Abstract
An efficient Agrobacterium-mediated durum wheat transformation system has been developed for the production of 121 independent transgenic lines. This improved system used Agrobacterium strain AGL1 containing the superbinary pGreen/pSoup vector system and durum wheat cv Stewart as the recipient plant. Acetosyringone at 400 μM was added to both the inoculation and cultivation medium, and picloram at 10 mg l−1 and 2 mg l−1 was used in the cultivation and induction medium, respectively. Compared with 200 μM in the inoculation and cultivation media, the increased acetosyringone concentration led to significantly higher GUS (β-glucuronidase) transient expression and T-DNA delivery efficiency. However, no evident effects of acetosyringone concentration on regeneration frequency were observed. The higher acetosyringone concentration led to an improvement in average final transformation efficiency from 4.7% to 6.3%. Furthermore, the concentration of picloram in the co-cultivation medium had significant effects on callus induction and regeneration. Compared with 2 mg l−1 picloram in the co-cultivation medium, increasing the concentration to 10 mg l−1 picloram resulted in improved final transformation frequency from 2.8% to 6.3%, with the highest frequency of 12.3% reached in one particular experiment, although statistical analysis showed that this difference in final transformation efficiency had a low level of significance. Stable integration of foreign genes, their expression, and inheritance were confirmed by Southern blot analyses, GUS assay, and genetic analysis. Analysis of T1 progeny showed that, of the 31 transgenic lines randomly selected, nearly one-third had a segregation ratio of 3:1, while the remainder had ratios typical of two or three independently segregating loci.
Keywords: Acetosyringone, Agrobacterium tumefaciens, durum wheat cv Stewart, picloram, transformation
Introduction
Genetic transformation is fundamental to wheat molecular genetics and improvement through genetic engineering. Agrobacterium-mediated transformation and microparticle bombardment are the two widely used methods for wheat genetic transformation. So far, the majority of wheat genetic transformation that has been reported utilized microparticle bombardment (Vasil et al., 1992, 1993; Weeks et al., 1993; Becker et al., 1994; Nehra et al., 1994; Zhou et al., 1995; Altpeter et al., 1996; Barro et al., 1998; Lazzeri and Jones, 2009). However, when transgene arrangement/copy number are scrutinized as part of the regulatory processes (EFSA, 2006, 2007), Agrobacterium-mediated transformation has advantages (Komari et al., 1996; Hiei et al., 1997; Hansen and Wright, 1999; Gheysen et al., 1998; Shibata and Liu, 2000; Dai et al., 2001; Matthews et al., 2001; Miller et al., 2002; Jones, 2005; Jones et al., 2005; Travella et al., 2005; Sparks and Jones, 2009).
There has been significant progress in Agrobacterium transformation of wheat (Triticum aestivum L.) in recent years, but it is still confined mainly to a few responsive varieties with quite different transformation frequencies such as the model spring genotype ‘Bobwhite’ (Cheng et al., 1997, 2003; Hu et al., 2003), other spring wheat varieties, Fielder, Cadenza, and Veery 5 (Weir et al., 2001; Khanna and Daggard, 2003; Wu et al., 2003; Jones et al., 2005), and the winter wheat variety Florida (Wu et al., 2003; Jones et al., 2005). Efficient Agrobacterium-mediated transformation of wheat via in planta inoculation was reported by Risacher et al. (2009). This method is used successfully by Biogemma in France and by NIAB in the UK, but it is still too early to say whether it will be adopted more widely due to both the patent (Risacher and Craze 1992) and the specific expertise required. Considerable efforts have also been devoted to the optimization of Agrobacterium-mediated transformation of mature embryos and the development of direct Agrobacterium-mediated transformation of germline cells in shoot meristem, seeds, or developing inflorescences (floral dip), with limited success (Zhao et al., 2006; Agarwal et al., 2009; Ding et al., 2009; Zale et al., 2009).
To date, there have been many studies examining factors affecting the efficiency of Agrobacterium-mediated transformation of cereals. Most commonly, these have examined factors such as medium composition, Agrobacterium inoculation procedure, Agrobacterium strain and vector, selective agent, and explant type and size (Bartlett et al., 2008). In wheat, factors influencing the efficiency of T-DNA delivery have been evaluated including different explant types, cell density of Agrobacterium for inoculation, inoculation period, co-culture medium, surfactants in the inoculation medium, and the induction agent, such as acetosyringone, in the inoculation and co-culture media (Cheng et al., 1997). Khanna and Daggard (2003) reported that the use of superbinary vectors carrying additional vir genes, the addition of 250 μM acetosyringone to the inoculation and co-cultivation media, and the modification of the polyamine ratio in the regeneration medium greatly improved the final wheat transformation efficiency to 3.9%. Using freshly isolated embryos as the target explant, Agrobacterium inoculation procedures such as the effect of acetosyringone on DNA delivery, the effect of Silwet L-77 on embryo survival, callus induction, and DNA delivery, and the effect of pre-culture, inoculation time, and length of co-cultivation were examined (Wu et al., 2003). During the inoculation and co-cultivation period, acetosyringone appeared to act together with specific temperature requirements and an acidic environment to promote the expression of Agrobacterium vir genes. The presence of 200 μM acetosyringone markedly increased T-DNA delivery in bread wheat transformation (Wu et al., 2003). However, 400 μM acetosyringone might have a possible harmful effect on T-DNA transfer and it is not an absolute requirement for all genotypes and explants for stable transformation (Cheng et al., 1997; Amoah et al., 2001; Weir et al., 2001).
In addition, the major constraint to the improvement of Agrobacterium-mediated transformation of wheat is the ability to regenerate plants successfully from transformed callus. Medium composition, especially the concentration of auxin, is one of the major factors influencing embryogenic response and regeneration of the explants after infection by Agrobacterium. Two auxins commonly used to induce somatic embryogenesis and allow regeneration from cereal tissues are 2,4-dichlorophenoxyacetic acid (2,4-D) and picloram (Wernicke and Milkovits, 1987). Both are synthetic auxins with distinct effects on the induction of cell division, proliferation, and further regeneration. They act by inducing auxin-sensitive non-dividing cells, arrested in G1, to re-enter S phase and mitosis. The timing of this process depends on the auxin type and on the concentration applied (Wernicke and Milkovits, 1987; Barcelo et al., 1992; Barro et al., 1998). 2,4-D is the most commonly used exogenous growth regulator added to the culture medium for cereals, whereas picloram in the induction medium gave rise to more regenerative cultures than 2,4-D in wheat, with the optimal range between 2 mg l−1 and 6 mg l−1 (Barro et al., 1999).
Durum wheat (Triticum turgidum L. var. durum) is a major food crop of the Mediterranean Basin, with a cultivated area of ∼9% of the world total wheat crop; it possesses an ultra hard endosperm classification and is mainly used for making pasta and semolina (Shewry et al., 1995). It is also a vital part of the development of wheat functional genomic resources due to its ancestral AABB genome composition. Whereas the transformation of bread wheat varieties remains at low frequency and genotype dependent, an efficient high throughput Agrobacterium-mediated durum wheat transformation system may contribute to furthering our understanding of important physiological processes and elucidation of wheat gene functions. However, it has received less attention than bread wheat for genetic transformation, and most of the transformation done so far was by the biolistic-mediated method. The first successful genetic transformation was achieved via bombardment of cv Medora (Bommineni et al., 1997). He et al. (1999) transformed three pasta wheat cultivars (L35, Ofanto, and Svevo) and one breeding line (Latino×Lira) with the high molecular weight glutenin subunit genes. Wiley et al. (2007) transformed durum wheat cv Ofanto with PinA and PinB to study their expression pattern and role in determining grain hardness. Gadaleta et al. (2006) studied the effect of pre-treatment and different explant types on durum transformation as well as reporting the use of phosphomannose isomerase (pmi) as a positive selectable marker for durum wheat variety Svevo, and achieved a higher transformation efficiency than when using bar as the selectable marker. Pellegrineschi et al. (2002) studied the effect of pre-treatment on immature embryos (IEs) for achieving good transformation efficiency of three elite durum wheat varieties Mexicali, D5c31YN S74, and D5c31YN S48. Patnaik et al. (2006) and Vishnudasan et al. (2005) tested mature embryos as starter explants for both bread and durum wheat with limited success. However, the first case of successful A. tumefaciens-mediated durum wheat transformation was reported by Wu et al. (2008) using Ofanto as donor plants. The effect of additional vir genes in the helper plasmid on T-DNA delivery and stable transformation efficiencies was compared (Wu et al., 2008). To date, unlike bread wheat transformation, which was dominated by using the model variety Bobwhite, researchers used diverse durum wheat cultivars or breeding lines for introducing genes of interest or for transformation methodology development (Jones et al., 2005).
High efficiency of T-DNA delivery and good regeneration ability after Agrobacterium inoculation remain crucial for the success of Agrobacterium-mediated wheat transformation. Based on empirical studies through manipulation of culture medium components, such as phenolic inducers and growth regulators, here the successful Agrobacterium-mediated transformation of durum wheat cultivar cv Stewart using the superbinary pGreen/pSoup vector system with improved transformation efficiency is reported. The effects of acetosyringone and picloram in the inoculation and co-cultivation media on embryogenesis, regeneration, and final transformation efficiency were also investigated.
