Abstract
Strigolactones are considered a new group of plant hormones. Their role as modulators of plant growth and signalling molecules for plant interactions first became evident in Arabidopsis, pea, and rice mutants that were flawed in strigolactone production, release, or perception. The first evidence in tomato (Solanum lycopersicon) of strigolactone deficiency is presented here. Sl-ORT1, previously identified as resistant to the parasitic plant Orobanche, had lower levels of arbuscular mycorrhizal fungus (Glomus intraradices) colonization, possibly as a result of its reduced ability to induce mycorrhizal hyphal branching. Biochemical analysis of mutant root extracts suggested that it produces only minute amounts of two of the tomato strigolactones: solanacol and didehydro-orobanchol. Accordingly, the transcription level of a key enzyme (CCD7) putatively involved in strigolactone synthesis in tomato was reduced in Sl-ORT1 compared with the wild type (WT). Sl-ORT1 shoots exhibited increased lateral shoot branching, whereas exogenous application of the synthetic strigolactone GR24 to the mutant restored the WT phenotype by reducing the number of lateral branches. Reduced lateral shoot branching was also evident in grafted plants which included a WT interstock, which was grafted between the mutant rootstock and the scion. In roots of these grafted plants, the CCD7 transcription level was not significantly induced, nor was mycorrhizal sensitivity restored. Hence, WT-interstock grafting, which restores mutant shoot morphology to WT, does not restore mutant root properties to WT. Characterization of the first tomato strigolactone-deficient mutant supports the putative general role of strigolactones as messengers of suppression of lateral shoot branching in a diversity of plant species.
Keywords: CCD7, CCD8, grafted plants, mycorrhiza, shoot branching, strigolactones, tomato
Introduction
Today, strigolactones—or their biosynthetic metabolites—are considered to be a new group of plant hormones. They are suggested to play a pivotal role in the regulation of above-ground plant architecture (Gomez-Roldan et al., 2008; Umehara et al., 2008).
Strigolactones are suggested to be synthesized mainly in the lower parts of the stem and in roots (reviewed by Dun et al., 2009), but their biosynthetic pathway in plants is not fully understood. Strigolactones are suggested to be derived from the carotenoid pathway (synthesized in the plastids; Naik et al., 2003; Matusova et al., 2005), and to involve novel branching of the carotenoid cleavage pathway, via the activity of different oxygenases (Gomez-Roldan et al., 2008; Umehara et al., 2008; Floss and Walter, 2009; reviewed by Dun et al., 2009); the involvement of additional, unknown enzymes in the synthesis of this complex molecule has been suggested.
Strigolactones have been shown to have a role below-ground as well, in plant interactions with the parasitic plants Orobanche and Striga; strigolactones have been suggested as important signalling factors for Orobanche and Striga seed germination (Joel et al., 1995, 2007; Yokota et al., 1998; Goldwasser et al., 2008; Matusova et al., 2005; Bouwmeester et al., 2007; Xie et al., 2007, 2008). Strigolactones have also been suggested to be inducers of hyphal branching and of mitochondrial metabolism and mitotic activity of arbuscular mycorrhizal fungi (AMF; Akiyama et al., 2005; Akiyama and Hayashi, 2006; Besserer et al., 2006, 2008; Gomez-Roldan et al., 2008; Yoneyama et al., 2008); AMF are soil micro-organisms that establish mutual symbioses with higher plants and promote plant growth under suboptimal growth conditions (reviewed by Koltai et al., 2009).
The role of strigolactones, both as modulators of plant growth and as signalling molecules for plant interactions, became evident from studies of Arabidopsis, pea, and rice mutants, flawed in strigolactone production or perception (Gomez-Roldan et al., 2008; Umehara et al., 2008). Additional studies have suggested two key hypotheses regarding the role that strigolactones may play in determining shoot architecture: either strigolactones serve as auxin-promoted secondary messengers that move up into buds to repress their outgrowth (Brewer et al., 2009; Ferguson and Beveridge, 2009; reviewed by Dun et al., 2009), or they act primarily by reducing the capability of polar auxin transport from the apical meristem, thereby inhibiting polar auxin transport from buds, resulting in restrained bud outgrowth (Bennett et al., 2006; Mouchel and Leyser, 2007; Ongaro and Leyser, 2008; Leyser, 2009).
