Abstract
DNA repair is a double-edged sword in stem cells. It protects normal stem cells in both embryonic and adult tissues from genetic damage, thus allowing perpetuation of intact genomes into new tissues. Fast and efficient DNA repair mechanisms have evolved in normal stem and progenitor cells. Upon differentiation, a certain degree of somatic mutations becomes more acceptable and, consequently, DNA repair dims. DNA repair turns into a problem when stem cells transform and become cancerous. Transformed stem cells drive growth of a number of tumours (e.g., high grade gliomas) and being particularly resistant to chemo- and radiotherapeutic agents often cause relapses. The contribution of DNA repair to resistance of these tumour-driving cells is the subject of intense research, in order to find novel agents that may sensitize them to chemotherapy and radiotherapy.
1. Introduction
Endogenous damage (e.g., oxidative metabolism linked) and external exposures (e.g., environmental pollution linked) all damage DNA causing a number of modifications including base and backbone alterations, single strand breaks (SSB) and double strand breaks (DSB) that may limit survival and the regenerative potential of both embryonic stem cells (ESC) and adult stem cells (ASC). ESC differentiate to all cell types in the mammalian body, including germ line cells. The maintenance of genomic stability in ESC must be stringent, any genetic alterations in those progenitor cells compromising the genomic stability and functionality of entire cell lineages. Consistently, the mutation rate and the frequency of mitotic recombination are lower in murine ESC than in adult somatic cells or isogenic mouse embryonic fibroblasts (MEF). For instance, the frequency of spontaneous mutation at the aprt gene is around 10−6 in ESC and 100-fold higher (~10−4) in MEF [1]. Mechanisms of mutagenesis differ as well. Most mutation events involve loss of heterozigosity (LOH) in both ESC and MEF but LOH is generated mainly through nondisjunction in ESC and through mitotic recombination in MEF [2]. Similarly, when spontaneous mutation is assessed at the X-linked locus hprt, it is undetectable in ESC (<10−8) and ~10−5 in MEF. Hence, robust mechanisms counteracting spontaneous mutagenesis may exist in ESC and DNA repair is likely one of them [2–6]. On the other hand, ASC are important in the long-term maintenance of tissues throughout life [7]. For instance, the effector cells of the blood have limited lifetimes and must be replenished continuously throughout life from a small reserve of hematopoietic stem cells in the bone marrow. Although the replicative potential of hematopoietic stem cells may be finite, studies conducted in murine genetic models indicate that DNA repair is critical to the longevity and stress response of the hematopoietic stem cell pool [8]. This likely applies to other ASC types including mesenchymal stem cells (MSC) [9].
2. The Bright Side: DNA Repair in Normal Stem Cells
2.1. ESC
Using single-cell gel electrophoresis (SCGE) Maynard and coworkers [5] found that human ESC have more efficient repair of different types of DNA damage [generated from H2O2, ultraviolet (UV)-C, ionizing radiation (IR) or psoralen] than human primary fibroblasts and, with the exception of UV-C damage, HeLa cells (Table 1). A microarray gene expression analysis showed that the mRNA levels of several DNA repair genes, including some involved in DNA base excision repair (BER) and interstrand crosslink (ICL) repair, were elevated in human ESC compared with their differentiated forms [embryoid bodies (EB)]. Hence, multiple DNA repair pathways are over-regulated in human ESC, relative to differentiated human cells [5]. Consistently, the expression of antioxidant and DNA repair genes was reduced and the DNA damage levels increased during spontaneous differentiation of two human ESC lines [10]. Replicating chromatin in ESC is particularly vulnerable to strand breaks [11]. Two pathways seal DSBs in mammalian cells, including ESC: non homologous end joining (NHEJ) that is the main pathway and homology-directed repair (HDR) that may act as a backup in the absence of NHEJ components [12]. The expression of strand break repair genes such as Rad51 fades when murine ESC differentiate [2, 10, 13] (Table 1). Despite their high expression of O6 methylGua DNA methyltransferase (MGMT), murine ESC undergo apoptosis at much higher frequency than differentiated cells after treatment with N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) [14] (Table 1). This is due to elevated expression in ESC of the mismatch repair (MMR) proteins MSH2 and MSH6 that trigger futile cycles of O6 methylguanine repair/replication [15]. It has been hypothesized that the high apoptotic response of murine ESC may contribute to reduce the mutational load in these cells [14, 16, 17] (Table 1). Embryonic Fanconi anemia (FA) neural stem cells (NSC) have a reduced capacity to self-renew in vitro [18, 19] and the expression of FA G and FA L (but not FA C) genes is enhanced in human ESC relative to human EB [5]. In summary, 7 out of 9 (78%) studies indicate that ESC possess more elevated DNA repair capacity than their differentiated derivatives (Table 1).
Table 1.
DNA repair capacity in embryonic, adult and mesenchymal stem versus differentiated cells.
| Stem cell system | DNA repair mechanism/enzyme | Higher | Similar | Lower | Remarks | Ref. |
|---|---|---|---|---|---|---|
| ESC | ||||||
| Human ESC | Expression of double strand break repair genes | √ | Compared to human embryoid bodies and fibroblasts | [5] | ||
| Human ESC | Expression of BER genes | √ | Compared to human embryoid bodies and fibroblasts | [5] | ||
| Human ESC | Expression of NER genes | √ | Compared to human embryoid bodies and fibroblasts | [5] | ||
| Human ESC | Expression of antioxidant and DNA repair genes | √ | Compared to differentiated cells | [10] | ||
| Murine ESC | Expression of strand break repair genes | √ | Compared to differentiated cells | [1, 2] | ||
| Murine ESC | Expression of antioxidant and strand break repair genes | √ | Compared to differentiated cells | [13] | ||
| Murine ESC | NER | √ | Strong apoptosis | [16] | ||
| Murine ESC | NER | √ | Strong apoptosisS-phase delay | [17] | ||
| Murine ESC | MGMTMMR | √ | Compared to differentiated cells Highly sensitive to methylation damageStrong apoptosis | [14] | ||
| ASC | ||||||
| Human CD34+ 38− hematopoietic stem cells | Removal of ENU or melphalan-induced DNA adducts | √ | Compared to progenitor or mature cellsResistant to DNA-reactive drugs | [20] | ||
| Human CD34+ 38− hematopoietic stem cells | Resealing of strand breaks and repair gaps | √ | Compared to progenitor or mature cellsResistant to DNA-reactive drugs | [20] | ||
| Human CD 34+ hematopoietic stem cells | KU70 expression | √ | Negatively correlated with donor age | [21] | ||
| Human cells of the monocytic lineage | NER | √ | Compared to macrophages | [22] | ||
| Human neural precursors | NER | √ | Compared to neurons | [23, 24] | ||
| Human cycling CD34+ hematopoietic stem cells | MGMTMMR | √ | Compared to mature CD34− cellsNormally sensitive to methylation damage | [25] | ||
| Murine neural precursors | √ | Sensitive to IR | [26] | |||
| Stem cells in the subventricular zone and oligodendrocyte precursor cells in rodent and human brain | √ | Sensitive to IR | [27] | |||
| Murine neural stem/progenitor cells | BER (OGG1) | √ | Compared to differentiated cells | [28] | ||
| Murine neural stem/progenitor cells | BER (NEIL3) | √ | Compared to differentiated cells | [29, 30] | ||
| Murine fetal hematopoietic cells | BER (DNA pol β) | √ | Compared to adult hematopoietic cellsLow point mutation frequency | [31] | ||
| Murine myoblasts | BER (DNA ligase I and XRCC1) | √ | Compared to myotubes | [32] | ||
| Murine keratinocytes | NER | √ | [33] | |||
| MSC | ||||||
| Human fetal MSC | DNA repair gene expression | √ | Compared to adult MSC | [34] | ||
| Human MSC from bone marrow transplantation patients | DSB repair | √ | Compared to lung or breast cancer cellsResistant to IR | [35] | ||
| Human MSC obtained from bone marrow transplantation patients | ROS-scavenging capacity | √ | Resistant to IR | [35] | ||
| Human MSC | Single and double-strand break repair | √ | Resistant to IRHigh telomerase activity | [36] | ||
| Rat MSC | DNA repair gene expression | √ | Compared to senescent MSC | [9] | ||
2.2. ASC
