Abstract
The absorption spectra of a series of inhibitors bound at the active site of Δ5-3-ketosteroid isomerase from Pseudomonas putida were found to exhibit substantial variations in the contributions of the protonated and deprotonated forms. Systematic variation of the inhibitor solution pKa combined with a method of quantifying the contributions of each protonation state showed the oxyanion hole in the active site of wild-type Δ5-3-ketosteroid isomerase to have a proton affinity equal to a solution pKa of 10.05 ± 0.03, which is similar to the measured pKa, 10.0, of the reaction intermediate. This observation supports the prediction of Cleland, Kreevoy, Frey, Gassman, and Gerlt that an enzyme utilizing a strong hydrogen bond for catalysis matches the proton affinity of the protein to the intermediate [Cleland, W.W. and Kreevoy, M.M. (1994) Science 264, 1887-189.], [Frey, P.A., Whitt, S., and Tobin, J. (1994) Science 264, 1927–1930], [Gerlt, J.A. and Gassman, P.G. (1993) Biochemistry 32, 11934 – 11952]. As the difference in proton affinity decreases, the strength of the hydrogen bond increases, and the closely matched proton affinity between the active site and the reaction intermediate supports the possibility that a short, strong hydrogen bond is catalytically relevant in Δ5-3-ketosteroid isomerase.
The origin(s) of catalytic rate enhancement in enzymes remains an open and important question. In some enzymes, short-strong hydrogen bonds (SSHBs) between the substrate and protein have been proposed to produce a rate enhancement by stabilizing high-energy reaction intermediates (1–4). In order to form a SSHB, the proton affinity of the substrate must approximate the proton affinity of its hydrogen-bonding partner. In this paper, we present evidence of a matched proton affinity between a reaction intermediate and its hydrogen-bonding partner in the active site of Δ5-3-ketosteroid isomerase (KSI) from Pseudomonas putida (pKSI) and Comamonas testosteroni (tKSI). The observation of a matched proton affinity suggests that the formation of a SSHB may be possible and catalytically relevant in KSI
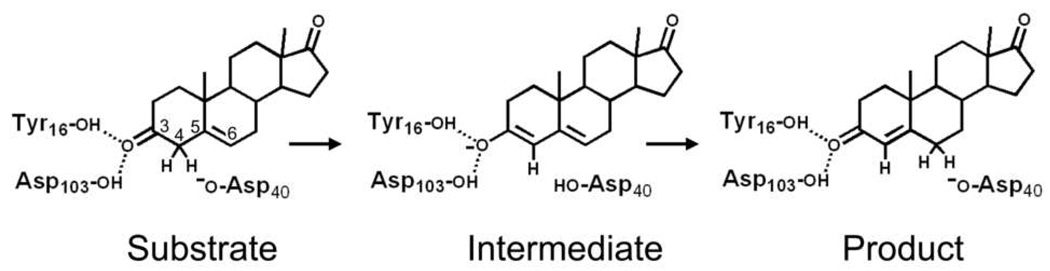
KSI catalyses a C-H bond cleavage and formation through an enolate intermediate at diffusion limited rate as depicted in Figure 1 (5). The general base Asp40 abstracts a proton from the 4 position of the steroid ring to form an enolate that is stabilized by the hydrogen bond donating Tyr16 and Asp103 (all residue numbers use the P. putida sequence unless specifically describing tKSI). Tyr16 and Asp103 are positioned deep within the hydrophobic active site and form a so-called oxyanion hole. Protonated Asp40 then transfers its proton to the 6 position of the steroid ring to complete the reaction. The hydrogen bonds from Tyr16 and Asp103 are known to significantly affect the rate of catalysis in KSI. Mutation of either Tyr16 or Asp103 in pKSI to non-polar residues reduces the catalytic efficiency by 103.4 and 102.1, respectively (6). In tKSI, mutation of Tyr14 (homologous to Tyr16 in pKSI) results in a 104.7 (7) reduction in catalytic efficiency while the mutation of Asp99 (homologous to Asp103 in pKSI) reduced catalytic efficiency by 103.6 (8). We note, however, that all amino acid residues in the active site interact strongly and contribute to catalysis, so site-directed mutagenesis can produce secondary effects that may lead to misinterpretation.
Figure 1.
Schematic of enzymatic isomerization of a Δ5-3-ketosteroid to a Δ4-3-ketosteroid as observed in KSI from Pseudomonas putida and Comamonas testosteroni. Residues are numbered according to the P. putida sequence. In C. testosteroni, Tyr16 is Tyr14, Asp103 is Asp99, and Asp40 is Asp38.
Gerlt and Gassman proposed the formation of unusually short, strong hydrogen bonds between the KSI oxyanion hole and the reaction intermediate as a means of catalytic rate enhancement 15 years ago (1,2). In their model, high-energy states along the reaction coordinate are specifically stabilized by the formation of these bonds. Since then, the catalytic role of SSHBs has been debated (9,10). Crystal structures of phenol, a reaction intermediate analog, bound to the active site of pKSI show the O-O distance between phenol and the oxyanion hole residues to be 2.49 Å for Tyr16 and 2.61 Å for Asp103 (11). Also, far downfield shifts of ~16–18 ppm associated with the oxyanion hole protons in NMR studies have been interpreted as evidence of SSHB activity. (12–15). The term “short, strong hydrogen bond” is general, but is sometimes used interchangeably with “low-barrier hydrogen bond” (LBHB). As originally conceived, the LBHB is a specific physical state where the proton involved in a hydrogen bond becomes delocalized between heavy atom centers, that is, the barrier between hydrogen bonding partners is low (16,17). The resulting potential energy surface is broader than for a normal hydrogen bond, the zero point vibrational energy is lowered, and this produces a dramatically stabilized hydrogen bond. LBHB’s have been proposed to be present in KSI based on NMR (12–14) and crystallographic evidence (18). In a related system, photoactive yellow protein, where such data likewise supports the presence of a LBHB (19,20), neutron scattering experiments have more directly identified a LBHB by observing a proton located equidistant from the two hydrogen bonding oxygen centers, not localized to either heavy atom (21).
