Abstract
Lung cancer is a complex spectrum of diseases characterized by extensive genomic instability, which can be detected among both histological subtypes and different foci within a tumor. Conventional and cutting edge investigative technologies have uncovered scores of genomic changes in individual specimens that have been used to characterize specific molecular subtypes. Oncogenes with predominant roles in lung cancer include EGFR, MYC and RAS family members, PIK3CA, NKX2-1 and ALK; tumor suppressor genes include TP53, RB1, CDKN2, and a cluster of genes mapped at 3p. MicroRNA regulators also have been linked to lung cancer. The functional role of the recurrent genomic changes in lung tumors has been explored, which has led to a better understanding of cell growth, differentiation and apoptotic pathways. Additionally, this knowledge has supported the development of novel therapeutics and translational tools for selection of patients for personalized therapy.
Key words: chromosomal aberrations, gene amplification, gene fusion, oncogene, tumor suppressor gene, microRNA
Introduction
Lung cancer, comprised of two major clinico-pathological categories small-cell (SCLC) and non-small-cell lung carcinoma (NSCLC), is the leading cause of cancer-related morbidity and mortality worldwide.1 SCLC accounts for less than 20% of lung tumors, displays neuroendocrine features and has a propensity for rapid growth and early metastasis. NSCLC represents the vast majority of these tumors and includes adenocarcinoma and squamous cell carcinoma, the two most common histological subtypes. Lung cancers are characterized by extensive genomic instability, which can be detected among both histological subtypes and among different foci within a tumor. The genomic changes occur at different levels, from mutations in single or few nucleotides to gains or losses of entire chromosomes. Some mutations are completely innocuous, but many of genomic events are responsible for dramatic functional changes and involve the core of lung carcinogenesis. In this article, we review relevant chromosomal and genomic alterations in lung cancer and discuss recent findings that have contributed to an understanding of their molecular profiles and the development of strategies for earlier diagnosis and more efficient therapies.
Chromosomal Rearrangements in Lung Cancer: What is Known and How it Impacts Gene Expression
Usually lung carcinomas are highly aneusomic, with gains and losses of entire chromosomes or large chromosome regions. These tumors also exhibit simple and complex structural rearrangements responsible for alterations in transcription and protein expression. Included are variations in gene copy number due to deletions, duplications or amplifications, and gene fusions driven by insertions, inversions and translocations. Conventional cytogenetic methods, such as G-banding, were fundamental for initial discoveries on molecular mechanisms of lung carcinogenesis, but had limited utility in instances of cryptic or very complex rearrangements. The advent of molecular cytogenetic strategies in the early 1990s, such as multiplex FISH (M-FISH),2 spectral karyotyping (SKY)3 and comparative genomic hybridization (CGH)4 have increased the accuracy of identifying chromosomal rearrangements (Fig. 1A and B), but these approaches were still limited by low resolution (5–10 megabases). New technological advances and the availability of genomic resources in the last decade have fostered the shift to microarray-based platforms, which has progressed from using only a few hundred DNA clones,5,6 to mining the entire genome for copy number variants at the 1 Mb resolution,7 and more recently selected analyses at the nucleotide level.8,9 Although high resolution platforms have been largely used to identify genomic rearrangements in lung cancer, intra-tumor heterogeneity still poses a challenge. Chromosomal abnormalities detected by these new technologies have been independently validated by other high-resolution laboratory approaches, such as fluorescence in situ hybridization (FISH) and polymerase chain reaction (PCR)-based techniques. Both are able to accurately define specific genomic regions involved in rearrangements and the PCR-based approach has high throughput. On the other hand, FISH has the critical advantage of investigating target phenomena in single cells “in situ” and of preserving the original tissue architecture. Ultimately, it is the combination of multiple technical approaches that provides the most powerful strategy for understanding the molecular pathways underlying the lung tumor development.
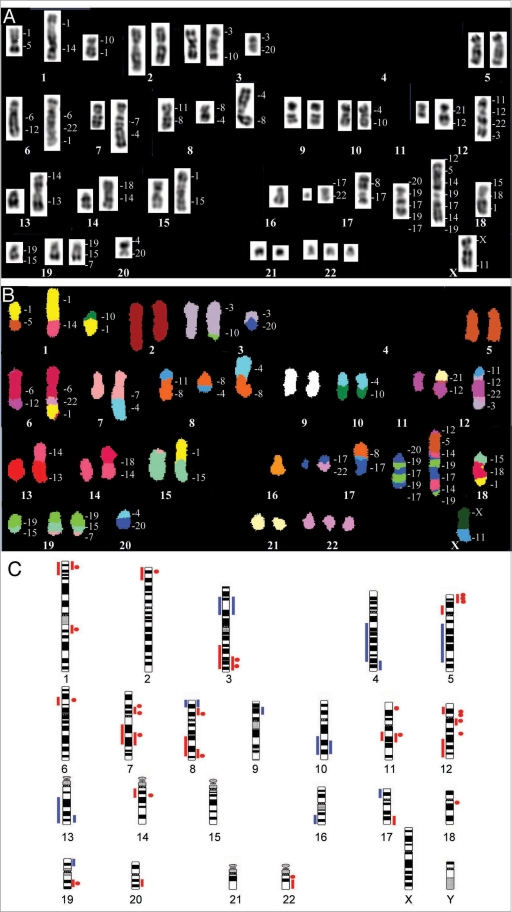
Figure 1.