Materials and methods
Donor plant material and explants
Durum wheat cv Stewart was grown in the field at the CAAS experimental station in mid February 2008 under normal field management conditions. Each plot consisted of 2 m rows spaced 0.3 m apart. The plants were kept free of weeds by 1–2 applications of the broad-range herbicide Tantizon (isomethiozin) (Baodai, Suzhou, China) in early March and/or before the jointing stage. To avoid mildew, the fungicide Triadimefon (Lvba, Jinan, China) was applied 2–3 times at the recommended concentration either as a preventative during the jointing period or whenever the first sign of disease appeared during later stages. Rogor (40%) (Fubang, Shandong, China) was used 2–3 times as a pesticide to manage aphids at heading, flowering, and grain-filling stages. Ears were harvested in late May at 12–14 days after pollination for isolation of fresh embryos. Immature caryopses were removed from the spikelet, surface-sterilized first with 70% ethanol for 1 min, followed by 10% (v/v) Domestos (Lever) for 15 min, and then rinsed three times with sterilized distilled water. IEs were isolated under aseptic conditions after removal of the entire axis. The length of IEs ranged from 0.8 mm to 1.5 mm.
Agrobacterium tumefaciens strain and plasmid vectors
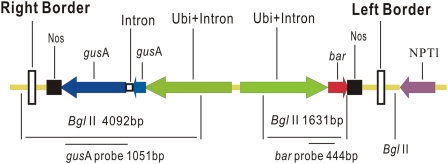
Agrobacterium tumefaciens strain AGL1 (Lazo et al., 1991) was used for all experiments. The plasmid combination pAL154 and pAL156 based on the pSoup/pGreen series (Hellens et al., 2000) was kindly provided by Drs Wendy Harwood and Mark Smedley (John Innes Centre). The pSoup-derived plasmid (pAL154) contained the 15 kb Komari fragment and functioned as a helper plasmid providing replication functions in trans for pAL156. The pGreen-based plasmid (pAL156) contained T-DNA incorporating the bar gene and a modified gusA gene [coding for β-glucuronidase (GUS) with a maize RPOT (T3/T7-like DNA-dependent RNA polymerase) intron inserted at nucleotide 385 to prevent expression in Agrobacterium (Bourdon et al., 2001; Ke et al., 2002). Both genes were driven by the maize Ubiquitin1 promoter plus the ubiquitin1 intron (Christensen and Quail, 1996). The bar gene was adjacent to the left border and gusA was adjacent to the right border (Fig 1). Kanamycin and carbenicillin at 100 mg l−1 and 200 mg l−1, respectively, were added to maintain the purity of the AGL1 strain harbouring the pAL154/pAL156 combination.
Fig. 1.
Schematic of the gusA and bar gene expression cassettes in the pAL156 plasmid, BglII restriction sites, and T-DNA borders. (This figure is available in colour at JXB online.)
Wheat transformation
Agrobacterium-mediated transformation was performed following the protocol described by Jones et al. (2005) with a few modifications. Agrobacterium tumefaciens was grown from glycerol stock in 10 ml of MG/L liquid medium (Tingay et al., 1997) supplemented with 1 mg l−1 biotin, 100 mg l−1 kanamycin, and 200 mg l−1 carbenicillin. The culture was incubated overnight at 27–29 °C with shaking (250 rpm) and, when the culture was at density of 1.0–1.5 OD600, cells were pelleted by centrifugation at 4 °C, 4500 g for 10 min and then resuspended in inoculation medium with 200 μM or 400 μM acetosyringone with continued shaking for 1–3 h until the IEs were ready for inoculation. The same concentration of acetosyringone was used in co-cultivation medium thereafter. The media used for inoculation, co-cultivation, and induction were as given in Wu et al. (2009) except for the picloram concentrations which were adjusted to 2, 4, and 10 mg l−1, in three concentration regimes in the co-cultivation medium, respectively. Freshly isolated IEs were pre-cultured with their scutella side up on co-cultivation medium, and then immersed immediately in Agrobacterium suspension for 1–3 h in the dark; Silwet L-77 (Lehle seeds, USA) was added during inoculation at a concentration of 0.015%.
Cultures were incubated at 24–25 °C in the dark. The explants were then transferred to fresh co-cultivation medium. Co-cultivation was carried out in the dark at 24–25 °C. After 3 d of co-cultivation in darkness, explants were transferred to induction medium containing 160 mg l−1 of the antibiotic timentin [Smithkline Beecham, UK; ticarcillin:clavulanic acid (15:1)]. All subsequent media plates contained timentin at this concentration. Explants were maintained on induction medium for 3–4 weeks, after which they were transferred to RDZ medium (R medium plus 5 mg l−1 zeatin and 0.1 mg l−1 2,4-D), and moved to light. After a further 3–4 weeks, regenerating tissues were transferred to RPPT2.5 medium [R medium containing 2.5 mg l−1 L-phosphinothricin (PPT)]. Approximately 3–4 weeks later, plantlets which survived the first round of selection were transferred to a second round on RPPT4.0 medium (R medium containing 4.0 mg l−1 PPT) and, if necessary, a third round of selection. Plants showing resistance to PPT were subjected to a PCR analysis and/or a GUS activity assay. Positive plants were transferred to soil in a 10 cm pot in the glasshouse.
Assay for GUS activity
Transient GUS expression was determined as described by Jones et al. (2005). Explants were sampled after 3 d on induction medium and incubated overnight at 37 °C in X-Gluc buffer containing 1 mM X-Gluc, 100 mM sodium phosphate buffer pH 7.0, 0.5 mM potassium ferricyanide, 0.5 mM potassium ferrocyanide, and 0.1% (v/v) Triton X-100. Blue foci were counted with the aid of a microscope. T-DNA delivery was assessed by counting embryos that had at least one GUS focus and then counting the number of foci per embryo. To assay for stable expression, calli, shoots, leaf fragments, and seeds from regenerating plants were also incubated in X-Gluc buffer overnight at 37 °C, and then chlorophyll was removed from tissues using 70% ethanol under light for 12–24 h.
Ammonium assay for bar gene expression
The ammonium assay, for qualitative detection of phosphinothricin acetyltransferase (PAT) activity was performed according to Wu et al. (2006). After imbibing leaf pieces from transgenic and control plants in the incubation solution, under light for 5 h, a 200 μl aliquot was transferred to a fresh tube and vortexed with 1 ml of the two reagent mixes followed by incubation at 37 °C for 15 min to 30 min in the dark. Finally, the colour of the solution was judged qualitatively as either positive or negative. Leaf samples with no bar gene expression gave a darker blue colour whereas these expressing the bar gene gave an obviously lighter colour of pale blue or white.
PCR and Southern blot analysis of transgenic plants
Genomic DNA was extracted from wheat leaves using the cetyltrimethylammonium bromide (CTAB) method as described by Sambrook et al. (1989). PCR was used to confirm the presence or the absence of transgenes in the primary transformants and their progeny (Pastori et al., 2001). The primer sequences were: bar, 5′-GTCTGCACCATCGTCAACC-3′ and 5′- GAAGTCCAGCTGCCAGAAAC-3′; and gusA, 5′-AGTGTACGTATCACCGTTTGTGTGAAC-3′ and 5′-ATCGCCGCTTTGGACATACCATCCGTA-3′. The annealing temperature was 57 °C and the approximate product length 444 bp for bar; and 62 °C, 1051 bp for gusA. At least two replicates were carried out for each PCR analysis.
For Southern blot analysis, 30 μg of genomic DNA was digested overnight with restriction enzyme BglII, which cut the plasmid as shown in Fig 1, fractionated on 0.8% agarose gel in 1× TAE buffer for 12 h at 60 V, and then blotted to Hybond-N+ membranes (Amersham, Inc.). Hybridization was performed in hybridization reagents (1× HSB, 0.2× Denhardt's III, 0.05 mg ml−1 salmon sperm DNA) at 65 °C for 16 h using the 1051 bp gusA gene fragment as probe, which was labelled by [α-32P]dCTP by a random priming method (Promega). Following hybridization, the membranes were washed twice at low stringency (2× SSC and 0.5% SDS) and then twice at high stringency (0.2× SSC and 0.5% SDS). All washes were done for 15 min at 65 °C. The membrane was then exposed to a storage phosphor screen for 2–3 d and image data were captured on the Molecular Dynamics Phosphor Imager (Amersham, Inc).
Segregation analysis
T0 plants with ∼30 seeds or more were harvested for segregation analysis. The seed kernel was cut transversely across the centre; the half with the embryo was used for planting in the greenhouse, whereas the other half was used for GUS assay. At the 3-/4-leaf stage, leaf material was harvested and subjected to bar gene ammonium assay (Wu et al., 2006) and GUS assay if necessary. Seeds with no histochemical GUS staining were confirmed to be null segregants by PCR.
Statistical analyses
To compare the effects of different concentrations of acetosyringone on embryogenesis, regeneration, and final transformation efficiency, a one-tailed Student's t-test was performed. To compare the effects of different concentrations of picloram on embryogenesis, regeneration, and final transformation efficiency, a one-way analysis of variance (ANOVA) was used on the full data set, with an F-test to assess the overall differences between the three treatments. At the same time, a t-test was also employed to clarify further the significance of the difference existing between the two treatments. Significance (P-value) was evaluated at the 5% level for all comparisons. For each treatment, the standard error of the mean (SE) was calculated based on at least three biological replicates.