Several conditions must be met to allow SSHB formation. First, the dielectric constant of the medium surrounding the hydrogen bond must be low. In the gas phase, the heat of formation of hydrogen bonds can reach −25 to −30 kcal mol−1 (22). As the dielectric constant of the medium increases to ~6, the maximum strength of the hydrogen bonds decreases to about half the gas phase value (23). The KSI active site around Tyr16 and Asp103 is often considered a low dielectric medium because the site is largely composed of hydrophobic amino acids, and intermediate analogs have been shown to bind with increasing affinity as analog hydrophobicity is increased. For example, equilenin binds approximately three orders of magnitude tighter than substituted phenols (11), and, as shown below, the Kd for substituted naphthols binding to KSI falls between that for the substituted phenols and equilenin (the larger analogs sample a much larger area of the active site than just the oxyanion hole which complicates simple interpretations of binding data). Other researchers have observed Tyr14 in tKSI has a perturbed pKa and enhanced fluorescence quantum yield compared to the tyrosine solution values, and these observations have been interpreted as evidence for a low dielectric constant in the active site (24). However, the pKa of Tyr14 is probably perturbed by its immediate hydrogen bonding partners as well as from the local dielectric (25). A more precise and local measure of the dielectric is provided by dynamic Stokes shift experiments, which are presented in a separate paper (26). The dynamic Stokes shift experiments demonstrate only a very limited response of the oxyanion hole to a sudden change in the pKa of a bound fluorophore indicating a limited capacity for solvating a charge shift, which can be interpreted as a low dielectric environment.
Another important condition for SSHB formation and the role of SSHBs in enzymatic catalysis is the matching of proton affinities between the hydrogen bond donor and acceptor groups. Formation of SSHBs to the high-energy species along the reaction coordinate contributes to catalysis by stabilizing the formation of these species. SSHB formation is governed by the changing proton affinities between the reaction ground state and high energy intermediate during the catalytic cycle. If SSHBs are active in KSI, then the SSHB must be formed to the intermediate, not the starting substrate. In this work, we compare the proton affinity of the oxyanion hole to that of the reaction intermediate and demonstrate that they are well matched.
Proton affinity is often described by a pKa value; however, pKa is a bulk solvent defined term. Comparing the proton affinities by comparing pKa values of two small molecules titrated in the same solvent is entirely reasonable; however, the oxyanion hole is deep within a hydrophobic pocket where the environment cannot be described as aqueous. Therefore, titrating the oxyanion hole may provide a pKa for the hydrogen bonding residues, but that pKa is not comparable to the solution values obtained for the reaction intermediate or its analogs. Instead of titrating the active site residues by changing the solution pH, we have compared the proton affinity of the oxyanion hole to that of substituted naphthols by determining the degree to which the naphthol is protonated while bound to the enzyme. This is quantified by measuring the contributions of protonated and deprotonated substituted naphthols whose electronic absorption spectra are sufficiently different to be observed directly. The proton affinity of the oxyanion hole is reported in terms of the solution pKa that would bind as a 50:50 mixture of protonated and deprotonated states. Wild-type KSI, especially pKSI, is found to have a proton affinity matched to the reaction intermediate, and the matched proton affinity between the intermediate and oxyanion hole is consistent with enzymatic rate enhancement utilizing SSHBs.
Methods
Protein expression and purification followed standard methods that have been described in detail previously (11). Electronic absorption measurements were conducted with either a PerkinElmer Lambda 25 or Cary 6000i spectrophotometer. Unless otherwise specified, absorption spectra of pKSI/naphthol complexes were obtained with 1 mM protein and 0.5 mM naphthol in pH 7, 40 mM potassium phosphate buffer and 1% DMSO, with protein-alone solutions serving as a blank. Experiments with tKSI were preformed with 1.6 mM protein and 62.5 µM naphthol with otherwise identical conditions as pKSI. High pH basis spectra were obtained in 0.5 M NaOH, 20% DMSO solution, and 0.5 M HCl, 20% DMSO was used for low pH basis spectra for all naphthols except equilenin, which required 50% DMSO for solubility. All spectra were obtained in a 1 mm quartz cuvette.
Protein concentration was determined by absorption at 280 nm. As determined by the Scripps protein calculator (27,28), pKSI and tKSI have molar extinction coefficients at 280 nm of 16,500 M−1cm−1 and 3840 M−1cm−1, respectively.
Binding constants were determined by monitoring changes in the fluorescence emission spectra of naphthols upon binding to KSI. Fluorescence emission spectra (330 nm excitation for all samples except 320 nm for 6-cyano-2-naphthol) were obtained for naphthols free in solution (16.5 µM substituted naphthol, pH 7, 40 mM potassium phosphate buffer with 20% DMSO) and bound to pKSI D40N (16.5 µM substituted naphthol, pH 7, 40 mM potassium phosphate buffer, 2 mM pKSI D40N). The D40N mutation is commonly used to improve intermediate analog binding to pKSI. Nearly complete binding of the substituted naphthol by pKSI in the previously described conditions was guaranteed by demonstrating that the emission lineshape from the 16.5 µM substituted naphthol was independent of pKSI D40N concentration from 2 mM to at least 0.5 mM. To determine Kds, spectra were then taken in conditions where both bound and unbound populations could be observed (16.5 µM substituted naphthol, 25 µM pKSI D40N, pH 7, 40 mM potassium phosphate buffer with 20% DMSO and 25 µM substituted naphthol, 25 µM pKSI D40N, pH 7, 40 mM potassium phosphate buffer with 20% DMSO). The spectra showing both populations were fit as a linear combination of the bound and free fluorescence spectra. The resulting fits yielded the concentration of bound and unbound naphthol, and the concentrations were used directly to determine the Kd by simple equilibrium theory.