In the present research, a tomato (Solanum lycopersicon) strigolactone-deficient mutant, Sl-ORT1, previously demonstrated to be resistant to the parasitic plant Orobanche was characterized; this is the first reported strigolactone-deficient mutant in tomato. The mutant's interaction with AMF, its shoot morphology, and its reversion to WT phenotype via exogenous addition of a synthetic strigolactone, GR24, to roots are characterized here. WT interstock grafting also led to a reduction in shoot branching; however, it did not lead to changes in the grafted plant roots, in either transcription of the key enzymes of strigolactone biosynthesis or AMF sensitivity. Hence, WT interstock grafting, which restores mutant shoot morphology to WT, does not restore mutant root properties to WT; rather, it may act on the shoot to suppress lateral branching.
Materials and methods
Plant mutant and WT strains
Tomato (Solanum lycopersicon) cv. M82 seeds (WT; Eshed et al., 1992) were obtained from Tarsis Agricultural Chemicals Ltd. (Petah Tikva, Israel). The mutant Sl-ORT1 was induced by irradiation with fast-neutron irradiation mutagenesis and selected for Orobanche resistance, as described in Dor et al. (2010). Sl-ORT1 was self-propagated for another five generations for homogeneity.
Arbuscular mycorrhizal strain and assays for mycorrhizal susceptibility
All mycorrhizal studies were conducted with the AMF Glomus intraradices (LPA 8). The initial screening studies were performed in the greenhouse. G. intraradices was used as a sand-based inoculum. The different types of inoculum, prepared as described previously by David-Schwartz et al. (2001), were: ‘spores only’ (200 spores per plant), and ‘whole inoculum’ (30 propagules per plant, including both spores and colonized roots); grafted plants were inoculated with ‘spores only’.
Experiments were carried out in a randomized block design with six replicates for each treatment, and in each replicate 10 plants were examined. Infectivity potential of these inocula was determined using the most probable number (MPN) method (Haas and Krikun, 1985). Means and standard error were determined for all replicates; means of replicates were subjected to statistical analysis by multiple-range test (P ≤0.05), using the JMP statistical package (SAS, Cary, NC).
Roots from all experiments were stained with trypan blue solution (Phillips and Hayman, 1970) and mycorrhization was enumerated using the gridline intersection method (Giovannetti and Mosse, 1980).
Determining the effects of root exudates on AMF hyphal branching
Roots of WT and Sl-ORT1 plants grown in pots were gently removed and transferred to sterilized Erlenmeyer flasks (500 ml capacity). The culture solution containing root exudates was decanted and filtered through a polycarbonate filter to remove sloughed root debris. The hyphal branching assay was performed as described previously by Nagahashi and Douds (2000) and Gadkar et al. (2003) using G. intraradices (DAOM 181602) as the test AMF. Hyphal branching was determined following 7 d of incubation with plant extract or controls (GR24—Johnson et al., 1981—as a positive control and sterile distilled water as a negative control). Two experiments were conducted, with 20 replicates each; in each replicate, branching of at least two spores was examined. Means and standard error were determined for all replicates; means of replicates were subjected to statistical analysis by multiple-range test (P ≤0.05), using the JMP statistical package.
LC-MS analysis
For LC-MS analysis of root extracts, whole roots were cut from 5 week hydroponically grown plants. For hydroponic plant growth, SL-ORT1 and WT seeds were surface-sterilized in 70% ethanol for 0.5 min and then in 1% sodium hypochlorite containing 0.02% (v: v) Tween 20 for 2.5 min. After rinsing three times with an excessive amount of sterile distilled water, the seeds were placed on moistened rock wool (approximately 500 tomato seeds per container). Two containers were used for the seeds of each line. The containers were placed in growth chamber at 25 °C with 16/8 h (day/night) and the nutrient solution was replaced twice a week. For root extraction, tomato roots were blended in a blender at top speed for 1 min in acetone (w/v=1:2). The acetone was evaporated under reduced pressure at 35 °C.