The repair capacity for DNA strand breaks diminishes during maturation of cells of the human lymphohematopoietic system. Bracker and coworkers [20] have studied the variation of DNA repair capacity and expression of DNA repair genes during maturation of hematopoietic cells treated with ENU or melphalan (Table 1). The removal of DNA adducts, the resealing of strand breaks and the resistance to DNA-reactive drugs were higher in stem (CD34+ 38−) than in mature (CD34−) or progenitor (CD34+ 38+) cells isolated from umbilical cord blood from the same individual [20]. Hence, slow dividing stem cells may be protected by extensive DNA repair while more mature and less valuable cells, if damaged, could be rather eliminated by apoptosis. The NHEJ components such as the Ku70 protein are downregulated during ageing of the hematopoietic stem cell donor. Ku70 expression shows highest levels in newborn, 2.6-fold lower levels in young (mean age: 30 years) and 6.3-fold lower levels in old (mean age: 87.6 years) donors [21]. Transcription-coupled repair (TCR) is a subpathway of nucleotide excision repair (NER) that preferentially repairs the transcribed strand of active genes, as compared to the non-transcribed strand, thus providing cells with a smart mechanism of safeguard of expressed genes [37]. Likewise strand break repair and BER, NER is strongly downregulated in several human cell types undergoing differentiation (e.g., in cells of the monocytic lineage when they differentiate to macrophages or NSC differentiating to neurons) [23, 24, 38] (Table 1). Attenuation of NER during differentiation results from lack of ubiquitination of NER proteins that in turn is linked to differences in phosphorylation of the ubiquitin-activating enzyme E1 [24]. To ensure proficient repair of active genes, besides TCR, a second specialized mechanism termed differentiation-associated repair (DAR) exists in differentiated cells. DAR recruits to transcribed genes of differentiated cells the remaining proficient NER enzymes not yet engaged in TCR that eventually repair both DNA strands [24]. Hence, DAR may be considered as a subpathway of global genome NER, focussing on the chromatin domains of differentiated cells within which transcription occurs [22, 24]. In human hematopoietic cord blood cells, Casorelli et al. [25] found no difference in sensitivity to methylating agents between cycling CD34+ (stem) and CD34− (mature) cells (Table 1). In this study, MGMT significantly protected against N-Methyl-N-Nitrosourea (MNU) toxicity while MMR enhanced as expected the MNU sensitivity of the cells by processing O6-methylguanine into a lethal lesion [15, 25]. The important role of DSB repair in maintenance of hematopoietic stem cell function has been recently emphasized in two murine studies [8, 39, 40]. Progressive loss of hematopoietic stem cells and a decrease of bone marrow cell count were observed in mice with defective DSB repair and stem cell function in tissue culture and transplantation was severely impaired [40]. Stem cells in the central nervous system (CNS) might behave differently. No resistance of neural precursors to IR was observed in two studies [26, 27] (Table 1). Nowak and coworkers [26] reported massive apoptosis of neural precursors but not of neurons in the developing mouse brain after gamma-irradiation while the level of induced damage was similar in the two cell types. In the second study, Panagiotakos et al. [27] have observed a specific association between radiation injury and irreversible damage to stem cells in the subventricular zone (SVZ) and loss of oligodendrocyte progenitor cells (OPC) in both rodent and human brain. Hence unlike hematopoietic precursors, DSB repair might be less efficient in the neural precursors in relation to their greater radiosensitivity. Expression of 8-oxoguanine DNA glycosylase (OGG1) is elevated in regions of the neonatal mouse brain that are rich in neural stem/progenitor cells, namely the medial wall of the lateral ventricle and the SVZ [28] (Table 1). Both the expression and the activity of OGG1 are high in neural stem/progenitor cells from newborn mice and decrease in adult animals and upon induction of cell differentiation. Enhanced expression of OGG1 and other BER enzymes may protect neural stem/progenitor cells from oxidatively damaged DNA [28]. Murine Nei endonuclease VIII-like 3 (Neil3) glycosylase follows as well an expression pattern involving brain regions harbouring stem cell populations [29, 30] (Table 1). The glycosylase activities are stable through prolonged in vitro culture required for expansion of stem cells to clinically relevant numbers [41]. Downregulation of oxidatively damaged DNA repair genes and a concomitant increase in 8-oxoguanine (8-oxoGua) DNA levels during differentiation of mouse proliferating (myoblasts) to terminally differentiated (myotubes) muscle cells have been described by Narciso and coworkers [32] (Table 1). Both short and long patch BER pathways were impaired in myotubes. The defect in BER was linked to the nearly complete lack of DNA ligase I and to the strong down-regulation of XRCC1 with consequent destabilization of DNA ligase III [32]. The FA pathway exerts a central role in neural stem and progenitor cells during developmental and adult neurogenesis [18, 19]. Reduced proliferation of neural progenitor cells and enhanced NSC depletion has been observed in ageing FA mice. Lower reactive oxygen species (ROS) levels may be achieved in NSC by higher expression of key antioxidant enzymes involved in basal mitochondrial metabolism (e.g., uncoupling protein 2 (UCP2) and glutathione peroxidase (GPX)) as compared to postmitotic neural cells [42]. Following exposure to the mitochondrial toxin 3-nitropropionic acid and unlike postmitotic cells, NSC fastly upregulate UCP2, GPX and superoxide dismutase 2 and successfully recover. Thus, a fast response of antioxidant enzymes may represent an important “vigilance” mechanism of NSC to counteract oxidatively damaged DNA in the CNS. Summary reckoning indicates that in 8 out of 13 (61%) studies, ASC display more elevated DNA repair capacity than mature cells (Table 1; [31, 33]).
2.3. MSC
The gene expression profiles of undifferentiated MSC derived from first trimester fetal liver and adult bone marrow were compared by serial analysis of gene expression, and validated by either reverse transcription polymerase chain reaction or immunoblotting of selected genes [34]. Transcripts implicated in cell cycle promotion, chromatin regulation and DNA repair were more abundant in fetal than in adult MSC (Table 1). Likewise, MSC obtained from bone marrow transplantation patients display increased DSB repair capacity and resistance to IR and possess elevated ROS-scavenging capacity as compared to lung cancer and breast cancer cells [35] (Table 1). Telomerase activity is an additional mechanism by which MSC may resist IR damage. Telomerase-immortalized derivatives of human MSC have been found IR-resistant as compared to primary stem cells while DNA repair capacity was similar in the two cell types [36] (Table 1). Considering the aforementioned study by Galderisi and coworkers on senescence of MSC [9], 4 out of 5 (80%) studies indicate that DNA repair is elevated in MSC although appropriate comparison to differentiated cells is not available.
3. The Dark Side: DNA Repair in Cancer Stem Cells
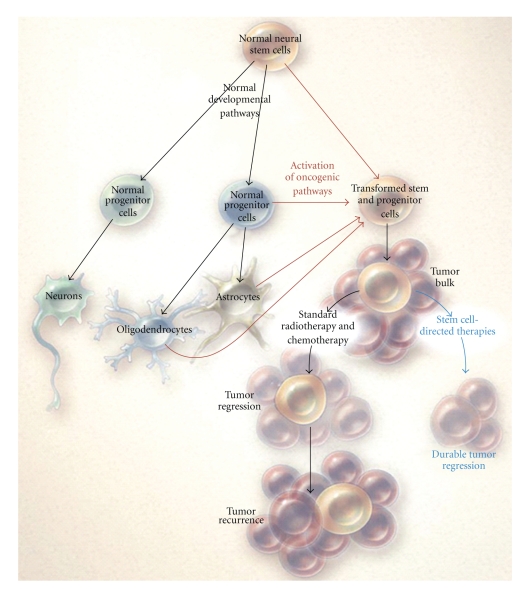
Stem cells and cancer cells share some features: similar signalling pathways may regulate self-renewal in stem cells and cancer cells, and cancer cells may include “cancer stem cells”—rare cells with indefinite potential for self renewal and differentiation that drive tumorigenesis [56]. A detailed knowledge of the biological distinctness of cancer stem cells may be crucial for the development of specific therapies aimed to tumour eradication [57–59]. In particular, the existence of cells endowed with features of primitive progenitor cells and tumor-initiating function has been demonstrated in high grade gliomas [60] (Figure 1).
Figure 1.
Resistance in GSC. Normal NSC self-renew and give rise to multipotential progenitor cells that form neurons, oligodendroglia, and astrocytes. GSC arise from the transformation of either NSC or progenitor cells (red) or, less likely, from de-differentiation of oligodendrocytes or astrocytes (thin red arrows) and lead to malignant gliomas. GSC are relatively resistant to standard treatments such as radiation and chemotherapy and lead to regrowth of the tumor after treatment. Therapies directed at stem cells can deplete these cells and potentially lead to more durable tumor regression (blue) (from [60] with permission).
3.1. High Grade Gliomas
Despite aggressive surgical resections using preoperative and intraoperative neuroimaging, along with recent advances in radiotherapy and chemotherapy, the prognosis for high grade glioma patients remains dismal, the median survival being 24–60 months for patients with anaplastic astrocytoma (World Health Organization (WHO) grade III) and 12–15 months for patients with glioblastoma multiforme (WHO grade IV) [60, 61]. Established prognostic factors are limited and include age at diagnosis, Karnofsky performance status, extent of surgery and possibly MGMT promoter methylation [60, 62]. Standard treatment includes resection of >95% of the tumor, followed by concurrent chemotherapy (usually performed with the alkylating agent temozolomide (TMZ)) and radiotherapy. Malignant gliomas are associated with such dismal prognoses in part because glioma cells can actively migrate through the brain, often travelling relatively long distances, making them elusive targets for effective surgical removal and almost invariable sources of relapse [63]. In children, the management of high grade gliomas remains an even greater challenge for neuro-oncologists in part because of the greater vulnerability of the developing brain to treatment-related toxicity [64].
