell is made white. Depending on the contrast of the images, sometimes a lighter area inside the cell is erroneously made white. With a pencil of the “phase contrast to BW” macro this white region can be painted black. The resulting file was used as input file for the Quimp3 analysis. The pseudopodia were detected using the default parameters of the macro. The pseudopod algorithm of Quimp3 identifies and annotates pseudopodia in a sequence of several steps, starting with determining active contours of the cell outlines.
Cell outline, extensions and curvature.
The original Quimp (Quantitative Imaging of Membrane Proteins) program uses an active contour method to automatically recognize the outline of a cell.10 The program creates a number of interconnected nodes, based on a user-defined minimal and maximal distance between nodes. In Quimp2 the nodes are bar-coded, which allows to assign spatial-temporal properties to each node.11 In this procedure a manual chain selection encompassing the cell of interest is required for the first frame. The BOA_TN plugin then automatically determines the contour of that cell in all subsequent frames. It first creates a number of interconnected nodes on this chain, based on a user-defined distance between nodes. Subsequently, this chain of nodes is shrunk towards the cell; if the distance between two nodes becomes closer than a user-defined distance, one of them is deleted. The nodes are fixed in space and given a number when a user-defined steepness in fluorescent signal (the cell boundary) is reached. The cell boundary of the first frame is thus described by the position of the nodes. For subsequent movie frames, the chain of nodes of the preceding frame is enlarged, and new nodes with a higher number are inserted between the old ones. This is followed by a new round of chain shrinkage and cell boundary detection; during this process nodes with higher tracking numbers are lost first when nodes come too close. The number of nodes that will be kept between frames depends on the extent of cell deformation. In retracting regions nodes will be lost permanently while in extending regions nodes will be added that are maintained until that region retracts. The chain is processed alternating clockwise and anti-clockwise to rules out a bias in the selection process of nodes to be inserted or deleted. The optimal distance between nodes is about two pixels;10 the distance and total number of nodes is 0.45 μm and 170 nodes for typical images created using a confocal fluorescent microscope (pixel size 0.2 μm), and 1 μm and 70 nodes for images by phase contrast microscope with 20× objective (pixel size is 0.49 μm).
Retracting and extending nodes are identified by comparing the position of a node in the succeeding contour relative to the contour of the cell in the previous contour. The magnitude of the retracting/extension is expressed as the area change, which was calculated as follows:9 for a specific node the points halfway the two adjacent nodes were determined by linear interpolation for the present, previous and next frame. Area covered by the polygon that connects these points was calculated. The area change (in μm2/s) is this surface area divided by the interval between previous and next frame. This extension/retraction parameter is used as a sign (minus for retraction, plus for extension) in front of the area change of the node.
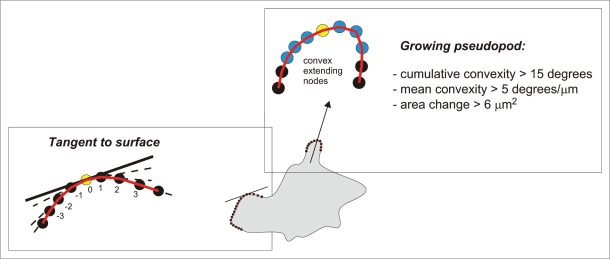
A pseudopod is an outward extension of a spherical cell. Therefore, we identified outward (convex) or inward (concave) deformations of a spherical cell as follows. For each node the program determines the outer angle α of the line segments pointing from a given node to its two neighbors. Since a perfect circle will yield angles α that are larger than 180 degrees, the obtained angles are corrected by subtracting 360/n because the chain of n nodes forms a closed polygon. Thus, the deformation from a spherical cell is defined as the shape curvature c, given by c = α − 180 − 360/n. Positive and negative values of c imply convex and concave nodes, respectively (see Fig. 1 of Bosgraaf et al.9). The slope of the tangent (βt) to the surface at a specific node was calculated as the weighted average of the angles between a node and its adjacent nodes up to three nodes away from the central convex node. βt = 1/12 [γ(−3,−2) + 2γ(−2,−1) + 3γ(−1,0) + 3γ(0,1) + 2γ(1,2) + γ(2,3)], where γ(a,a+1) indicates the angle between node a and node a+1 (see below in Fig. 2).