(A and B) Spectral karyotyping (SKY) of a lung adenocarcinoma showing numerous numerical and structural chromosome changes. The inverted-DAPI image is shown in (A) and the classified image with the pseudo-colors is shown in (B). The specimen was near-diploid, with rearrangements involving most chromosomes. In translocations, the origin of the material is listed on the right of the chromosomes. (C) Summary of genomic imbalances reported in lung cancer (reviewed in ref. 11). Small cell lung cancer (SCLC) is represented on the left of the chromosome idiograms, non-small cell lung cancer (NSCLC) is represented on the right. Copy number gain is represented by red bars, focal amplification by red dots and copy number loss by blue bars.
The first recurrent chromosomal abnormalities to be recognized in lung cancer were 3p deletions, identified by classical karyotyping in SCLC.10 For more than a decade, little new data were reported. It was only with the advent of M-FISH, SKY and CGH that cryptic rearrangements were detected, marker chromosomes were recognized and breakpoints were refined, providing a basis for the search of genes potentially deregulated and associated with tumor initiation and progression. A more detailed picture of genomic copy number variation in lung cancer was achieved recently with the array-based analyses and a summary of current data on gains and losses is presented in Figure 1C (reviewed in ref. 11). Analyses in more than 70 SCLC and 800 NSCLC cell lines and primary tumors identified important recurrent genomic changes, such as high-amplitude focal amplicons involving members of the MYC family (MYCL1, MYCN and MYC), participants in EGFR pathways (EGFR, PIK3CA, KRAS), and other genes controlling cell proliferation, such as FGFR1, TP63, TERT, CCND1, CCNE1 and NKX2-1. These data have contributed to a growing body of evidence supporting the hypothesis that multiple cooperating oncogenes are involved in amplification events, apparently in non-random frequency. Importantly, several studies have shown that the expression of genes located in chromosomal regions involved in gains or losses varies consistently with the DNA copy number.12,13 Altogether, these findings have important implications for the design of functional genomic studies aimed at identifying cancer-relevant genes, since single-gene assays will not uncover activities that rely on interactions among multiple collaborating genes.
Growth Signaling and Apoptotic Pathways: The Balance of Stimulatory and Inhibitory Genes
In clinically evident lung cancer, genomic changes involve both tumor suppressor genes and oncogenes. Tumor suppressor genes are commonly inactivated by a combination of genetic mechanisms such as point mutations, chromosomal rearrangements and mitotic recombinations, and by epigenetic events like methylation of promoter regions.14 The major tumor suppressor genes involved in lung cancer are TP53 (17p13.1), RB1 (13q14.11), CDKN2 (p16INK4a or MST1, 9p21), and several genes located at 3p. TP53 is well known for its key role in the negative regulation of the cell cycle G1/S phase transition and for being a gatekeeper for apoptosis.14,15 Mutations and overexpression of TP53 are almost universal in lung cancer and associated with smoking and more aggressive tumors.16–18 RB1 controls the G1/S transition through E2F19,20 and may also be inactivated by nonsense mutations or splicing abnormalities, most commonly in SCLC. CDKN2/p16/MTS1 encodes a CDK4 inhibitor and is frequently abnormal in NSCLC (16% to 100%).21 CDKN2 hypermethylation predicts a poor 5-year survival rate in resectable NSCLC22 and early recurrence in resected stage I NSCLC.23 Partial deletion of 3p occurs in almost all analyzed SCLCs and NSCLCs24 and encompasses numerous genes identified as tumor suppressors including FHIT (3p14.2), RASSF1 (3p21.3), TUSC2 (FUS1, 3p21.3), SEMA3B (3p21.3), SEMA3F (3p21.3) and MLH1 (3p22.3). Allelic imbalance of FHIT is associated with chromosomal deletions25,26 while RASSF1 and the mismatch repair gene MLH1 are inactivated by promoter hypermethylation.27–29 TUSC2,30 SEMA3F and SEMA3B transcripts31 are recurrently underrepresented in lung cancers and the SEMA3s were found to be targets of TP53,32 which suggests they could be activated during DNA damage or other stress responses.
Numerous proto-oncogenes contribute to lung cancer pathogenesis when constitutively activated, such as the members of the EGFR (ERBB), MYC and RAS families, as well as PIK3CA, NKX2-1 and ALK. The activation of proto-oncogenes frequently occurs by genetic mutations (KRAS, EGFR, and PIK3CA), amplifications (MYC, EGFR, HER2, PIK3CA, NKX2-1), and chromosomal rearrangements, such as translocations and inversions that place these genes under the regulation of constitutively activated genes (MYC) or create chimeric proteins (ALK-EML4).