3.2. Origins of Glioma Stem Cells
Although the brain is completely formed and structured few weeks after birth, it maintains a degree of plasticity throughout life, including axonal remodeling, synaptogenesis, but also neural cell birth, migration and integration [65]. The SVZ and the dentate gyrus of the hippocampus are the two main neurogenic sites in the adult brain. NSC reside in these structures and produce progenitors that possess migratory ability. One current fashionable model suggests that gliomas may arise from the transformation of neural stem or progenitor cells, originating cancer cells that are undifferentiated, self-renewing, with the capacity for driving tumor development and designated glioma stem cells (GSC), because of their stem-like properties. The origin of GSC has been investigated by activating oncogenic K-RAS in mouse neuronal precursor cells and adult SVZ cells [66]. K-RAS-activated mice showed a marked expansion of stem cell populations in the SVZ and developed intermediate grade, infiltrating glioma with 100% penetrance. Tumors were consistently located in the amygdalohippocampal region and nearby cortex and tumor cells expressed markers associated with neural progenitor cells, including OLIG2, BMI1, and platelet-derived growth factor (PDGF) receptor-alpha. Therefore, infiltrating tumor cells arose in this study from NSC transformed by oncogenic activation in vivo [66]. In a different approach, a mouse strain where tumor induction would be restricted to myelinating OPC was generated [67]. PDGF-B transfer to OPC could induce gliomas with an incidence of 33%. The majority of tumors resembled human WHO grade II oligodendroglioma based on close similarities in histopathology and expression of cellular markers. Thus, in this system OPC could act as cell of origin for experimental glioma [67]. The generation of mouse gliomas following the overexpression of PDGF-B in embryonic neural progenitors has been described by Appolloni et al. as well [68]. Histopathological, immunohistochemical and genome-wide expression features of PDGF-B induced tumors were surprisingly uniform, despite they were generated by transducing a highly heterogeneous population of progenitor cells known for their ability to produce all the cell types of CNS. This uniformity is likely due to the ability of PDGF-B overexpression to respecify competent embryonic NSC toward the oligodendroglial lineage. PDGF-B-induced tumors harbored different proliferating cell populations but only PDGF-B-overexpressing cells were tumorigenic [68]. The possibility that GSC originate in some cases from de-differentiation of tumoural cells cannot yet be ruled out [69, 70]. It has been recently observed that the reversion of mature astrocytes to an embryonic state is sufficient to sensitize them to oncogenic stress [71]. Prolonged exposure of astrocytes to transforming growth factor (TGF)-alpha is sufficient to trigger their reversion to a neural progenitor-like state (de-differentiated astrocytes). When dedifferentiated astrocytes were grafted intra-cerebrally, they showed the same cytogenomic profile as astrocytes, survived in vivo and did not give birth to tumors. After exposure to IR yet, they acquired cancerous properties: they were immortalized, showed cytogenomic abnormalities, and formed high-grade glioma-like tumors after brain grafting [71]. Anyway, whatever the GSC origin, malignancy of gliomas often correlates with the stem phenotype [43–47, 50, 51] (Table 2) and accordingly, if only a subset of glioma cells drives tumour progression it is important to target it specifically (Figure 1) [60, 72, 73].
Table 2.
Stem cell involvement in high grade glioma patient's outcome.
| Stem cell marker | Clinical model | Animal model | Association to poor outcome | Ref. | ||
|---|---|---|---|---|---|---|
| CD133 | Human bulk gliomas | Yes | [43] | |||
| CD133 | Gliomas of various grade and histology | Yes | [44] | |||
| CD133 | Low-grade and high-grade glioma specimens | Yes | [45] | |||
| CD133 | Cell lines from GBM specimens established under neural stem cell conditions | Yes | [46] | |||
| CD133 | U251 human glioma cells with knocked-down CD133 | Yes | [47] | |||
| CD133 | Two types of GSC within different regions of the same human GBM | No | [48] | |||
| Expression signature dominated by HOX genes, which comprises CD133 | GBM from initial surgery or resected at recurrence | Yes | [49] | |||
| CD133, Nestin, Sox-2, Musashi-1, CXCR4, Flt-4/VEGFR-3 and CD105/Endoglin | Astrocytomas of different WHO grades | Yes | [50] | |||
| Sox2, Musashi-1, nestin | Surgical specimens of human gliomatosis cerebri | Yes | [51] | |||
| Renewable neurosphere formation | Cultured human gliomas | Yes | [52] | |||
| CD133 | Mouse line whose CD133-expressing cells can be eliminated conditionally | No | [53] | |||
| CD133 | Rat C6 cell line | No | [54] | |||
| CD133, nestin | Rat N29 and N32 experimental gliomas | Yes | [55] | |||
3.3. What are GSC?
Although methods of deriving GSC from glioma tumours and sorting them according to certain marker expression (CD133, Nestin, Sox-2 and Musashi-1) have been described [50, 75, 76], the exact identity of GSC remain elusive [70]. The most extensive of the limited data on this topic relate to GSC identified by the surface marker CD133 (Table 2). The stem cell marker CD133/prominin-1 is a a five transmembrane domain glycoprotein which has been identified as a cancer stem cell marker in several solid tumor types, including those of the brain. CD133 is often expressed on the surface of human GSC and CD133-positive cells may represent >85% of certain human and animal glioma cell lines [55, 77]. To verify if CD133-expressing cells are essential for tumorigenesis, a glioma-initiating mouse cell line whose CD133-positive cells can be eliminated conditionally by a Cre-inducible diphtheria toxin fragment A (DTA) gene on the CD133 locus, was generated by Nishide and coworkers [53]. After induction of the DTA gene, the cell line maintained the capacity to form tumour spheres culture and transplantable multiform glioblastoma (GBM) in vivo. Hence at least in this mouse model, CD133-expressing cells are dispensable for gliomagenesis [53]. The presence of two types of GSC within different regions of the same human GBM supports this conclusion [48]. Cytogenetic and molecular analysis showed that the two types of GSC bore quite diverse tumorigenic potential and distinct genetic anomalies, and yet, CD133 expression was similar. Those two GSC populations might represent distinct cell targets, with a differential therapeutic importance independently of CD133 expression [48]. CD133 failed to identify the total cancer stem-like cell population in the rat C6 glioma cell line, since both CD133-positive and CD133-negative cells displayed cancer stem-like cell fractions showing characteristics of self-renewal, multilineage differentiation potentials in vitro, and tumorigenic capacity in vivo [54]. Methodological problems concerning purification of CD133-expressing cells may exist. It has been recently reported that the specific expression and enrichment of CD133 can be obtained in fresh human gliomas and gliomasphere cultures purified by fluorescence activating cell sorting while purification of CD133-positive GSC using the widely used CD133-microbeads may be affected by lack of specificity and lead to mixed populations [78]. In conclusion, CD133-positive GSC likely drive only an as yet unquantified subset of malignant gliomas, the remainder deriving from CD133-negative GSC with distinct phenotypical features [46, 79]. The neurosphere formation ability and tumorigenic capacity of cultured glioma cells have been recently explored as independent predictors of patient's outcome [52]. Those features of cultured glioma cells significantly correlated with an increased hazard of patient's death and more rapid tumor progression independently of Ki67 proliferation index [52]. These results suggest that the ability to propagate brain tumor stem cells in vitro is associated with clinical outcome although the lengthy duration of this assay may preclude direct clinical application. The ultrastructural features of GSC isolated from both a primary glioma tissue and the human glioma cell line SHG-44 have been described [80]. The ultrastructural features of the two kinds of GSC were similar, with relatively developed mitochondria, Golgi apparatuses, ribosomes, undeveloped rough endoplasmic reticula, rare lysosomes and no typical autophagosomes, and high nuclear-cytoplasmic ratio. Their nuclei, frequently containing huge amounts of euchromatin and a small quantity of heterochromatin, were mostly globular and the majority of them had only one nucleole. Typical apoptotic cells could hardly be found in tumor spheres, and between adjacent cells there were cell junctions, which probably were incompletely developed desmosomes or intermediate junctions [80]. A definite (and sometimes disregarded) feature of GSC is their ability to differentiate. This is the only parameter that unequivocally permitted, in our experience, to distinguish stem from nonstem glioma cells [77]. In this study, stem cells cultured as adherent monolayers conveniently allowed morphologic analysis of differentiation under the inverted light microscope. Removal of growth factors and addition of foetal calf serum (FCS-differentiating conditions) resulted after 3 weeks in acquisition of a typical astroglial morphology by genuine GSC. Recent evidence shows that even single GSC possess multilineage potential [81]. Analysis of marker expression usually confirms the morphologic analysis. For instance glial fibrillary acidic protein (GFAP) is an intermediate filament protein that is typical, although not exclusive, of cells of astroglial lineage and may indicate that differentiation of stem cultures is mostly oriented to astroglial commitment. In a sense, nothing is better than visual inspection for morphological changes upon growth factors removal to determine whether cells are bona fide stem or not. Tumourigenic glioma cells unable to acquire any of the astrocyte, neuron, or oligodendrocyte morphology upon growth factors removal may be called “tumour initiating cells” or “tumour-driving cells” but not tumour stem cells, even if they express so-called “stemness” markers.