Figure 2.
Pseudopod and tangent to surface. The pseudopod algorithm identifies growing pseudopodia by three steps: (i) it identifies a series of adjacent convex nodes, (ii) evaluates the ‘bending’ of the convex nodes, and (iii) sets a minimal growth area. Then the algorithm searches the central convex node (yellow) in the two frames where the area change became positive for the first time (start) and was positive for the last time (end), respectively. The arrow connects these points, and represents the growing pseudopod as a vector with length, timing and directionality. The slope of the tangent to the surface at a specific node (yellow node) was calculated as the weighted average of the angles between a node and its adjacent node up to three nodes away from the central convex node.
Pseudopodia.
A protrusion of the membrane contains a convex region (a number of consecutive convex nodes) at the tip. This feature was exploited in Quimp3 to build an automated detection algorithm for pseudopodia and the uropod (Fig. 1). To prevent local minor convexities from being detected as a protrusion, the convexity of the nodes is smoothed first over user-defined distance and frames (default is three nodes and three frames, using ‘edge-preserving’ smoothing. It calculates the average position of the node of interest and two nodes at the left, the node of interest and two nodes at the right, and the node of interest and one node at the right and one at the left; the value is chosen that is closest to the position of the node of interest. The method effectively removes small local fluctuations of the cell outline but preserves the edges of concave or convex areas). Subsequently, convex areas are identified that consist of adjacent convex nodes.
A protrusion fulfils minimal requirements for the strength of the curvature of the convex area (Figs. 1 and 2): The cumulative convexity (Σc) of the adjacent convex nodes is above a user-defined minimum (default 15 degrees) and the mean curvature of the nodes has to be above a user-defined minimum (default 5 degrees per μm). Growing pseudopodia are subsequently identified as protrusions with an uninterrupted series of frames with a positive area gain, and the cumulative area gain in this period is above a user-defined minimum (default 6 μm2; for a pseudopod with a width of ∼3 μm this implies a minimal extending length of ∼2 μm). In these frames the central node of the convex area is identified as the tip of the pseudopod. After the pseudopod has been identified in a series of frames, the macro searches back and forward to find the frames where the positive area change of the pseudopod area started and stopped, respectively; there is no requirement for convexity in this process, only positive area change. As a result of that process, the pseudopod vector connects the x,y,t coordinates of the node where a pseudopod starts and ends during its growing phase.
The aforementioned criteria identify most growing pseudopodia “correctly” as they were identified by eye by an investigator. The macro contains some other criteria that improve the detection, especially in noisy movies, or irregular protrusions. These sections of the macro incorporate criteria how to deal with minor concavities in a convex area, a rather flat tip on a broad pseudopod, or a short transient arrest of area change (see help file).
Uropod.
The uropod is the protrusion at the trailing end of the cell. The uropod is used to identify the polarity of the cell. The front of the cell is not easily defined, because cells extend pseudopodia to the right, left, or sometimes backwards. In contrast, considerably less fluctuation is observed in the uropod, and in most cases the uropod retains approximately the same position. Therefore, the central uropod node (see below for identification) is defined as the ‘rear’ of the cell. The ‘front’ of the cell is then loosely defined as the node farthest away (along the perimeter) from the central convex node of the uropod. Dictyostelium cells, as many other cells, move with persistence, meaning that they tend to extend pseudopodia in a similar direction as previous pseudopodia.12,13 Therefore, cells rarely make ‘head to tail’ changes of direction, the uropod retains its approximate position, and most pseudopodia are extended in the front, i.e., the area surrounding the front node.