Root extracts were then fractionated to isolate strigolactones according to Yoneyama et al. (2007) and analysed by LC–MS as described in Yoneyama et al. (2007) with modifications. Briefly, HPLC separation was conducted with a U980 HPLC instrument (Jasco, Tokyo, Japan) fitted with an ODS (C18) column (Mightysil RP-18, 2×250 mm, 5 μm; Kanto Chemicals, Tokyo, Japan). The mobile phase was 60% methanol in water (v/v) and was switched to 100% methanol 15 min after injection. The column was then washed with 100% methanol for 20 min at a flow rate of 0.2 ml min−1 and the column temperature set to 40 °C.
MS was performed with a Quattro LC mass spectrometer (Micromass, Manchester, UK) equipped with an electrospray source. Both the drying and nebulizing gas was nitrogen generated from pressurized air in an N2G nitrogen generator (Parker-Hanifin Japan, Tokyo, Japan). The nebulizing gas flow was set to approximately 100 l h−1, and the desolvation gas flow was set to 500 l h−1. The interface temperature was set to 400 °C, and the source temperature to 150 °C. MS/MS experiments were conducted using argon as the collision gas and a collision energy of 16 eV. The collision gas pressure was 0.15 Pa. The following m/z transitions were monitored: 367>270 for didehydro-orobanchol and 365>268 for solanacol. Data acquisition and analysis were performed with MassLynx software (ver. 3.2). Solanacol was quantified using a standard purified from cowpea root exudates (Xie et al., 2007). The root extract sample was dissolved in 50% aqueous methanol and filtered through a spin column (Ultra-free MC, 0.45-μm pore size, Millipore, Billerica, MA). An aliquot of the filtered solution was diluted with a volume of either pure 50% methanol or 50% methanol containing known amounts of solanacol. The increase in peak area on the chromatogram corresponded to the amounts of solanacol added, enabling estimation of the amounts of this compound in a sample. The identity of didehydro-orobanchol was not verified using a standard. Determination of this compound was based on the m/z 367>270 transition and retention time. All peaks corresponding to known strigolactones were confirmed by bioassay on broomrape seeds. Two replicates were performed for each line, WT or Sl-ORT1. Each replicate contained approximately 500 individual plants.
Determination of shoot branching
WT and Sl-ORT1 seeds were surface-sterilized and allowed to germinate and grow in styrofoam seedling trays in soil:vermiculite (1:1, v/v). GR24 was added at a concentration of 0.027 μM as water solution to the styrofoam trays for 3 weeks. Four-week-old seedlings were transferred to 3.0 l pots (one plant per pot) with a 1:1 mixture of soil and vermiculite. Plants were grown in a greenhouse under natural light conditions supplemented with artificial light (100 μmol m−2 s−1) to maintain a 16 h photoperiod at 28/24 °C (day/night). Number and weight of lateral shoots were measured for each plant after 45 d: eight plants were examined for each WT and Sl-ORT1 strain. Unsuppressed branching was counted as lateral branches of more than 0.3 g FW and more than 5 cm length. The experiment was repeated four times. Means and standard deviations were determined for all replicates; means of replicates were subjected to statistical analysis by multiple-range test (P ≤0.05), using the JMP statistical package.
Grafting experiments
Grafted plants were prepared by Hishtil nursery (Nehalim, Israel). Briefly, sterile-grown seedlings, 21 d post-germination, were sectioned within the hypocotyl region and the combinations of scion and rootstock were aligned to form a graft union.