Results
Effect of acetosyringone concentration in the inoculation and co-cultivation media on T-DNA transfer, regeneration, and final transformation efficiency
The effect of acetosyringone on T-DNA delivery may vary depending on the genotypes of the donor wheat tested. To investigate the effect of acetosyringone concentration on T-DNA delivery, callus regeneration, and final transformation efficiency in the transformation of cv Stewart, 200 μM or 400 μM acetosyringone, in both the inoculation and co-cultivation medium, was tested. As indicated in the Materials and methods, a two-component binary pGreen/pSoup system was used in this study. The pSoup plasmid pAL154 included the 15 kb Komari fragment supplying extra vir genes to enhance the T-DNA transfer to plant cells (Komari, 1990). The pGreen plasmid pAL156 contained the gusA gene interrupted by an intron which prevented its expression in Agrobacterium cells. Hence, visualization of GUS loci per embryo inoculated could reflect the efficiency of T-DNA transfer to plant cells. Following inoculation, 3 d of co-cultivation (Fig. 2A) and 3 d of culture on induction medium, 15–25 IE calli were sacrificed for GUS assay, with three replicates for each treatment (Fig. 2B). Efficiency of T-DNA delivery was measured as the percentage of embryos with at least one GUS blue spot on the surface of the scutellum and the mean number of blue foci per scutellum. While the concentration of picloram was maintained at 10 mg l−1 in the co-cultivation medium, 12 and seven treatments were performed with the concentration of acetosyringone at 400 μM and 200 μM in the inoculation and cultivation medium, respectively. As indicated in Table 1, Figs. 3 and 4A, B, following the increase in acetosyringone concentration from 200 μM to 400 μM, enhanced GUS transient expression and T-DNA delivery efficiency, in terms of both the embryos that displayed at least one locus of GUS staining on the scutellum surface and the average number of GUS foci per explant, was observed; the embryos that displayed at least one locus of GUS staining increased from 76.5% to 100% (P=0.0185, P <0.05) (Table 1, Fig. 4A), whereas the average number of blue spots per explant increased from 40.3 to 95.8 (P=0.0001, P <0.01) (Table 1, Fig. 4B). Additionally, no obvious sign of necrosis caused by the enhanced infection of Agrobacterium cells between each treatment was found (data not shown).
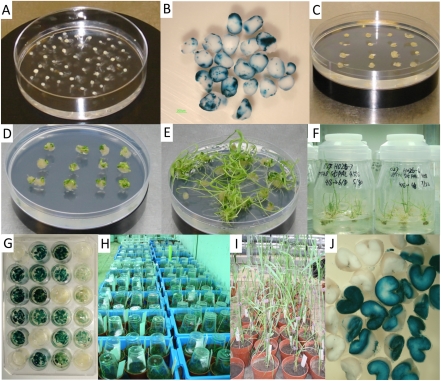
Fig. 2.
The whole process of Agrobacterium-mediated durum wheat cv Stewart transformation. (A) The growth of inoculated embryos after 3 d on co-cultivation medium. (B) GUS transient assay after another 3 d on induction medium. (C) The embryogenesis of callus on induction medium. (D) Regenerated shoots from calli on RDZ medium. (E) The growth of shoots and roots from the calli after 3 weeks on RDZ medium. (F) The growth of surviving plants after two rounds of selection. (G) Stable GUS expression in the leaves of some transgenic lines. (H) Positive plants transferred to soil. (I) The growth of some transgenic lines in the greenhouse. (J) Segregation analysis of GUS expression in T1 seeds.
Table 1.
Acetosyringone concentration in the inoculation and co-cultivation media and its effects on GUS transient expression, T-DNA transfer, regeneration, and final transformation efficiency
| Exp. no. | Treatmentsa | Embryos inoculated | Proportion of embryos with GUS spots (%) | No. of GUS foci per embryo | Frequency of embryogenesis (%) | Frequency of regeneration (%) | GUS-positive plants | PCR gusA | PCR bar | Final transformation efficiency (%) |
| C022-1 | As4P10 | 84 | NDb | ND | 85.7 | 66.7 | 2 | 2 | 2 | 2.4 |
| H026-5 | As4P10 | 84 | ND | ND | 84.5 | 58.1 | 2 | 4 | 4 | 4.8 |
| H025-3 | As4P10 | 64 | ND | ND | 60.9 | 43.8 | 3 | 3 | 3 | 4.7 |
| H027-3 | As4P10 | 87 | 100.0 | 85 | 94.3 | 74.7 | 5 | 5 | 5 | 5.8 |
| H027-4 | As4P10 | 88 | ND | ND | 88.6 | 75.0 | 2 | 3 | 3 | 3.4 |
| H027-5 | As4P10 | 83 | 100.0 | 115 | 89.2 | 69.9 | 4 | 4 | 5 | 6.0 |
| H027-6 | As4P10 | 86 | ND | ND | 97.7 | 58.1 | 3 | 4 | 4 | 4.7 |
| C024-2 | As4P10 | 98 | 100.0 | 86 | 73.5 | 57.8 | 3 | 4 | 4 | 4.1 |
| C024-4 | As4P10 | 102 | ND | ND | 92.2 | 49.0 | 10 | 11 | 12 | 11.8 |
| CL01-2 | As4P10 | 62 | ND | ND | 83.9 | 50.0 | 3 | 3 | 3 | 4.8 |
| X015-1 | As4P10 | 106 | ND | ND | 90.6 | 42.5 | 13 | 13 | 13 | 12.3 |
| X015-2 | As4P10 | 98 | 100.0 | 97 | 71.4 | 40.8 | 11 | 11 | 11 | 11.2 |
| Mean ±SE | 100.0±0.00 | 95.8±6.97 | 84.4±3.08 | 57.2±3.55 | 6.3±0.99 | |||||
| H027-2 | As2P10 | 76 | ND | ND | 69.5 | 62.9 | 1 | 1 | 1 | 1.3 |
| H027-7 | As2P10 | 60 | 77.8 | 35 | 75.0 | 55.0 | 4 | 4 | 4 | 6.7 |
| C024-1 | As2P10 | 115 | ND | ND | 88.7 | 63.5 | 6 | 6 | 6 | 5.2 |
| CL01-1 | As2P10 | 60 | 100.0 | 42 | 81.7 | 58.3 | 2 | 3 | 3 | 5.0 |
| H028-1 | As2P10 | 83 | 58.3 | 46 | 53.0 | 39.8 | 5 | 5 | 5 | 6.0 |
| H028-3 | As2P10 | 43 | n.d. | n.d. | 88.4 | 67.4 | 0 | 1 | 1 | 2.3 |
| H028-7 | As2P10 | 61 | 70.0 | 38 | 73.8 | 39.3 | 2 | 3 | 4 | 6.6 |
| Mean ±SE | 76.5±8.79 | 40.3±2.39 | 75.7±4.69 | 55.2±4.30 | 4.7±0.79 |
As4P10 and As2P10 indicate that the concentrations of acetosyringone and picloram in the co-cultivation medium were 400 μM and 10 mg l−1, and 200 μM and 10 mg l−1, respectively.
ND not determined.
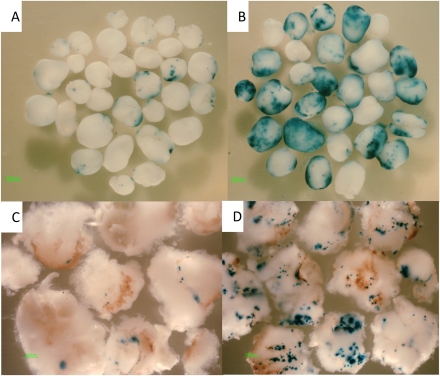
Fig. 3.
Comparison of GUS transient expression in the embryos on the co-cultivation medium containing 200 μM or 400 μM acetosyringone after 6 d and 20 d of Agrobacterium inoculation. (A) Six days after inoculation and co-cultivation on medium containing 200 μM acetosyringone. (B) Six days after inoculation and co-cultivation on medium containing 400 μM acetosyringone. (C) Twenty days after inoculation and co-cultivation on medium containing 200 μM acetosyringone. (D) Twenty days after inoculation and co-cultivation on medium containing 400 μM acetosyringone.
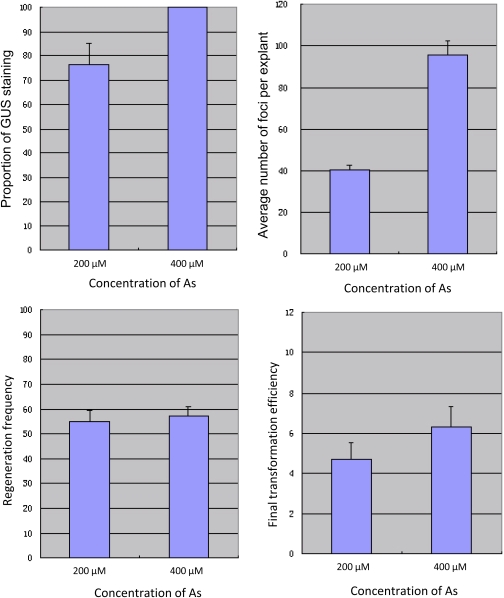
Fig. 4.
Acetosyringone concentration in inoculation and co-cultivation media and its effect on GUS transient expression, T-DNA delivery, regeneration, and final transformation efficiency. (A and B) The effect of acetosyringone concentration on T-DNA delivery, measured as the proportion of embryos that showed GUS expression (A) and the mean number of GUS foci per embryo (B). (C) The effect of acetosyringone concentration on regeneration frequency and (D) its effect on final transformation efficiency. As2 and As4 indicate that the concentration of acetosyringone in the inoculation and co-cultivation medium was 200 μM and 400 μM, respectively. Each assay was repeated at least three times, and the data were presented as means of measurements ±SE. (This figure is available in colour at JXB online.)