As described below, some of the protein-bound electronic absorption spectra of naphthols bound at the active site of pKSI are clearly a mixture of the protonated and deprotonated form of the naphthol that depends on the particular naphthol. The contributions from these forms cannot be obtained simply by taking a linear combination of the protonated and deprotonated basis spectra obtained in aqueous solution due to additional shifts in the absorption bands of both the neutral and anionic forms of the ligands when they are bound in the protein active site. In order to deal with the shifts and still obtain quantitative information on the populations of protonated and deprotonated naphthols, we start with a linear combination of the aqueous solution basis spectral lineshapes and band positions, then the basis spectra are allowed to shift in energy in 10 wavenumber steps, retaining their line shape to determine if shifting the spectra produces a better fit. If a better fit is found, then a new linear combination is determined with the shifted basis spectra. The process is repeated until the algorithm converges, usually about 10 iterations. Thus, each fit yields the ratio of the protonated and deprotonated forms, which are used to describe the proton affinity of the oxyanion hole, plus two band shifts. These shifts reflect the interaction of the polarizable chromophore with the protein environment, which is expected to be substantially different from the aqueous environment used to obtain the basis spectra, but they do not bear on the protonation state of the naphthol, and we do not attempt to interpret these shifts.
Results and Discussion
Binding constants for substituted naphthols
Substituted naphthols bind well to pKSI D40N, less well to wild-type. The mutation D40N is a commonly used as it improves intermediate analog binding (29), for example, in pKSI D40N, the Kd of equilenin is less than 1 nM (11). The kd’s for the other naphthols were determined as described above to be 20 ± 6 µM for 2,6-dihydroxynaphthalene, 4 ± 1 µM for 6-bromo-2-naphthol, 10 ± 5 µM for 6-methoxy-2-naphthol, and 2 ± 1 µM for 6-cyano-2-naphthol (data not shown). Solvent conditions were similar to those used in the absorption experiments (pH 7, 40 mM potassium phosphate) except a 1:5 mixture of DMSO and pH 7, 40 mM potassium phosphate buffer was used to facilitate naphthol solubility. As the DMSO increases the solubility of the naphthols in solution, we regard the measured Kd values to be upper limits for the solvent conditions used in the absorption experiments. Even for the poorest binding, 2,6-dihydroxynaphthalene, we calculate greater than 96% of the naphthol is bound to pKSI at the concentrations used for the absorption experiments.
Electronic absorption spectra of substituted naphthols bound to pKSI
Figure 2 shows the absorption spectra of equilenin, an intermediate analog with a solution pKa of 9.7, at high and low pH (the basis spectra), and the spectrum of equilenin bound to the active site of pKSI D40N. The absorption spectrum of equilenin bound in the protein is clearly different from either of the basis spectra, and the three poorly resolved peaks observed in the protein-bound spectrum are consistent with both protonation states being present.
Figure 2.
There is conflicting data in the literature on the protonation state of substituted naphthols bound to tKSI. Pollack and co-workers argue that the napthols are completely anionic when bound to tKSI (30) while others have observed a mixture of protonation states (31). We show below that the protonation state of the bound naphthol depends on its solution pKa. Also, many of the previous experiments designed to determine the protonation state of the reaction intermediate used tKSI with its common D38N mutation for enhanced binding affinity. However, as discussed below, the proton affinity of the tKSI D38N oxyanion hole is significantly higher than in wild-type tKSI or pKSI. As a result, observations of a fully anionic substrate in tKSI D38N may not reflect the protonation state of the intermediate during catalysis.
Proton affinity of the oxyanion hole
The acid/base equilibrium between the protonation of the oxyanion hole, A, and the substrate, B, is described by Equation (1):
| (1) |
and the modified Boltzmann distribution by Equation (2).
| (2) |
A is the titratable protein group, which may be influenced by contributions from several residues in the oxyanion hole. It is expected that the proton transfer originates from either of the residues that form hydrogen bonds to the substrate hydroxyl, Tyr16 or Asp103; however, we need not be more specific about which of these two centers transfers a proton to the substrate, and the data presented here does not directly address this question (see (25) for more discussion of the identity of the proton donor). The left side of Equation (2) can be rearranged as shown in (3) where [C]T is the total concentration of the complex and %A− is the percent of A deprotonated at equilibrium.
| (3) |
Rearranging (2) and (3) leads directly to Equation (4).
| (4) |
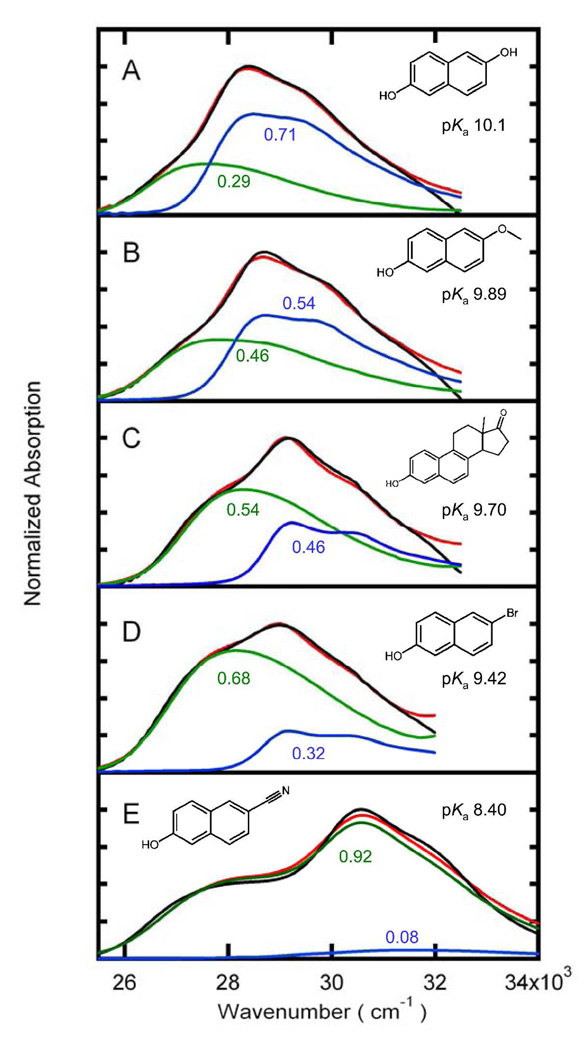
Figure 3 shows the absorption spectra of substituted naphthols of varying pKa bound to the KSI active site. The protein-bound absorption spectrum of each naphthol is fitted to a combination of high and low pH solution spectra. As described in the Methods section, fitting the high and low pH spectra to the protein-bound spectrum requires shifting the transition energies of the solution spectra. The fraction of protonated form bound to KSI and the transition energy shifts are listed in Table 1.