For interstock grafting, 5 cm long epicotyl segments with slanted cuts were grafted to the rootstock and, simultaneously, to the scion. Seedlings were incubated in a controlled chamber at 25/20 °C (day/night temperatures), with relative humidity of 75% for 3 weeks. Following healing of the graft union, seedlings were transferred to 1.5 l pots filled with a mixture of sterile sand and vermiculite (1:1 v/v), with or without the presence of AMF spores (as described above) and allowed to grow under greenhouse conditions, with 16 h light. Plants were examined following 8 weeks of greenhouse growth. Unsuppressed branching was counted as lateral branches with more than 0.3 g FW and more than 5 cm length.
For hypocotyl grafting experiments, WT and Sl-ORT1 were reciprocally grafted. For interstock grafting, four combinations of WT and Sl-ORT1 plants were used—WT/WT/WT: WTscion-WTinterstock-WTstock; O/O/O: Sl-ORT1scion-Sl-ORT1interstock-Sl-ORT1stock; WT/O/WT: WTscion-Sl-ORT1interstock-WTstock; O/WT/O: Sl-ORT1scion-WTinterstock-Sl-ORT1stock. Non-grafted WT and Sl-ORT1 plants were used as controls. Shoot lateral branching, AMF colonization and Sl-CCD7 and Sl-CCD8 transcription levels were determined as described above and below, respectively.
For all grafting experiments, eight replicates (plants) were used for each grafting combination and controls, and the experiment was repeated three times. Means of replicates were subjected to statistical analysis by multiple-range test (P ≤0.05), using the JMP statistical package.
Isolation of Sl-CCD8 gene fragment and determination of Sl-CCD8 and Sl-CCD7 transcription levels using semi-quantitative PCR
The Sl-CCD8 gene fragment was isolated from WT cDNA (H Koltai, unpublished results; see Supplementary Fig. S3 at JXB online) using primers designed based on conserved gene regions. A sequence of a fragment of Sl-CCD7 gene was kindly provided by Drs Klee and Vogel. RNA was extracted from whole roots of plants that were allowed to grow under greenhouse conditions, with 16/8 h (day/night) for 8 weeks, as described in Gal et al. (2006). Semi-quantitative PCR was performed by amplifying 140 bp and 150 bp fragments from each of the genes; Sl-CCD7 and Sl-CCD8, respectively, using forward primer 5′-TGGGAAGGTGGTGATCCTTA-3′ and reverse primer 5′-TAGCTGAGCAGCAACATCCA-3′ for Sl-CCD7 and forward 5′-CAATCACAGCGGTAACTCTTCCA-3′ and reverse primer 5′-GCATCCTGATTCTAAAGCATTT-3′ for Sl-CCD8. Tomato ribosomal 18S (accession no. AY552528) served as the reference gene for the amount of RNA, and was amplified using the forward primer 5′-TTGATTACGTCCCTGCCCTTTGTACAC-3′ and the reverse primer 5′-AGGTTCACACCTACGGAAACCTTGTTAC-3′. PCR amplification was performed for 30 cycles under the following conditions: 94 °C denaturation for 30 s, 58.4 °C annealing for 30 s and 72 °C extension for 30 s. The resulting PCR products were separated by electrophoresis on a 1.4% agarose gel, and quantified using TotalLab TL120 (Nonlinear Dynamics Ltd., Newcastle, UK). The amount of amplification of the gene of interest was determined relative to that of the 18S ribosomal gene. The experiment was performed in eight replicates; in each replicate, the resulting amount-of-amplification product values were normalized to that of the WT. Means and standard error were determined from all replicates; means of replicates were subjected to statistical analysis by multiple-range test (P ≤0.05), using the JMP statistical package.
Results
The Sl-ORT1 mutant is impaired in its interaction with the arbuscular mycorrhizal fungus Glomus intraradices
The Solanum lycopersicon Orobanche-Resistant Trait 1 (Sl-ORT1) mutant was isolated as described in Dor et al. (2010); Sl-ORT1 was demonstrated to be resistant to Orobanche under both field and greenhouse conditions as compared to its parental WT line.