To evaluate the possible harmful effect of an increased concentration of acetosyringone on callus regeneration, after 3 weeks on induction medium and another 3 weeks on regeneration medium (Fig. 2D, E), embryogenic calli with shoots and/or roots were used to assess the regeneration frequency (scored as the percentage of IEs that produced shoots and/or roots). As indicated in Table 1 and Fig. 4C, when the concentration of acetosyringone was at 400 μM, the frequencies of regeneration ranged from 40.8% to 75%, with an average of 57.2±3.55%, whereas at 200 μM, the frequencies ranged from 39.3% to 67.4%, with an average of 55.2±4.30%. Thus, while the increased acetosyringone concentration markedly enhanced T-DNA delivery, no statistically significant differences (at the 0.05 level) were observed for its effect on embryogenesis or regeneration.
The effect of acetosyringone concentration on the final transformation efficiency was also evaluated. After regeneration, two rounds of selection were used to produce putative transgenic plants (Fig. 2F). For the confirmation of transgenic plants, leaf material from the surviving plants was taken for PCR analysis (data not shown). After PCR analysis and confirmation of the presence of bar and/or gusA in plants, the GUS assay was used to confirm the expression of the gusA gene integrated into the wheat genome (Fig. 2G). Transgenic plants derived from the same single explants were treated as sister lines unless they were proven to be independent integration events by Southern blot analysis. These positive plants were then transferred into soil and grown to maturity in a greenhouse (Fig. 2H–J). For each treatment, the final transformation efficiency was calculated as the percentage of positive plants produced from the total number of IEs inoculated with Agrobacterium. As indicated in Table 1 and Fig. 4D, when the concentration of acetosyringone was at 400 μM, following the enhanced T-DNA transfer, the final transformation efficiency ranged from 2.4% to 12.3%, with an average of 6.3±0.99%, whereas that with 200 μM acetosyringone treatment was from 1.3% to 6.7%, and the average was 4.7±0.79%. The means transformation efficiency for 200 μM and 400 μM acetosyringone treatments indicated that an increased acetosyringone concentration also resulted in improved final transformation efficiency. However, the statistical significance of this observation was at a low level (P=0.142, P >0.05), due to the relatively low number of transgenic lines obtained in each experiment and the variance between replicate experiments. In subsequent experiments, acetosyringone was used at a concentration of 400 μM in both the inoculation and cultivation medium.
Effect of increased picloram concentration in the co-cultivation medium on callus embryogenesis, regeneration, and final transformation efficiency
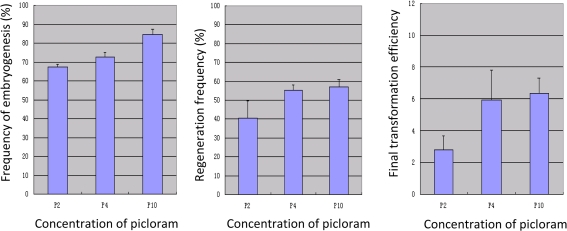
It has been demonstrated previously that co-cultivation of explant tissues with Agrobacterium cells decreases embryogenesis and regeneration frequency compared with non-inoculated controls. The concentration of auxin at the start of this tissue culture process might play an important role in mitigating this effect. A different concentration regime of picloram in the co-cultivation medium was examined for its influence on embryogenesis, regeneration, and final transformation efficiency, whereas its concentration in the induction medium was fixed at 2 mg l−1 for all treatments. The frequency of embryogenesis was scored as the percentage of IEs with good embryogenesis. Picloram in the co-cultivation medium was used at 2, 4, and 10 mg l−1, respectively. For all treatments, the non-inoculated control showed nearly a 100% embryogenesis and regeneration frequency (data not shown), whereas after co-cultivation with Agrobacterium various picloram treatments showed different levels of embryogenesis and regeneration frequencies. As indicated in Table 2, the frequencies of embryogenesis were 67.4±1.40, 72.7±2.39, and 84.4±3.08% when the concentration of picloram in the co-cultivation medium was at 2, 4 and 10 mg l−1, respectively (Fig. 5A). The frequencies of regeneration also improved, with mean values of 40.3±9.52, 55.5±2.80, and 57.2±3.55% following the same increasing picloram concentrations. As predicted, improved embryogenesis and regeneration performance resulted in increased final mean transformation efficiencies of 2.8±0.92, 5.9±1.86, and 6.3±0.99% for picloram treatments of 2, 4, and 10 mg l−1, respectively (Fig. 5A). ANOVA confirmed that the differences in embryogenesis and regeneration frequency between the 2 mg l−1 and 10 mg l−1 treatments were strongly significant (P <0.05). However, despite the higher mean values for transformation efficiency in the 10 mg l−1 treatment, the difference between the 2 mg l−1 and 10 mg l−1 treatments was statistically significant at a relatively low level of P=0.0552 (P >0.05) and the difference between 4 mg l−1 and 10 mg l−1 was not significantly different. The relatively high mean transformation rate at 10 mg l−1 picloram was skewed by three particularly high values from replicates C024-4, X015-1, and X015-2 (Table 2), which indicated that other factors were also influencing the final transformation efficiency.
Table 2.
Picloram concentration in the co-cultivation medium and its effects on embryogenesis, regeneration, and final transformation efficiency
| Exp no. | Treatmentsa,b | Embryos inoculated | Frequency of embryogenesis (%) | Frequency of regeneration (%) | GUS-positive plants | PCR gusA | PCR bar | Final transformation efficiency (%) |
| C023-3 | As4P2 | 113 | 66.4 | 39.8 | 4 | 4 | 4 | 3.5 |
| C022-4 | As4P2 | 107 | 65.7 | 57.0 | 0 | 1 | 1 | 0.9 |
| C023-1 | As4P2 | 104 | 70.2 | 24.0 | 3 | 4 | 4 | 3.9 |
| Mean ±SE | 67.4±1.40 | 40.3±9.52 | 2.8±0.92 | |||||
| H025-1 | As4P4 | 59 | 74.5 | 55.9 | 6 | 6 | 6 | 10.2 |
| H026-3 | As4P4 | 73 | 78.1 | 64.0. | 6 | 7 | 7 | 9.6 |
| H026-6 | As4P4 | 52 | 67.3 | 57.3 | 1 | 1 | 1 | 1.9 |
| H027-1 | As4P4 | 73 | 76.7 | 53.4 | 1 | 1 | 1 | 1.4 |
| H028-6 | As4P4 | 60 | 66.7 | 46.7 | 4 | 4 | 4 | 6.7 |
| Mean± SE | 72.7±2.39 | 55.5±2.80 | 5.9±1.86 | |||||
| C022-1 | As4P10 | 84 | 85.7 | 66.7 | 2 | 2 | 2 | 2.4 |
| H026-5 | As4P10 | 84 | 84.5 | 58.1 | 2 | 4 | 4 | 4.8 |
| H025-3 | As4P10 | 64 | 60.9 | 43.8 | 3 | 3 | 3 | 4.7 |
| H027-3 | As4P10 | 87 | 94.3 | 74.7 | 5 | 5 | 5 | 5.8 |
| H027-4 | As4P10 | 88 | 88.6 | 75.0 | 2 | 3 | 3 | 3.4 |
| H027-5 | As4P10 | 83 | 89.2 | 69.9 | 4 | 4 | 5 | 6.0 |
| H027-6 | As4P10 | 86 | 97.7 | 58.1 | 3 | 4 | 4 | 4.7 |
| C024-2 | As4P10 | 98 | 73.5 | 57.8 | 3 | 4 | 4 | 4.1 |
| C024-4 | As4P10 | 102 | 92.2 | 49.0 | 10 | 11 | 12 | 11.8 |
| CL0l-2 | As4P10 | 62 | 83.9 | 50.0 | 3 | 3 | 3 | 4.8 |
| X015-1 | As4P10 | 106 | 90.6 | 42.5 | 13 | 13 | 13 | 12.3 |
| X015- | As4P10 | 98 | 71.4 | 40.8 | 11 | 11 | 11 | 11.2 |
| Mean ±SE | 84.4±3.08 | 57.2±3.55 | 6.3±0.99 |
As4 indicates that the concentration of acetosyringone in the co-cultivation medium was 400 μM.
P2, P4, and P10 indicate that the concentration of picloram was at 2, 4, and 10 mg l−1, respectively.
Fig. 5.
Picloram concentration in the co-cultivation medium and its effects on callus embryogenesis, regeneration, and final transformation efficiency. (A) The effect of picloram concentration in the co-cultivation medium on callus embryogenesis. (B) The effect of picloram concentration in the co-cultivation medium on regeneration frequency. (C) The effect of picloram concentration in the co-cultivation medium on final transformation efficiency. P2, P4, and P10 indicate that the concentration of picloram in the co-cultivation medium was at 2, 4, and 10 mg l−1, respectively. Each assay was repeated at least three times, and the data were presented as means of measurements ±SE. (This figure is available in colour at JXB online.)
Production and molecular characterization of transgenic plants
After two rounds of selection, PCR analysis was performed to confirm the presence of bar and/or gusA genes in genomic DNA isolated from the putative transgenic plants, and GUS assay was used to detect the expression of the gusA gene integrated into the wheat genome (Fig. 2G). These positive plants were then transferred into soil and grown to maturity in a greenhouse (Fig. 2H, I). In total, 121 bar PCR-positive, transgenic plants were obtained from all treatments, with most of them being morphologically normal, except for five plants showing reduced height and setting very few seeds. Of these 121 plants, 118 were also gusA PCR positive although only 106 plants showed clear blue staining indicative of GUS expression (Table 3). Due to the strict selection pressure applied, no plants were found with only the gusA gene integrated in this experiment.
Table 3.
