Abstract
Vegetative growth and flowering initiation are two crucial developmental processes in the life cycle of annual plants that are closely associated. The timing of both processes affects several presumed adaptive traits, such as flowering time (FT), total leaf number (TLN), or the rate of leaf production (RLP). However, the interactions among these complex processes and traits, and their mechanistic bases, remain largely unknown. To determine the genetic relationships between them, the natural genetic variation between A. thaliana accessions Fei-0 and Ler has been studied using a new population of 222 Ler×Fei-0 recombinant inbred lines. Temporal analysis of the parental development under a short day photoperiod distinguishes two vegetative phases differing in their RLP. QTL mapping of RLP in consecutive time intervals of vegetative development indicates that Ler/Fei-0 variation is caused by 10 loci whose small to moderate effects mainly display two different temporal patterns. Further comparative QTL analyses show that most of the genomic regions affecting FT or TLN also alter RLP. In addition, the partially independent genetic bases observed for FT and TLN appear determined by several genomic regions with two different patterns of phenotypic effects: regions with a larger effect on FT than TLN, and vice versa. The distinct temporal and pleiotropic patterns of QTL effects suggest that natural variation for flowering time is caused by different genetic mechanisms involved in vegetative and/or reproductive phase changes, most of them interacting with the control of leaf production rate. Thus, natural selection might contribute to maintain this genetic variation due to its phenotypic effects not only on the timing of flowering initiation but also on the rate of vegetative growth.
Keywords: Arabidopsis thaliana, flowering locus C (FLC), flowering time, growth, HUA2, natural variation, plastochron, quantitative trait locus (QTL), rate of leaf production, vegetative development
Introduction
The transition from vegetative to reproductive development is a major phase change in the life cycle of annual plants known as reproductive phase change, flowering transition, floral induction, or flowering initiation. This phase change qualitatively modifies the fate of shoot meristem primordia from leaf to flower primordia (Poethig, 2003). The temporal control of this transition is an important adaptive trait in natural and agricultural environments since it synchronizes the reproductive phase with the most favourable season for seed development and maturation. In addition, the temporal control of flowering initiation determines the time invested in vegetative growth and, consequently, the vegetative resources available during reproduction. Although flowering initiation and vegetative growth are closely associated, the relationship between both processes is not straightforward and the mechanistic bases of their co-ordination remain largely unknown.
The process of flowering transition has been extensively studied in the past two decades at the developmental, environmental, genetic, and molecular levels, mainly in the annual model plant Arabidopsis thaliana (reviewed by Ausín et al., 2005; Corbesier and Coupland, 2005; Kobayashi and Weigel, 2007; Kim et al., 2009). In this species, environmentally or genetically induced changes in the time committed from germination to flowering initiation (so-called flowering time) correlate with the number of leaves produced during vegetative development (Koornneef et al., 1991; Martinez-Zapater and Somerville, 1990; Steynen et al., 2001). Induced mutations in numerous genes have been isolated that strongly increase (Koornneef et al., 1991) or decrease (Pouteau et al., 2004) flowering time (FT), nearly all of them also altering total leaf number (TLN) and, accordingly, the final vegetative biomass. However, although these mutations affect both traits, they do not always alter them proportionally. It is expected that mutations affecting the rate of leaf production will lead to differential effects on FT and TLN, whereas mutations that do not alter this rate might show similar effects on FT and TLN. Thus, some of the so-called flowering time mutations such as fca, fve (Koornneef et al., 1991) or early bolting from versailles-1 (ebv-1; Pouteau et al., 2004) also show certain effects on the rate of leaf production, which is a major component of vegetative growth. Furthermore, two consecutive phases named as juvenile and adult phases, with several subphases, are distinguished within vegetative development, their transition being referred to as vegetative phase change (Poethig, 2003). These vegetative phases quantitatively differ in leaf size and shape, vascular pattern, and the presence of abaxial trichomes (Telfer et al., 1997; Steynen et al., 2001). Since flowering time genes do not affect similarly the duration of the various vegetative phases in a similar way, these genes may also alter the average leaf size (Martinez-Zapater et al., 1995; Telfer et al., 1997; Soppe et al., 1999; Steynen et al., 2001), which is the second component of vegetative growth. Understanding the genetic relationship between flowering initiation and vegetative growth is a relevant question from basic and applied perspectives. On the one hand, determining the extent of shared genetic bases between FT and TLN will reveal the interactions among the various mechanisms and pathways that regulate flowering transition and leaf primordia production (Poethig, 2003). On the other hand, understanding the genetic variation with differential effects on both traits, FT and TLN, will enable independent genetic manipulation of two major components of yield, flowering time and the rate of vegetative growth.
The number of leaf primordia initiated per unit of time (leaves d−1) defines the rate of leaf production (RLP), which is the inverse of the plastochron (PLR) measuring the time between the initiation of two successive leaf primordia (d leaf−1) (reviewed in Lamoreaux et al., 1978). In A. thaliana and most dicotyledonous plant species, PLR and RLP are often estimated by macroscopic evaluation of leaf primordia appearance. However, given the distinct internode elongation and vegetative architecture of monocotyledonous cereals, the time for the macroscopic appearance of leaf primordia is called phyllochron in these species, and it is distinguished from plastochron measurements of leaf initiation at the microscopic level (Klepper et al., 1982). Classical descriptions show that PLR and RLP are not constant through vegetative development and, particularly, increases in the rate of leaf production have been found previous to flowering initiation in many but not all species (see references in Groot and Meicenheimer, 2000; Bernier et al., 1981). Various genes have recently been identified in A. thaliana, rice, and maize that strongly affect RLP and PLR, including: PHYTOCHROME B (PHYB; Koornneef et al., 1995); gibberellin biosynthesis and sensitivity genes (Wilson et al., 1992); genes involved in carbon metabolism like PHOSPHOGLUCOMUTASE (PGM; Caspar et al., 1985); the cytochrome P450 genes PLASTOCHRON1 (Miyoshi et al., 2004), CYP78A5 and CYP78A7 (Wang et al., 2008); the glutamate carboxipeptidase genes ALTERED MERISTEM PROGRAM 1 (AMP1; Helliwell et al., 2001) and PLASTOCHRON3 (PLA3; Kawakatsu et al., 2009); genes encoding the RNA binding proteins TERMINAL EAR1 (TE1; Veit et al., 1998), PLASTOCHRON2 (PLA2; Kawakatsu et al., 2006), and SERRATE (SE; Prigge and Wagner, 2001); and the microRNA genes MIR156 (Wu and Poethig, 2006; Wang et al., 2008) and MIR172 (Aukerman and Sakai, 2003; Wu et al., 2009) that target other genes involved in vegetative phase changes or flowering initiation. All these genes also affect FT and/or TLN differently, which further shows the complex interaction between the control of phase changes and the rate of vegetative growth. In addition, it has been shown that genetic modifications of cell division rate through a cell cycle gene correlate with changes in the rate of leaf initiation (Cockcroft et al., 2000). Hence, it has been suggested that variation in the rate of leaf initiation is largely determined by changes in cell division rate in the apical shoot meristem (Poethig, 2003).
In addition to artificially induced genetic variation, a wealth of natural variation has been described for flowering time, among wild or cultivated accessions of A. thaliana, rice, and cereals (reviewed in Alonso-Blanco et al., 2009). In the past few years, the genetic and molecular bases of this variation have been partly elucidated by identifying about 20 different genes that underlie large effect quantitative trait loci (QTL) (Alonso-Blanco et al., 2009). By contrast, very few studies have approached the natural genetic variation for the rate of leaf production or plastochron. Current analyses have shown that there is also considerable genetic variation for these traits among varieties of different grasses, which interacts significantly with environmental factors such as temperature or photoperiod (Padilla and Otegui, 2005; Clerget et al., 2008). Nevertheless, the genetic dissection of this quantitative variation has been only initiated in rice (Miyamoto et al., 2004).
In this study, the aim was to determine the genetic basis of the natural variation that exists for the rate of leaf production among two A. thaliana accessions. In particular, a wild accession, Feira-0 (Fei-0), was identified that shows a faster rate of leaf production than the laboratory strain Landsberg erecta (Ler). These two accessions also differ in their flowering time and total leaf number, which enables the analysis of the relationship between the genetic bases of these traits. To achieve these goals, a new population of recombinant inbred lines (RILs) was developed from the Ler×Fei-0 cross, and this was used for QTL analyses of the various traits. Temporal analysis of the rate of leaf production indicates that 10 QTL affect this trait, their additive effects showing two main temporal patterns approximately corresponding to both halves of vegetative development. Comparative QTL mapping indicates that most genomic regions affecting flowering time also affect the rate of leaf production, but two different patterns of pleiotropic effects account for the partially independent genetic bases of FT and TLN. The temporal and pleioitropic patterns of these QTL suggest that natural variation for flowering time in the Ler×Fei-0 cross is determined by different mechanisms involved in the regulation of several phase changes, which, in most cases, interact with the control of the rate of leaf production.