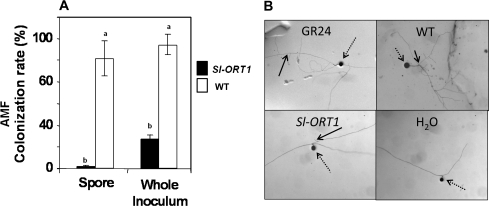
Since it has previously been suggested that the parasitic Orobanche and the symbiotic AMF respond to similar signals for their interaction with host plants (Akiyama and Hayashi, 2006; Bouwmeester et al., 2007; Yoneyama et al., 2008), the ability of the Sl-ORT1 mutant to be infected by AMF was determined. Sl-ORT1 plants were shown to have reduced G. intraradices colonization rates, by either whole inoculum or spores, relative to WT plants. AMF whole inoculum was significantly more infective to Sl-ORT1 than spores, whereas both spores and whole inoculum were similarly infective to the WT (Fig. 1A).
Fig. 1.
(A) Colonization rate by arbuscular mycorrhizal fungi of WT (Solanum lycopersicon cv. M82) and Sl-ORT1 plants. Six replicates were done for each treatment, and in each replicate 10 plants were examined. Means and standard error were determined for all replicates; means of replicates were subjected to statistical analysis by multiple-range test. (B) Examples of arbuscular mycorrhiza hyphal branching following exposure to WT or Sl-ORT1 root exudates, or to GR24 as a positive control or sterile distilled water as a negative control. Two experiments were conducted, with 20 replicates each; in each replicate, branching of at least two spores was examined. Means and standard error were determined for all replicates; means of replicates were subjected to statistical analysis by multiple-range test. Different lowercase letters (a, b) above bars represent significantly different means (P ≤0.05). Dashed arrows point to AMF spores; black arrows point to AMF hyphal branching sites.
To determine a possible cause of Sl-ORT1 resistance to AMF, the ability of Sl-ORT1 and WT root exudates to induce AMF hyphal branching was determined. A reduced level of hyphal branching was evident in the presence of Sl-ORT1 versus WT exudates; GR24 (Johnson et al., 1981), which is a synthetic strigolactone analogue previously shown to have biological activities (Gomez-Roldan et al., 2008; Umehara et al., 2008) served as a positive control, and distilled sterile water served as a negative control (Fig. 1B; Table 1). Hence, the resistance of Sl-ORT1 to AMF may result from its inability to promote hyphal branching.
Table 1.
Results of two experiments studying hyphal branching of arbuscular mycorrhiza (Glomus intradices) following exposure to WT or mutant (Sl-ORT1) or to GR24 as a positive control or sterile distilled water as a negative control
| 2° branching | 3° branching | 4° branching | 5° branching | Total branching | ||
| WT | 7.02±1.08 | 3.99±0.61 | 0.83±0.28 | 0.19±0.02 | 12.03 | |
| Sl-ORT1 | 4.02±0.74 | 1.76±0.45 | 0.13±0.13 | 5.91 | ||
| GR24 | 4.81±0.52 | 2.92±0.63 | 0.40±0.31 | 8.13 | ||
| H2O | 2.71±0.39 | 0.88±0.36 | 0.25±0.25 | 3.84 | ||
| 2° branching | 3° branching | 4° branching | 5° branching | 6° branching | Total branching | |
| WT | 6.23±0.88 | 3.84±1.73 | 1.93±0.94 | 0.65±0.40 | 0.23±0.23 | 12.89 |
| Sl-ORT1 | 2.46±0.53 | 2.46 | ||||
| GR24 | 3.68±0.71 | 1.66±0.43 | 0.31±0.20 | 0.14±0.14 | 5.79 | |
| H2O | 3.28±0.58 | 0.68±0.29 | 3.96 |
Six replicates were done for each treatment, in each replicate 10 plants were examined. Means and standard error were determined for all replicates.