Figure 3.
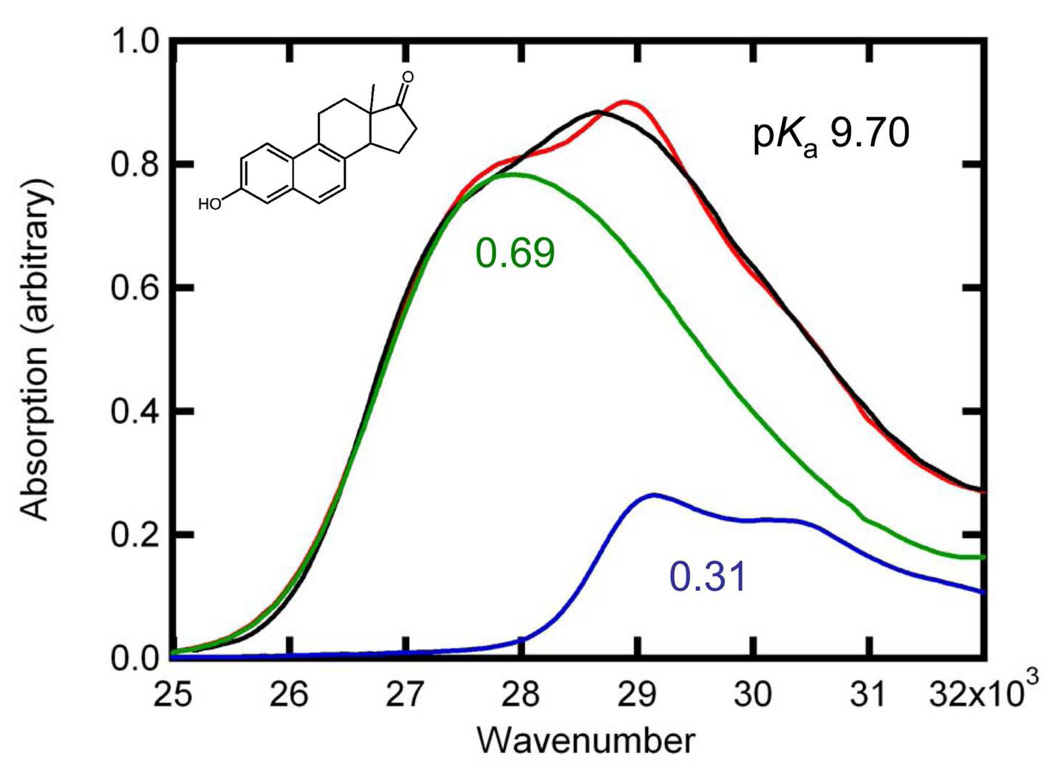
The absorption spectra of substituted naphthols (Black, normalized at the absorption maximum) fit to a linear combination (Red) of the protonated (Blue) and deprotonated (Green) species, each shifted as described in text and in Table 1. The protonated and deprotonated spectra are scaled by their relative contributions to the fit (Table 1). The scaling factors are shown in the color corresponding to the curve they describe. (A) 2,6-dihydroxynaphthalene, solution pKa 10.1; (B) 6-methoxy-2-naphthol, solution pKa 9.89; (C) equilenin, solution pKa 9.7; (D) 6-bromo-2-naphthol, solution pKa 9.42; and (E) 6-cyano-2-naphthol, solution pKa 8.4.
Table 1.
Shifts in absorption band maxima in cm−1 relative to those in solution resulting from binding of naphthols in the pKSI active site and the fraction of the neutral species observed when bound. Errors indicate one standard deviation.
| Ligand | Protein | Neutral shift | Anionic shift | Fraction neutral |
|---|---|---|---|---|
| Equilenin | WT | −280 ±17 | 446 ± 28 | 0.31 ± 0.02 |
| Equilenin | D40N | −180 ± 14 | 730 ± 56 | 0.456 ± 0.005 |
| 6-bromo-2-naphthol | D40N | −475 ± 7 | 190 ± 28 | 0.32 ± 0.02 |
| 2,6-hydroxynaphthalene | D40N | −405 ± 63 | 1230 ± 212 | 0.71 ± 0.01 |
| 6-cyano-2-naphthol | D40N | −1480 ± 280 | 275 ± 7 | 0.08 ± 0.03 |
| 6-methoxy-2-naphthol | D40N | −390 ± 14 | 235 ± 7 | 0.54 ± 0.03 |
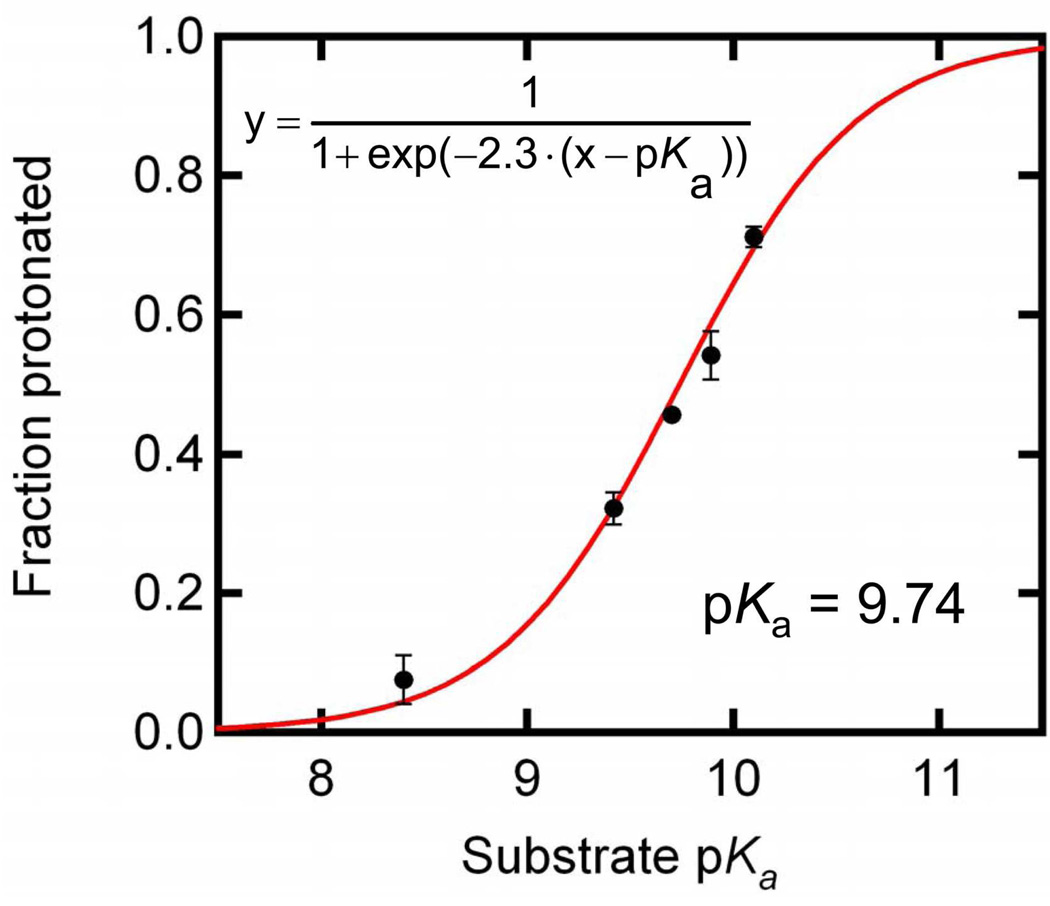
It is evident from the spectra in Figure 3 that substituted naphthols with different solution pka’s bind to the pKSI D40N active site with varying degrees of protonation. A comparison of the degree of protonation while bound to pKSI (Table 1) with the solution pKa of each substituted naphthol yields a proton affinity curve (analogous to a titration curve) for the pKSI D40N active site, as shown in Figure 4. This proton affinity curve cannot be used to determine the actual pKa of titratable groups in the oxyanion hole. The pKa of any titratable group is a solvent defined quantity; usually, the solvent is water, and the pKa values listed for the naphthols are the aqueous values. However, the active site constitutes a very specific solvent environment highly perturbed from water. By