3.4. Deregulated Pathways in GSC
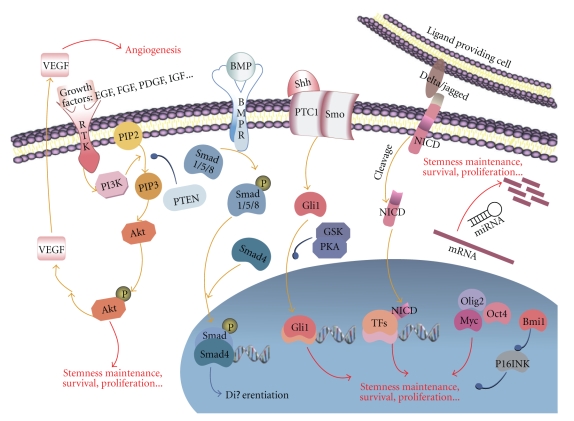
Invasive malignant glioma cells often show a decrease in their proliferation rates and a relative resistance to apoptosis that may underlie their resistance to conventional chemotherapy and radiotherapy [61]. Invasive growth and resistance to apoptosis results from changes at the genomic, transcriptional and post-transcriptional level of a number of cellular factors involved in complex signal pathways (reviewed in [74]) (Figure 2). For instance, the proliferation of normal stem and progenitor cells in the brain is under control of p53 [84]. The altered expression of several cell cycle regulators, in particular a pronounced downregulation of p21, has been observed in p53-mutant NSC indicating that p53 may act as a growth suppressor of GSC [85]. Deregulation of a number of additional cell cycle control pathways, including the p16-CDK4-RB pathway may underlie the generation of GSC in the brain [86, 87]. Activation of signaling pathways like the PDGF pathway [88], often accompanied by Ras inactivation [89], has been implicated in transformation of SVZ NSC (Figure 2). Another contributor is the Sonic Hedgehog (Shh) pathway that regulates the patterning, proliferation and survival of NSC within the CNS [90]. Shh signalling is mediated by Gli1 [91] that likely serves as a protective mechanism against premature mitosis in normal NSC. Deregulation of Gli1 has been observed in GSC [91] (Figure 2). Similarly, the WNT Notch and TGF-beta/Bone Morphogenetic Protein (BMP) developmental pathways have been also found aberrantly expressed in GSC [92]. Secretion of the angiogenic factor vascular endothelial growth factor (VEGF) by GSC has been observed and this phenomenon is further induced by hypoxia [93, 94] (Figure 2). Monoclonal antibodies and low molecular-weight kinase inhibitors of some of the above pathways may be of help in targeting GBM. For instance, the anti-VEGF neutralizing antibody bevacizumab limits the proangiogenic effects of GSC and may suppress the growth of GSC-derived xenografts in some cases [95]. However, most clinical trials of these agents as monotherapies have failed to demonstrate significant survival benefit, likely linked to the complexity of GBM biology that is characterized by a constantly changing microenvironment that greatly influences both tumor growth and response to therapy [63, 96].
Figure 2.
Complex signal pathways and cellular factors regulate GSC. GSC are controlled at multiple levels by complicated regulatory networks. Signals initiated by receptor tyrosine kinases (RTK), bone morphogenetic protein receptors (BMPR), Hedgehog, and Notch result in complicated intracellular events to help balance self-renewal and differentiation of GSC as well as the promotion of cell survival and proliferation. Intracellular regulators including transcriptional factors (Olig2, Myc, Oct4, etc.), epigenetic modifiers (Bmi1), and microRNAs are also highly potent of maintaining GSC populations due to their ability to regulate massive downstream targets simultaneously (from [74] with permission).
3.5. DNA Repair as a Resistance Mechanism in GSC
Whether DNA repair is a major mechanism of resistance to apoptosis in cancer stem cells, likewise normal stem cells, still is an open question [6, 97]. For instance, myeloid progenitor bone marrow cells derived from BER-defective mice display unexpected bone marrow alkylation resistance as compared to progenitor cells from wild-type mice [98]. Hence, in this case, repairing the damaged base seems more lethal than leaving it unprocessed. The phosphatidylinositol-3-kinase (PI3K)-Akt pathway, a protumorigenic signaling cascade involved in several human cancers, is frequently up regulated in gliomas (Figure 2) (reviewed in [99]). Hyperactivation of the PI3K-Akt pathway occurs in gliomas through a variety of mechanisms, including loss of the inhibitory effects of the phosphatase and tensin homolog (PTEN) tumor suppressor [100]. Enhanced Akt signaling may cause a tumourigenic phenotype with increased cell proliferation, metastasis and angiogenesis. Akt inhibitors may significantly reduce viability of GSC relative to matched non stem cells [101] and sensitize them to chemotherapeutic agents [102]. Inhibiton of the Akt pathway further causes delayed repair of IR-induced DSB detected by gamma-H2AX foci formation and radiosensitization [103]. Hence activation of the Akt signaling may underlie at least some cases of radioresistance in GSC. Another route involves the activation of the DNA damage checkpoint response. Tumour cells expressing CD133 increase in gliomas treated with IR. CD133-positive GSC are more resistant than CD133-negative cells to IR in both in vitro experiments or when expanded in the brains of NOD/SCID mice and activate the DNA damage checkpoint response with unusual intensity [104]. Increased DNA repair capacity, evaluated by the SCGE assay has been also reported [104, 105]. Debromohymenialdisine (DBH), a specific inhibitor of the Chk1 and Chk2 checkpoint kinases, delayed the checkpoint response and reversed the resistant phenotype [104]. Similarly, IR-mediated phosphorylation of Chk1 and Chk2 have been found distinctly higher in undifferentiated human embryonal carcinoma cells compared with differentiated cells [106]. G2 and G1-arrested NSC rapidly increase after IR in the mouse, with concomitant phosphorylation of cdc2 and p53 and inhibition of Notch [107]. Hence, although the selective resistance of GSC has been questioned [108], CD133-positive cells may represent the cellular population that resists to IR and chemotherapy at least in a subset of glioma tumours and activation of the DNA damage checkpoint response may be a major underlying mechanism in this regard [49, 77, 104]. On the contrary, we could not confirm enhanced DNA repair capacity in GSC [77]. It should be pointed out that the concept “enhanced DNA repair capacity” implies “per time unit”. Five GSC lines isolated from primary grade IV gliomas were examined for their cell cycle and DNA repair features [77]. The population doubling time of the GSC lines was significantly longer as compared to nonstem cell lines while no enhanced DNA repair examined by an in vitro BER assay, the SCGE assay or resolution of pH2AX nuclear foci, could be observed. The DNA damage checkpoint response was constitutively activated in CD133-positive but not in CD133-negative cells, with Chk1 and Chk2 kinases phosphorylated in the absence of treatment. Recent studies indicate cell cycle restriction as a mechanism of resistance in leukaemia as well [109]. Hence, constitutive activation of the DNA damage checkpoint response may confer GSC increased time for the removal of radio- and chemotherapeutic- induced DNA lesions before arrival of the replication fork while whether improved efficacy of repair (per time unit) is a common resistance mechanism in GSC still requires further studies.
3.6. Targeting the Checkpoint Response in GSC
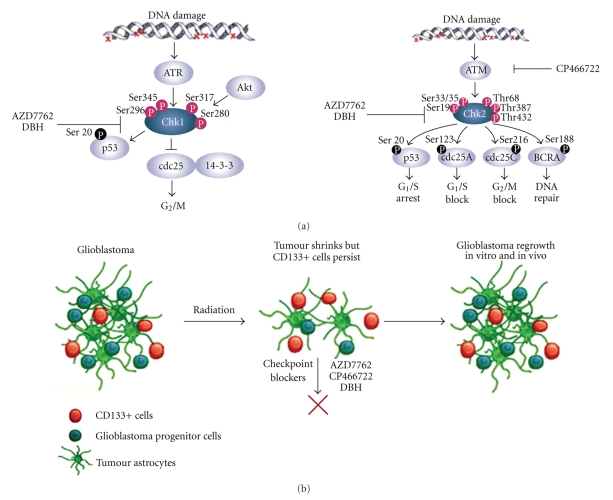
Once DNA damage is identified with the aid of sensors, the checkpoint transducers ATM (ataxia-telangiectasia mutated) and ATR (ATM and Rad3-related) proteins undergo conformational change and/or localisation, resulting in their activation. ATM and ATR phosphorylate the downstream effectors checkpoint kinase-2 (Chk2) and checkpoint kinase-1 (Chk1) respectively, two serine/threonine kinases that serve as functional analogues (reviewed in [110, 111]) (Figure 3(a)). Significant crosstalk exists between the two pathways [112]. Sites of Chk2 phosphorylation by ATM include Ser 33/35, Thr68, Ser19, Thr387 and Thr432 while ATR phosphorylates Chk1 mainly at serines 280, 296, 317 and 345. The first and second-level transducers ATM, ATR, Chk1 and Chk2 phosphorylate a number of effector molecules, such as p53 and CDC25 phosphatases. In particular, CDC25 phosphatases are key checkpoint kinase targets for controlling cell cycle transitions. Human cells have three CDC25 proteins that regulate cell cycle transitions by removing the inhibitory phosphorylation from cyclin-dependent kinases (CDKs). Chk1 and Chk2 phosphorylation of the CDC25 proteins inhibits their activity through either ubiquitin-mediated degradation or cytoplasmic sequestration and prevents CDK activation. This negative regulation of the CDC25 phosphatases is a major checkpoint mechanism for entry of cells into mitosis.