The uropod is defined as follows: After the movie is loaded, the user is asked to identify approximately where the uropod is at the beginning of the movie with a mouse click. The macro identifies the nearest protrusion as the uropod, and its central node as the rear node. In subsequent frames, the rear node is identified by the central node of this protrusion. This method works very well if the uropod remains in the “back” half of the cell. In case the cell does make head to tail conversions (which does happen in shortly starved cells) the user can choose an alternative, fully automatic and dynamic method of uropod tracking. With this alternative method activated, the macro iterates back and forth over the entire length of the movie. The longest living protrusion with a negative area gain (i.e., it loses more area than it gains) is defined as the uropod at the start of the movie. As the movie proceeds, the algorithm notifies when the uropod area exhibits area gain in stead of retraction above a user-defined setting (default 20 μm2). The algorithm will then select another protrusion as the new uropod using the aforementioned method.
Pseudopod annotation.
Split or de novo.
Pseudopodia are very often formed near the tip of a previous pseudopod. These pseudopodia are termed “split” because they split off from a preceding protrusion.14 In contrast to this type of pseudopod, cells also produce “de novo” pseudopodia, which are protrusions that appear in a region of the cell body that was not previously part of a protrusion. The investigator identifies split pseudopodia by observing different frames of a movie and judging whether the start position of a new pseudopod is within or outside the domain of an existing pseudopod; this domain is identified by combining convexity (the convex tip is usually flanked by two concave areas at the base of the pseudopod forming the boundary with the cell), pseudopod start (a pseudopod usually grows out from a position that remains the base of the pseudopod), and structure (pseudopodia are generally contain few organelles). Although this judgment is subjective, we noticed that three independent observers annotated ∼90% of the pseudopodia identically as split or de novo.
The computer algorithm does not use these human-based rules, but decides whether a pseudopod is a split of an existing pseudopod by making use of the convex area of the two pseudopodia as follows: The new pseudopod is considered a split of an existing pseudopod, if at the start of the extension period of the new pseudopod, its convex region includes nodes that also belong to the convex region of an existing pseudopod, OR lay within a user-defined distance from a node of that convex region of the existing pseudopod (default 1 μm). The convex area of an existing pseudopod is restricted to the front of the pseudopod. The second criterion was introduced to correctly assign pseudopodia that split at the side of an existing pseudopod. Visual inspection reveals that the first criterion (overlapping convex nodes) always correctly assigns split pseudopodia, and that ∼80% of split pseudopodia are covered by this criterion. The second criterion with a default setting of 1 μm provides the optimum between missing splitting pseudopodia and incorporating de novo pseudopodia; judged by visual inspection, ∼95% of the pseudopodia are correctly annotated as split versus de novo by these automated pseudopod tracking criteria (see also Table 1).
Table 1.
The output file for pseudopod tracking (summary of .dat8 file)
| Output | Explanation |
| ID | Pseudopod identification number: Each pseudopod is assigned a unique number. |
| Parent ID | If a pseudopod emerges from, or very close to an existing pseudopod, it is considered a “child” of that “parent” pseudopod (0 if pseudopod emerged de novo). |
| gained area | The total net area that the protrusion gained during its extension period (in µm2). See note in help file for precaution. |
| Fs | The number of the frame at which the pseudopod started extending. |
| Fe | The number of the frame that was the final frame of the extension period. |
| xs | x coordinate at start of extension: |
| ys | y coordinate at start of extension: |
| xe | x coordinate at end of extension: |
| ye | y coordinate at end of extension: |
| Role of pseudopod | Pseudopod is maintained (1) or retracted (−1): When a pseudopod starts extending, the positions of the two nodes flanking the pseudopod are stored. The nodes that pass between these points are tracked. If it reaches a user defined threshold, the pseudopod is considered “maintained” and assigned a 1 in this column. If the pseudopod disappears before the threshold is met or if it merges with the uropod, it is considered “retracted” and assigned −1 in this column. |
| Fate of pseudopod | The fate of the pseudopod after its extension period has finished. One of seven coded options is possible; negative numbers indicate retraction of the pseudopod. |