Among the most important factors for lung tumor growth and proliferation are the tyrosine kinase receptors of the ERBB family, which are coded by the genes epidermal growth factor receptor (EGFR, 7p12), ERBB2 (HER2/neu, 17q12), ERBB3 (12q13) and ERBB4 (2q33.3). The EGFR protein is overexpressed in the majority of lung carcinomas.32,33 Activating mutations in the EGFR tyrosine kinase domain prevail in lung cancer patients of East Asian ethnicity, never-smokers, females, and NSCLC with adenocarcinoma histology.34–37 The EGFR gene is amplified in approximately 10% to 15% of advanced NSCLC.34,38–42 Phosphorylation of EGFR activates signaling to cell proliferation and survival via RAS/MAPK and PIK3CA/AKT pathways.43 Both EGFR protein overexpression and gene amplification have shown a trend towards poor prognosis33,42 while activating mutations have been associated with better prognosis and indolent disease.44,45 The other members of the EGFR family are also important, although less critical. Overexpression of ERBB2 ranges from 10 to 30% in NSCLC;46 ERBB2 gene amplification is less common (6 to 20%)47,48 and activating mutations are rare.47 These features are associated with poor survival and resistance to EGFR tyrosine kinase inhibitors (TKIs) in cases with clinical and biological features of sensitivity to such treatment.47 ERBB3 is overexpressed in 20 to 60% of lung tumors, especially squamous cell carcinomas,49 is genomically amplified in 5% without histology subtype specification,50 and is also correlated with shorter survival.51 ERBB4 is still poorly understood and seems to infrequently (<3%) harbor mutations in NSCLC.52
The genes of the RAS family (HRAS at 11p15.1, KRAS at 12p12.1, and NRAS at 1p13.2) encode for highly homologous G-proteins located at the inner surface of the cell membrane with essential roles in signal transduction pathways involved in differentiation, proliferation and survival. In lung cancer, KRAS is more frequently mutated than HRAS and NRAS.53 The mutant proteins permanently fixed in the active position and constitutively activate downstream signaling pathways, including BRAF, MAPK and PI3K/AKT.54 KRAS mutations prevail in large-cell carcinomas and adenocarcinomas (20–30%). KRAS and EGFR mutations are almost completely mutually exclusive.37 KRAS mutation has been reported as a negative prognostic factor for survival in NSCLC55 and for not responding to EGFR tyrosine kinase inhibitors. 56 Other downstream effectors of the RAS pathway, such as BRAF, which encodes a serine-threonine kinase activated by point mutation, are infrequently mutated in lung cancer (<5%) and are likely to have a lesser relevant role in the pathogenesis of these carcinomas.57
The MYC family of genes (MYC at 8q24.1, MYCN at 2p24 and MYCL1 at 1p34) encodes basic-helix-loop-helix zipper (bHLHz) transcription factors that, after dimerization with MYC-associated factor X (Max), binds to E-box motifs (CACGTG, CANNTG) and stimulates the transcription of various target genes relevant for cell growth, differentiation and apoptosis.58 Additionally, there is increasing evidence that the MYC genes bind ubiquitously throughout the genome, apparently to genomic sites at up to 15% of all cellular genes, which hints at a potential non-transcriptional function.59 The alternative model for the role of MYC in cell growth and tumorigenesis is corroborated by studies showing that MYC promotes DNA replication via non-transcriptional mechanisms and its deregulation causes DNA damage predominantly during the S-phase.60 MYC was shown to be the most frequently amplified oncogene in lung cancer cell lines (28% of 53 investigated lines).61 Amplification and overexpression of MYC genes occurs in more than 20% of SCLCs and NSCLCs in association with resistance to chemotherapy, tumor progression and worse prognosis.62
The PI3K-PTEN-AKT signaling pathway transmits a strong cell survival signal through interactions between cell surface receptors (IGF1R, PDGF, EGFR), extracellular ligands (EGF, TGFα), and the recruitment of class I PI3Ks and specific intracellular proteins (PDK-1, Akt/PKB) by mechanisms regulated by PTEN and AKT.63 The p110α catalytic subunit of PI3Ks is coded by the PI3KCA gene (3q26) and there is increasing evidence that constitutive activation of the PI3K pathways in lung cancer occurs as a consequence of PIK3CA mutation or amplification. PIK3CA genomic gain detected by FISH was reported in 43% of lung cancers with a preference for squamous cell carcinoma64 and overexpression of phosphorylated Akt has been observed in approximately 50% of advanced NSCLC.34
It has been postulated that genetic alterations that directly interfere with transcriptional networks regulating lung development may be a more common feature of lung cancer than previously realized.65 Supporting this was the recent finding of amplification of the homeobox transcription factor, NKX2-1 (14q13.3),65,66 which plays a master role in induction and maintenance of lung and thyroid morphogenesis and differentiation of epithelial cell lineages.67 Gain at 14q13.3 was present in more than 10% of lung cancer specimens and was significantly more frequent in adenocarcinomas.68
An interesting example of activation of a tyrosine kinase by gene fusion due to structural chromosomal rearrangements involves ALK. The EML4-ALK fusion (Fig. 2A) resulting from inversion in chromosome 2p and the TFG-ALK and KIF5B-ALK fusions resulting from the translocations t(2;3)(p23;q21) and t(2;10)(p23.2;p11.22), respectively, occur in approximately 4% of NSCLC and comprise a newly defined molecular subtype.69–71 In these gene rearrangements, the promoter of the 5′ partner gene controls transcription of the resulting fusion gene. The fusion partner typically contains an oligomerization domain that mediates constitutive dimerization, subsequent autophosphorylation and activation of the ALK kinase in the absence of ligand, which is important for both tumorigenesis and tumor maintenance. These ALK gene rearrangements appear to be more common in lung adenocarcinomas from never or light smokers whose tumors are wild-type for EGFR and KRAS.72,73
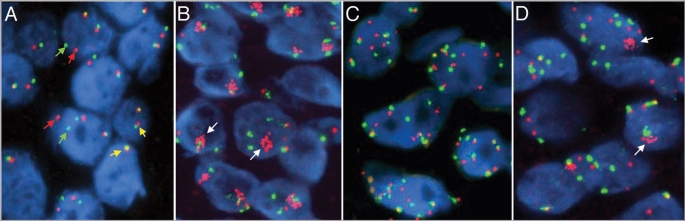
Figure 2.