Materials and methods
Plant materials
The laboratory strain Ler and the wild genotype Fei-0 collected in Santa Maria da Feira (Portugal) (Picó et al., 2008) were studied in this work. A population of 222 recombinant inbred lines (RILs) was developed by single seed descent from a single F1 (Ler×Fei-0) plant obtained using Ler as the mother parent (Alonso-Blanco et al., 2006). A single F8 plant from each Ler×Fei-0 RIL was genotyped, and seeds from 10 F9 plants per line were bulked to obtain the final F10 generation phenotypically analysed. The population is available through the Nottingham Arabidopsis Stock Centre (http://arabidopsis.info).
The Landsberg erecta (Ler) strain used as a parent for the Ler×Fei-0 RILs carries the loss-of-function mutant alleles erecta-1 (er-1; Rédei, 1962) and hua2-5 (Doyle et al., 2005). To evaluate ERECTA and HUA2 as candidate genes for QTL identified in this work, two near isogenic lines carrying active wild-type alleles of these genes in a Landsberg genetic background were analysed: line Ler-1, carrying wild-type alleles of HUA2 (obtained from the Cold Spring Harbor Laboratory, New York, USA) and line Landsberg (La) carrying wild-type alleles in ERECTA and HUA2 (Rédei, 1962). In addition, lines bearing the following mutations were also studied: phyB-1 in Ler-1, hua2-5 genetic background (Koorneef et al., 1980); er-105 (Torii et al., 1996) and pgm-1 (Caspar et al., 1985) in Columbia (Col) background.
Growth conditions and measurements of quantitative traits
Seeds were sown in Petri dishes containing a filter paper soaked with demineralized water, and then stored for 4 d at 4 °C to break seed dormancy. Thereafter, seeds were transferred to a growth chamber at 21 °C, where they remained for 4 d for germination. Subsequently, only germinated seedlings were planted to avoid the effects of temporal variation for seed germination on the temporal analysis of developmental traits. Plants were grown in pots with soil/vermiculite mix at 3/1 v/v proportion, in growth chambers at 21 °C. The short day (SD) photoperiod was provided as 8/16 h light/darkness in a small growth cabinet (parental, hybrids, and mutant evaluations) or a large growth room (RIL population evaluation) illuminated with cool-white fluorescent lamps. The long-day (LD) photoperiod was provided as 16/8 h light/darkness in a growth cabinet. For the evaluation of the RIL population, all lines and the parents were grown simultaneously in a single experiment organized in a two-complete-blocks design. Five plants per RIL were grown in one pot per block and lines were completely randomized within blocks. Twenty plants were grown per parent and block in four pots.
Flowering time (FT) was measured as the number of days from germination until macroscopic visualization of flower buds (∼1–2 mm) by the naked eye. This estimate of flowering initiation did not include the subsequent time for bolting and flower anthesis, hence, reducing variation due to factors acting on those processes. Total leaf number (TLN) was estimated as the total number of rosette and cauline leaves, which were counted after anthesis of the first flowers.
Two reciprocal growth parameters were estimated based on ∼1–2 mm leaf primordia: the rate of leaf production (RLP; leaves per day) and plastochron (PLR; days per leaf). For each plant, the number of leaf primordia that are visible by the naked eye was counted every 3–15 d after germination, depending on the experiment, until flowering initiation. For detailed parental analyses, data were collected every 3–4 d. For the analysis of the RIL population, the times for data collection were selected based on the parental study. Thus, the first measurement was taken 31 d after germination and, subsequently, at approximate intervals of 10–15 d. From these data, the average RLP was estimated for each consecutive time interval by dividing the number of leaf primordia that appeared in that interval by the corresponding number of days. Average RLP during the first 52 d of vegetative development (RLP1-52) was similarly estimated, and average RLP throughout vegetative development (Total RLP) was calculated by the ratio TLN/FT. In addition, average RLP during the first 52 d or for the complete vegetative development were also estimated as slopes from linear regressions of leaf number on time, using all the individual data points available for each genotype for the first 52 d or before flower bud visualization (sRLP1-52 and Total sRLP, respectively). Linear regressions consistently fitted temporal leaf data since R2 values were >0.95 for each of the RILs and parents. The reciprocal plastochron variables PLR1-52, Total PLR, sPLR1-52, and Total sPLR were inversely calculated.
Genotyping and genetic map construction
DNA was isolated as previously described (Bernartzky and Tanksley, 1986) without mercaptoethanol. RILs were genotyped with 90 markers selected from different sources and covering 96% of the A. thaliana physical map (see Supplementary Table S1 at JXB online). In a first step, 45 microsatellites previously reported (Bell and Ecker, 1994; Loudet et al., 2002; Clauss et al., 2002; http://www.arabidopsis.org) and 36 indels developed in this work based on Ler/Col sequence polymorphisms (Jander et al., 2002) were evenly selected at approximate physical intervals of 1.5 Mb. Thereafter, six CAPS markers were developed to fill large physical or genetic intervals using already described sequence polymorphisms between Ler and Fei-0 (Nordborg et al., 2005). New indel and CAPS markers were named according to the BAC clone containing the corresponding sequence. Two additional PCR markers corresponding to known indel polymorphisms in FRI (Johanson et al., 2000) and FLC (Michaels et al., 2003), as well as the ERECTA morphological marker segregating in the Ler/Fei-0 population, were also genotyped. Genotypes of the RILs for the 90 markers are provided in Supplementary Table S2 at JXB online.
Most microsatellites (42 out of 45) and 14 indels were PCR amplified in 10 mixed reactions of two to nine markers using a forward primer labelled with one of the Perkin-Elmer Applied Biosystems fluorochromes 6-FAM, NED, PET, and VIC (see Supplementary Table S1 at JXB online). PCRs were carried out in 15 μl volume reactions containing 5 ηg of DNA and a mix of primers for the corresponding markers. Amplifications were performed with one step at 95 °C for 5 min, followed by 35 cycles of 94 °C for 1 min, 55 °C for 1 min, 72 °C for 2 min, and a final step at 72 °C for 40 min. Simultaneously amplified fragments of each marker mix were separated in an ABI PRISM 3700 DNA analyser using GeneScan-500-LIZ (Applied Biosystems) as the internal size standard. Electropherograms were visually inspected and manually scored for Ler and Fei-0 alleles using GeneScan 3.7 software (Applied Biosystems). The remaining PCR markers were analysed on standard agarose gels as previously described (Konieczny and Ausubel, 1993). The 90×222 Ler/Fei-0 RIL dataset contained an average of 0.43% missing data per marker, the largest proportion of missing data corresponding to 2.7% (see Supplementary Table S2 at JXB online).
The Ler/Fei-0 linkage map was constructed using the JOINMAP 3.0 software package (Van Ooijen and Voorrips, 2001) and applying the RI8 mapping population type. Markers were assigned to linkage groups with the grouping module, markers consistently remaining on the same linkage group from LOD values of 3 to 15. Markers were arranged within linkage groups using the following mapping thresholds: REC of 0.45, LOD of 1, and JUMP of 5. Recombination frequencies were converted to genetic distances in cM using Kosambi′s mapping function (Kosambi, 1944).
QTL analyses
QTL mapping of each trait was carried out separately using mean RIL values that were previously log transformed to improve the normality of the variables. Phenotypic values of all RILs are provided in Supplementary Table S3 at JXB online. Five RILs were discarded from the quantitative analyses because not enough plants survived for their phenotypic evaluation. QTL were mapped by the multiple-QTL-model method (MQM) implemented in the software package MapQTL v. 4.0 (Van Ooijen, 2000) as described in its reference manual (http://www.mapqtl.nl). LOD threshold values for QTL detection were estimated for each trait with the permutation test implemented in MapQTL, using 10 000 permutations. LOD values of 2.5–2.6, corresponding to a genome-wide significance α=0.05, were used for the detection of most QTL. However, a few putative QTL were also declared using thresholds of 1.7–1.9, corresponding to a chromosome-wide significance of 0.05. Two-LOD support intervals were established as ≈95% QTL confidence intervals (Van Ooijen, 1992). The additive allele effect and the percentage of variance explained by each QTL, as well as the total variance explained by all QTL detected for each trait, were obtained from the MQM models. QTL additive allele effects correspond to half the differences between the estimated means of the two homozygous genotypic groups of RILs. Two-way genetic interactions at the genome-wide level were searched by testing all pair-wise combinations of 90 markers using EPISTAT (Chase et al., 1997) with log-likelihood ratio thresholds corresponding to a significance of P <0.0001. One hundred thousand trials were used in Monte Carlo simulations performed with EPISTAT to establish the statistical significance. The percentage of variance explained by significant interactions was estimated by ANOVA type III variance components analysis. This and other statistical tests were performed with the statistical package SPSS v13. Most statistical comparisons shown in the results were based on log-transformed data but none of the outcomes and conclusions were changed when using the original data. Hence, phenotypic descriptions, histograms, and approximate additive effects are presented as estimated from the original scales. Since RLP and PLR are reciprocal measurements they identified the same genomic regions, and these variables are referred to as RLP/PLR.