challenging the active site of pKSI with naphthols whose solution pka’s are quite different, we can compare the proton affinity of the oxyanion hole to that of the substituted naphthol. Therefore, the value determined by the proton affinity curve in Figure 4 is not the pKa of the oxyanion hole, rather the oxyanion hole has a proton affinity equal to a naphthol with a solution pKa of that value.
Figure 4.
Fraction of substrate bound as the protonated species (Table 1) versus the substrate pKa (black dots) is fit to Equation 4 (red line) to determine the proton affinity of the active site. Error bars indicate one standard deviation.
The proton affinity of the oxyanion hole can be found with Equation (4) by varying the inhibitor solution pKa and measuring the degree of protonation. As shown in the fit in Figure 4, we observe the proton affinity of the oxyanion hole in pKSI D40N to match that of a naphthol with solution pKa of 9.76 ± 0.03. The high precision of this measurement is possible because the fit of Equation 4 to the data in Figure 4 requires only one adjustable parameter. The shape of the sigmoid is determined by acid-base theory, as described above in the derivation of Equation 4 with the endpoints set to 0% and 100% protonated by definition.
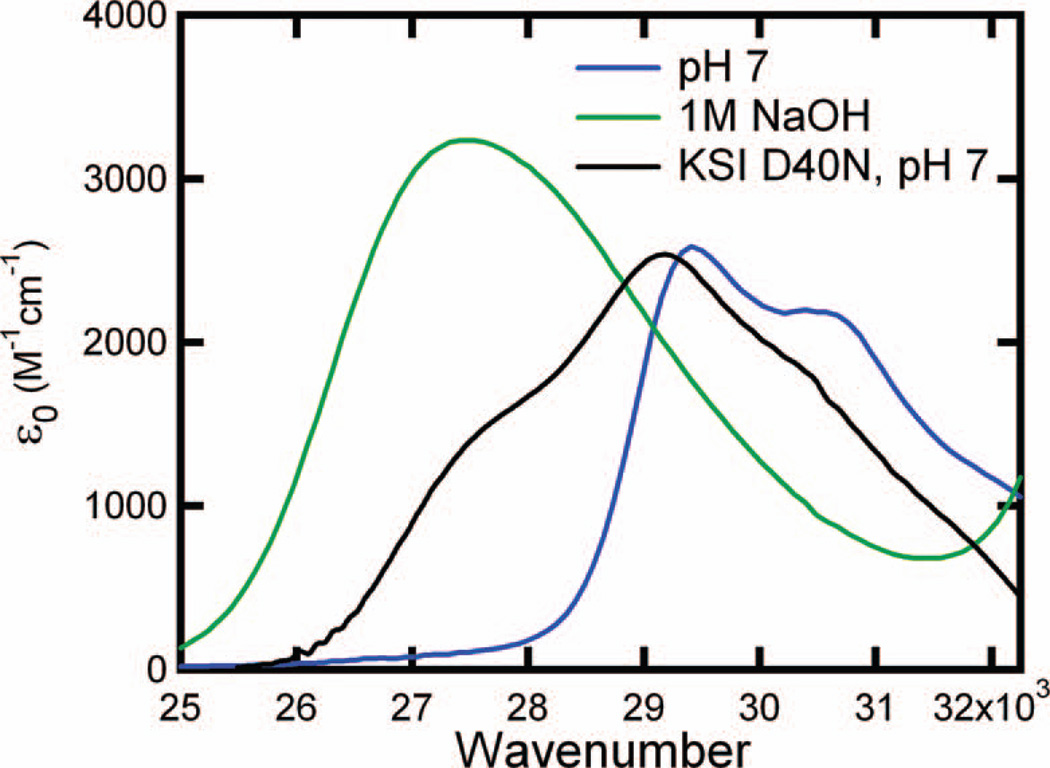
Absorption spectra of equilenin bound to wild-type pKSI also show a mixture of protonation states, as shown in Figure 5. Wild-type pKSI binds the substituted naphthols more weakly than pKSI D40N, and this weaker binding significantly limits our ability to probe the active site with multiple substituted naphthols as most bind with Kd greater than 1 mM. However, equilenin binds to wild-type pKSI with a 1.8 µM Kd (32), which is sufficient to assure nearly 100% binding under the conditions of the experiment. Based only on the contributions from the protonated and deprotonated forms of equilenin, the proton affinity of the oxyanion hole of wild-type pKSI is observed to match a solution pKa of 10.05 ± 0.03. As discussed below, the solution pKa of the reaction intermediate has been measured to be 10.0 (33), and the matched proton affinity between the protein and reaction intermediate suggests SSHBs may form during the catalytic cycle.