Fluorescence in situ hybridization (FISH) images of sections of non-small cell lung cancer hybridized with the ALK Break-Apart (A), the EGFR/CEP 7 (B and C) and the MET/CEP 7 (C) probe sets. ALK Break-Apart and the EGFR/CEP 7 probe are commercially available (Abbott Molecular), MET/CEP 7 is a “homebrew” probe. In each panel, the chromatin from the nuclei is stained with DAPI (blue color). In (A), the ALK Break Apart FISH probe includes DNA sequences contiguous to the 3′ end of ALK labeled in red and sequences of the 5′ end of ALK labeled in green. (A) Shows an adenocarcinoma specimen harboring the EML4-ALK fusion that is detected as split red and green signals (red and green arrows). The fused red/green signals (yellow arrows) indicate native status of the ALK gene. In (B and C), DNA sequences encompassing the EGFR gene are labeled in red and the centromere 7 control is labeled in green. In normal copies of chromosome 7, these two signals are physically close since EGFR maps at 7p12. (B) Shows a lung adenocarcinoma specimen harboring amplification of the EGFR gene (clusters of red spots indicated by the white arrows). (C) Shows a squamous cell carcinoma exhibiting copy number gain for both the EGFR gene and the control CEP 7. In (D), sequences encompassing the MET gene were labeled in red and the centromere 7 control is in green. (D) Shows an adenocarcinoma specimen exhibiting amplification of the MET gene (clusters of red spots indicated by the white arrows).
Genomic Changes and Target Therapy to NSCLC
The genomic changes in proto-oncogenes are important drivers for therapeutic strategies. Observations that inactivation of a few or even a single oncogene was sufficient to induce a sustained tumor regression have supported the “oncogene-addiction” hypothesis. The model proposes that tumors may become irrevocably addicted to the oncogene that initiated tumorigenesis and a sudden interruption of its activity shifts the balance towards proliferative arrest and apoptosis.74 The ALK-driven cancers, for example, have show excellent response to specific ALK inhibitors in Phase I clinical trials.72 The status of the EGFR gene has also proved to be a powerful predictive marker for targeted therapy. Not surprisingly, patients with EGFR activating mutations and high copy numbers are more sensitive to EGFR tyrosine kinase inhibitors, such as gefitinib and erlotinib.34–36,40,43,75 NSCLC patients with EGFR gene amplification or high-level genomic gain by chromosomal aneusomy (Fig. 2B and C) have also shown higher sensitivity to the monoclonal anti-EGFR antibody, cetuximab.39 Amplification of other genes, such as MYC and EIF3H, has also been associated with better response to EGFR TKIs76 although the mechanisms involved are still to be determined. Another interesting example implicates MET (7q31.2). Enhanced MET regulation leads to oncogenic changes including cell proliferation, reduced apoptosis, angiogenesis, altered cytoskeletal function and metastasis. Mutations in the tyrosine kinase domain and MET gene amplification are uncommon in unselected NSCLC77,78 but MET gene amplification (Fig. 2D) was found to be a major mechanism by which lung tumors overcome EGFR inhibition and develop resistance to EGFR TKIs.77,79
MicroRNAs as Novel Regulators in Lung Cancer
Interesting new players in carcinogenesis are the microRNAs (miRNAs), a recently identified class of highly conserved, endogenous, non-coding RNAs that regulate gene expression in a sequence-specific manner.80,81 In their mature form, miRNAs are 19 to 25 nucleotides in length and are predicted to regulate as many as 300 to 400 messenger RNA (mRNA) targets. These molecules are of particular importance in cancer biology because many have been shown to be altered by amplification or deletion, a hallmark of the cancer genome.82 Furthermore, miRNAs are better classifiers of tissue origin for cancer cell lines or tumor tissues than are mRNA biomarkers83 and signatures of miRNA expression can define molecular subsets of tumors84,85 and predict outcome.82,86,87 The tissue specificity of miRNA expression, their incredible stability and their ability to regulate multiple mRNA targets make them attractive as a novel class of biomarkers in lung cancer.
There are numerous studies focusing on miRNAs and lung cancer with relevant results. Overexpressed microRNAs are expected to function as oncogenes and one such example involves the hsa-mir-17-92 cluster. This cluster comprises more than forty distinct miRNAs residing in an intron of MIRHG1 at 13q31.3, a gene that is markedly overexpressed and occasionally amplified in lung cancer.88 The predicted targets for this miRNA cluster comprise a large number of genes, including the tumor suppressors PTEN and RB2.89 Under the same principle, some mi-RNAs function as tumor suppressor genes. Downregulation of miRNA hsa-let-7g in 3p21.2 and miRNA hsa-mir-128b in 3p22 was recently found to be associated with overexpression of RAS and EGFR, respectively.90 These findings provide a functional link between the first recurrent abnormality detected in lung cancer, deletion of 3p sequences, and deregulation of KRAS and EGFR. Hsa-mir-128b was demonstrated to directly regulate EGFR and, most importantly, its loss had a favorable impact in the sensitivity to EGFR TKIs, which was comparable to EGFR copy number gain, associated with significantly better disease control and longer survival. Amplification of oncogenes also regulates signaling pathways through miRNAs. For instance, MYC activates expression of the miRNA cluster on chromosome 13, and two of these miRNAs (hsa-mir-17-5p and hsa-mir-20A) negatively regulate E2F.91 This association reveals the tightly controlled mechanism for activation of transcription and limitation of translation exert by MYC on E2F.