Figure 3.
Cell cycle checkpoint pathways, possible targets in GSC. (a) Once DNA damage is identified with the aid of sensors, the checkpoint transducers ATM and ATR undergo conformational change and/or localisation, resulting in their activation. ATM and ATR activate a series of downstream molecules, including the checkpoint kinases Chk1 and Chk2. The latter inactivate CDC25 phosphatases, culminating in cell cycle arrest. AZD7762 (AstraZeneca) and DBH are specific inhibitors of Chk1 and Chk2 kinases. CP466722 (Pfizer) is a specific inhibitor of ATM (modified from [82] with permission). (b) Targeting GSC may yield durable tumor regression. Glioblastomas are heterogeneous tumours containing CD133-positive GSC among other, more differentiated, CD133-negative cells, including glioblastoma progenitor cells. Following radiation, the bulk glioblastoma responds and the tumour shrinks but CD133-positive cells activate checkpoint controls for DNA repair more strongly than CD133-negative cells, resist radiation and prompt the tumour to regrow. These cells could be targeted with DNA-checkpoint blockers (e.g., AZD7762, CP466722 and DBH) to render them radiosensitive (modified from [83] with permission).
Releasing the constitutively-pressed brake that prevents GSC from dividing may push them into cell cycle and sensitize them to IR and chemotherapeutic agents such as TMZ that primarily act by damaging DNA [82]. To this purpose, specific inhibitors for the key actors of the checkpoint response namely ATM, ATR, Chk1 and Chk2 are the object of intense academical and industrial research [113]. One possible candidate could be AZD7762 developed by Zabludoff and coworkers at AstraZeneca [114]. AZD7762 is a potent ATP-competitive checkpoint kinase inhibitor that was shown to potentiate the cytotoxicity of DNA-damaging drugs towards different types of tumours cultivated in vitro, by abrogating the DNA damage checkpoint response. Importantly, the potentiation was observed in vivo as well, using multiple xenograft models and several DNA-damaging agents, indicating that the drug could be worth exploring in the clinical setting to increase patient's response rates. Another potentially interesting drug is CP466722 developed by Rainey and coworkers [115]. These authors identified CP466722 as a potent and specific ATM inhibitor after screening a targeted compound library. Inhibition by CP466722 abrogated the ATM-dependent phosphorylation activity and the cell cycle checkpoint response and could be reversed by removing the drug. HeLa and AT GM02052 cells were sensitized to IR in the presence of CP466722 in vitro. No in vivo experiments were reported in this study. A number of additional cell cycle checkpoint inhibitors are available (e.g., the aforementioned DBH) or under development [113]. Their use could permit significant sensitization of GSC to radiotherapy and chemotherapy.
4. Conclusions
Enhanced DNA repair capacity is often observed in normal (embryonic or adult; human or murine) stem cells as compared to differentiated cells, suggesting that normal stem cells often protect their genome through enhanced DNA repair. This may not be the case for cancer stem cells. At least in gliomas, DNA repair rates are normal but low proliferation and constitutive activation of the DNA damage checkpoint response confer increased time for lesion removal or bypass before arrival of the replication fork. Hence, GSC do not repair DNA better. They just have more time to do that. Those features may be common to stem cells from other tumour types as well [109]. Drugs targeting cell cycle restriction in GSC could be of help for complete eradication of the tumor and several novel agents of this kind are under development. In particular ATM and Chk1 and Chk2 kinase inhibitors may effectively sensitize GSC to IR and alkylating agents by stimulating their proliferation.
Acknowledgments
The work is partially supported by Compagnia di S. Paolo, Turin, Italy (Progetto “Sensibilizzare i tumori cerebrali alla radio- e chemioterapia con inibitori dei checkpoint del ciclo cellulare”) and Istituto Superiore Sanità, Rome, Italy (Programma Italia-USA “Malattie Rare”).
Abbreviations
- ASC:
Adult stem cell(s)
- ATM:
Ataxia-telangiectasia mutated
- ATR:
ATM and Rad3-related kinase
- BER:
DNA base excision repair
- BMP:
Bone morphogenetic protein
- BMPR:
Bone morphogenetic protein receptor
- CDK:
Cyclin-dependent kinase
- Chk1:
Checkpoint kinase-1
- Chk2:
Checkpoint kinase-2
- CNS:
Central nervous system
- DAR:
Differentiation-associated repair
- DBH:
Debromohymenialdisine
- DSB:
Double-strand break(s)
- DTA:
Diphtheria toxin fragment A
- EB:
Embryoid body
- ESC:
Embryonic stem cell(s)
- FA:
Fanconi anemia
- FCS:
Foetal calf serum
- GBM:
Multiform glioblastoma
- GFAP:
Glial fibrillary acidic protein
- GPX:
Glutathione peroxidise
- GSC:
Glioma stem cell(s)
- HDR:
Homology-directed repair
- ICL:
Interstrand crosslink
- IR:
Ionizing radiation
- LOH:
Loss of heterozigosity
- MEF:
Mouse embryonic fibroblast(s)
- MGMT:
O6 methylGua DNA methyltransferase
- MMR:
Mismatch repair
- MNNG:
N-methyl-N′-nitro-N-nitrosoguanidine
- MNU:
N-methyl-N-nitrosourea
- MSC:
Mesenchymal stem cell(s)
- Neil3:
Nei endonuclease VIII-like 3 glycosylase
- NER:
Nucleotide excision repair
- NHEJ:
Non homologous end joining
- NSC:
Neural stem cell(s)
- OGG1:
8-oxoGua DNA glycosylase
- 8-oxoGua:
8-oxoguanine
- OPC:
Oligodendrocyte progenitor cell(s)
- pol:
DNA polymerase
- PI3K:
Phosphatidylinositol-3-kinase
- PDGF:
Platelet-derived growth factor
- PTEN:
Phosphatase and tensin homologue
- ROS:
Reactive oxygen species
- RTK:
Receptor tyrosine kinase
- SCGE:
Single cell gel electrophoresis
- Shh:
Sonic Hedgehog
- SSB:
Single strand break(s)
- SVZ:
Subventricular zone
- TCR:
Transcription-coupled repair
- TGF:
Transforming growth factor
- TMZ:
Temozolomide
- UCP2:
Uncoupling protein 2
- UV:
Ultraviolet
- VEGF:
Vascular endothelial growth factor
- WHO:
World Health Organization.