Figure 5.
The absorption spectrum of equilenin bound to WT-pKSI (Black) fit to a linear combination (Red) of the protonated (Blue) and deprotonated (Green) species. The protonated and deprotonated spectra are scaled by their relative contributions to the fit, and the scaling factors are shown below each curve.
Electronic absorption spectra of equilenin bound to tKSI
If the matched proton affinity found in pKSI is observed in KSI from other species, then the matched proton affinity is likely an important, conserved feature of the active site, and the proton affinity would not be expected to vary much from species to species that share the same natural substrate. There is insufficient evidence in the literature to guarantee that tKSI and pKSI utilize exactly the same catalytic mechanism; however, they share 34% sequence identity, and the active site of tKSI has analogs of the most catalytically relevant residues, Asp40, Asp103, Tyr16, and Tyr57, in pKSI. The structural similarities between tKSI and pKSI suggest at least a similar catalytic mechanism takes place in both enzymes.
Electronic absorption spectra of substituted naphthols bound to tKSI with the active site mutation D38N (analogous to D40N in pKSI) show more of the anionic species than when bound to pKSI. The most basic of the naphthols, 2,6-dihydroxynaphthalene, bound as only ~30% protonated to tKSI D38N (data not shown), while in pKSI D40N 2,6-dihydroxynaphthalene was found to be 71% protonated. Because most of the substituted naphthols fall near 0% protonated on the proton affinity curve, the proton affinity of the tKSI D38N oxyanion hole cannot be measured as accurately as in pKSI D40N. Nonetheless, from this much more limited data a proton affinity curve was created for tKSI and the proton affinity was estimated to be equal to a solution pKa of ~10.5 for tKSI D38N. Like wild-type pKSI, wild-type tKSI does not bind the substituted naphthols well enough to construct a proton affinity curve. However, equilenin binds to tKSI with a 3 µM Kd (29), and experiments with equilenin like those described above for wild-type pKSI show a proton affinity of wild-type tKSI to be equal to a solution pKa of 10.3 ± 0.1.
The proton affinity observed in wild-type tKSI is similar, but not identical to wild-type pKSI. The 0.3 pKa unit difference may indicate the two enzymes utilize hydrogen bonds of different strength during catalysis, or possibly, tKSI has been optimized to catalyze the isomerization of steroids with slightly different proton affinity than the canonical steroid shown in Figure 1. However, the D40N (pKSI) and D38N (tKSI) mutations indicate that a small number of mutations can affect the oxyanion hole proton affinity, and the similarity of the two wild-type proton affinities suggests a proton affinity near 10 is functionally relevant. As described above, if the proton affinity were not functionally relevant, then the two proteins could have oxyanion hole proton affinities that diverge over time.
Connection between active site proton affinity and catalytic efficiency
The proton affinity of the oxyanion hole in pKSI and tKSI is similar to the proton affinity of the intermediate shown in Figure 1. The intermediate in Figure 1 is widely considered to be representative of intermediates produced in vivo. Zeng and Pollack synthesized the intermediate shown in Figure 1 and determined the solution pKa to be 10.0 (33). We have presented evidence that the proton affinity of the oxyanion hole is equal to a solution pKa of 10.05 ± 0.03 in pKSI and 10.3 ± 0.1 in tKSI. A matched proton affinity between the oxyanion hole and reaction intermediate has previously been proposed by Cleland, Kreevoy, Frey, Gassman, and Gerlt (1–4) to enhance the catalytic rate in KSI, and the observation of the matched proton affinity supports this claim.
Matching the proton affinity between the reaction intermediate and oxyanion hole is believed to produce a catalytic rate enhancement by stabilizing the intermediate via the formation of strong hydrogen bonds. Other labs have examined the dependence of hydrogen bond strength versus the difference in proton affinity between the hydrogen bonding groups (34,35). Decreasing the difference in proton affinity between two hydrogen bonding groups does lower the heat of formation for a hydrogen bond. The degree to which the heat of formation decreases depends on other factors as well, particularly the solvent. Shan et al (34) have shown log KHB, where KHB is the equilibrium constant of hydrogen bond formation, to vary linearly with ΔpKa in both water (dielectric constant of 80) and dimethyl sulfoxide (dielectric constant of 47.2) with slopes 0.05 and 0.73, respectively. Taking the value of 0.73 to be more relevant for the hydrophobic KSI active site, the hydrogen bond becomes 0.95 kcal/mol less stable for every pKa unit mismatch between hydrogen-bonding partners. Ab initio calculations of LBHB systems in vacuum have determined a 1.8 kcal/mol penalty to hydrogen bond stability for every unit of pKa mismatch between hydrogen-bonding partners (35). Either value suggests a large stabilization of the intermediate in KSI. Given the 0.95 kcal/mol value, the difference in ΔG° for hydrogen bond formation between the oxyanion hole and the ketone starting material with conjugate acid pKa ~ −2 versus the intermediate with matched pKa would be 11.5 kcal/mol. However, we note that the assumption of linearity for the log KHB versus ΔpKa is not necessarily valid over the 12 pKa unit range.
Whether the hydrogen bonds in KSI are of the low-barrier variety or not, there is a significant amount of stabilization energy available to produce a rate enhancement. Åqvist and Feierberg (36) have used empirical valence bond simulations to determine that stabilization of the intermediate accounts for 60% of the rate enhancement observed in pKSI with electrostatic preorganization of the transition state accounting for the balance. However, they find no evidence of LBHB activity.