Summary of the Agrobacterium-mediated durum wheat transformation treatments in developing the improved system
| Treatmentsa,b | Total no of embryos inoculated | Mean frequency of embryogenesis ±SE (%) | Mean frequency of regeneration ±SE (%) | Total no. of GUS-positive plants | Total no. of PCR gusA | Total no. of PCR bar | Mean final transformation efficiency ±SE (%) |
| As2P10 | 498 | 75.7±4.69 | 55.2±4.30 | 20 | 23 | 24 | 4.7±0.79 |
| As4P10 | 1042 | 84.4±3.08 | 57.2±3.55 | 61 | 67 | 69 | 6.3±0.99 |
| As4P4 | 317 | 72.7±2.39 | 55.5±2.80 | 18 | 19 | 19 | 5.9±1.86 |
| As4P2 | 324 | 67.4±1.40 | 40.3±9.52 | 7 | 9 | 9 | 2.8±0.92 |
| Total | 2181 | – | – | 106 | 118 | 121 | – |
As2 and As4 indicate that the concentration of acetosyringone in the co-cultivation medium was 200 μM and 400 μM, respectively.
P2, P4, and P10 indicate that the concentrations of picloram were at 2, 4, and 10 mg l−1, respectively.
‘–’, not determined.
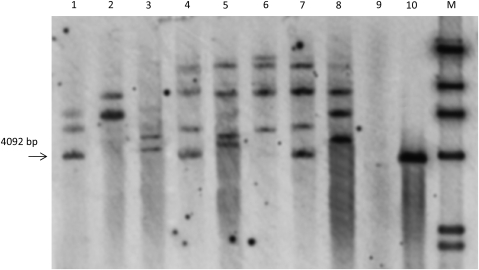
The integration of the gusA gene was tested in eight randomly selected lines by Southern blot analysis. Both plasmid and genomic DNA were digested by BglII and hybridized with the 1051 bp gusA gene probe. As indicated in Fig. 1, BglII cuts five times in the pAL156 plasmid and releases fragments of 4092 bp and 1631 bp containing the gusA and bar sequences, respectively. Therefore, BglII-digested pAL156 showed the expected band of 4092 bp in size when hybridized with the gusA gene probe. In genomic DNA from transgenic plants, where it would be expected for only the T-DNA to be integrated, BglII cuts only three times between the border sequences, and makes other cuts randomly located in the flanking wheat genomic DNA. Therefore, it would be expected that BglII-digested Southerns, probed with the 4092 bp gusA gene probe, would give hybridization bands of at least 4000 bp, with the number of bands indicating approximately the number of transgene copies integrated. As indicated in Fig. 6, gusA-probed filters containing DNA from eight transgenic lines possessed bands of a size ≥4092 bp, and seven of the eight lines had unique banding patterns indicative of independent integration events. Although lanes 4 and 7 had the same band pattern, they were not from the same treatment; therefore, these two were not sister lines. Furthermore, two lines H027-3-6 and H028-7-3 (lanes 2 and 3) displayed two bands, two lines C023-3-1 and H027-2-1 (lanes 1 and 8) had three bands, and four lines C024-1-5, X015-2-1, H027-7-1, and H027-7-2 (lanes 4, 5, 6, and 7) possessed four bands, suggesting a range of between two and four copies of the gusA gene integrated into the durum wheat genome at the same time.
Fig. 6.
Southern blot analysis of eight randomly selected transgenic T0 lines. Genomic and plasmid DNA was digested with BglII and hybridized with a 1051 bp gusA gene fragment. 1, C023-3-1; 2, H027-3-6; 3, H028-7-3; 4, C024-1-5; 5, X015-2-1; 6, H027-7-1; 7, H027-7-2; 8, H027-2-1; 9, non-transformed control; 10, pAL156; M, λ-HindIII ladder.
Transgene segregation and expression in the T1 generation
Transgene segregation was analysed in the seeds or seedlings of 31 randomly selected T0 lines. For each line, 17–87 seeds or seedlings were used for GUS assay. The PAT protein assay was conducted to confirm the expression of the bar gene in the T1 generation plants. Among these selected lines, as shown in Table 4, 29% (9/31) displayed a typical 3:1 segregation ratio for the expression of the gusA gene, indicating one or more copies of the gusA gene integrated at a single segregating locus. The most common segregation ratio was 15:1, indicating the presence of two independently segregating gusA gene loci, as shown by 15 of the 31 lines tested (48.9%). Five lines (16.1%) possessed a 63:1 segregation ratio typical of three loci. Two lines showed a non-Mendelian segregation ratio of 1:1, presumably due to aberrant gamete or seed formation. PCR detection was also performed, which confirmed the GUS expression results. In all of the 31 selected lines, as calculated from PAT activity assays, the bar gene segregated at the same ratio and at the same locus as that of the gusA gene (data not shown).
Table 4.
Segregation analysis of GUS expression in T1 seeds of transgenic lines
| Exp. batch no. | Treatments | Observed GUS (positive:negative) | Expected segregation ratio (positive:negative) | χ2 value | P-value |
| H027-2-1 | As2P10 | 13:6 | 3:1 | 0.4386 | 0.5078** |
| H027-7-1 | As2P10 | 49:1 | 63:1 | 0.0622 | 0.8030** |
| H027-7-2 | As2P10 | 36:3 | 15:1 | 0.1384 | 0.7098** |
| H028-1-3 | As2P10 | 17:8 | 3:1 | 0.6533 | 0.4189* |
| H028-7-3 | As2P10 | 16:1 | 15:1 | 0.0039 | 0.9501** |
| H028-7-11 | As2P10 | 34:4 | 15:1 | 1.1860 | 0.2761* |
| C024-1-4 | As2P10 | 36:2 | 15:1 | 0.1385 | 0.7098** |
| C024-1-5 | As2P10 | 25:8 | 3:1 | 0.0101 | 0.9199** |
| C024-1-6 | As2P10 | 17:1 | 15:1 | 0.0148 | 0.9031** |
| C024-1-7 | As2P10 | 18:1 | 15:1 | 0.0316 | 0.8590** |
| CL01-1-2 | As2P10 | 15:2 | 15:1 | 0.8824 | 0.3476* |
| H027-3-6 | As4P10 | 18:1 | 15:1 | 0.0316 | 0.8590** |
| H027-3-10 | As4P10 | 17:9 | 3:1 | 1.2821 | 0.2575* |
| H027-5-1 | As4P10 | 37:13 | 3:1 | 0.0267 | 0.8702** |
| H027-5-2 | As4P10 | 17:1 | 15:1 | 0.0148 | 0.9031** |
| H027-6-1 | As4P10 | 9:6 | 1:1 | 0.6000 | 0.4385* |
| X015-1-1 | As4P10 | 26:2 | 15:1 | 0.0381 | 0.8453** |
| X015-1-8 | As4P10 | 24:10 | 3:1 | 0.3529 | 0.5524** |
| X015-1-11 | As4P10 | 33:2 | 15:1 | 0.0171 | 0.8958** |
| X015-2-1 | As4P10 | 57:1 | 63:1 | 0.0099 | 0.9209** |
| X015-2-8 | As4P10 | 18:1 | 15:1 | 0.0316 | 0.8590** |
| X015-2-11 | As4P10 | 24:1 | 15:1 | 0.0923 | 0.7613** |
| X015-2-16 | As4P10 | 15:1 | 15:1 | 0.0000 | 1.0000** |
| C024-2-3 | As4P10 | 19:4 | 3:1 | 0.7101 | 0.3994* |
| C024-4-1 | As4P10 | 86:1 | 63:1 | 0.0965 | 0.7561** |
| C024-4-4 | As4P10 | 44:1 | 63:1 | 0.1273 | 0.7212** |
| C024-4-11 | As4P10 | 40:1 | 63:1 | 0.2048 | 0.6509** |
| C024-4-15 | As4P10 | 23:1 | 15:1 | 0.1778 | 0.6733** |
| CL01-2-1 | As4P10 | 10:11 | 1:1 | 0.0476 | 0.8273** |
| C023-3-1 | As4P2 | 15:9 | 3:1 | 2.0000 | 0.1573* |
| H025-1-20 | As4P4 | 24:4 | 3:1 | 1.7143 | 0.1904* |
χ2 results are shown for the closest Mendelian ratios to be observed. χ2 values indicate that all observed frequencies were not significantly different from those expected.
aAs2 and As4 indicate that the concentration of acetosyringone in the co-cultivation medium was 200 μM and 400 μM, respectively.
bP2, P4, and P10 indicate that the concentration of picloram was at 2, 4, and 10 mg l−1, respectively.
P >0.5, in very good agreement with 3:1, 15:1, or 1:1.
*0.1 < P < 0.5, in good agreement with 3:1, 15:1, 63:1, or 1:1.
Discussion
The influence of picloram concentration in the co-cultivation medium on regeneration and transformation efficiency
In this study, the effects of different concentrations of picloram used in the co-cultivation medium on induction of embryogenesis, regeneration, and final efficiency of Agrobacterium-mediated durum wheat cv Stewart transformation were investigated for the first time. The co-cultivation step marks the beginning of the callus induction phase with de-differentiation and cell division of the immature scutella epithelial cells. It is thought that auxin levels applied at this stage are likely to affect subsequent tissue culture steps. Compared with 2 mg l−1 picloram in the co-cultivation medium, the present results strongly suggested that an increase in the picloram concentration to 4 mg l−1 or 10 mg l−1 led to improved embryogenesis and regeneration performance, and the efficiency of final transformation was greatly increased in some replicates. However, no statistically significant difference was observed in the recovery of T0 transformants from the 4 mg l−1 and 10 mg l−1 picloram treatments (Table 2) and, in order to keep the auxin levels as low as possible to avoid somaclonal mutations and other unwanted effects, 4 mg l−1 seems a good compromise.