Results
Rate of leaf production and flowering initiation of Ler and Fei-0
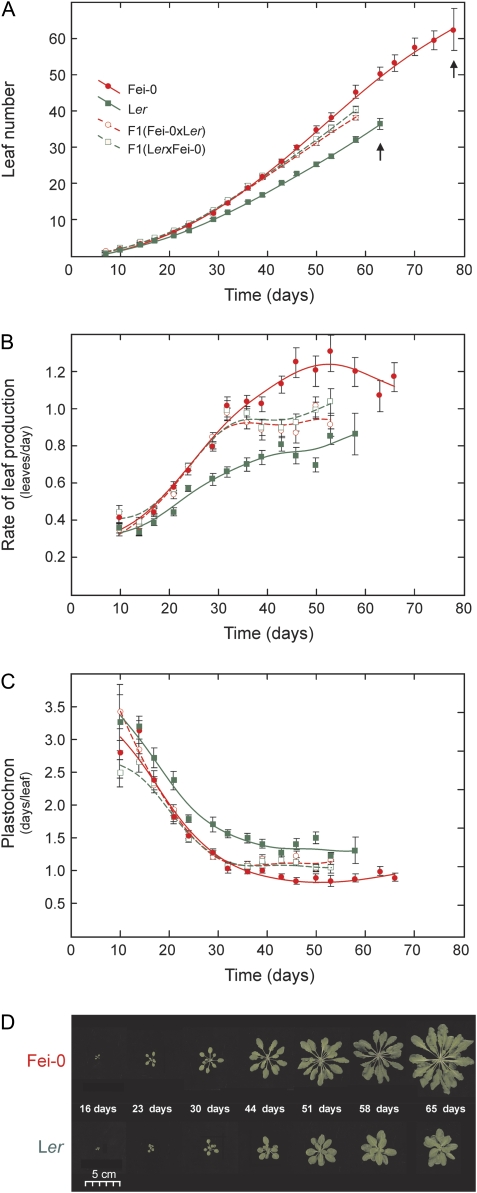
Preliminary analysis of several wild genotypes (Picó et al., 2008) identified the Fei-0 accession as showing faster vegetative development than the reference strain Landsberg erecta (Ler) because under LD photoperiod conditions it only flowers slightly later (33.6±2.9 d versus 26.4±3.0 d) but it produces proportionally more leaves (20.6±2.9 versus 7.9±0.9). To characterize in detail this genetic variation, Fei-0 and Ler accessions were grown under a SD photoperiod and newly developed leaf primordia were counted every 3 or 4 d. Under these conditions, Fei-0 flowered 26% later than Ler but it produced 68% more leaves (Table 1). As shown in Fig. 1, Fei-0 showed faster development of leaf primordia from germination to flowering initiation, although differences between both accessions were significant from day 20 onwards (P <0.001). The temporal dynamics of this variation was further characterized by estimating the average rate of leaf production (RLP) and plastochron (PLR) in consecutive time intervals. Both accessions showed a similar temporal pattern of RLP and PLR variation characterized by a continuous acceleration of vegetative development in two distinct temporal phases, which approximately corresponds to the two halves of vegetative development (Fig. 1B, C). During the first half (∼30 d and ∼35 d for Ler and Fei-0, respectively), RLP rapidly increased from its lowest value right after germination to reach a nearly steady level, while PLR shows the opposite behaviour. In the second phase, RLP and PLR follow the same trends but at substantially lower rates. Fei-0 and Ler significantly differed in both phases (P <0.001), Fei-0 always showing higher RLP and lower PLR than Ler. Given this ontogenetic variation of developmental rates and the consequent influence of flowering time genetic variation on the total average estimations of RLP and PLR, these parameters were also calculated during the first 52 d, which is the common time interval of vegetative development for both accessions. Thus, Fei-0 consistently showed an average RLP1-52 40% larger than Ler in different experiments (Table 1). Similar behaviours of RLP and PLR during vegetative development were found under LD photoperiod conditions, although both parents showed faster RLP values in LD than SD photoperiod (see Supplementary Fig. S1 at JXB online). In addition, phenotypic differences between genotypes were smaller under LD, independently of the generally shorter times of vegetative development observed in these conditions.
Table 1.
Flowering time, total leaf number and general rates of leaf production and plastochrons of Ler, Fei-0, reciprocal hybrid plants and the RIL population
| FT | TLN | RLP1-52 | PLR1-52 | Total RLP | Total PLR | |
| (d) | (leaves) | (leaves d−1) | (d leaf−1) | (leaves d−1) | (d leaf−1) | |
| Ler | 62.8±2.5 | 43.1±2.9 | 0.52±0.04 | 1.95±0.17 | 0.69±0.05 | 1.46±0.10 |
| Fei-0 | 79.4±8.9 | 72.4±6.9 | 0.72±0.07 | 1.41±0.15 | 0.92±0.13 | 1.11±0.17 |
| F1 (Ler×Fei-0) | 60.7±3.0 | 52.6±3.2 | 0.66±0.05 | 1.52±0.13 | 0.87±0.07 | 1.16±0.11 |
| F1 (Fei-0×Ler) | 58.9±2.5 | 50.3±3.1 | 0.65±0.06 | 1.54±0.14 | 0.86±0.07 | 1.18±0.09 |
| Ler1 | 70.7±8.9 | 29.6±6.4 | 0.34±0.03 | 2.96±0.30 | 0.42±0.06 | 2.44±0.33 |
| Fei-01 | 96.6±11.6 | 89.7±14.8 | 0.48±0.08 | 2.13±0.35 | 0.93±0.12 | 1.10±0.14 |
| RIL mean | 81.8±27.2 | 53.6±27.5 | 0.39±0.06 | 2.71±0.41 | 0.62±0.15 | 1.71±0.41 |
| Min–max RIL mean | 34.1–179.0 | 12.0–131.8 | 0.24–0.53 | 1.91–4.56 | 0.33–1.02 | 0.98–3.03 |
Values are mean±SD.
Parental lines grown in the same experiment than the Ler×Fei-0 RIL population.
Fig. 1.
Rate of vegetative development of Ler, Fei-0 and their reciprocal hybrids. (A) Leaf number, (B) rate of leaf production (RLP), (C) plastochron (PLR), and (D) vegetative phenotypes, in relation to time after germination. Data are mean ±SE of 15 plants per genotype grown under short day photoperiod conditions. RLP and PLR values are estimated in 1 week intervals around each time point. Curves were fitted by distance-weighted least squares. In (A), leaf number is estimated until the appearance of flower buds, which is indicated with arrows for the two parental lines.
To determine the degree of dominance of the various traits, the behaviour of hybrid plants obtained from reciprocal crosses between Fei-0 and Ler (Fig. 1; Table 1) were analysed simultaneously. Both hybrids did not differ for any trait indicating that maternal effects are not significant (P >0.01). In addition, hybrid plants flowered at nearly the same time as Ler but they produced an intermediate leaf number between the two parents, which indicates that the later FT of Fei-0 behaves as recessive but TLN as semidominant. In addition, hybrids showed different RLP and PLR behaviours in the two temporal phases, the faster development of Fei-0 appearing as dominant during the first phase but as semidominant during the second one (Fig. 1). Thus hybrid analyses supported the distinction of two phases within vegetative development differentiated by their leaf production rate. In addition, the change of dominance observed in the course of ontogenetic development suggested that different loci account for the variation between Fei-0 and Ler in each phase or that dominance of the loci involved is conditioned by a changing developmental environment.