Conclusions
Lung cancer is a spectrum of diseases with numerous alterations in expression patterns resulting from acquired genetic and epigenetic mechanisms. Conventional and cutting-edge investigative technologies have detected scores of genomic changes in individual specimens. However, few of those changes are recurrent among large numbers of tumors, a characteristic that poses a challenge for the precise definition of molecular subtypes.
Studies focusing on the functional role of genomic changes in lung cancer are in dramatic expansion. Ultimately, these genomic discoveries are expected to contribute to a better understanding of cell growth, differentiation and death pathways, and to the development of novel therapeutics and translational tools for assessment of risk, early diagnosis and selection of patients for personalized therapy.
Footnotes
Previously published online: www.landesbioscience.com/journals/celladhesion/article/10884
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:38–42. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Speicher MR, Gwyn BS, Ward DC. Karyotyping human chromosomes by combinatorial multi-fluor FISH. Nat Genet. 1996;12:368–375. doi: 10.1038/ng0496-368. [DOI] [PubMed] [Google Scholar]
- 3.Schröck E, du Manoir S, Veldman T, Schoell B, Wienberg J, Ferguson-Smith MA, et al. Multicolor spectral karyotyping of human chromosomes. Science. 1996;273:494–497. doi: 10.1126/science.273.5274.494. [DOI] [PubMed] [Google Scholar]
- 4.Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, et al. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 5.Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 6.Solinas-Toldo S, Lampel S, Stilgenbauer S, Nickolenko J, Benner A, Döhner H, et al. Matrix-based comparative genomic hybridization: biochips to screen for genomic imbalances. Genes Chromosomes Cancer. 1997;20:399–407. [PubMed] [Google Scholar]
- 7.Chung YJ, Jonkers J, Kitson H, Fiegler H, Humphray S, Scott C, et al. A whole-genome mouse BAC microarray with 1-Mb resolution for analysis of DNA copy number changes by array comparative genomic hybridization. Genome Res. 2004;14:188–196. doi: 10.1101/gr.1878804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang YT, Heist RS, Chirieac LR, Lin X, Skaug V, Zienolddiny S, et al. Genome-wide analysis of survival in early-stage non-small-cell lung cancer. J Clin Oncol. 2009;27:2660–2667. doi: 10.1200/JCO.2008.18.7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blons H, Pallier K, Le Corre D, Danel C, Tremblay-Gravel M, Houdayer C, et al. Genome wide SNP comparative analysis between EGFR and KRAS mutated NSCLC and characterization of two models of oncogenic cooperation in non-small cell lung carcinoma. BMC Med Genomics. 2008;1:25. doi: 10.1186/1755-8794-1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whang-Peng J, Kao-Shan CS, Lee EC, Bunn PA, Carney DN, Gazdar AF, et al. Specific chromosome defect associated with human small-cell lung cancer; deletion 3p(14-23) Science. 1982;215:181–182. doi: 10.1126/science.6274023. [DOI] [PubMed] [Google Scholar]
- 11.Varella-Garcia M. Genomic Alterations in Lung Cancer. In: Pass HI, editor. Principles and Practice of Lung Cancer. 4th ed. Lippincott Williams & Wilkins; 2009. pp. 77–96. [Google Scholar]
- 12.Heidenblad M, Lindgren D, Veltman JA, Jonson T, Mahlamäki EH, Gorunova L, et al. Microarray analyses reveal strong influence of DNA copy number alterations on the transcriptional patterns in pancreatic cancer: implications for the interpretation of genomic amplifications. Oncogene. 2005;24:1794–1801. doi: 10.1038/sj.onc.1208383. [DOI] [PubMed] [Google Scholar]
- 13.Wang Q, Diskin S, Rappaport E, Attiyeh E, Mosse Y, Shue D, et al. Integrative genomics identifies distinct molecular classes of neuroblastoma and shows that multiple genes are targeted by regional alterations in DNA copy number. Cancer Res. 2006;66:6050–6062. doi: 10.1158/0008-5472.CAN-05-4618. [DOI] [PubMed] [Google Scholar]
- 14.Knudson AG. Chasing the cancer demon. Annu Rev Genet. 2000;34:1–19. doi: 10.1146/annurev.genet.34.1.1. [DOI] [PubMed] [Google Scholar]
- 15.Levine AJ, Momand J, Finlay CA. The p53 tumour suppressor gene. Nature. 1991;351:453–456. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- 16.Subramanian J, Govindan R. Lung cancer in never smokers: a review. J Clin Oncol. 2007;25:561–570. doi: 10.1200/JCO.2006.06.8015. [DOI] [PubMed] [Google Scholar]
- 17.Toyooka S, Tsuda T, Gazdar AF. The TP53 gene, tobacco exposure, and lung cancer. Hum Mutat. 2003;21:229–239. doi: 10.1002/humu.10177. [DOI] [PubMed] [Google Scholar]