The effect of the higher concentrations of picloram on the final transformation efficiency might not be due simply to its influence on embryogenesis and regeneration as suggested by Barro et al. (1998). As indicated in the present results, the increase in the final transformation efficiencies was not proportional to the changes of frequency of embryogenesis and regeneration (Table 3). It is known that auxins play a role in the activation of genes involved in cell de-differentiation and division (Dudits et al., 1991), and cells in the S phase (DNA synthesis phase) of the cell cycle are more predisposed to the integration of foreign DNA (Villemont et al., 1997). The type and concentration of auxin determined the cell division, proliferation and further regeneration, and the recovery of transformed plants (Wernicke and Milkovits, 1987; Barcelo et al., 1992; Barro et al., 1998). For example, for scutellum cultures of durum wheat cv Desf, picloram can significantly increase the frequency of regeneration compared with 2,4-D, with the optimized concentration of 2 mg l−1 in the induction medium (He et al., 2001). In wheat biolistic transformation, picloram in the induction medium gave rise to more regenerative cultures and produced more transgenic plants from scutellum cultures than 2,4-D, with the highest transformation frequency reached at 2.5% from cultures induced on medium containing 4 mg l−1 picloram (Barro et al., 1999). Wu et al. (2008) reported that the presence of 2 mg l−1 picloram in the co-cultivation medium for the Agrobacterium-mediated transformation of durum wheat cv Ofanto, using the same superbinary pGreen/pSoup system and transformation procedure as applied here, resulted in final transformation efficiencies of between 0.6% and 9.7%, with an average of 3.1%. For the different genotype used in this study, compared with the result with 2 mg l−1 picloram treatments, it is concluded that increased concentrations of picloram in the co-cultivation medium for Agrobacterium-mediated transformation had a tendency to improve the final transformation efficiency but were probably acting in combination with other, as yet unidentified, factors to give unusually high transformation rates in some replicates. Considering that different wheat genotypes might have various degrees of responses to picloram treatments, in practice it is suggested that different concentrations of picloram in the co-cultivation and induction media should be tested for a given wheat variety.
The influence of acetosyringone concentration on regeneration and transformation efficiency
It is accepted that the concentration of acetosyringone in the inoculation and co-cultivation media plays an important role in Agrobacterium-mediated wheat transformation and needs to be optimized for each explant type and genotype. For freshly isolated embryos, the presence of acetosyringone (200 μM) and glucose in the inoculation and co-culture media was crucial for efficient T-DNA delivery (Cheng et al., 1997; Wu et al., 2003). Enhanced transient green fluorescent protein (GFP) expression was observed in wheat cell clusters with acetosyringone at 400 μM in the co-cultivation medium but not the inoculation medium (Weir et al., 2001). While no gusA expression was observed when acetosyringone was excluded from the inoculation and co-culture media using wheat inflorescence tissue as explant, an increase in the concentration of acetosyringone from 200 μM to 400 μM produced fewer responding explants and fewer GUS spots per explant, indicating a possible harmful effect on T-DNA transfer (Amoah et al., 2001). The need for acetosyringone has been reported for a variety of wheat explant types (Cheng et al., 1997; McCormac et al., 1998; Amoah et al., 2001), but not for wheat cell suspension cultures where exogenous induction agents were not necessary for stable transformation (Cheng et al., 1997).
In this study, enhanced GUS transient expression and T-DNA delivery efficiency, in terms of both the embryos that displayed at least one locus of GUS staining on the scutellum surface and the average number of GUS foci per explant, were observed following the increase in acetosyringone concentration from 200 μM to 400 μM in the inoculation and co-cultivation media. However, no strongly significant differences of its effect on regeneration were observed. The mean transformation efficiency of 200 μM treatments was 4.7%, lower than that of 400 μM treatments (6.3%) (Table 1), but the statistical significance of this difference was weak (P=0.1422, P >0.05), due to the variation between replicate batches. This variation might be caused by the different physiological status of donor plants which may play an important role in determining the final transformation efficiency, although additional work will be needed to clarify this unequivocally.
The efficiency of genetic transformation of durum wheat varieties
The efficiency reported here for Agrobacterium-mediated transformation of durum wheat cv Stewart in the improved protocol (average 6.3%, with a maximum in one batch of 12.3%) was higher than that reported for bread wheat and other durum wheat varieties. For example, Khanna and Daggard (2003) reported 1.2–3.9% for the variety Veery-5 using the LBA4404/pHK21 strain and vector combination (pHK21 is also a superbinary vector containing additional vir genes), while Cheng et al. (1997) achieved 1.4–4.3% efficiency for the variety Bobwhite using C58ABI/pMON18365. Wu et al. (2008) used the same superbinary pGreen/pSoup system (pAL154/pAL156) for the Agrobacterium-mediated transformation of durum wheat cv Ofanto and obtained final transformation efficiencies of between 0.6% and 9.7%, with an average of 3.1%, but transformation of bread wheat varieties Florida and Cadenza with the same Agrobacterium strain and plasmid combinations gave only 0.3–3.0% efficiency (Wu et al., 2003). When Chinese bread wheat varieties (Xinchun 9 and Kenong 199) were transformed with the same protocol as used in this report, the efficiency was almost at the same level as reported, ranging from 0.5% to 3.3% (data not shown). The numbers of transformed lines and efficiencies of transformation using the protocol described here were also significantly higher than those reported for durum wheat using biolistics as observed by Wu et al. (2008). For example, Wiley et al. (2005) obtained an efficiency of 0.21%; He et al. (1999) produced four lines at 0.6% efficiency; and Bell (2003), 10 lines at 0.06% efficiency. Pellegringechi et al. (2002) used three elite CIMMYT durum wheat cultivars to generate an overall efficiency of 1.7%. Therefore, it could be concluded that for the few varieties tested so far, for durum wheat genetic transformation, the Agrobacterium-mediated method would be a better choice than biolistics.
Transgene integration, expression, and inheritance in regenerated transformed plants
In the present study, all the eight lines randomly selected for Southern blot analysis showed 2–4 copies, in agreement with the observation that Agrobacterium-mediated transformation tends to produce transgenic lines with a low copy number of transgenes integrated (Hiei et al., 1997; Gheysen et al., 1998; Hansen and Wright 1999; Shibata and Liu, 2000; Dai et al., 2001; Jones, 2005; Jones et al., 2005; Travella et al., 2005; Sparks and Jones, 2009). Furthermore, some lines showed a typical 3:1 segregation ratio, for example lines C-023-3-1 and C024-1-5, but they had three and four gusA genes integrated, respectively (Fig. 6), suggesting that all the gusA genes had integrated into the wheat genome as a single segregating locus. Other lines had segregation ratios indicative of two or more loci segregating independently. Integration of transgenes at a single locus and often as concatomers, regardless of the copy number, is typically seen in transgenic lines produced using both direct DNA delivery and the Agrobacterium-mediated method of transformation (Deroles et al., 1988; Spencer et al., 1992). In addition, of the eight lines selected for Southern blot analysis, none with a single copy of gusA was observed, presumably due to the use of additional vir genes to aid transformation in recalcitrant species which may result in more transgene copies at one or more loci as suggested by Wu et al. (2006). Moreover, as indicated in Table 3, of 121 bar PCR-positive transformants obtained in the development of this system, 118 were gusA PCR positive, among which only 106 plants showed GUS expression. The latter events could be indicative of the frequencies of incomplete T-DNA transfers or rearrangements or silencing of the unselected gene. Incomplete T-DNA integration was also observed by Wu et al. (2006) in that 44% of wheat transformants contained an incomplete T-DNA in which either the gusA gene or the bar gene was lost. It would be expected that only bar-positive transformants would survive the whole selection process. However, some gusA PCR-positive or GUS-positive transgenic plants without the bar gene were observed in other experiments (data not shown). The structure and characterization of the selected bar gene and unselected transgene (gusA) in the progeny of the transgenic plants obtained need to be further clarified in understanding the integration process. In addition, rearrangements or silencing of the unselected gene can be a challenge for transgenic research in general. In maize, the transgene (gusA) expression cassette was more likely to be rearranged if expression of that gene was not selected for during callus growth (Register et al., 1994). It remains unclear whether this is specific to the gusA gene or whether other genes are subject to this rearrangement or silencing during Agrobacterium-mediated transformation.
Compared with the previous reports on analysis of transgene loci produced by Agrobacterium-mediated wheat transformation, the present data showed a shift towards more loci as indicated by higher segregation ratios. As shown in Table 4, among the 31 randomly selected lines, 29.0% displayed a typical 3:1 segregation ratio for the expression of both bar and gusA genes, indicating the complete linkages of these two genes and segregation together as a single functional locus, although there were one or more copies of the gusA gene integrated, for example in lines C023-3-1 and C024-1-5, as indicated in Southern blot analysis (Fig. 6); 48.4% of all lines selected possessed two loci on which the gusA gene was integrated; 16.1% segregated at a 63:1 ratio, suggesting three or more copies of the gusA gene integrated into at least three loci on the chromosome (Table 4). These ratios were higher than those of other reports and indicated that the transformation treatments adopted here generated transgene insertions at more loci than previously reported. For example, Khanna and Daggard (2003) demonstrated that 35% of transgenic lines showed 3:1 segregation ratios, whereas Hu et al. (2003) reported 46% of 3:1 ratios. Analysis of 26 plants revealed single gene copies in 35% of the lines tested, two or three copies in 50%, and four to five copies in only 15% (Cheng et al., 1997). Using the same strain and vector combination as reported here, Wu et al. (2006) detected that 38% of 34 lines analysed had the expected 3:1 ratio of segregation, while only 18% possessed two or more transgene loci and the remaining 44% contained an incomplete T-DNA in which either the gusA gene or the bar gene was lost. Therefore, it might be concluded that although Agrobacterium-mediated transformation generally produces fewer loci and a lower copy number in transgenic wheat plants, the integration pattern depends on the specific methods used for transformation, with the presence of additional vir genes probably playing a part in determining copy and locus number.