Flowering initiation and rates of leaf production of the Ler×Fei-0 RIL population
To determine the genetic bases of the variation for the rate of leaf production and its relationship with flowering time and leaf number, a population of 222 recombinant inbred lines (RILs) between Fei-0 and Ler was developed. Analysis of this population under SD conditions showed nearly the same amount of phenotypic variation for FT than for TLN (Table 1; Fig. 2), but FT displayed larger transgression in both directions than TLN. Both traits were highly correlated (r=0.96; P <0.001), but substantial TLN variation was observed for each FT value, which indicates partly independent genetic bases for FT and TLN (see Supplementary Fig. S2 at JXB online).
Fig. 2.
Frequency distributions of flowering time, total leaf number, and general rates of leaf production in the Ler×Fei-0 RIL population. Arrows and horizontal bars depict mean ±SD of parental lines. RLP1-52: average rate of leaf production between days 1 to 52. Total RLP: average rate of leaf production during the complete vegetative development of each RIL.
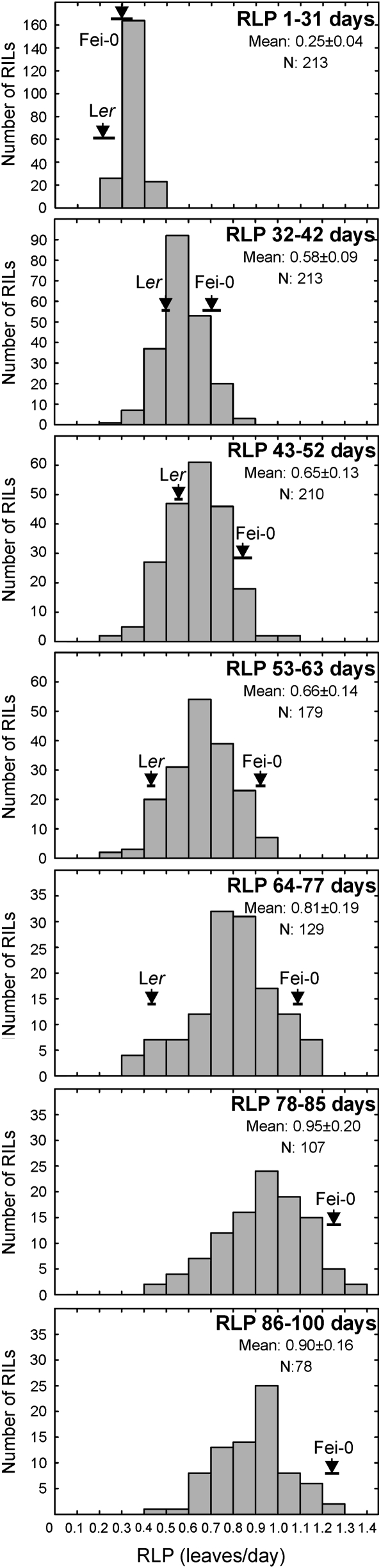
The number of leaf primordia developed by each line was estimated at 31, 42, 52, 63, 77, 85, and 100 d after germination, and average RLP and PLR values of each RIL were calculated for successive time intervals. Similar temporal patterns of RLP and PLR variation were observed in the RIL population (Fig. 3) than those described in the parents. Overall, a rapid increase of RLP was found during the first 42 d to reach a nearly steady level at 52–63 d. Thereafter, the average RLP of the population increased slowly until Fei-0 flowered. Significant genetic variation was found for RLP/PLR at each time interval, as well as during the first 52 d when most RILs remained vegetative (only 25 RILs flowered before 52 d), or along the total vegetative development of each line (Figs 2, 3). As expected from the ontogenetic pattern of vegetative development, FT and TLN values correlated with Total RLP and PLR rates (0.78 <r <0.93, P <0.0001; Supplementary Table S4 at JXB online). However, FT did not correlate with average RLP estimated in the first 52 d (P >0.1), while TLN weakly correlated (r=0.29; P <0.0001) showing partly independent genetic components for these traits. In addition, RLP values estimated in the early time periods did not correlate or showed only low correlations with RLP values estimated in later intervals, suggesting partly independent genetic bases for RLP variation along vegetative development (see Supplementary Table S4 at JXB online).
Fig. 3.
Frequency distributions of rates of leaf production along vegetative development in the Ler×Fei-0 RIL population. Each panel shows the average RLP estimated in one of seven successive time intervals. Inside each panel is indicated the time interval, population mean, and the number of included RILs showing vegetative development (N). Arrows and horizontal bars depict mean ±SD of parental lines. The Ler parent flowered in 70 d and, therefore, it is not included in subsequent panels.
Genetic map of Ler×Fei-0 RIL population
A genetic map including 90 markers evenly distributed at an average distance of 4.9 cM was developed from the 222 RILs (see Materials and methods). This map showed a total length of 416 cM, the largest genetic interval between adjacent markers corresponding to 12.3 cM (Fig. 4). The genetic order of all markers was similar to that of the Col physical map (http://www.arabidopsis.org/) with the exception of pair T1J24-T26N6 located in the centromeric region of chromosome 4, which appeared as inverted. A comparison of physical and genetic maps (see Supplementary Fig. S3A at JXB online) indicated that the recombination rate is homogeneously distributed along the five chromosomes with an average value of 391 kb cM−1. However, recombination was substantially lower in all five pericentromeric regions. In total, the 222 RILs provide 1700 recombination events on this genetic map, with an average of 7.7 breakpoints per line. As expected for the F8 generation, heterozygosity of most markers was below 1%. Only eight markers located in three genomic regions presented higher values ranging from 1.8% to 2.7% (Supplementary Fig. S3B at JXB online). Moreover, only seven markers mapping in the upper arm of chromosome 1 showed distortion from the expected 1:1 segregation of homozygous genotypes (P <0.01; Supplementary Fig. S3B at JXB online). These results indicate that the statistical power for QTL mapping in this population is nearly similar for most of the genome.
Fig. 4.
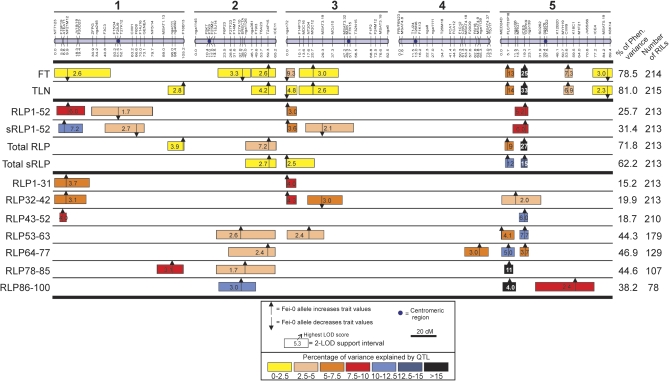
QTL mapping of traits related with flowering initiation and rates of leaf production in the Ler/Fei-0 RIL population. Bars on the top represent the genetic map of the five linkage groups. Thick horizontal lines separate three groups of traits analysed: flowering initiation related traits; general measurements of the rate of leaf production; and temporal measurements of the rate of leaf production in seven consecutive time intervals (indicated in the trait name as days after germination). Columns on the right show the total percentage of phenotypic variance explained by the additive effects of all detected QTL, and the number of RILs used for QTL mapping (see text for details). For each trait, the locations of all QTL identified are shown as 2-LOD support intervals, the position of arrows corresponding to the maximum LOD score values. Numbers inside the boxes indicate the highest LOD score. Colours of the QTL boxes correspond to the different ranges of QTL explained variances as described in the legend. Upper and lower arrows indicate that the additive effect of Fei-0 alleles increase or decrease the trait values, respectively, compared with the Ler allele. (This figure is available in colour at JXB online.)
QTL mapping for flowering time and total leaf number
This genetic map was used to identify and locate QTL involved in the variation for the various traits in the Ler×Fei-0 RIL population (Fig. 4). Nine and eight loci were detected affecting FT and TLN, respectively, located on all chromosomes except chromosome 4, and together they accounted for 79–81% of the phenotypic variance (see Supplementary Table S5 at JXB online). Five individual loci involved in each trait show small additive effects explaining less than 2.5% of the variation. Only two QTL linked in the upper arm of chromosome 5 displayed large relative effects, each accounting for 7–20% of the phenotypic variance. These QTL identified conservatively a total of 10 genomic regions, two of them (MQC2 and MSAT5.19, on chromosomes 3 and 5, respectively) showing a similar relative effect on FT and TLN (the difference in relative effect was smaller than 0.5%). However, the remaining eight regions had differential relative effects on both traits (Fig. 4; see Supplementary Table S5 at JXB online). Fei-0 alleles in six of these 10 regions increased FT and/or TLN compared with Ler alleles, which is in agreement with the parental values and the transgressive variation observed in the RIL population. Only the two large effect loci located on chromosome 5 interacted genetically, lines carrying Fei-0 alleles in both regions flowering significantly later than expected from their additive effects. This synergy accounted for 15–17% of the phenotypic variance for these traits, and no other significant QTL by QTL interaction was detected throughout the genome (see Supplementary Table S5 at JXB online).