- 18.Vega FJ, Iniesta P, Caldes T, et al. p53 exon 5 mutations as a prognostic indicator of shortened survival in non-small-cell lung cancer. Br J Cancer. 1997;76:44–51. doi: 10.1038/bjc.1997.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ewen ME. The cell cycle and the retinoblastoma protein family. Cancer Metastasis Rev. 1994;13:45–66. doi: 10.1007/BF00690418. [DOI] [PubMed] [Google Scholar]
- 20.Sherr CJ. The INK4a/ARF network in tumour suppression. Nat Rev Mol Cell Biol. 2001;2:731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- 21.Singhal S, Vachani A, ntin-Ozerkis D, Kaiser LR, Albelda SM. Prognostic implications of cell cycle, apoptosis, and angiogenesis biomarkers in non-small cell lung cancer: a review. Clin Cancer Res. 2005;11:3974–3986. doi: 10.1158/1078-0432.CCR-04-2661. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, Lee JJ, Wang L, Liu DD, Lu C, Fan YH, et al. Value of p16INK4a and RASSF1A promoter hypermethylation in prognosis of patients with resectable non-small cell lung cancer. Clin Cancer Res. 2004;10:6119–6125. doi: 10.1158/1078-0432.CCR-04-0652. [DOI] [PubMed] [Google Scholar]
- 23.Brock MV, Hooker CM, Ota-Machida E, Han Y, Guo M, Ames S, et al. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med. 2008;358:1118–1128. doi: 10.1056/NEJMoa0706550. [DOI] [PubMed] [Google Scholar]
- 24.Kok K, Naylor SL, Buys CH. Deletions of the short arm of chromosome 3 in solid tumors and the search for suppressor genes. Adv Cancer Res. 1997;71:27–92. doi: 10.1016/s0065-230x(08)60096-2. [DOI] [PubMed] [Google Scholar]
- 25.Croce CM, Sozzi G, Huebner K. Role of FHIT in human cancer. J Clin Oncol. 1999;17:1618–1624. doi: 10.1200/JCO.1999.17.5.1618. [DOI] [PubMed] [Google Scholar]
- 26.Sard L, Accornero P, Tornielli S, Delia D, Bunone G, Campiglio M, et al. The tumor-suppressor gene FHIT is involved in the regulation of apoptosis and in cell cycle control. Proc Natl Acad Sci USA. 1999;96:8489–8492. doi: 10.1073/pnas.96.15.8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burbee DG, Forgacs E, Zöchbauer-Müller S, Shivakumar L, Fong K, Gao B, et al. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J Natl Cancer Inst. 2001;93:691–699. doi: 10.1093/jnci/93.9.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaira K, Sunaga N, Tomizawa Y, Yanagitani N, Ishizuka T, Saito R, et al. Epigenetic inactivation of the RAS-effector gene RASSF2 in lung cancers. Int J Oncol. 2007;31:169–173. [PubMed] [Google Scholar]
- 29.Wang YC, Lu YP, Tseng RC, Lin RK, Chang JW, Chen JT, et al. Inactivation of hMLH1 and hMSH2 by promoter methylation in primary non-small cell lung tumors and matched sputum samples. J Clin Invest. 2003;111:887–895. doi: 10.1172/JCI15475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji L, Roth JA. Tumor suppressor FUS1 signaling pathway. J Thorac Oncol. 2008;3:327–330. doi: 10.1097/JTO.0b013e31816bce65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Potiron VA, Roche J, Drabkin HA. Semaphorins and their receptors in lung cancer. Cancer Lett. 2009;273:1–14. doi: 10.1016/j.canlet.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Futamura M, Kamino H, Miyamoto Y, Kitamura N, Nakamura Y, Ohnishi S, et al. Possible role of semaphorin 3F, a candidate tumor suppressor gene at 3p21.3, in p53-regulated tumor angiogenesis suppression. Cancer Res. 2007;67:1451–1460. doi: 10.1158/0008-5472.CAN-06-2485. [DOI] [PubMed] [Google Scholar]
- 33.Hirsch FR, Varella-Garcia M, Cappuzzo F, McCoy J, Bemis L, Xavier AC, et al. Combination of EGFR gene copy number and protein expression predicts outcome for advanced non-small-cell lung cancer patients treated with gefitinib. Ann Oncol. 2007;18:752–760. doi: 10.1093/annonc/mdm003. [DOI] [PubMed] [Google Scholar]
- 34.Cappuzzo F, Hirsch FR, Rossi E, Bartolini S, Ceresoli GL, Bemis L, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst. 2005;97:643–655. doi: 10.1093/jnci/dji112. [DOI] [PubMed] [Google Scholar]
- 35.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 36.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 37.Shigematsu H, Lin L, Takahashi T, Nomura M, Suzuki M, Wistuba II, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 38.Felip E, Rojo F, Reck M, Heller A, Klughammer B, Sala G, et al. A phase II pharmacodynamic study of erlotinib in patients with advanced non-small cell lung cancer previously treated with platinum-based chemotherapy. Clin Cancer Res. 2008;14:3867–3874. doi: 10.1158/1078-0432.CCR-07-5186. [DOI] [PubMed] [Google Scholar]
- 39.Hirsch FR, Herbst RS, Olsen C, Chansky K, Crowley J, Kelly K, et al. Increased EGFR gene copy number detected by fluorescent in situ hybridization predicts outcome in non-small-cell lung cancer patients treated with cetuximab and chemotherapy. J Clin Oncol. 2008;26:3351–3357. doi: 10.1200/JCO.2007.14.0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu CQ, da Cunha Santos G, Ding K, Sakurada A, Cutz JC, Liu N, et al. Role of KRAS and EGFR As Biomarkers of Response to Erlotinib in National Cancer Institute of Canada Clinical Trials Group Study BR.21. J Clin Oncol. 2008;26:4268–4275. doi: 10.1200/JCO.2007.14.8924. [DOI] [PubMed] [Google Scholar]