| −4) the pseudopod retracts but is still present until the movie ends. This code should only occur towards the end of the movie; | |
| −3) The pseudopod retracts and eventually disappears. | |
| −2) The pseudopod retracts and eventually merges with either the uropod or another pseudopod. This often occurs for lateral pseudopodia that are retracted when the uropod region gets close. | |
| −1) The pseudopod retracts fully but its parental pseudopod persists (i.e., a protrusion remains) and a new pseudopod emerges from its parent. | |
| +1) The pseudopod persists and a new pseudopod emerges from it (i.e., it serves as a parent for another pseudopod). | |
| +2) The pseudopod does not retract, but merges with another pseudopod. This is a rare case for most cell lines. | |
| +4) The pseudopod extends till the end of the movie. Should only occur towards the end of the movie. | |
| Type of split | If the pseudopod arose from a parent, the pseudopod may be part of a ‘split series’. |
| Step (3): The second pseudopod in a pair of alternating left-right or right-left pseudopodia. | |
| Hop (2): The second pseudopod in a pair of pseudopodia that both have the same direction (i.e., right-right or left-left). | |
| Y-split (1): Two pseudopodia that grow simultaneously from a parent pseudopod | |
| (0): If neither of the previously mentioned criteria are met, the value 0 is returned. These are de novo pseudopodia and pseudopodia that are the first in a split series. | |
| Intensity | Mean intensity during extension: The average membrane intensity of the nodes belonging to the pseudopod during its extension period. (relevant for cells expressing fluorescent markers) |
Parental pseudopod.
If a pseudopod is recognized as a “split” pseudopod (see above), it means per definition that it arose out of a previously existing protrusion. The newly appearing pseudopod is termed the ‘child’ of the previous, which is termed the ‘parent’ of the subsequent pseudopod. A pseudopod may have multiple children, but can have only one parent (or none in the case of a de novo pseudopod or at the start of the movie). In most cases, the parental pseudopod belonging to a newly appearing one is readily recognized by the algorithm.
Step/hop/Y-split.
If a pseudopod has a parental pseudopod, the history of the protrusion may be further characterized. Amoeboid cells such as Dictyostelium often produce split pseudopodia that originate alternating from the left/right side of the preceding pseudopodia. This series of split pseudopodia leads to a typical zig-zag path of the leading edge. If a given pseudopod is the second pseudopod of a left/right or right/left pair, it is termed a “step.” Alternatively, if it is the second pseudopod of a left/left or right/right pair, it is termed a “hop.” Note that at least two previous generations of parental pseudopodia are required for this definition; i.e., when a de novo pseudopod is the parent of a family of splitting pseudopodia, the grandchild of the parent is the first pseudopod that can be a step or a hop.
Two newly appearing pseudopodia may extend simultaneously from a preexisting pseudopod. These two newly appearing pseudopodia are termed “Y-split” pseudopodia. The criterion is based on the algorithm that assigns parenthood of pseudopodia. When a split pseudopod emerges (child), the algorithm investigates whether the presumed parent had grown above a threshold level (default setting is 1.5 μm). If the presumed parent did not meet this criterion and if it continues to grow, both pseudopodia are assigned as Y-split children of the previous pseudopod. (If the presumed parent does not grow beyond 1.5 μm, the child is allocated to the parent of the presumed parent; see help file for details).
Maintained or lost.
Some pseudopodia make large contributions to the movement of the cell and are termed “maintained.” Other pseudopodia are retracted before they merge with the cell body, or the cell makes a new dominant pseudopod in another direction by which the nodes of the old pseudopod merge with the cell body and eventually with the uropod, and are retracted. Thus, every extending pseudopod is assigned as ‘maintained’ or ‘lost’ as follows. At the start of a pseudopod the two nodes that flank the convex area are marked as ‘guard’ nodes, which are followed during the entire movie. When a pseudopod (or its children) grows out, new nodes will be added in the growing pseudopod. For a pseudopod that contributes to cell movement, many of these new nodes lay in the region between the guard nodes. In contrast, very few new nodes will be introduced between guard nodes of a pseudopod that is not extended in the major direction of cell movement (a lateral pseudopod, or a pseudopod that will be retracted). If a certain percentage of cell perimeter lies between these guard nodes, the pseudopod is assigned ‘maintained.’ The default value is 40%.