QTL mapping for rates of leaf production
To identify loci that affect the rate of leaf production, RLP/PLR variables estimated in Ler×Fei-0 population were used for QTL mapping. Average rates were estimated during the first 52 d of vegetative development (RLP1-52) and through the total vegetative development of each RIL (Total RLP) in two ways: as ratios of leaf number and time, or as slopes from linear regressions of leaf number on time (see Materials and methods). Nearly the same loci were identified by both methods, although some small effect loci were significantly detected only in one analysis (Fig. 4). In total, five genomic regions were found affecting RLP1-52/sRLP1-52, which accounted for 26–31% of the phenotypic variance. Three of these loci showed a moderate additive effect (>5% of explained variance), while two other QTL presented smaller effects (see Supplementary Table S5 at JXB online). Also five genomic regions affected Total RLP/Total sRLP, their additive effects explaining 62–72% of the variation. Comparison of the QTL found for RLP1-52 and Total RLP showed that only two genomic regions affected both sets of traits (the top of chromosome 3, marker nga172; the top of chromosome 5, marker ICE5), while the six remaining regions appeared as significant only in one of these two RLP variables. In addition, comparison of RLP QTL with the genomic regions affecting FT or TLN shows that seven out of 10 regions affecting the latter traits also significantly affected the rate of vegetative development (Fig. 4). The previously described synergistic interaction between the two large effect QTL on chromosome 5 was only significant on Total RLP, where Fei-0 alleles on both loci interact to increase this rate. One additional genomic region was detected on chromosome 1 (around marker F6D8) affecting RLP traits but not FT or TLN. Overall, these results suggest that most FT and TLN loci also affect RLP, but their relative effect is not steady during vegetative development.
To determine the genetic bases of the RLP/PLR temporal variation during the vegetative phases directly, QTL mapping was carried out with the average rates estimated in the seven consecutive time intervals, from germination to day 100 (RLP1-31, RLP32-42, RLP43-52, RLP53-63, RLP64-77, RLP78-85, and RLP86-100). A total of two to four QTL were detected at each time interval, their additive effects accounting for 15–47% of the phenotypic variance (Fig. 4; see Supplementary Table S6 at JXB online). In contrast to previous analyses, most individual loci explained more than 5% of the phenotypic variance, hence showing a moderate-to-large relative effect. Comparisons among the seven analyses identified a conservative minimum number of ten genomic regions affecting RLP in at least one time interval. Nine of these regions overlap with the above-described FT or TLN loci, whereas only one additional QTL on chromosome 4 appeared affecting RLP64-77.
Analysis of the temporal patterns of QTL effects shows that different loci affected RLP in the various time intervals (Fig. 4). Four genomic regions appeared as significant mainly in the first three time intervals spanning from day 1 to 52, which had been previously mapped for RLP1-52 (chromosome 1, marker NF21M12; chromosome 3, markers nga172 and MQC2; chromosome 5, marker ICE5). By contrast, other four genomic regions detected in time intervals from days 53 to 100 overlapped with the specific QTL affecting Total RLP but not RLP1-52 (bottom of chromosome 1, marker F18B13; chromosome 2, markers ERECTA and T24P15; top of chromosome 5, marker T31P16/FLC). Two additional regions were also significant in a single time interval from 53–100 d, which were not detected in other RLP analyses (chromosome 4, marker nga1139; chromosome 5, marker CIW9). No significant genetic interaction was found among these 10 regions, and a single two-way interaction was detected for RLP1-31 and RLP53-63 involving markers from other genomic regions (see Supplementary Table S6 at JXB online).
In nine of these 10 genomic regions (all except QTL on chromosome 3, marker MQC2) Fei-0 alleles increased RLP, in agreement with the high Fei-0 RLP and the limited transgression observed in the RIL population. However, analysis of the absolute allelic effects of these regions on FT and TLN showed two distinct genetic behaviours (Fig. 4; Supplementary Table S5 at JXB online). In three of these regions (top of chromosome 1, marker NF21M12; chromosome 2, marker ERECTA; top of chromosome 3, marker nga172) Fei-0 alleles reduced FT more than TLN. By contrast, Fei-0 alleles in five other regions (bottom of chromosome 1, marker F18B13; bottom of chromosome 2, marker T24P15; chromosome 5, markers T31P16/FLC, ICE5, and CIW9) increased TLN more than FT. Thus, the fast Fei-0 RLP is determined by at least 10 loci acting mainly on one of two consecutive temporal periods of vegetative development, and showing two major patterns of, presumably, pleiotropic effects on FT and TLN.
Effects of HUA2 and ERECTA on rates of leaf production and flowering initiation
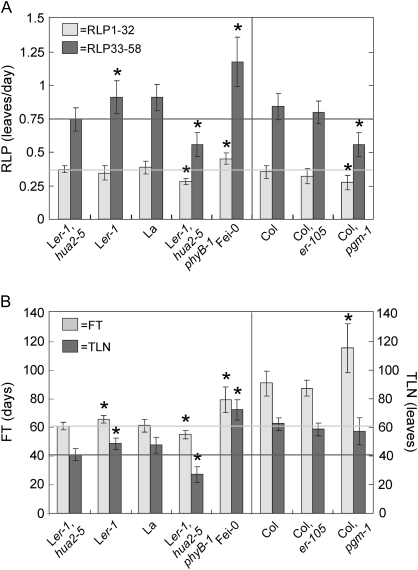
The Ler strain used as parent of the Ler×Fei-0 RILs carries the loss-of-function mutant alleles er-1 (Rédei, 1962) and hua2-5 (Doyle et al., 2005), which overlap with QTL regions identified on chromosomes 2 (ERECTA) and 5 (closely linked to ICE5), respectively. These genes have previously been shown to affect flowering time and/or various leaf developmental traits (Doyle et al., 2005; Van Zanten et al., 2009) suggesting that they might underlie some QTL identified in this population. To test if HUA2 and ERECTA also affect the rate of leaf production, genotypes carrying mutant and wild-type alleles of these genes were compared in Landsberg (Ler-1, hua2-5; Ler-1 and accession La) and Columbia (er-105 and Col) genetic backgrounds (Fig. 5). In addition, the phyB-1 and pgm-1 mutations, known to affect the rate of vegetative development (Koornneef et al., 1995; Caspar et al., 1985), were included as positive controls. As shown in Fig. 5, the genotype Ler-1, hua2-5 not only flowers earlier and with fewer leaves than the Ler-1 line, but also shows significantly reduced RLP from days 33 to 58 (RLP33-58). These phenotypic differences were similar to the allelic effects estimated for QTL located on chromosome 5 (ICE5), indicating that HUA2 underlies those QTL. By contrast, lines carrying er-1 or er-105 mutations did not differ significantly from their corresponding wild-type genotypes for any of the traits analysed (P >0.01), which suggests that genes other than ERECTA account for the QTL on chromosome 2.
Fig. 5.
Rates of leaf production, flowering time, and leaf number of mutant lines. (A) Average rate of leaf production from day 1 to 32 (RLP1-32) and from days 33 to 58 (RLP33-58). (B) Flowering time (FT) and total leaf number (TLN) of the same genotypes. Each panel shows the mean ±SD of 10–15 plants grown under SD photoperiod of the following genotypes: Landsberg parental line of the Ler×Fei-0 RILs carrying the er-1 and hua2-5 mutant alleles (Ler-1, hua2-5); Landsberg line carrying wild-type alleles of HUA2 (Ler-1); Landsberg accession with wild-type ERECTA and HUA2 alleles (La); phyB-1 mutation in Ler-1, hua2-5 genetic background (Ler-1, hua2-5, phyB-1); Fei-0 parental accession; Col accession; er-105 and the pgm-1 mutants in a Col genetic background. Statistical comparisons were carried out between the following pairs of genotypes in the Ler background: Ler-1 and Ler-1, hua2-5; La and Ler-1; Ler-1, hua2-5, phyB-1 and Ler-1, hua2-5; Fei-0 and Ler-1, hua2-5. Col background mutants were compared to Col accession. Significant phenotypic differences are indicated with asterisk on the first genotype of those pair comparisons (P <0.01). Horizontal lines mark the phenotypic levels of the Ler-1, hua2-5 parental line.