- 41.Sone T, Kasahara K, Kimura H, Nishio K, Mizuguchi M, Nakatsumi Y, et al. Comparative analysis of epidermal growth factor receptor mutations and gene amplification as predictors of gefitinib efficacy in Japanese patients with nonsmall cell lung cancer. Cancer. 2007;109:1836–1844. doi: 10.1002/cncr.22593. [DOI] [PubMed] [Google Scholar]
- 42.Hirsch FR, Varella-Garcia M, Bunn PA, Jr, Di Maria MV, Veve R, Bremmes RM, et al. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol. 2003;21:3798–3807. doi: 10.1200/JCO.2003.11.069. [DOI] [PubMed] [Google Scholar]
- 43.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 44.Cappuzzo F. EGFR FISH versus mutation: different tests, different end-points. Lung Cancer. 2008;60:160–165. doi: 10.1016/j.lungcan.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 45.Kobayashi N, Toyooka S, Ichimura K, Soh J, Yamamoto H, Matsuo K, et al. Non-BAC component but not epidermal growth factor receptor gene mutation is associated with poor outcomes in small adenocarcinoma of the lung. J Thorac Oncol. 2008;3:704–710. doi: 10.1097/JTO.0b013e31817c6080. [DOI] [PubMed] [Google Scholar]
- 46.Mitsudomi T, Kosaka T, Endoh H, Horio Y, Hida T, Mori S, et al. Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non-small-cell lung cancer with postoperative recurrence. J Clin Oncol. 2005;23:2513–2520. doi: 10.1200/JCO.2005.00.992. [DOI] [PubMed] [Google Scholar]
- 47.Cappuzzo F, Bemis L, Varella-Garcia M. HER2 mutation and response to trastuzumab therapy in non-small-cell lung cancer. N Engl J Med. 2006;354:2619–2621. doi: 10.1056/NEJMc060020. [DOI] [PubMed] [Google Scholar]
- 48.Hirsch FR, Varella-Garcia M, Franklin WA, Veve R, Chen L, Helfrich B, et al. Evaluation of HER-2/neu gene amplification and protein expression in non-small cell lung carcinomas. Br J Cancer. 2002;86:1449–1456. doi: 10.1038/sj.bjc.6600286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hilbe W, Dirnhofer S, Oberwasserlechner F, Eisterer W, Ammann K, Schmid T, et al. Immunohistochemical typing of non-small cell lung cancer on cryostat sections: correlation with clinical parameters and prognosis. J Clin Pathol. 2003;56:736–741. doi: 10.1136/jcp.56.10.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cappuzzo F, Toschi L, Domenichini I, Bartolini S, Ceresoli GL, Rossi E, et al. HER3 genomic gain and sensitivity to gefitinib in advanced non-small-cell lung cancer patients. Br J Cancer. 2005;93:1334–1340. doi: 10.1038/sj.bjc.6602865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lai WW, Chen FF, Wu MH, Chow NH, Su WC, Ma MC, et al. Immunohistochemical analysis of epidermal growth factor receptor family members in stage I non-small cell lung cancer. Ann Thorac Surg. 2001;72:1868–1876. doi: 10.1016/s0003-4975(01)03207-6. [DOI] [PubMed] [Google Scholar]
- 52.Soung YH, Lee JW, Kim SY, Wang YP, Jo KH, Moon SW, et al. Somatic mutations of the ERBB4 kinase domain in human cancers. Int J Cancer. 2006;118:1426–1429. doi: 10.1002/ijc.21507. [DOI] [PubMed] [Google Scholar]
- 53.Aviel-Ronen S, Blackhall FH, Shepherd FA, Tsao MS. K-ras mutations in non-small-cell lung carcinoma: a review. Clin Lung Cancer. 2006;8:30–38. doi: 10.3816/CLC.2006.n.030. [DOI] [PubMed] [Google Scholar]
- 54.Molina JR, Adjei AA. The Ras/Raf/MAPK pathway. J Thorac Oncol. 2006;1:7–9. [PubMed] [Google Scholar]
- 55.Marks JL, Broderick S, Zhou Q, Chitale D, Li AR, Zakowski MF, et al. Prognostic and therapeutic implications of EGFR and KRAS mutations in resected lung adenocarcinoma. J Thorac Oncol. 2008;3:111–116. doi: 10.1097/JTO.0b013e318160c607. [DOI] [PubMed] [Google Scholar]
- 56.Massarelli E, Varella-Garcia M, Tang X, Xavier AC, Ozburn NC, Liu DD, et al. KRAS mutation is an important predictor of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. Clin Cancer Res. 2007;13:2890–2896. doi: 10.1158/1078-0432.CCR-06-3043. [DOI] [PubMed] [Google Scholar]
- 57.Ueda M, Toji E, Nunobiki O, Izuma S, Okamoto Y, Torii K, et al. Mutational analysis of the BRAF gene in human tumor cells. Hum Cell. 2008;21:13–17. doi: 10.1111/j.1749-0774.2008.00046.x. [DOI] [PubMed] [Google Scholar]
- 58.Dang TP, Gazdar AF, Virmani AK, Sepetavec T, Hande KR, Minna JD, et al. Chromosome 19 translocation, overexpression of Notch3, and human lung cancer. J Natl Cancer Inst. 2000;92:1355–1357. doi: 10.1093/jnci/92.16.1355. [DOI] [PubMed] [Google Scholar]