References
- 1.Hong Y, Cervantes RB, Tichy E, Tischfield JA, Stambrook PJ. Protecting genomic integrity in somatic cells and embryonic stem cells. Mutation Research. 2007;614(1-2):48–55. doi: 10.1016/j.mrfmmm.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 2.Tichy ED, Stambrook PJ. DNA repair in murine embryonic stem cells and differentiated cells. Experimental Cell Research. 2008;314(9):1929–1936. doi: 10.1016/j.yexcr.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park Y, Gerson SL. DNA repair defects in stem cell function and aging. Annual Review of Medicine. 2005;56:495–508. doi: 10.1146/annurev.med.56.082103.104546. [DOI] [PubMed] [Google Scholar]
- 4.Lin Q, Donahue SL, Ruley HE. Genome maintenance and mutagenesis in embryonic stem cells. Cell Cycle. 2006;5(23):2710–2714. doi: 10.4161/cc.5.23.3517. [DOI] [PubMed] [Google Scholar]
- 5.Maynard S, Swistowska AM, Lee JW, et al. Human embryonic stem cells have enhanced repair of multiple forms of DNA damage. Stem Cells. 2008;26(9):2266–2274. doi: 10.1634/stemcells.2007-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frosina G. DNA repair in normal and cancer stem cells, with special reference to the central nervous system. Current Medicinal Chemistry. 2009;16(7):854–866. doi: 10.2174/092986709787549253. [DOI] [PubMed] [Google Scholar]
- 7.Kenyon J, Gerson SL. The role of DNA damage repair in aging of adult stem cells. Nucleic Acids Research. 2007;35(22):7557–7565. doi: 10.1093/nar/gkm1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warren LA, Rossi DJ. Stem cells and aging in the hematopoietic system. Mechanisms of Ageing and Development. 2009;130(1-2):46–53. doi: 10.1016/j.mad.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galderisi U, Helmbold H, Squillaro T, et al. In vitro senescence of rat mesenchymal stem cells is accompanied by downregulation of stemness-related and DNA damage repair genes. Stem Cells and Development. 2009;18(7):1033–1041. doi: 10.1089/scd.2008.0324. [DOI] [PubMed] [Google Scholar]
- 10.Saretzki G, Walter T, Atkinson S, et al. Downregulation of multiple stress defense mechanisms during differentiation of human embryonic stem cells. Stem Cells. 2008;26(2):455–464. doi: 10.1634/stemcells.2007-0628. [DOI] [PubMed] [Google Scholar]
- 11.Banáth JP, Bañuelos CA, Klokov D, MacPhail SM, Lansdorp PM, Olive PL. Explanation for excessive DNA single-strand breaks and endogenous repair foci in pluripotent mouse embryonic stem cells. Experimental Cell Research. 2009;315(8):1505–1520. doi: 10.1016/j.yexcr.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 12.Weinstock DM, Jasin M. Alternative pathways for the repair of RAG-induced DNA breaks. Molecular and Cellular Biology. 2006;26(1):131–139. doi: 10.1128/MCB.26.1.131-139.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saretzki G, Armstrong L, Leake A, Lako M, von Zglinicki T. Stress defense in murine embryonic stem cells is superior to that of various differentiated murine cells. Stem Cells. 2004;22(6):962–971. doi: 10.1634/stemcells.22-6-962. [DOI] [PubMed] [Google Scholar]
- 14.Roos WP, Christmann M, Fraser ST, Kaina B. Mouse embryonic stem cells are hypersensitive to apoptosis triggered by the DNA damage O6-methylguanine due to high E2F1 regulated mismatch repair. Cell Death and Differentiation. 2007;14(8):1422–1432. doi: 10.1038/sj.cdd.4402136. [DOI] [PubMed] [Google Scholar]
- 15.Casorelli I, Russo MT, Bignami M. Role of mismatch repair and MGMT in response to anticancer therapies. Anti-Cancer Agents in Medicinal Chemistry. 2008;8(4):368–380. doi: 10.2174/187152008784220276. [DOI] [PubMed] [Google Scholar]
- 16.van der Wees C, Jansen J, Vrieling H, van der Laarse A, Van Zeeland A, Mullenders L. Nucleotide excision repair in differentiated cells. Mutation Research. 2007;614(1-2):16–23. doi: 10.1016/j.mrfmmm.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 17.de Waard H, Sonneveld E, de Wit J, et al. Cell-type-specific consequences of nucleotide excision repair deficiencies: embryonic stem cells versus fibroblasts. DNA Repair. 2008;7(10):1659–1669. doi: 10.1016/j.dnarep.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 18.Sii-Felice K, Etienne O, Hoffschir F, et al. Fanconi DNA repair pathway is required for survival and long-term maintenance of neural progenitors. The EMBO Journal. 2008;27(5):770–781. doi: 10.1038/emboj.2008.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sii-Felice K, Barroca V, Etienne O, et al. Role of fanconi DNA repair pathway in neural stem cell homeostasis. Cell Cycle. 2008;7(13):1911–1915. doi: 10.4161/cc.7.13.6235. [DOI] [PubMed] [Google Scholar]
- 20.Bracker TU, Giebel B, Spanholtz J, et al. Stringent regulation of DNA repair during human hematopoietic differentiation: a gene expression and functional analysis. Stem Cells. 2006;24(3):722–730. doi: 10.1634/stemcells.2005-0227. [DOI] [PubMed] [Google Scholar]
- 21.Prall WC, Czibere A, Jäger M, et al. Age-related transcription levels of KU70, MGST1 and BIK in CD34+ hematopoietic stem and progenitor cells. Mechanisms of Ageing and Development. 2007;128(9):503–510. doi: 10.1016/j.mad.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 22.Nouspikel TP, Hyka-Nouspikel N, Hanawalt PC. Transcription domain-associated repair in human cells. Molecular and Cellular Biology. 2006;26(23):8722–8730. doi: 10.1128/MCB.01263-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nouspikel T, Hanawalt PC. Terminally differentiated human neurons repair transcribed genes but display attenuated global DNA repair and modulation of repair gene expression. Molecular and Cellular Biology. 2000;20(5):1562–1570. doi: 10.1128/mcb.20.5.1562-1570.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nouspikel T. DNA repair in differentiated cells: some new answers to old questions. Neuroscience. 2007;145(4):1213–1221. doi: 10.1016/j.neuroscience.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 25.Casorelli I, Pelosi E, Biffoni M, et al. Methylation damage response in hematopoietic progenitor cells. DNA Repair. 2007;6(8):1170–1178. doi: 10.1016/j.dnarep.2007.03.024. [DOI] [PubMed] [Google Scholar]
- 26.Nowak E, Etienne O, Millet P, et al. Radiation-induced H2AX phosphorylation and neural precursor apoptosis in the developing brain of mice. Radiation Research. 2006;165(2):155–164. doi: 10.1667/rr3496.1. [DOI] [PubMed] [Google Scholar]
- 27.Panagiotakos G, Alshamy G, Chan B, et al. Long-term impact of radiation on the stem cell and oligodendrocyte precursors in the brain. PLoS ONE. 2007;2(7, article e588) doi: 10.1371/journal.pone.0000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hildrestrand GA, Diep DB, Kunke D, et al. The capacity to remove 8-oxoG is enhanced in newborn neural stem/progenitor cells and decreases in juvenile mice and upon cell differentiation. DNA Repair. 2007;6(6):723–732. doi: 10.1016/j.dnarep.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 29.Rolseth V, Rundén-Pran E, Luna L, McMurray C, Bjørås M, Ottersen OP. Widespread distribution of DNA glycosylases removing oxidative DNA lesions in human and rodent brains. DNA Repair. 2008;7(9):1578–1588. doi: 10.1016/j.dnarep.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hildrestrand GA, Neurauter CG, Diep DB, et al. Expression patterns of Neil3 during embryonic brain development and neoplasia. BMC Neuroscience. 2009;10, article 45 doi: 10.1186/1471-2202-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang L, Deng L, Mendonca MS, et al. X-rays induce distinct patterns of somatic mutation in fetal versus adult hematopoietic cells. DNA Repair. 2007;6(9):1380–1385. doi: 10.1016/j.dnarep.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narciso L, Fortini P, Pajalunga D, et al. Terminally differentiated muscle cells are defective in base excision DNA repair and hypersensitive to oxygen injury. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(43):17010–17015. doi: 10.1073/pnas.0701743104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Backendorf C, de Wit J, Van Oosten M, et al. Repair characteristics and differentiation propensity of long-term cultures of epidermal keratinocytes derived from normal and NER-deficient mice. DNA Repair. 2005;4(11):1325–1336. doi: 10.1016/j.dnarep.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 34.Gotherstrom C, West A, Liden J, Uzunel M, Lahesmaa R, Le Blanc K. Difference in gene expression between human fetal liver and adult bone marrow mesenchymal stem cells. Haematologica. 2005;90(8):1017–1026. [PubMed] [Google Scholar]
- 35.Chen M-F, Lin C-T, Chen W-C, et al. The sensitivity of human mesenchymal stem cells to ionizing radiation. International Journal of Radiation Oncology Biology Physics. 2006;66(1):244–253. doi: 10.1016/j.ijrobp.2006.03.062. [DOI] [PubMed] [Google Scholar]