Discussion
Natural genetic variation for the rate of leaf production in different vegetative phases
The rate of leaf production (or the plastochron) is a major component of vegetative growth whose genetic mechanisms have just begun to be elucidated. Our analysis of Ler and Fei-0 accessions shows that, in A. thaliana, this rate is not constant throughout vegetative development from germination to flowering initiation, and that such a rate depends on photoperiod. Under short day photoperiods, two distinct phases are distinguished during vegetative development by their RLP/PLR estimated from the appearance of 1–2 mm leaf primordia. In agreement with a previous report of Col background (Groot and Meicenheimer, 2000) the first temporal half of vegetative development shows a low average rate of leaf production, together with a rapid increase in this rate, while the second half is characterized by a high and nearly stable rate of leaf production. These phases partly overlap with the juvenile and adult vegetative phases previously described according to leaf morphological features (Telfer et al., 1997; Steynen et al., 2001), suggesting that the rate of leaf initiation is specific to each developmental phase. In addition, substantial natural genetic variation has been identified among these three accessions for the rate of leaf production in both temporal vegetative phases, Ler and Fei-0 differing as much as induced mutants with strong effects on these rates (Fig. 5). However, Ler/Fei-0 natural variation appears determined by the additive effects of at least 10 loci with a small to moderate effect.
Temporal effects of QTL controlling the rate of leaf production
As shown in this work, the rate of leaf production is a complex trait that varies during vegetative development by the effects of a large number of loci. Therefore, estimation and dissection of the average total rate of leaf production show inevitable limitations for QTL mapping, despite the use of different analytical methods (e.g. TLN/FT ratios or slopes from linear regressions). For instance, QTL identified with the average Total RLP/PLR but not with RLP1-52 are not necessarily phase-specific RLP loci. Since RLP/PLR is changing along vegetative phases, loci affecting the developmental timing of particular phase changes might not alter the rate of leaf production of those phases but they will still affect the average rate estimated on total vegetative development. This limitation is further illustrated by the detection of RLP1-52-specific QTL that are not found with Total RLP. This is probably a consequence of the high RLP in the second vegetative phase, which would strongly reduce the contribution of first-phase-specific loci on the total leaf number and average Total RLP. Hence, a systematic temporal dissection is necessary to decipher the complex genetic bases of this quantitative variation.
Several results from the temporal analyses indicate that the Ler/Fei-0 natural variation for the rate of leaf production is determined by different loci along vegetative development. First, the analysis of F1 hybrid plants shows different dominance behaviour of RLP/PLR in the two plastochron-based vegetative phases. Second, different QTL are detected in the Ler×Fei-0 RIL population when analysing the average RLP/PLR estimated in consecutive time intervals. Genetic interactions changing with the ontogeny of vegetative phases could limit QTL detection power in these analyses. However, this possibility is not supported because two-way QTL interactions only accounted for a small proportion of the phenotypic variance. Thus, the Ler/Fei-0 variation for the rate of leaf production is mostly determined by 10 QTL whose additive effects appear under temporal control. The two main temporal patterns of these loci further suggest that they might be involved mainly in two vegetative phases, the early and late acting QTL roughly corresponding to juvenile and adult phase loci, respectively.
Genetic relationship between flowering initiation and the rate of leaf production
Comparative QTL analyses show that all except one genomic region (the bottom of chromosome 5, marker MSAT5.19) that affect flowering time or leaf number in the Ler×Fei-0 population also affect the rate of leaf production in some but not all periods of vegetative development. This result explains the small independent genetic bases observed for FT and TLN, which is mainly determined by eight genomic regions with two distinct patterns of phenotypic effects on the various traits (Fig. 4). Three of them affect RLP/PLR but show larger absolute effect on FT than TLN, whereas the remaining five regions also affect RLP/PLR but present a larger effect on TLN than FT. The Fei-0 accession carries alleles increasing the rate of leaf production in those eight genomic regions, but it only bears alleles reducing this rate in another locus (middle of chromosome 3, marker MQC2), which explains its faster vegetative development compared to Ler. Assuming that QTL colocations are due to a single locus, these patterns of phenotypic effects can be interpreted as distinct pleiotropic behaviours. Thus, in three loci, Fei-0 alleles increase RLP but accelerate flowering initiation, two of these loci acting early in vegetative development while one seems to function consistently later. In contrast, in five loci, Fei-0 alleles increase RLP but delay flowering initiation. Only one of these loci acts in the first half of vegetative development, while the remaining four loci function in different periods of later vegetative development.
The differential effects of QTL on FT and TLN might reflect different functions of those loci in two distinct processes: developmental phase change and vegetative growth (Poethig, 2003). Taking into account that alterations of the developmental timing of phase changes involve modifications in the leaf number produced in a particular phase, it can be hypothesized that the five loci with a stronger effect on TLN than FT might participate in the regulation of phase changes. Accordingly, the early and late acting loci can be interpreted as affecting the vegetative or reproductive phase changes, respectively. In contrast, those three loci with an opposite behaviour could participate more directly in the control of RLP/PLR growth components since they mainly alter the time invested for leaf production. Alternatively, the differential QTL effects on FT and TLN might be a consequence of their temporal patterns. For instance, due to the slow RLP during early vegetative development, loci involved in the early regulation of vegetative phase change might show a larger effect on FT than TLN. On the contrary, QTL involved in the regulation of late vegetative development or flowering transition might show a smaller effect on FT than TLN due to the faster RLP. Therefore, given the quantitative effects of most of these loci on FT, TLN, and RLP/PLR, and their temporal variation, it is not possible conclusively to speculate on the specific developmental processes affected by these loci.
Genetic and molecular mechanisms underlying the natural variation for flowering time and the rate of vegetative growth
Most QTL detected in this work overlap with QTL identified in some of the large number of quantitative analyses of flowering time carried out in A. thaliana (for recent studies see El-Lithy et al., 2006; Simon et al., 2008; O'Neill et al., 2008). However, the differential temporal and pleiotropic patterns of the QTL described in this study indicate that reported flowering time QTL affect several developmental mechanisms that, directly or indirectly, alter the rate of leaf initiation. Currently, there are nine genes that have been isolated underlying A. thaliana flowering time QTL (reviewed in Alonso-Blanco et al., 2009). CRY2, FLM, FRL1, MAF2-MAF5, FLC, and HUA2, located on chromosomes 1 and 5, overlap with Ler/Fei-0 QTL suggesting that they might correspond to some of those loci (Michaels and Amasino, 1999; El-Assal et al., 2001; Doyle et al., 2005; Werner et al. 2005; Schlappi et al. 2006; Caicedo et al., 2009). By contrast, no QTL was found colocating with the FRIGIDA (FRI) gene, which is a major determinant of the natural variation for flowering time in many crosses (Johanson et al., 2000; Alonso-Blanco et al., 2009). Since Ler carries a loss-of-function allele of FRI (Johanson et al., 2000), this result indicates that Fei-0 also bears FRI defective alleles with similar effects to those of Ler. FRI sequencing has shown that an insertion/deletion, possibly generating a truncated protein without 220 C-terminal amino acids, causes FRI dysfunction of Fei-0 (data not shown). In addition, the Ler parental line of the Ler×Fei-0 population carries loss-of-function alleles of FLC and HUA2 colocating with the two large effect QTL mapped on chromosome 5 (Michaels et al., 2003; Doyle et al., 2005). Supporting that these two genes cause part of the Ler/Fei-0 natural variation, it is found that: (i) Fei-0 carries functional alleles of FLC and HUA2 (sequence data not shown); and (ii) QTL in these regions interact synergistically to delay flowering, in a similar way to that previously described for FLC and HUA2 active alleles (Poduska et al., 2003). In addition, it has been shown that HUA2 also affects the rate of leaf production, demonstrating that it contributes to the overlapping QTL affecting the various traits. Furthermore, HUA2 QTL affects the rate of leaf production earlier during vegetative development than FLC-colocating QTL, in agreement with its role as a positive regulator of FLC and other MADS-box genes like AG and FLM (Doyle et al., 2005; Wang et al., 2007). A gain-of-function natural allele of HUA2 has also been described in the Sy-0 accession that is not present in Fei-0 (Wang et al., 2007), which might be expected to increase the rate of leaf production further. Thus, the pleiotropic effects of HUA2 and the genes underlying the rest of the QTL suggest that they are targets of natural selection acting not only through their phenotypic effects on the timing of flowering initiation but also on the rate of leaf production during vegetative development.