The Sl-ORT1 mutant is deficient in strigolactone production
Since strigolactones are prominent signalling molecules for both plant–AMF and plant–Orobanche interactions, the Sl-ORT1 mutant's ability to produce strigolactones was analysed by determining the level of strigolactones in the WT and Sl-ORT1 mutant root extracts.
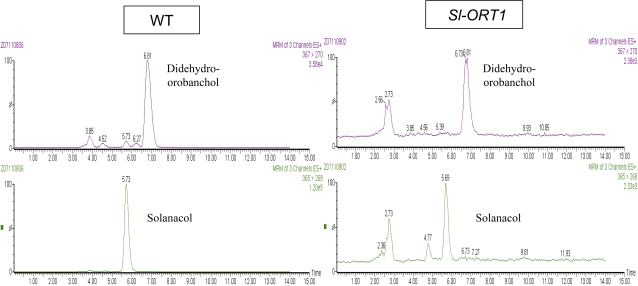
In the WT root extracts, two peaks corresponding to previously identified tomato strigolactones were examined (López-Ráez et al., 2008); these were detected in different channels of tandem mass spectrometry (MS/MS) analysis of WT root extracts (Fig. 2; Table 2). One, corresponding to solanacol, was detected in the 365 >268 transition with a retention time of 5.73 min (Fig. 2; Table 2). The MS/MS analysis and the addition of solanacol as an internal standard, together with its ability to induce broomrape seed germination in a bioassay (not shown), confirmed its identity as solanacol. The second peak, corresponding to a putative didehydro-orobanchol isomer(s), was detected in the 367 >270 transition with a retention time of 6.81 min (Fig. 2). The identity of this compound was not verified using a standard (due to its unavailability), but was confirmed by MS/MS analysis and broomrape seed-germination bioassay (not shown).
Fig. 2.
Results of LC-MS analysis of strigolactones in WT (Solanum lycopersicon cv. M82) and Sl-ORT1 tomato root extracts. Two replicates were performed for each line, WT or Sl-ORT1. Each replicate contained approximately 500 individual plants. One of the replicates is presented in the figure, whereas the second replicate demonstrated similar data. (This figure is available in colour at JXB online.)
Table 2.
Content of strigolactones solanacol and didehydro-orobanchol in root extracts obtained from hydroponically grown tomato plants following 5 weeks of growth
| Tomato line | WT | Sl-ORT1 |
| Solanacol | ||
| concentration (ng g−1 root) | 6.76 | 0.61 |
| Didehydro-orobanchol | ||
| peak area | 13 048 | 408 |
Two replicates were performed for each line, WT or Sl-ORTI. Each replicate contained approximately 500 individual plants; means for two replicates are presented.
In Sl-ORT1 root extracts the two peaks detected in the different channels correspond to strigolac-335 tones were demonstrated. Solanacol was detected as a peak in the 365 >268 transition with a retention time of 5.69 min, as confirmed by the addition of solanacol as an internal standard. Didehydro-orobanchol isomer(s) was detected as a peak in the 367 >270 transition at a retention time of 6.81 min (Fig. 2). However, both solanacol and didehydro-orobanchol isomer(s) were found to be reduced in Sl-ORT1 as compared to the WT (Table 2). These results suggested that the Sl-ORT1 mutant produces only minute amounts of the two strigolactones relative to the WT.
Mutant characterization
Sl-ORT1 mutant has aberrant shoot morphology compared to the WT
Shoot morphology of Sl-ORT1 and WT plants was examined. In tomato WT, apical dominance is maintained up until the formation of 7–11 metamers (reviewed by McSteen and Leyser, 2005; Schmitz and Theres, 2005). Then the primary shoot apical meristem is converted into an inflorescence, and in determined plants (such as WT, the progenitor of Sl-ORT1; Pnueli et al., 1998), shoots acquire a sympodial architecture.