Hydrogen bonds exist between the oxyanion hole and substrate during the course of the reaction. We have shown the proton affinity of a reaction intermediate is similar to the proton affinity of the oxyanion hole. This observation suggests catalytic rate enhancement is at least partially achieved by varying the proton affinity between the substrate and oxyanion hole. As the reaction intermediate is formed, the difference in proton affinity between the substrate and oxyanion hole decreases, and the increasingly stable hydrogen bonds stabilize the reaction intermediate. Our observations therefore support predictions from Cleland, Kreevoy, Frey, Gassman, and Gerlt (1–4) who theorized a hydrogen bond could produce a catalytic rate enhancement if the proton affinities between protein and substrate are matched for the high energy states along a reaction coordinate.
Conclusion
Substituted naphthols with solution pKa’s ~10 bind to the active site of pKSI and tKSI as a mixture of protonation states. By quantifying the degree of protonation at the active site as a function of solution pKa, a proton affinity curve was obtained for the oxyanion hole. It was determined from this proton affinity curve that pKSI D40N from Pseudomonas putida has a proton affinity equal to a solution pKa value of 9.76 ± 0.03, and wild-type pKSI has a value of 10.05 ± 0.03. The same experiment performed on tKSI from Comamonas testosteroni yields proton affinities equal to ~10.5 and 10.3 ± 0.1 for the D38N mutant and wild-type, respectively. The similarity of the wild-type KSI proton affinities to the measured solution pKa of the reaction intermediate shown in Figure 1 supports theories of a catalytic rate enhancement via changes in hydrogen bond stabilization energy along the reaction coordinate.
Catalytic rate enhancement in KSI probably results from a combination of active site characteristics including electrostatic preorganization, geometrical constraints and SSHBs. We provide evidence that KSI is particularly well suited to form SSHBs between the high-energy intermediate and oxyanion hole as they appear to have equal proton affinity. The formation of SSHBs results in a catalytically relevant amount of stabilization energy, which is only available to chemical species whose proton affinity is matched to that of the oxyanion hole. Such an oxyanion hole whose proton affinity is matched to the reaction intermediate allows KSI to stabilize the intermediate relative to the starting materials through a SSHB.
Acknowledgments
This work was supported in part by a grant from the National Institute of Health GM27738. Plasmids containing the tKSI and pKSI genes were graciously provided by the Daniel Herschlag lab at Stanford University. We are grateful for extended discussions with members of the Herschlag lab as well as Aaron Fafarman in our lab.
Abbreviations
- DMSO
dimethyl sulfoxide
- KSI
ketosteroid isomerase
- LBHB
low-barrier hydrogen bond
- NMR
nuclear magnetic resonance
- pKSI
ketosteroid isomerase from Pseudomonas putida
- tKSI
ketosteroid isomerase from Comamonas testosteroni
- SSHB
short-strong hydrogen bond
Footnotes
This work was supported in part by a grant from the National Institute of Health GM27738.
REFERENCES
- 1.Gerlt JA, Gassman PG. Understanding the rates of certain enzyme-catalyzed reactions: Proton abstraction from carbon acids, acyl transfer reactions, and displacement reactions of phosphodiesters. Biochemistry. 1993;32:11934–11952. doi: 10.1021/bi00096a001. [DOI] [PubMed] [Google Scholar]
- 2.Gerlt JA, Gassman PG. An explanation for rapid enzyme-catalyzed proton abstraction from carbon acids: importance of late transition states in concerted mechanisms. J. Am. Chem. Soc. 1993;115:11552–11568. [Google Scholar]
- 3.Cleland WW, Kreevoy MM. Low-barrier hydrogen bonds and enzymic catalysis. Science. 1994;264:1887–1890. doi: 10.1126/science.8009219. [DOI] [PubMed] [Google Scholar]
- 4.Frey PA, Whitt S, Tobin J. A low-barrier hydrogen bond in the catalytic triad of serine proteases. Science. 1994;264:1927–1930. doi: 10.1126/science.7661899. [DOI] [PubMed] [Google Scholar]
- 5.Pollack RM. Enymatic mechanisms for catalysis of enolization: ketosteroid isomerase. Bioorg. Chem. 2004;32:341–353. doi: 10.1016/j.bioorg.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 6.Choi G, Ha NC, Kim SW, Kim DH, Park S, Oh BH, Choi KY. Asp-99 donates a hydrogen bond not to Tyr-14 but to the steroid directly in the catalytic mechanism of Δ5-3-ketosteriod isomerase from Pseudomonas putida Biotype B. Biochemistry. 2000;39:903–909. doi: 10.1021/bi991579k. [DOI] [PubMed] [Google Scholar]
- 7.Kuliopulos A, Mildvan AS, Shortle D, Talalay P. Kinetic and ultraviolet spectroscopic studies of active-site mutants of Δ5-3-ketosteroid isomerase. Biochemistry. 1989;28:149–159. doi: 10.1021/bi00427a022. [DOI] [PubMed] [Google Scholar]
- 8.Pollack RM, Thornburg LD, Wu LR, Summers MF. Mechanistic insights from the three-dimensional structure of 3-oxo-Δ5-steroid isomerase. Arch. Biochem. Biophys. 1999;370:9–15. doi: 10.1006/abbi.1999.1384. [DOI] [PubMed] [Google Scholar]
- 9.Warshel A, Papazyan A, Kollman PA. On low-barrier hydrogen bonds and enzyme catalysis. Science. 1995;269:102–104. doi: 10.1126/science.7661987. [DOI] [PubMed] [Google Scholar]
- 10.Guthrie JP. Short strong hydrogen bonds: can they explain enzymic catalysis? Chem. Biol. 1996;3:163–170. doi: 10.1016/s1074-5521(96)90258-6. [DOI] [PubMed] [Google Scholar]