- 59.Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. EMBO J. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, et al. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448:445–451. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- 61.Lockwood WW, Chari R, Coe BP, Girard L, Macaulay C, Lam S, Gazdar AF, et al. DNA amplification is a ubiquitous mechanism of oncogene activation in lung and other cancers. Oncogene. 2008;27:4615–4624. doi: 10.1038/onc.2008.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brennan J, O’Connor T, Makuch RW, Simmons AM, Russell E, Linnoila RI, et al. myc family DNA amplification in 107 tumors and tumor cell lines from patients with small cell lung cancer treated with different combination chemotherapy regimens. Cancer Res. 1991;51:1708–1712. [PubMed] [Google Scholar]
- 63.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 64.Massion PP, Taflan PM, Shyr Y, Rahman SM, Yildiz P, Shakthour B, et al. Early involvement of the phosphatidylinositol 3-kinase/Akt pathway in lung cancer progression. Am J Respir Crit Care Med. 2004;170:1088–1094. doi: 10.1164/rccm.200404-487OC. [DOI] [PubMed] [Google Scholar]
- 65.Kendall J, Liu Q, Bakleh A, Krasnitz A, Nguyen KC, Lakshmi B, et al. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc Natl Acad Sci USA. 2007;104:16663–16668. doi: 10.1073/pnas.0708286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kimura S, Hara Y, Pineau T, Fernandez-Salguero P, Fox CH, Ward JM, et al. The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain and pituitary. Genes Dev. 1996;10:60–69. doi: 10.1101/gad.10.1.60. [DOI] [PubMed] [Google Scholar]
- 68.Berghmans T, Mascaux C, Haller A, Meert AP, Van HP, Sculier JP. EGFR, TTF-1 and Mdm2 expression in stage III non-small cell lung cancer: a positive association. Lung Cancer. 2008;62:35–44. doi: 10.1016/j.lungcan.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 69.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 70.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 71.Takeuchi K, Choi YL, Togashi Y, Soda M, Hatano S, Inamura K, et al. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin Cancer Res. 2009;15:3143–3149. doi: 10.1158/1078-0432.CCR-08-3248. [DOI] [PubMed] [Google Scholar]
- 72.Shaw AT, Yeap BY, Mino-Kenudson M, Digumarthy SR, Costa DB, Heist RS, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27:4247–4253. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Solomon B, Varella-Garcia M, Camidge DR. ALK gene rearrangements: A new therapeutic target in a molecularly-defined subset of non-small cell lung cancer. J Thorac Oncol. 2009;4:1450–1454. doi: 10.1097/JTO.0b013e3181c4dedb. [DOI] [PubMed] [Google Scholar]
- 74.Weinstein IB, Joe A. Oncogene addiction. Cancer Res. 2008;68:3077–3080. doi: 10.1158/0008-5472.CAN-07-3293. [DOI] [PubMed] [Google Scholar]
- 75.Hirsch FR, Varella-Garcia M, McCoy J, West H, Xavier AC, Gumerlock P, et al. Increased epidermal growth factor receptor gene copy number detected by fluorescence in situ hybridization associates with increased sensitivity to gefitinib in patients with bronchioloalveolar carcinoma subtypes: a Southwest Oncology Group Study. J Clin Oncol. 2005;23:6838–6845. doi: 10.1200/JCO.2005.01.2823. [DOI] [PubMed] [Google Scholar]
- 76.Cappuzzo F, Varella-Garcia M, Rossi E, Gajapathy S, Valente M, Drabkin H, et al. MYC and EIF3H Coamplification significantly improve response and survival of non-small cell lung cancer patients (NSCLC) treated with gefitinib. J Thorac Oncol. 2009;4:472–478. doi: 10.1097/JTO.0b013e31819a5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bean J, Brennan C, Shih JY, Riely G, Viale A, Wang L, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci USA. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cappuzzo F, Marchetti A, Skokan M, Rossi E, Gajapathy S, Felicioni L, et al. Increased MET gene copy number negatively affects survival of surgically resected non-small-cell lung cancer patients. J Clin Oncol. 2009;27:1667–1674. doi: 10.1200/JCO.2008.19.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 80.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 81.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 82.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, Zepeniuk M, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26:462–469. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- 84.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 85.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 87.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 88.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 89.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 90.Weiss GJ, Bemis LT, Nakajima E, Sugita M, Birks DK, Robinson WA, et al. EGFR regulation by microRNA in lung cancer: correlation with clinical response and survival to gefitinib and EGFR expression in cell lines. Ann Oncol. 2008;19:1053–1059. doi: 10.1093/annonc/mdn006. [DOI] [PubMed] [Google Scholar]
- 91.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]