Data analysis.
The pseudopod macro exports a result table (.dat8) that contains quantitative and qualitative information on the detected pseudopodia (see Table 1). The result tables of the manual and automatic pseudopod tracking were analyzed using Excel. Primary calculations are size, extension period and direction of each individual pseudopod. Secondary calculations were made on the connection between subsequent pseudopodia, and include time period between pseudopodia, angle between present and previous pseudopod(s), and distance between start of present pseudopod and end of previous pseudopod.
A typical database contains information from 200–300 pseudopodia obtained from 6–10 cells from two independent movies. To select cells for pseudopod analysis, we first determined the displacement during 15 min of all ∼20–30 cells in the field of observation, and then selected the 3–5 cells that have a displacement closest to the mean displacement. The data are presented as the means and standard deviation (SD), or standard error of the means (SEM) where n represents the number of pseudopodia or number of cells analyzed. From mutant analysis we have learned that the optimal protocol is first to analyze a few cells by the manual procedure, and only then employ the fully automated method. This has the important advantage that during the intense manual tracking the investigator learns about the basic properties of the mutant cell line. In addition, by tracking the same cells manually and automatically, it may provide information to adjust the default settings. We observed that the defold settings of the parameters are satisfactory for wild type cells at different stages of development, and many mutant cells with signalling defects, such as pi3k1/2-null cells, sgc-null cells, pla2A-null cells. Mutants such as pten-null and GbpD-overexpressor cells have a very broad front with multiple pseudopodia require adjustment of the parameters that are used to discriminate between nearby pseudopodia.
The methodology of data collection and analysis assumes two-dimensional cells and pseudopodia, which is obviously incorrect. Cells move on a 2D agar surface, which implies that the movement in the plane of the agar surface is more important for understanding cell translocation than movement of the pseudopod in the z-direction. In addition it is extremely difficult, if not impossible, to obtain 3D information on pseudopod extension with a 1 second time resolution, and we suspect it will be difficult to extract and analyze pseudopod vectors in 3D. We are aware that data are obtained and discussed in 2D, and their relevance in 3D should be evaluated; for instance we may draw conclusions on pseudopod length, which is similar in 2D and 3D, but not on pseudopod area, because this has a completely different meanings in 2D and 3D.
Conclusions
We developed an automatic pseudopod tracking algorithm that yields very similar data as determined manually. This is observed for both quantitative properties as size and period, and qualitative properties as split or de novo pseudopodia. From a single movie we routinely obtain quantitative information on a few hundred pseudopodia from about six cells. Since cell movement is largely stochastic in nature large databases are essential to deduce the fundaments of pseudopod extension and the consequences for cell movement. We expect that the method is applicable to other extending convex protrusions such as filopodia, and protrusions in other cells such as growth cone and lamellipodia. We have started to analyze pseudopod extension and cell movement in the absence of external and internal cues, demonstrating that pseudopodia are not extended randomly, but preferentially by splitting in an alternating right/left fashion leading to strong persistence of the direction of movement.8 We suppose that external cues such as chemoattractants induce a bias of one or multiple properties of pseudopodia, such that pseudopodia that are extended in the direction of the gradient differ from pseudopodia that are extended in other directions. A cell may also respond to internal cues, such as starvation. Dictyostelium cells starved for 5 h move faster than growing cells. Do they extend more pseudopodia, or are the pseudopodia extended by growing and starved cell different? Finally, the pseudopod algorithm may be very useful to characterize the altered movement of mutant cells with signaling defects or altered cytoskeleton.