Acknowledgments
This project is funded by the Research Initiatives on Development of Precise and Efficient Transformation Technologies (2008ZX08010-003) and Development of Disease and Insect Resistance Transgenic Wheat Plants (2008ZX08002-001) supported by the Chinese Government. Rothamsted Research receives grant-aided support from the Biotechnology and Biological Sciences Research Council (BBSRC) of the UK. The plasmids pAL154 and pAL156 were kindly provided by Dr Wendy Harwood and Dr Mark Smedley (John Innes Centre).
References
- Agarwal S, Loar S, Steber C, Zale J. Floral transformation of wheat. Methods in Molecular Biology. 2009;478:105–114. doi: 10.1007/978-1-59745-379-0_6. [DOI] [PubMed] [Google Scholar]
- Altpeter F, Vasil V, Srivastava V, Stoger E, Vasil IK. Accelerated production of transgenic wheat (Triticum aestivum L.) plants. Plant Cell Reports. 1996;16:12–17. doi: 10.1007/BF01275440. [DOI] [PubMed] [Google Scholar]
- Amoah BK, Wu H, Sparks CA, Jones HD. Factors influencing Agrobacterium-mediated transient expression of uidA in wheat inflorescence tissue. Journal of Experimental Botany. 2001;52:1135–1142. doi: 10.1093/jexbot/52.358.1135. [DOI] [PubMed] [Google Scholar]
- Barcelo P, Lazzeri PA, Martin A, Lörz H. Competence of leaf cells II. Influence of auxin, ammonium and explants age on regeneration. Journal of Plant Physiology. 1992;139:448–454. [Google Scholar]
- Barro F, Cannell ME, Lazzeri PA, Barcelo P. The influence of auxins on transformation of wheat and tritordeum and analysis of transgene integration patterns in transformants. Transgenic Research. 1998;97:684–695. [Google Scholar]
- Barro F, Martin A, Lazzeri PA, Barcelo P. Medium optimization for efficient somatic embryogenesis and plant regeneration from immature inflorescences and immature scutella of élite cultivars of wheat, barley and tritordeum. Euphytica. 1999;108:161–167. [Google Scholar]
- Bartlett JG, Alves SC, Smedley M, Snape JW, Harwood W. High-throughput Agrobacterium-mediated barley transformation. Plant Methods. 2008;22:1–12. doi: 10.1186/1746-4811-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker D, Brettschneider R, Lorz H. Fertile transgenic wheat from microprojectile bombardment of scutellar tissue. The Plant Journal. 1994;5:299–307. doi: 10.1046/j.1365-313x.1994.05020299.x. [DOI] [PubMed] [Google Scholar]
- Bell P. Manipulation of lipoxygenase activity in durum wheat for the improvement of pasta colour. PhD thesis, Department of Biological Sciences and CPI Division. Bristol University and Rothamsted Research; 2003. [Google Scholar]
- Bommineni VR, Jauhar PP, Peterson TS. Transgenic durum wheat by microprojectile bombardment of isolated scutella. Journal of Heredity. 1997;88:475–481. [Google Scholar]
- Bourdon V, Harvey A, Lonsdale DM. Introns and their positions affect the translational activity of mRNA in plant cells. EMBO Reports. 2001;2:394–398. doi: 10.1093/embo-reports/kve090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M, Fry JE, Pang SZ, Zhou HP, Hironaka CM, Duncan DR, Conner TW, Wan YC. Genetic transformation of wheat mediated by Agrobacterium tumefaciens. Plant Physiology. 1997;115:971–980. doi: 10.1104/pp.115.3.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M, Hu TC, Layton J, Liu CN, Fry JE. Desiccation of plant tissues post- Agrobacterium infection enhances T-DNA delivery and increases stable transformation efficiency in wheat. In Vitro Cellular and Developmental Biology-Plant. 2003;39:595–604. [Google Scholar]
- Christensen AH, Quail PH. Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Research. 1996;5:213–218. doi: 10.1007/BF01969712. [DOI] [PubMed] [Google Scholar]
- Dai SH, Zheng P, Marmey P, Zhang SP, Tian WZ, Chen SY, Beachy RN, Fauquet C. Comparative analysis of transgenic rice plants obtained by Agrobacterium-mediated transformation and particle bombardment. Molecular Breeding. 2001;7:25–33. [Google Scholar]
- Deroles SC, Gardner RC. Expression and inheritance of kanamycin resistance in a large number of transgenic petunias generated by Agrobacterium-mediated transformation. Plant Molecular Biology. 1988;11:355–364. doi: 10.1007/BF00027392. [DOI] [PubMed] [Google Scholar]
- Ding LP, Li SC, Gao JM, Wang YS, Yang GX, He GY. Optimization of Agrobacterium-mediated transformation conditions in mature embryos of elite wheat. Molecular Biology Reporter. 2009;36:29–36. doi: 10.1007/s11033-007-9148-5. [DOI] [PubMed] [Google Scholar]
- Dudits D, Bögre L, Gyögyey J. Molecular and cellular approaches to the analysis of plant embryo development from somatic cells in vitro. Journal of Cell Science. 1991;99:475–484. [Google Scholar]
- EFSA. Guidance document of the Scientific Panel on Genetically Modified Organisms for the risk assessment of genetically modified plants and derived food and feed. The EFSA Journal. 2006;99:1–100. http://www.efsa.europa.eu/EFSA/efsa_locale-1178620753812_1178620775747.htm. [Google Scholar]
- EFSA. Guidance document of the Scientific Panel on Genetically Modified Organisms for the risk assessment of genetically modified plants containing stacked transformation events. The EFSA Journal. 2007;512:1–5. http://www.efsa.europa.eu/cs/BlobServer/Scientific_Document/gmo_guidance_ej512_GM_stacked_events_en.pdf. [Google Scholar]
- Gadaleta A, Giancaspro A, Blechl A, Blanco A. Phosphomannose isomerase, pmi, as a selectable marker gene for durum wheat transformation. Journal of Cereal Science. 2006;43:31–37. [Google Scholar]
- Gheysen G, Angenon G, Van Montague M. Agrobacterium-mediated plant transformation: a scientifically intriguing story with significant application. In: Lindsey K, editor. Transgenic Plant Research. The Netherlands: Harwood Academic; 1998. Press, 1–33. [Google Scholar]
- Hansen G, Wright MS. Recent advances in the transformation of plants. Trends in Plant Science. 1999;4:226–231. doi: 10.1016/s1360-1385(99)01412-0. [DOI] [PubMed] [Google Scholar]
- He GY, Lazzeri PA, Cannell ME. Fertile transgenic plants obtained from tritordeum inflorescences by tissue electroporation. Plant Cell Reports. 2001;20:67–72. doi: 10.1007/s002990000285. [DOI] [PubMed] [Google Scholar]
- He GY, Rooke L, Steele S, Bekes F, Gras P, Tatham AS, Fido R, Barcelo P, Shewry PR, Lazzeri PA. Transformation of pasta wheat (Triticum turgidum L-var. durum) with high-molecular-weight glutenin subunit genes and modification of dough functionality. Molecular Breeding. 1999;5:377–386. [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Molecular Biology. 2000;42:819–832. doi: 10.1023/a:1006496308160. [DOI] [PubMed] [Google Scholar]
- Hiei Y, Komari T, Kubo T. Transformation of rice mediated by Agrobacterium tumefaciens. Plant Molecular Biology. 1997;35:205–218. [PubMed] [Google Scholar]
- Hu T, Metz S, Chay C, et al. Agrobacterium-mediated large-scale transformation of wheat (Triticum aestivum L.) using glyphosate selection. Plant Cell Reports. 2003;21:1010–1019. doi: 10.1007/s00299-003-0617-6. [DOI] [PubMed] [Google Scholar]
- Jones HD. Wheat transformation: current technology and applications to grain development and composition. Journal of Cereal Science. 2005;41:137–147. [Google Scholar]
- Jones HD, Doherty A, Wu H. Review of methodologies and a protocol for the Agrobacterium-mediated transformation of wheat. Plant Methods. 2005 doi: 10.1186/1746-4811-1-5. 2005, 1:5(doi:10.1186/1746-4811-1-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke XY, McCormac AC, Harvey A, Lonsdale D, Chen DF, Elliott MC. Manipulation of discriminatory T-DNA delivery by Agrobacterium into cells of immature embryos of barley and wheat. Euphytica. 2002;126:333–343. [Google Scholar]
- Khanna HK, Daggard GE. Agrobacterium tumefaciens-mediated transformation of wheat using a superbinary vector and a polyamine-supplemented regeneration medium. Plant Cell Reports. 2003;21:429–436. doi: 10.1007/s00299-002-0529-x. [DOI] [PubMed] [Google Scholar]
- Komari T. Transformation of cultured-cells of Chenopodium quinoa by binary vectors that carry a fragment of DNA from the virulence region of pTiBo542. Plant Cell Reports. 1990;9:303–306. doi: 10.1007/BF00232856. [DOI] [PubMed] [Google Scholar]