In spite of the high correlation between FT and TLN described in the Ler×Fei-0 cross, our results identify a diversity of natural alleles that might enable partly independent manipulation of flowering time and the rate of vegetative growth. In particular, QTL on top of chromosomes 1 and 3 carry Fei-0 alleles reducing flowering time and increasing leaf production rate. However, understanding the potential usefulness of these alleles for the genetic manipulation of those traits awaits its molecular isolation and identification of the cellular mechanisms involved. In addition, understanding this genetic variation requires determining the extent of shared environmental regulation between the rate of leaf production and flowering time. Both complex traits integrate in their regulation the signals of environmental factors like photoperiod and temperature, but they are also probably regulated by multiple unknown clues (Granier et al., 2002; Ausín et al., 2005; Corbesier and Coupland, 2005; Kim et al., 2009). Since, under natural environments, life cycle phases and environmental factors vary simultaneously, these signals are likely to be differentially perceived and integrated during vegetative development to regulate growth and phase changes. Thus, it is expected that further characterization of existing natural genetic variation under diverse environments will elucidate the role of environmental factors on its temporal regulation, and on the co-ordination of vegetative growth and the various developmental phase changes.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. S1. Rate of vegetative development of Ler, Fei-0, and their reciprocal hybrids under LD photoperiod.
Supplementary Fig. S2. Relationship between flowering time and total leaf number in the Ler×Fei-0 RIL population.
Supplementary Fig. S3. Ler/Fei-0 linkage map.
Supplementary Table S1. Markers used to construct the Ler×Fei-0 genetic map.
Supplementary Table S2. Genotypes of the Ler×Fei RILs for 90 markers used to develop the genetic map.
Supplementary Table S3. Quantitative data used for QTL mapping for flowering time, total leaf number and the various estimates of the rate of leaf production.
Supplementary Table S4. Correlations among traits measured in the Ler×Fei-0 RIL population.
Supplementary Table S5. QTL affecting flowering time (FT), total leaf number (TLN), and rates of leaf production during the first 52 d of vegetative development (RLP1-52/PLR1-52, sRLP1-52/sPLR1-52) or during the complete vegetative development of each RIL (Total RLP/Total PLR, Total sRLP/Total sPLR).
Supplementary Table S6. QTL affecting average rates of leaf production in seven consecutive time intervals from day 1 to 100 after germination (RLP1-31, RLP32-42, RLP43-52, RLP53-63, RLP64-77, RLP78-85, and RLP86-100).
Supplementary Material
Acknowledgments
The authors thank Jenifer Pozas and Ana Ibañez for excellent technical assistance. This work was funded by grant GEN2006-27786-E/VEG from the Ministerio de Ciencia and Innovación of Spain to CA-B.
References
- Alonso-Blanco C, Aarts MG, Bentsink L, Keurentjes JJ, Reymond M, Vreugdenhil D, Koornneef M. What has natural variation taught us about plant development, physiology, and adaptation? The Plant Cell. 2009;21:1877–1896. doi: 10.1105/tpc.109.068114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, Koornneef M, van Ooijen JW. QTL analysis. Methods in Molecular Biology. 2006;323:79–99. doi: 10.1385/1-59745-003-0:79. [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. The Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausin I, Alonso-Blanco C, Martinez-Zapater JM. Environmental regulation of flowering. International Journal of Developmental Biology. 2005;49:689–705. doi: 10.1387/ijdb.052022ia. [DOI] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Bernartzky R, Tanksley S. Genetics of acting-related sequences in tomato. Theoretical and Applied Genetics. 1986;72:314–324. doi: 10.1007/BF00288567. [DOI] [PubMed] [Google Scholar]
- Bernier G, Kinet J-M, Sachs RM. The physiology of flowering. Boca Raton, Florida: CRC Press; 1981. [Google Scholar]
- Caicedo AL, Richards C, Ehrenreich IM, Purugganan MD. Complex rearrangements lead to novel chimeric gene fusion polymorphisms at the Arabidopsis thaliana MAF2–5 flowering time gene cluster. Molecular Biology and Evolution. 2009;26:699–711. doi: 10.1093/molbev/msn300. [DOI] [PubMed] [Google Scholar]
- Caspar T, Huber SC, Somerville C. Alterations in growth, photosynthesis, and respiration in a starchless mutant of Arabidopsis thaliana L. deficient in chloroplast phosphoglucomutase activity. Plant Physiology. 1985;79:11–17. doi: 10.1104/pp.79.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase K, Adler FR, Lerk KG. EPISTAT: a computer program for identifying and testing interactions between pairs of quantitative trait loci. Theoretical and Applied Genetics. 1997;242:81–89. [Google Scholar]
- Clauss MJ, Cobban H, Mitchell-Olds T. Cross-species microsatellite markers for elucidating population genetic structure in Arabidopsis and Arabis (Brassiceae) Molecular Ecology. 2002;11:591–601. doi: 10.1046/j.0962-1083.2002.01465.x. [DOI] [PubMed] [Google Scholar]
- Clerget B, Dingkuhn M, Goze E, Rattunde HF, Ney B. Variability of phyllochron, plastochron and rate of increase in height in photoperiod-sensitive sorghum varieties. Annals of Botany. 2008;101:579–594. doi: 10.1093/aob/mcm327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockcroft CE, den Boer BG, Healy JM, Murray JA. Cyclin D control of growth rate in plants. Nature. 2000;405:575–579. doi: 10.1038/35014621. [DOI] [PubMed] [Google Scholar]
- Corbesier L, Coupland G. Photoperiodic flowering of Arabidopsis: integrating genetic and physiological approaches to characterization of the floral stimulus. Plant, Cell and Environment. 2005;28:54–66. [Google Scholar]
- Doyle MR, Bizzell CM, Keller MR, Michaels SD, Song J, Noh YS, Amasino RM. HUA2 is required for the expression of floral repressors in Arabidopsis thaliana. The Plant Journal. 2005;41:376–385. doi: 10.1111/j.1365-313X.2004.02300.x. [DOI] [PubMed] [Google Scholar]
- El-Assal SE-D, Alonso-Blanco C, Peeters AJ, Raz V, Koornneef M. A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nature Genetics. 2001;29:435–440. doi: 10.1038/ng767. [DOI] [PubMed] [Google Scholar]
- El-Lithy ME, Bentsink L, Hanhart CJ, Ruys GJ, Rovito D, Broekhof JL, van der Poel HJ, van Eijk MJ, Vreugdenhil D, Koornneef M. New Arabidopsis recombinant inbred line populations genotyped using SNPWave and their use for mapping flowering-time quantitative trait loci. Genetics. 2006;172:1867–1876. doi: 10.1534/genetics.105.050617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granier C, Massonnet C, Turc O, Muller B, Chenu K, Tardieu F. Individual leaf development in Arabidopsis thaliana: a stable thermal-time-based programme. Annals of Botany. 2002;89:595–604. doi: 10.1093/aob/mcf085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot EP, Meicenheimer RD. Comparison of leaf plastochron index and allometric analyses of tooth development in Arabidopsis thaliana. Journal of Plant Growth Regulation. 2000;19:77–89. doi: 10.1007/s003440000008. [DOI] [PubMed] [Google Scholar]
- Helliwell CA, Chin-Atkins AN, Wilson IW, Chapple R, Dennis ES, Chaudhury A. The Arabidopsis AMP1 gene encodes a putative glutamate carboxypeptidase. The Plant Cell. 2001;13:2115–2125. doi: 10.1105/TPC.010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL. Arabidopsis map-based cloning in the post-genome era. Plant Physiology. 2002;129:440–450. doi: 10.1104/pp.003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science. 2000;290:344–347. doi: 10.1126/science.290.5490.344. [DOI] [PubMed] [Google Scholar]
- Kawakatsu T, Itoh J, Miyoshi K, Kurata N, Alvarez N, Veit B, Nagato Y. PLASTOCHRON2 regulates leaf initiation and maturation in rice. The Plant Cell. 2006;18:612–625. doi: 10.1105/tpc.105.037622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T, Taramino G, Itoh J, et al. PLASTOCHRON3/GOLIATH encodes a glutamate carboxypeptidase required for proper development in rice. The Plant Journal. 2009;58:1028–1040. doi: 10.1111/j.1365-313X.2009.03841.x. [DOI] [PubMed] [Google Scholar]
- Kim D-H, Doyle MR, Sung S, Amasino RM. Vernalization in plants. Annual Review of Cell Developmental Biology. 2009;25:21–44. [Google Scholar]
- Klepper B, Rickman RW, Peterson CM. Quantitative characterization of vegetative development in small cereal grains. Agronomy Journal. 1982;74:789–793. [Google Scholar]