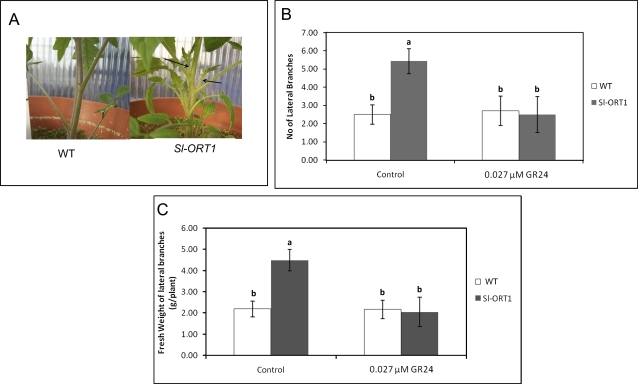
A higher number of unsuppressed side branches emerged in Sl-ORT1 plants than in the WT, prior to acquisition of sympodial architecture (Fig. 3A, B, C). Lack of suppression of side branches was evident in Sl-ORT1 from the first to seventh (±2) metamer (n=48). Further up in the metamers and inflorescence, a WT-like architecture resumed. In Sl-ORT1, primary side branches did not display multiplicity of unsuppressed secondary side branches and were similar to those of the WT. Upon exogenous application of 0.027 μM GR24, Sl-ORT1 shoot morphology was restored to that of the WT: the hyper-branching phenotype was suppressed, as evidenced by both the number of lateral branches and their fresh weight (Fig. 3B, C).
Fig. 3.
Morphological analysis of shoots of WT (Solanum lycopersicon cv. M82) and Sl-ORT1 plants. (A) An example of Sl-ORT1 and WT shoots. Arrows denote sites of lateral shoot branching (B) Lateral shoot number per plant, with or without (control) exogenous application of GR24 (0.027 μM). (C) Lateral shoot fresh weight, with or without (control) exogenous application of GR24 (0.027 μM). Number and weight of lateral shoots were measured for eight plants for each WT and Sl-ORT1 strains. The experiment was repeated four times. Means and standard deviations are shown; means of replicates were subjected to statistical analysis by multiple-range test. Different lowercase letters (a, b) above bars represent significantly different means (P ≤0.05). (This figure is available in colour at JXB online.)
Phenotype complementation in WT and mutant grafted plants
Sl-ORT1 shoots grafted to WT roots exhibited reduced lateral branching, and were similar to WT/WT (WTscion-WTstock) plants. Also WT shoots grafted to Sl-ORT1 roots exhibited a phenotype similar to that of WT/WT (Fig. 4). Notably, in addition to a lower level of lateral shoot branching, O/WT (Sl-ORT1scion-WTstock) and WT/O (WTscion-Sl-ORT1stock) plants exhibited a postponement in the appearance of lateral branching, such that unlike in Sl-ORT1 or O/O (Sl-ORT1scion-Sl-ORT1stock) plants, and similarly to WT or WT/WT (WTscion-WTstock) plants, nodes 1–3 did not develop lateral branches (see Supplementary Fig. S1 at JXB online).
Fig. 4.
Morphological analysis of WT (Solanum lycopersicon cv. M82), Sl-ORT1 and hypocotyl grafted plants. (A) Lateral shoot number per plant. (B) Lateral shoot fresh weight. (C) Total (main and lateral) shoot fresh weight. WT/WT: WTscion-WTstock; O/O: Sl-ORT1scion-Sl-ORT1stock; WT/O: WTscion-Sl-ORT1stock; O/WT: Sl-ORT1scion-WTstock. Number and weight of lateral shoots were measured for eight plants for each WT and Sl-ORT1 strain. The experiment was repeated three times. Means and standard deviations are shown; means of replicates were subjected to statistical analysis by multiple-range test. Different lowercase letters (a, b, c) above bars represent significantly different means (P ≤0.05).
However, mycorrhizal sensitivity was not restored to that of the WT in WT-scion grafted to Sl-ORT1 roots (WT/O), whereas WT roots grafted to Sl-ORT1 scion (O/WT) were sensitive to mycorrhizae (Table 3). No differences were observed in shoot fresh weights of the different grafted plant combinations (Fig. 4).