- 11.Kraut DA, Sigala PA, Pybus B, Liu CW, Ringe D, Petsko GA, Herschlag D. Testing electrostatic complementarity in enzyme catalysis: hydrogen bonding in the ketosteroid isomerase oxyanion hole. PLoS Biol. 2006;4:e99. doi: 10.1371/journal.pbio.0040099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao Q, Abeygunawardana C, Talalay P, Mildvan A. NMR evidence for the participation of a low-barrier hydrogen bond in the mechanism of Δ5-3-ketosteroid isomerase. Proc. Natl. Acad. Sci. USA. 1996;93:8220–8224. doi: 10.1073/pnas.93.16.8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho HS, Ha NC, Choi G, Kim HJ, Lee D, Oh KS, Kim KS, Lee W, Choi KY, Oh BH. Crystal Structure of Δ5-3-ketosteroid isomerase from Pseudomonas testosteroni in complex with equilenin settles the correct hydrogen bonding scheme for transition state stabilization. J. Bio. Chem. 1999;274:32863–32868. doi: 10.1074/jbc.274.46.32863. [DOI] [PubMed] [Google Scholar]
- 14.Cleland WW. Low-barrier hydrogen bonds and enzymatic catalysis. Arch. Biochem. Biophys. 2000;382:1–5. doi: 10.1006/abbi.2000.2011. [DOI] [PubMed] [Google Scholar]
- 15.Steiner T. The hydrogen bond in the solid state. Angew. Chem. Int. Ed. 2002;41:48–76. doi: 10.1002/1521-3773(20020104)41:1<48::aid-anie48>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 16.Kreevoy MM, Liang TM, Chang KC. Structures and isotopic fractionation factors of complexes AHA−1. J. Am. Chem. Soc. 1977;99:5207–5209. [Google Scholar]
- 17.Kreevoy MM, Liang TM. Structures and isotopic fractionation factors of complexes, A1HA2−1. J. Am. Chem. Soc. 1980;102:3315–3322. [Google Scholar]
- 18.Kim SW, Cha SS, Cho HS, Kim JS, Ha NC, Cho MJ, Joo S, Kim KK, Choi KY, Oh BH. High-resolution crystal structures of delta5-3-ketosteroid isomerase with and without a reaction intermediate analogue. Biochemistry. 1997;36:14030–14036. doi: 10.1021/bi971546+. [DOI] [PubMed] [Google Scholar]
- 19.Anderson S, Crosson S, Moffat K. Short hydrogen bonds in photoactive yellow protein. Acta Crystallog. 2004;D 60:1008–1016. doi: 10.1107/S090744490400616X. [DOI] [PubMed] [Google Scholar]
- 20.Sigala PA, Tsuchida MA, Herschlag D. Hydrogen bond dynamics in the active site of photoactive yellow protein. Proc. Natl. Acad. Sci. USA. 2009;106:9232–9237. doi: 10.1073/pnas.0900168106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamaguchi S, Kamikubo H, Kurihara K, Kuroki R, Niimura N, Shimizu N, Yamazaki Y, Kataoka M. Low-barrier hydrogen bond in photoactive yellow protein. Proc. Natl. Acad. Sci. USA. 2009;106:440–444. doi: 10.1073/pnas.0811882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Viloca M, Gonzalez-Lafont A, Llunch JM. Theoretical study of the low-barrier hydrogen bond in the hydrogen maleate anion in the gas phase. Comparison with normal hydrogen bonds. J. Am. Chem. Soc. 1997;119:1081–1086. [Google Scholar]
- 23.Pan Y, McAllisster MA. Characterization of low-barrier hydrogen bond. 6. Cavity polarity effects on the formic acid-formate anion model system. An ab initio and DFT investigation. J. Am. Chem. Soc. 1997;120:166–169. [Google Scholar]
- 24.Li YK, Kuliopulos A, Mildvan AS, Talalay P. Environments and mechanistic roles of the tyrosine residues of Δ5-3-ketosteroid isomerase. Biochemistry. 1993;32:1816–1824. doi: 10.1021/bi00058a016. [DOI] [PubMed] [Google Scholar]
- 25.Fafarman AT, Sigala PA, Herschlag D, Boxer SG. in preparation. [Google Scholar]
- 26.Childs W, Boxer SG. in preparation. [Google Scholar]
- 27. http://www.scripps.edu/~cdputnam/protcalc.html.
- 28.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 29.Hawkinson DC, Pollack RM, Ambulos NP., Jr Evaluation of the internal equilibrium constant for 3-oxo-delta 5-steroid isomerase using the D38E and D38N mutations: the energetic basis for catalysis. Biochemistry. 1994;33:12172–12183. doi: 10.1021/bi00206a021. [DOI] [PubMed] [Google Scholar]
- 30.Petrounia IP, Blotny G, Pollack RM. Binding of 2-napthols to D38E mutants of 3-oxo-Δ5-steroid isomerase: variation of ligand ionization state with the nature of the electrophilic component. Biochemistry. 2000;39:110–116. doi: 10.1021/bi9917838. [DOI] [PubMed] [Google Scholar]
- 31.Xue L, Kuliopulos A, Mildvan AS, Talalay P. Catalytic mechanism of an active-site mutant (D38N) of Δ 5-3-ketosteroid isomerase. Biochemistry. 1991;30:4991–4997. doi: 10.1021/bi00234a022. [DOI] [PubMed] [Google Scholar]
- 32.Kim DH, Nam GH, Jang DS, Choi G, Joo S, Kim JS, Oh BH, Choi KY. Roles of active site aromatic residues in catalysis by ketosteroid isomerase from Pseudomonas putida Biotype B. Biochemistry. 1999;38:13810–13819. doi: 10.1021/bi991040m. [DOI] [PubMed] [Google Scholar]
- 33.Zeng B, Pollack RM. Microscopic rate constants for the acetate ion catalyzed isomerization of 5-androstene-3,17-dione to 4-androstene-3,17-dione: a model for steroid isomerase. J. Am. Chem. Soc. 1991;113:3838–3842. [Google Scholar]
- 34.Shan SO, Loh S, Herschlag D. The energetics of hydrogen bonds in model systems: Implications for enzymatic catalysis. Science. 1996;272:97–101. doi: 10.1126/science.272.5258.97. [DOI] [PubMed] [Google Scholar]
- 35.Kumar GA, McAllister MA. Characterization of low-barrier hydrogen bonds. 8. Substituent effects on the strength and geometry of the formic acid-formate anion model system. An ab initio and DFT investigation. J. Am. Chem. Soc. 1998;120:3159–3165. [Google Scholar]
- 36.Feierberg I, Åqvist J. The catalytic power of ketosteroid isomerase investigated by computer simulation. Biochemistry. 2002;41:15728–15735. doi: 10.1021/bi026873i. [DOI] [PubMed] [Google Scholar]