- Kobayashi Y, Weigel D. Move on up, it's time for change: mobile signals controlling photoperiod-dependent flowering. Genes and Development. 2007;21:2371–2384. doi: 10.1101/gad.1589007. [DOI] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. The Plant Journal. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Hanhart C, Loenen-Martinet P, Blankestijn de Vries H. The effect of daylength on the transition to flowering in phytochrome-deficient, late-flowering and double mutants of Arabidopsis thaliana. Physiologia Plantarum. 1995;95:260–266. [Google Scholar]
- Koornneef M, Hanhart CJ, van der Veen JH. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Molecular and General Genetics. 1991;229:57–66. doi: 10.1007/BF00264213. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Rolff E, Spruit CJP. Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana L. Heynh. Zeitschrift für Pflanzenphysiologie. 1980;100:147–160. [Google Scholar]
- Kosambi DD. The estimation of map distances from recombination values. Annals of Eugenics. 1944;12:172–175. [Google Scholar]
- Lamoreaux RJ, Chaney WR, Brown KM. The plastochron index: a review after two decades of use. American Journal of Botany. 1978;65:586–593. [Google Scholar]
- Loudet O, Chaillou S, Camilleri C, Bouchez D, Daniel-Vedele F. Bay-0×Shahdara recombinant inbred line population: a powerful tool for the genetic dissection of complex traits in Arabidopsis. Theoretical and Applied Genetics. 2002;104:1173–1184. doi: 10.1007/s00122-001-0825-9. [DOI] [PubMed] [Google Scholar]
- Martínez-Zapater JM, Jarillo JA, Cruz-Alvarez M, Roldán M, Salinas J. Arabidopsis late-flowering fve mutants are affected in both vegetative and reproductive development. The Plant Journal. 1995;7:543–541. [Google Scholar]
- Martinez-Zapater JM, Somerville CR. Effect of light quality and vernalization on late-flowering mutants of Arabidopsis thaliana. Plant Physiology. 1990;92:770–776. doi: 10.1104/pp.92.3.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. The Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, He Y, Scortecci KC, Amasino RM. Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2003;100:10102–10107. doi: 10.1073/pnas.1531467100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto N, Goto Y, Matsui M, Ukai Y, Morita M, Nemoto K. Quantitative trait loci for phyllochron and tillering in rice. Theoretical and Applied Genetics. 2004;109:700–706. doi: 10.1007/s00122-004-1690-0. [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Ahn BO, Kawakatsu T, Ito Y, Itoh J, Nagato Y, Kurata N. PLASTOCHRON1, a timekeeper of leaf initiation in rice, encodes cytochrome P450. Proceedings of the National Academy of Sciences, USA. 2004;101:875–880. doi: 10.1073/pnas.2636936100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M, Hu TT, Ishino Y, et al. The pattern of polymorphism in. Arabidopsis thaliana. PLoS Biology. 2005;3:e196. doi: 10.1371/journal.pbio.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill CM, Morgan C, Kirby J, et al. Six new recombinant inbred populations for the study of quantitative traits in Arabidopsis thaliana. Theoretical and Applied Genetics. 2008;116:623–634. doi: 10.1007/s00122-007-0696-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla JM, Otegui ME. Co-ordination between leaf initiation and leaf appearance in field-grown maize Zea mays: genotypic differences in response of rates to temperature. Annals of Botany London. 2005;96:997–1007. doi: 10.1093/aob/mci251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pico FX, Mendez-Vigo B, Martinez-Zapater JM, Alonso-Blanco C. Natural genetic variation of Arabidopsis thaliana is geographically structured in the Iberian peninsula. Genetics. 2008;180:1009–1021. doi: 10.1534/genetics.108.089581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poduska B, Humphrey T, Redweik A, Grbić V. The synergistic activation of FLOWERING LOCUS C by FRIGIDA and a new flowering gene AERIAL ROSETTE 1 underlies a novel morphology in Arabidopsis. Genetics. 2003;163:1457–1465. doi: 10.1093/genetics/163.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig RS. Phase change and the regulation of developmental timing in plants. Science. 2003;301:334–336. doi: 10.1126/science.1085328. [DOI] [PubMed] [Google Scholar]
- Pouteau S, Ferret V, Gaudin V, Lefebvre D, Sabar M, Zhao G, Prunus F. Extensive phenotypic variation in early flowering mutants of Arabidopsis. Plant Physiology. 2004;135:201–211. doi: 10.1104/pp.104.039453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge MJ, Wagner DR. The arabidopsis serrate gene encodes a zinc-finger protein required for normal shoot development. The Plant Cell. 2001;13:1263–1279. doi: 10.1105/tpc.13.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rédei GP. Single locus heterosis. Zeitschrift für Vererbungslehre. 1962;93:164–170. [Google Scholar]
- Schlappi MR. FRIGIDA LIKE 2 is a functional allele in Landsberg erecta and compensates for a nonsense allele of FRIGIDA LIKE 1. Plant Physiology. 2006;142:1728–1738. doi: 10.1104/pp.106.085571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M, Loudet O, Durand S, Berard A, Brunel D, Sennesal FX, Durand-Tardif M, Pelletier G, Camilleri C. Quantitative trait loci mapping in five new large recombinant inbred line populations of Arabidopsis thaliana genotyped with consensus single-nucleotide polymorphism markers. Genetics. 2008;178:2253–2264. doi: 10.1534/genetics.107.083899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppe WJ, Bentsink L, Koornneef M. The early-flowering mutant efs is involved in the autonomous promotion pathway of Arabidopsis thaliana. Development. 1999;126:4763–4770. doi: 10.1242/dev.126.21.4763. [DOI] [PubMed] [Google Scholar]
- Steynen QJ, Bolokoski DA, Schultz EA. Alteration in flowering time causes accelerated or decelerated progression through Arabidopsis vegetative phases. Canadian Journal of Botany. 2001;79:657–665. [Google Scholar]
- Telfer A, Bollman KM, Poethig RS. Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development. 1997;124:645–654. doi: 10.1242/dev.124.3.645. [DOI] [PubMed] [Google Scholar]
- Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y. The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. The Plant Cell. 1996;8:735–746. doi: 10.1105/tpc.8.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ooijen JW. Accuracy of mapping quantitative trait loci in autogamous species. Theoretical and Applied Genetics. 1992;84:803–811. doi: 10.1007/BF00227388. [DOI] [PubMed] [Google Scholar]
- Van Ooijen JW. MapQTL Version 4.0: user friendly power in QTL mapping: Addendum to the Manual of version 3.0. Wageningen, The Netherlands: Plant Research International; 2000. [Google Scholar]
- Van Ooijen JW, Voorrips RE. JoinMap® 3.0, Software for the calculation of genetic linkage maps. Wageningen, the Netherlands: Plant Research International; 2001. [Google Scholar]
- Van Zanten M, Snoek LB, Proveniers MC, Peeters AJ. The many functions of ERECTA. Trends in Plant Science. 2009;14:214–218. doi: 10.1016/j.tplants.2009.01.010. [DOI] [PubMed] [Google Scholar]
- Veit B, Briggs SP, Schmidt RJ, Yanofsky MF, Hake S. Regulation of leaf initiation by the terminal ear 1 gene of maize. Nature. 1998;393:166–168. doi: 10.1038/30239. [DOI] [PubMed] [Google Scholar]
- Wang JW, Schwab R, Czech B, Mica E, Weigel D. Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. The Plant Cell. 2008;20:1231–1243. doi: 10.1105/tpc.108.058180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Sajja U, Rosloski S, Humphrey T, Kim MC, Bomblies K, Weigel D, Grbic V. HUA2 caused natural variation in shoot morphology of A. thaliana. Current Biology. 2007;17:1513–1519. doi: 10.1016/j.cub.2007.07.059. [DOI] [PubMed] [Google Scholar]
- Werner JD, Borevitz JO, Warthmann N, Trainer GT, Ecker JR, Chory J, Weigel D. Quantitative trait locus mapping and DNA array hybridization identify an FLM deletion as a cause for natural flowering-time variation. Proceedings of the National Academy of Sciences, USA. 2005;102:2460–2465. doi: 10.1073/pnas.0409474102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RN, Heckman JW, Somerville CR. Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiology. 1992;100:403–408. doi: 10.1104/pp.100.1.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. 2009;138:750–759. doi: 10.1016/j.cell.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Poethig RS. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development. 2006;133:3539–3547. doi: 10.1242/dev.02521. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.