- Komari T, Hiei Y, Saito Y, Murai N, Kumashiro T. Vectors carrying two separate T-DNAs for co-transformation of higher plants mediated by Agrobacterium tumefaciens and segregation of transformants free from selection markers. The Plant Journal. 1996;10:165–174. doi: 10.1046/j.1365-313x.1996.10010165.x. [DOI] [PubMed] [Google Scholar]
- Lazo GR, Stein PA, Ludwig RA. A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Bio-Technology. 1991;9:963–967. doi: 10.1038/nbt1091-963. [DOI] [PubMed] [Google Scholar]
- Lazzeri PA, Jones HD. Transgenic wheat, barley and oats: production and characterization. Methods in Molecular Biology. 2009;478:3–22. doi: 10.1007/978-1-59745-379-0_1. [DOI] [PubMed] [Google Scholar]
- Matthews PR, Wang MB, Waterhouse PM, Thornton S, Fieg SJ, Gubler F, Jacobsen JV. Marker gene elimination from transgenic barley, using co-transformation with adjacent ‘twin T-DNAs’ on a standard Agrobacterium transformation vector. Molecular Breeding. 2001;7:195–202. [Google Scholar]
- McCormac AC, Wu H, Bao M, Wang Y, Xu R, Elliott MC, Chen DF. The use of visual marker genes as cell-specific reporters of Agrobacterium-mediated T-DNA delivery to wheat (Triticum aestivum L.) and barley (Hordeum vulgare L.) Euphytica. 1998;99:17–25. [Google Scholar]
- Miller M, Tagliani L, Wang N, Berka B, Bidney D, Zhao ZY. High-efficiency transgene segregation in co-transformed maize plants using an Agrobacterium tumefaciens 2 T-DNA binary system. Transgenic Research. 2002;11:381–396. doi: 10.1023/a:1016390621482. [DOI] [PubMed] [Google Scholar]
- Nehra NS, Chibbar RN, Leung N, Caswell K, Mallard C, Steinhauer L, Baga M, Kartha KK. Self-fertile transgenic wheat plants regenerated from isolated scutellar tissues following microprojectile bombardment with 2 distinct gene constructs. The Plant Journal. 1994;5:285–297. [Google Scholar]
- Pastori GM, Wilkinson MD, Steele SH, Sparks CA, Jones HD, Parry MAJ. Age-dependent transformation frequency in elite wheat varieties. Journal of Experimental Botany. 2001;52:857–863. doi: 10.1093/jexbot/52.357.857. [DOI] [PubMed] [Google Scholar]
- Patnaik D, Vishnudasan D, Khurana P. Agrobacterium-mediated transformation of mature embryos of Triticum aestivum and Triticum durum. Current Science. 2006;91:307–317. [Google Scholar]
- Pellegrineschi A, Brito RM, Velazquez L, Noguera LM, Pfeiffer W, McLean S, Hoisington D. The effect of pretreatment with mild heat and drought stresses on the explant and biolistic transformation frequency of three durum wheat cultivars. Plant Cell Reports. 2002;20:955–960. [Google Scholar]
- Register JC, Peterson DJ, Bell PJ, et al. Structure and function of selectable and non-selectable transgenes in maize after introduction by bombardment. Plant Molecular Biology. 1994;25:951–961. doi: 10.1007/BF00014669. [DOI] [PubMed] [Google Scholar]
- Risacher T, Craze M. Plant transformation method. 1992 WO 00/63398. [Google Scholar]
- Risacher T, Craze M, Bowden S, Paul W, Barsby T. Highly efficient Agrobacterium-mediated transformation of wheat via in planta inoculation. Methods in Molecular Biology. 2009;478:115–124. doi: 10.1007/978-1-59745-379-0_7. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Shewry PR, Tatham AS, Barro F, Barcelo P, Lazzeri P. Biotechnology of breadmaking—unraveling and manipulating the multi-protein gluten complex. Bio-Technology. 1995;13:1185–1190. doi: 10.1038/nbt1195-1185. [DOI] [PubMed] [Google Scholar]
- Shibata D, Liu YG. Agrobacterium-mediated plant transformation with large DNA fragments. Trends in Plant Science. 2000;5:354–357. doi: 10.1016/s1360-1385(00)01689-7. [DOI] [PubMed] [Google Scholar]
- Sparks CA, Jones HD. Biolistic transformation of wheat. Methods in Molecular Biology. 2009;478:71–92. doi: 10.1007/978-1-59745-379-0_4. [DOI] [PubMed] [Google Scholar]
- Spencer TM, O'Brien JV, Start WG, Adams TR, Gordon-kamm WJ, Lemaux PG. Segregation of transgenes in maize. Plant Molecular Biology. 1992;18:201–210. doi: 10.1007/BF00034949. [DOI] [PubMed] [Google Scholar]
- Tingay S, McElroy D, Kalla R, Fieg S, Wang MB, Thornton S, Brettell R. Agrobacterium tumefaciens-mediated barley transformation. The Plant Journal. 1997;11:1369–1376. [Google Scholar]
- Travella S, Ross SM, Harden J, Everett C, Snape JW, Harwood WA. A comparison of transgenic barley lines produced by particle bombardment and Agrobacterium-mediated techniques. Plant Cell Reports. 2005;23:780–789. doi: 10.1007/s00299-004-0892-x. [DOI] [PubMed] [Google Scholar]
- Vasil V, Castillo AM, Fromm ME, Vasil IK. Herbicide resistant fertile transgenic wheat plants obtained by microprojectile bombardment of regenerable embryogenic callus. Bio-Technology. 1992;10:667–674. [Google Scholar]
- Vasil V, Srivastava V, Castillo AM, Fromm ME, Vasil IK. Rapid production of transgenic wheat plants by direct bombardment of cultured immature embryos. Bio-Technology. 1993;11:1553–1558. [Google Scholar]
- Villemont E, Dubois F, Sangwan RS, Vasseur G, Bourgeois Y, Sangwan-Norreel BS. Roles of the host cell cycle in Agrobacterium-mediated transformation of Petunia: evidence of an S-phase control mechanism for T-DNA transfer. Planta. 1997;201:160–172. [Google Scholar]
- Vishnudasan D, Tripathi MN, Rao U, Khurana P. Assessment of nematode resistance in wheat transgenic plants expressing potato proteinase inhibitor (Pin2) gene. Transgenic Research. 2005;14:665–675. doi: 10.1007/s11248-005-5696-4. [DOI] [PubMed] [Google Scholar]
- Weeks JT, Anderson OD, Blechl AE. Rapid production of multiple independent lines of fertile transgenic wheat (Triticum aestivum L.) Plant Physiology. 1993;102:1077–1084. doi: 10.1104/pp.102.4.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir B, Gu X, Wang MB, Upadhyaya N, Elliott AR, Brettell RIS. Agrobacterium tumefaciens-mediated transformation of wheat using suspension cells as a model system and green fluorescent protein as a visual marker. Australian Journal of Plant Physiology. 2001;28:807–818. [Google Scholar]
- Wernicke W, Milkovitz L. Effect of auxin on mitotic cell cycle in cultured leaf segments at different stages of development in wheat. Plant Physiology. 1987;69:16–22. [Google Scholar]
- Wiley PR, Tosi P, Evrard A, Lovegrove A, Jones HD, Shewry PR. Promoter analysis and immunolocalisation show that puroindoline genes are exclusively expressed in starchy endosperm cells of wheat grain. Plant Molecular Biology. 2007;64:125–136. doi: 10.1007/s11103-007-9139-x. [DOI] [PubMed] [Google Scholar]
- Wiley PR. The use of genetic transformation to determine the molecular basis for grain texture in wheat. PhD thesis, Department of Biological Sciences and CPI Division. Bristol University and Rothamsted Research; 2005. [Google Scholar]
- Wu H, Doherty A, Jones HD. Efficient and rapid Agrobacterium-mediated transformation of durum wheat using additional virulence genes. Transgenic Research. 2008;17:425–436. doi: 10.1007/s11248-007-9116-9. [DOI] [PubMed] [Google Scholar]
- Wu H, Doherty A, Jones HD. Agrobacterium-mediated transformation of bread and durum wheat using freshly isolated immature embryos. Methods in Molecular Biology. 2009;478:93–103. doi: 10.1007/978-1-59745-379-0_5. [DOI] [PubMed] [Google Scholar]
- Wu H, Sparks CA, Amoah B, Jones HD. Factors influencing successful Agrobacterium-mediated genetic transformation of wheat. Plant Cell Reports. 2003;21:659–668. doi: 10.1007/s00299-002-0564-7. [DOI] [PubMed] [Google Scholar]
- Wu H, Sparks CA, Jones HD. Characterisation of T-DNA loci and vector backbone sequences in transgenic wheat produced by Agrobacterium-mediated transformation. Molecular Breeding. 2006;18:195–208. [Google Scholar]
- Zale JM, Agarwal S, Loar S, Steber CM. Evidence for stable transformation of wheat by floral dip in Agrobacterium tumefaciens. Plant Cell Reports. 2009;28:903–915. doi: 10.1007/s00299-009-0696-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao TJ, Zhao SY, Chen HM, Zhao QZ, Hu ZM, Hou BK, Xia GM. Transgenic wheat progeny resistant to powdery mildew generated by Agrobacterium inoculums to the basal portion of wheat seedling. Plant Cell Reports. 2006;25:1199–1204. doi: 10.1007/s00299-006-0184-8. [DOI] [PubMed] [Google Scholar]
- Zhou H, Arrowsmith JW, Fromm ME, Hironaka CM, Taylor ML, Rodriguez D, Pajeau ME, Brown SM, Santino CG, Fry JE. Glyphosate-tolerant CP4 and GOX genes as a selectable marker in wheat transformation. Plant Cell Reports. 1995;15:159–163. doi: 10.1007/BF00193711. [DOI] [PubMed] [Google Scholar]