- 36.Serakinci N, Christensen R, Graakjaer J, et al. Ectopically hTERT expressing adult human mesenchymal stem cells are less radiosensitive than their telomerase negative counterpart. Experimental Cell Research. 2007;313(5):1056–1067. doi: 10.1016/j.yexcr.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 37.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nature Reviews Molecular Cell Biology. 2008;9(12):958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 38.Hsu P-H, Hanawalt PC, Nouspikel T. Nucleotide excision repair phenotype of human acute myeloid leukemia cell lines at various stages of differentiation. Mutation Research. 2007;614(1-2):3–15. doi: 10.1016/j.mrfmmm.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 39.Rossi DJ, Bryder D, Seita J, Nussenzweig A, Hoeijmakers J, Weissman IL. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 2007;447(7145):725–729. doi: 10.1038/nature05862. [DOI] [PubMed] [Google Scholar]
- 40.Nijnik A, Woodbine L, Marchetti C, et al. DNA repair is limiting for haematopoietic stem cells during ageing. Nature. 2007;447(7145):686–690. doi: 10.1038/nature05875. [DOI] [PubMed] [Google Scholar]
- 41.Hildrestrand GA, Duggal S, Bjørås M, Luna L, Brinchmann JE. Modulation of DNA glycosylase activities in mesenchymal stem cells. Experimental Cell Research. 2009;315(15):2558–2567. doi: 10.1016/j.yexcr.2009.05.017. [DOI] [PubMed] [Google Scholar]
- 42.Madhavan L, Ourednik V, Ourednik J. Increased “vigilance” of antioxidant mechanisms in neural stem cells potentiates their capability to resist oxidative stress. Stem Cells. 2006;24(9):2110–2119. doi: 10.1634/stemcells.2006-0018. [DOI] [PubMed] [Google Scholar]
- 43.Thon N, Damianoff K, Hegermann J, et al. Presence of pluripotent CD133+ cells correlates with malignancy of gliomas. Molecular and Cellular Neuroscience. 2010;43(1):51–59. doi: 10.1016/j.mcn.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 44.Zeppernick F, Ahmadi R, Campos B, et al. Stem cell marker CD133 affects clinical outcome in glioma patients. Clinical Cancer Research. 2008;14(1):123–129. doi: 10.1158/1078-0432.CCR-07-0932. [DOI] [PubMed] [Google Scholar]
- 45.Rebetz J, Tian D, Persson A, et al. Glial progenitor-like phenotype in low-grade glioma and enhanced CD133-expression and neuronal lineage differentiation potential in high-grade glioma. PLoS ONE. 2008;3(4, article e1936) doi: 10.1371/journal.pone.0001936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Günther HS, Schmidt NO, Phillips HS, et al. Glioblastoma-derived stem cell-enriched cultures form distinct subgroups according to molecular and phenotypic criteria. Oncogene. 2008;27(20):2897–2909. doi: 10.1038/sj.onc.1210949. [DOI] [PubMed] [Google Scholar]
- 47.Yao J, Zhang T, Ren J, Yu M, Wu G. Effect of CD133/prominin-1 antisense oligodeoxynucleotide on in vitro growth characteristics of Huh-7 human hepatocarcinoma cells and U251 human glioma cells. Oncology Reports. 2009;22(4):781–787. doi: 10.3892/or_00000500. [DOI] [PubMed] [Google Scholar]
- 48.Piccirillo SGM, Combi R, Cajola L, et al. Distinct pools of cancer stem-like cells coexist within human glioblastomas and display different tumorigenicity and independent genomic evolution. Oncogene. 2009;28(15):1807–1811. doi: 10.1038/onc.2009.27. [DOI] [PubMed] [Google Scholar]
- 49.Murat A, Migliavacca E, Gorlia T, et al. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. Journal of Clinical Oncology. 2008;26(18):3015–3024. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- 50.Ma Y-H, Mentlein R, Knerlich F, Kruse M-L, Mehdorn HM, Held-Feindt J. Expression of stem cell markers in human astrocytomas of different WHO grades. Journal of Neuro-Oncology. 2008;86(1):31–45. doi: 10.1007/s11060-007-9439-7. [DOI] [PubMed] [Google Scholar]
- 51.Kong D-S, Kim MH, Park W-Y, et al. The progression of gliomas is associated with cancer stem cell phenotype. Oncology Reports. 2008;19(3):639–643. [PubMed] [Google Scholar]
- 52.Laks DR, Masterman-Smith M, Visnyei K, et al. Neurosphere formation is an independent predictor of clinical outcome in malignant glioma. Stem Cells. 2009;27(4):980–987. doi: 10.1002/stem.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nishide K, Nakatani Y, Kiyonari H, Kondo T. Glioblastoma formation from cell population depleted of prominin1-expressing cells. PLoS ONE. 2009;4(8, article e6869) doi: 10.1371/journal.pone.0006869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen G, Shen F, Shi Z, et al. Identification of cancer stem-like cells in the C6 glioma cell line and the limitation of current identification methods. In Vitro Cellular & Developmental Biology—Animal. 2008;44(7):280–289. doi: 10.1007/s11626-008-9115-z. [DOI] [PubMed] [Google Scholar]
- 55.Bexell D, Gunnarsson S, Siesjö P, Bengzon J, Darabi A. CD133+ and nestin+ tumor-initiating cells dominate in N29 and N32 experimental gliomas. International Journal of Cancer. 2009;125(1):15–22. doi: 10.1002/ijc.24306. [DOI] [PubMed] [Google Scholar]
- 56.Cho RW, Clarke MF. Recent advances in cancer stem cells. Current Opinion in Genetics and Development. 2008;18(1):48–53. doi: 10.1016/j.gde.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 57.Dirks PB. Brain tumor stem cells: bringing order to the chaos of brain cancer. Journal of Clinical Oncology. 2008;26(17):2916–2924. doi: 10.1200/JCO.2008.17.6792. [DOI] [PubMed] [Google Scholar]
- 58.Regenbrecht CRA, Lehrach H, Adjaye J. Stemming cancer: functional genomics of cancer stem cells in solid tumors. Stem Cell Reviews. 2008;4(4):319–328. doi: 10.1007/s12015-008-9034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou J, Zhang Y. Cancer stem cells: models, mechanisms and implications for improved treatment. Cell Cycle. 2008;7(10):1360–1370. doi: 10.4161/cc.7.10.5953. [DOI] [PubMed] [Google Scholar]
- 60.Wen PY, Kesari S. Malignant gliomas in adults. The New England Journal of Medicine. 2008;359(5):492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 61.Kanu OO, Mehta A, Di C, et al. Glioblastoma multiforme: a review of therapeutic targets. Expert Opinion on Therapeutic Targets. 2009;13(6):701–718. doi: 10.1517/14728220902942348. [DOI] [PubMed] [Google Scholar]
- 62.Casartelli G, Dorcaratto A, Ravetti JL, et al. Survival of high grade glioma patients depends on their age at diagnosis. Cancer Biology and Therapy. 2009;8(18):1719–1721. doi: 10.4161/cbt.8.18.9209. [DOI] [PubMed] [Google Scholar]
- 63.Lefranc F, Rynkowski M, DeWitte O, Kiss R. Present and potential future adjuvant issues in high-grade astrocytic glioma treatment. Advances and Technical Standards in Neurosurgery. 2009;34:3–35. doi: 10.1007/978-3-211-78741-0_1. [DOI] [PubMed] [Google Scholar]
- 64.Lafay-Cousin L, Strother D. Current treatment approaches for infants with malignant central nervous system tumors. Oncologist. 2009;14(4):433–444. doi: 10.1634/theoncologist.2008-0193. [DOI] [PubMed] [Google Scholar]
- 65.Cayre M, Canoll P, Goldman JE. Cell migration in the normal and pathological postnatal mammalian brain. Progress in Neurobiology. 2009;88(1):41–63. doi: 10.1016/j.pneurobio.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abel TW, Clark C, Bierie B, et al. GFAP-Cre-mediated activation of oncogenic K-ras results in expansion of the subventricular zone and infiltrating glioma. Molecular Cancer Research. 2009;7(5):645–653. doi: 10.1158/1541-7786.MCR-08-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lindberg N, Kastemar M, Olofsson T, Smits A, Uhrbom L. Oligodendrocyte progenitor cells can act as cell of origin for experimental glioma. Oncogene. 2009;28(23):2266–2275. doi: 10.1038/onc.2009.76. [DOI] [PubMed] [Google Scholar]
- 68.Appolloni I, Calzolari F, Tutucci E, et al. PDGF-B induces a homogeneous class of oligodendrogliomas from embryonic neural progenitors. International Journal of Cancer. 2009;124(10):2251–2259. doi: 10.1002/ijc.24206. [DOI] [PubMed] [Google Scholar]
- 69.Stiles CD, Rowitch DH. Glioma stem cells: a midterm exam . Neuron. 2008;58(6):832–846. doi: 10.1016/j.neuron.2008.05.031. [DOI] [PubMed] [Google Scholar]
- 70.Park DM, Rich JN. Biology of glioma cancer stem cells. Molecules and Cells. 2009;28(1):7–12. doi: 10.1007/s10059-009-0111-2. [DOI] [PubMed] [Google Scholar]
- 71.Dufour C, Cadusseau J, Varlet P, et al. Astrocytes reverted to a neural progenitor-like state with transforming growth factor alpha are sensitized to cancerous transformation. Stem Cells. 2009;27(10):2373–2382. doi: 10.1002/stem.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rubin LL. Stem cells and drug discovery: the beginning of a new era? Cell. 2008;132(4):549–552. doi: 10.1016/j.cell.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 73.Ischenko I, Seeliger H, Schaffer M, Jauch K-W, Bruns CJ. Cancer stem cells: how can we target them? Current Medicinal Chemistry. 2008;15(30):3171–3184. doi: 10.2174/092986708786848541. [DOI] [PubMed] [Google Scholar]