Acknowledgements
We thank Ineke Keizer-Gunnink for recording movies, and Till Bretschneider for suggestions on the design and implementation of Quimp3.
Footnotes
Previously published online: www.landesbioscience.com/journals/celladhesion/article/9953
References
- 1.Le Clainche C, Carlier MF. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol Rev. 2008;88:489–513. doi: 10.1152/physrev.00021.2007. [DOI] [PubMed] [Google Scholar]
- 2.Van Haastert PJM, Devreotes PN. Chemotaxis: signalling the way forward. Nat Rev Mol Cell Biol. 2004;5:626–634. doi: 10.1038/nrm1435. [DOI] [PubMed] [Google Scholar]
- 3.Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- 4.Arrieumerlou C, Meyer T. A local coupling model and compass parameter for eukaryotic chemotaxis. Dev Cell. 2005;8:215–227. doi: 10.1016/j.devcel.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Costantino S, Kent CB, Godin AG, Kennedy TE, Wiseman PW, Fournier AE. Semi-automated quantification of filopodial dynamics. J Neurosci Methods. 2008;171:165–173. doi: 10.1016/j.jneumeth.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 6.Wessels D, Voss E, Von Bergen N, Burns R, Stites J, Soll DR. A computer-assisted system for reconstructing and interpreting the dynamic three-dimensional relationships of the outer surface, nucleus and pseudopods of crawling cells. Cell Motil Cytoskeleton. 1998;41:225–246. doi: 10.1002/(SICI)1097-0169(1998)41:3<225::AID-CM4>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 7.Machacek M, Danuser G. Morphodynamic profiling of protrusion phenotypes. Biophys J. 2006;90:1439–1452. doi: 10.1529/biophysj.105.070383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bosgraaf L, van Haastert PJM. The Ordered Extension of Pseudopodia by Amoeboid Cell in the Absence of external Cues. PLoS ONE. 2009;4:5253. doi: 10.1371/journal.pone.0005253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bosgraaf L, Van Haastert PJM, Bretschneider T. Analysis of cell movement by simultaneous quantification of local membrane displacement and fluorescent intensities using Quimp2. Cell Motil Cytoskel. 2009;66:156–165. doi: 10.1002/cm.20338. [DOI] [PubMed] [Google Scholar]
- 10.Dormann D, Libotte T, Weijer CJ, Bretschneider T. Simultaneous quantification of cell motility and protein-membrane-association using active contours. Cell Motil Cytoskel. 2002;52:221–230. doi: 10.1002/cm.10048. [DOI] [PubMed] [Google Scholar]
- 11.Bosgraaf L, Van Haastert PJM, Bretschneider T. Analysis of cell movement by simultaneous quantification of local membrane displacement and fluorescent intensities using Quimp2. Cell Motil Cytoskel. 2009;66:156–165. doi: 10.1002/cm.20338. [DOI] [PubMed] [Google Scholar]
- 12.Potel MJ, Mackay SA. Preaggregative cell motion in Dictyostelium. J Cell Sci. 1979;36:281–309. doi: 10.1242/jcs.36.1.281. [DOI] [PubMed] [Google Scholar]
- 13.Li L, Norrelykke SF, Cox EC. Persistent cell motion in the absence of external signals: a search strategy for eukaryotic cells. PLoS ONE. 2008;3:2093. doi: 10.1371/journal.pone.0002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andrew N, Insall RH. Chemotaxis in shallow gradients is mediated independently of PtdIns 3-kinase by biased choices between random protrusions. Nat Cell Biol. 2007;9:193–200. doi: 10.1038/ncb1536. [DOI] [PubMed] [Google Scholar]
- 15.Wessa P. Maximum-likelihood Gamma Distribution Fitting (v1.0.2) in Free Statistics Software (v1.1.23-r3) 2008. Office for Research Development and Education, URL www.wessa.net/rwasp_fitdistrgamma.wasp/