- 74.Li. Z, Wang H, Eyler CE, Hjelmeland AB, Rich JN. Turning cancer stem cells inside out: an exploration of glioma stem cells signalling pathways. Journal of Biological Chemistry. 2009;284(25):16705–16709. doi: 10.1074/jbc.R900013200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fael Al-Mayhani TM, Ball SLR, Zhao J-W, et al. An efficient method for derivation and propagation of glioblastoma cell lines that conserves the molecular profile of their original tumours. Journal of Neuroscience Methods. 2009;176(2):192–199. doi: 10.1016/j.jneumeth.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 76.Pollard SM, Yoshikawa K, Clarke ID, et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell. 2009;4(6):568–580. doi: 10.1016/j.stem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 77.Ropolo M, Daga A, Griffero F, et al. Comparative analysis of DNA repair in stem and nonstem glioma cell cultures. Molecular Cancer Research. 2009;7(3):383–392. doi: 10.1158/1541-7786.MCR-08-0409. [DOI] [PubMed] [Google Scholar]
- 78.Clément V, Dutoit V, Marino D, Dietrich P-Y, Radovanovic I. Limits of CD133 as a marker of glioma self-renewing cells. International Journal of Cancer. 2009;125(1):244–248. doi: 10.1002/ijc.24352. [DOI] [PubMed] [Google Scholar]
- 79.Bidlingmaier S, Zhu X, Liu B. The utility and limitations of glycosylated human CD133 epitopes in defining cancer stem cells. Journal of Molecular Medicine. 2008;86(9):1025–1032. doi: 10.1007/s00109-008-0357-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhao Y, Huang Q, Zhang T, et al. Ultrastructural studies of glioma stem cells/progenitor cells. Ultrastructural Pathology. 2008;32(6):241–245. doi: 10.1080/01913120802289165. [DOI] [PubMed] [Google Scholar]
- 81.Borovski T, Vermeulen L, Sprick MR, Medema JP. One renegade cancer stem cell? Cell Cycle. 2009;8(6):803–808. doi: 10.4161/cc.8.6.7935. [DOI] [PubMed] [Google Scholar]
- 82.Frosina G. DNA repair and resistance of gliomas to chemotherapy and radiotherapy. Molecular Cancer Research. 2009;7(7):989–999. doi: 10.1158/1541-7786.MCR-09-0030. [DOI] [PubMed] [Google Scholar]
- 83.Dirks PB. Cancer: stem cells and brain tumours. Nature. 2006;444(7120):687–688. doi: 10.1038/444687a. [DOI] [PubMed] [Google Scholar]
- 84.Medrano S, Burns-Cusato M, Atienza MB, Rahimi D, Scrable H. Regenerative capacity of neural precursors in the adult mammalian brain is under the control of p53. Neurobiology of Aging. 2009;30(3):483–497. doi: 10.1016/j.neurobiolaging.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Meletis K, Wirta V, Hede S-M, Nistér M, Lundeberg J, Frisén J. p53 suppresses the self-renewal of adult neural stem cells. Development. 2006;133(2):363–369. doi: 10.1242/dev.02208. [DOI] [PubMed] [Google Scholar]
- 86.Hulleman E, Helin K. Molecular mechanisms in gliomagenesis. Advances in Cancer Research. 2005;94(1):1–27. doi: 10.1016/S0065-230X(05)94001-3. [DOI] [PubMed] [Google Scholar]
- 87.Boyer MJ, Cheng T. The CDK inhibitors: potential targets for therapeutic stem cell manipulations? Gene Therapy. 2008;15(2):117–125. doi: 10.1038/sj.gt.3303064. [DOI] [PubMed] [Google Scholar]
- 88.Jackson EL, Alvarez-Buylla A. Characterization of adult neural stem cells and their relation to brain tumors. Cells Tissues Organs. 2008;188(1-2):212–224. doi: 10.1159/000114541. [DOI] [PubMed] [Google Scholar]
- 89.Lee J-S, Gil J-E, Kim J-H, et al. Brain cancer stem-like cell genesis from p53-deficient mouse astrocytes by oncogenic Ras. Biochemical and Biophysical Research Communications. 2008;365(3):496–502. doi: 10.1016/j.bbrc.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 90.Cai C, Thorne J, Grabel L. Hedgehog serves as a mitogen and survival factor during embryonic stem cell neurogenesis. Stem Cells. 2008;26(5):1097–1108. doi: 10.1634/stemcells.2007-0684. [DOI] [PubMed] [Google Scholar]
- 91.Galvin KE, Ye H, Erstad DJ, Feddersen R, Wetmore C. Gli1 induces G2/M arrest and apoptosis in hippocampal but not tumor-derived neural stem cells. Stem Cells. 2008;26(4):1027–1036. doi: 10.1634/stemcells.2007-0879. [DOI] [PubMed] [Google Scholar]
- 92.Clark PA, Treisman DM, Ebben J, Kuo JS. Developmental signaling pathways in brain tumor-derived stem-like cells. Developmental Dynamics. 2007;236(12):3297–3308. doi: 10.1002/dvdy.21381. [DOI] [PubMed] [Google Scholar]
- 93.Yao X-H, Ping Y-F, Chen J-H, et al. Glioblastoma stem cells produce vascular endothelial growth factor by activation of a G-protein coupled formylpeptide receptor FPR. Journal of Pathology. 2008;215(4):369–376. doi: 10.1002/path.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Knizetova P, Darling JL, Bartek J. Vascular endothelial growth factor in astroglioma stem cell biology and response to therapy. Journal of Cellular and Molecular Medicine. 2008;12(1):111–125. doi: 10.1111/j.1582-4934.2007.00153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wong ET, Brem S. Antiangiogenesis treatment for glioblastoma multiforme: challenges and opportunities. The Journal of the National Comprehensive Cancer Network. 2008;6(5):515–522. doi: 10.6004/jnccn.2008.0039. [DOI] [PubMed] [Google Scholar]
- 96.Bleau AM, Huse JT, Holland EC. The ABCG2 resistance network of glioblastoma. Cell Cycle. 2009;8(18):2936–2944. [PubMed] [Google Scholar]
- 97.Johannessen T-CA, Bjerkvig R, Tysnes BB. DNA repair and cancer stem-like cells—potential partners in glioma drug resistance? Cancer Treatment Reviews. 2008;34(6):558–567. doi: 10.1016/j.ctrv.2008.03.125. [DOI] [PubMed] [Google Scholar]
- 98.Roth RB, Samson LD. 3-methyladenine DNA glycosylase-deficient Aag null mice display unexpected bone marrow alkylation resistance. Cancer Research. 2002;62(3):656–660. [PubMed] [Google Scholar]
- 99.Hambardzumyan D, Squatrito M, Carbajal E, Holland EC. Glioma formation, cancer stem cells, and Akt signaling. Stem Cell Reviews. 2008;4(3):203–210. doi: 10.1007/s12015-008-9021-5. [DOI] [PubMed] [Google Scholar]
- 100.Kwon C-H, Zhao D, Chen J, et al. Pten haploinsufficiency accelerates formation of high-grade astrocytomas. Cancer Research. 2008;68(9):3286–3294. doi: 10.1158/0008-5472.CAN-07-6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Eyler CE, Foo W-C, LaFiura KM, McLendon RE, Hjelmeland AB, Rich JN. Brain cancer stem cells display preferential sensitivity to Akt inhibition. Stem Cells. 2008;26(12):3027–3036. doi: 10.1634/stemcells.2007-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Westhoff M-A, Kandenwein JA, Karl S, et al. The pyridinylfuranopyrimidine inhibitor, PI-103, chemosensitizes glioblastoma cells for apoptosis by inhibiting DNA repair. Oncogene. 2009;28(40):3586–3596. doi: 10.1038/onc.2009.215. [DOI] [PubMed] [Google Scholar]
- 103.Kao GD, Jiang Z, Fernandes AM, Gupta AK, Maity A. Inhibition of phosphatidylinositol-3-OH kinase/Akt signaling impairs DNA repair in glioblastoma cells following ionizing radiation. Journal of Biological Chemistry. 2007;282(29):21206–21212. doi: 10.1074/jbc.M703042200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 105.Rich JN. Cancer stem cells in radiation resistance. Cancer Research. 2007;67(19):8980–8984. doi: 10.1158/0008-5472.CAN-07-0895. [DOI] [PubMed] [Google Scholar]
- 106.Wang X, Lui VCH, Poon RTP, Lu P, Poon RYC. DNA damage mediated s and G2 checkpoints in human embryonal carcinoma cells. Stem Cells. 2009;27(3):568–576. doi: 10.1634/stemcells.2008-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kato T, Kanemura Y, Shiraishi K, Miyake J, Kodama S, Hara M. Early response of neural stem/progenitor cells after X-ray irradiation in vitro. NeuroReport. 2007;18(9):895–900. doi: 10.1097/WNR.0b013e3281053c34. [DOI] [PubMed] [Google Scholar]
- 108.Beier D, Röhrl S, Pillai DR, et al. Temozolomide preferentially depletes cancer stem cells in glioblastoma. Cancer Research. 2008;68(14):5706–5715. doi: 10.1158/0008-5472.CAN-07-6878. [DOI] [PubMed] [Google Scholar]
- 109.Viale A, De Franco F, Orleth A, et al. Cell-cycle restriction limits DNA damage and maintains self-renewal of leukaemia stem cells. Nature. 2009;457(7225):51–56. doi: 10.1038/nature07618. [DOI] [PubMed] [Google Scholar]
- 110.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nature Reviews Molecular Cell Biology. 2008;9(8):616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Reinhardt HC, Yaffe MB. Kinases that control the cell cycle in response to DNA damage: Chk1, Chk2, and MK2. Current Opinion in Cell Biology. 2009;21(2):245–255. doi: 10.1016/j.ceb.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Enders GH. Expanded roles for Chk1 in genome maintenance. Journal of Biological Chemistry. 2008;283(26):17749–17752. doi: 10.1074/jbc.R800021200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bucher N, Britten CD. G2 checkpoint abrogation and checkpoint kinase-1 targeting in the treatment of cancer. British Journal of Cancer. 2008;98(3):523–528. doi: 10.1038/sj.bjc.6604208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zabludoff SD, Deng C, Grondine MR, et al. AZD7762, a novel checkpoint kinase inhibitor, drives checkpoint abrogation and potentiates DNA-targeted therapies. Molecular Cancer Therapeutics. 2008;7(9):2955–2966. doi: 10.1158/1535-7163.MCT-08-0492. [DOI] [PubMed] [Google Scholar]
- 115.Rainey MD, Charlton ME, Stanton RV, Kastan MB. Transient inhibition of ATM kinase is sufficient to enhance cellular sensitivity to ionizing radiation. Cancer Research. 2008;68(18):7466–7474. doi: 10.1158/0008-5472.CAN-08-0763. [DOI] [PMC free article] [PubMed] [Google Scholar]