Table 3.
Colonization rate by arbuscular mycorrhizal fungus (AMF) Glomus intraradices of WT, mutant (Sl-ORT1) and hypocotyl grafted plants
| Planta | AMF colonization rateb |
| WT | 80.3±10.0 a |
| Sl-ORT1 | 4.1±1.6 c |
| WT/WT | 57.0±7.4 b |
| O/O | 6.0±5.8 c |
| WT/O | 5.9±5.1 c |
| O/WT | 67.3±10.7 ab |
Six replicates were done for each treatment, in each replicate 10 plants were examined. Means and standard error were determined for all replicates; means of replicates were subjected to statistical analysis by multiple-range test.
WT/WT: WTscion-WTstock; O/O: Sl-ORT1scion-Sl-ORT1stock; O/WT: Sl-ORT1scion-WTstock; WT/O: WTscion-Sl-ORT1stock.
Different lowercase letters (a, b, c) represent significantly different means (P ≤0.05).
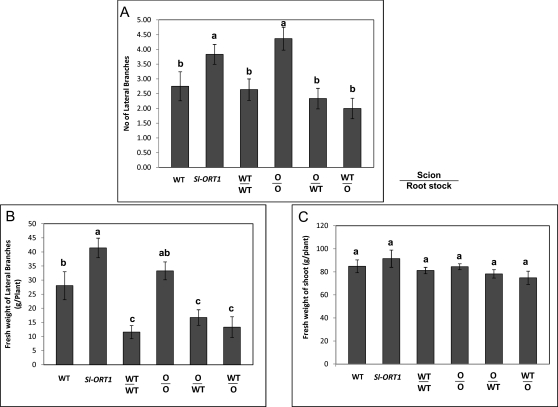
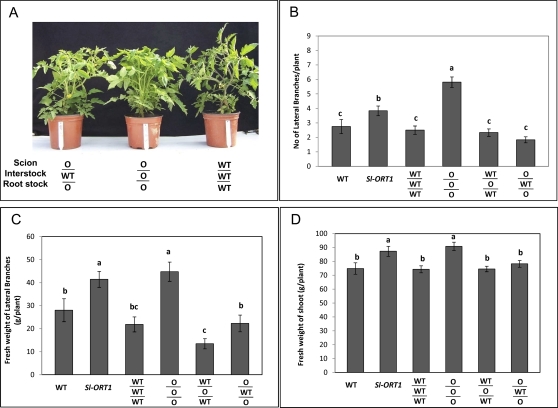
Restoration of Sl-ORT1 shoot morphology to that of the WT also appeared in interstock-grafted plants: WT interstock that was grafted between Sl-ORT1 stock and Sl-ORT1 scion (O/WT/O plants) suppressed Sl-ORT1 hyper-branching and restored the WT phenotype, as evidenced by both the number and fresh weight of the lateral branches (Fig. 5). Similar to hypocotyl grafting, in addition to the lower level of lateral shoot branching, both WT/O/WT (WTscion-Sl-ORT1interstock-WTstock) and O/WT/O (Sl-ORT1scion-WTinterstock-Sl-ORT1stock) plants exhibited a postponement in lateral branch appearance, such that unlike in Sl-ORT1 or O/O/O (Sl-ORT1scion-Sl-ORT1interstock-Sl-ORT1stock) plants, and similar to WT and WT/WT/WT (WTscion-WTinterstock-WTstock) plants, nodes 1–3 did not develop lateral branches (see Supplementary Fig. S2 at JXB online). Shoot fresh weight, however, was higher in Sl-ORT1 and O/O/O plants, probably reflecting the marked increase in lateral branching compared to the WT (Fig. 5). However, in O/WT/O roots, mycorrhizal sensitivity was not restored to that of the WT (Table 4). Hence, also with regard to mycorrhizal sensitivity, in O/WT/O plants, WT interstock may not have restored WT-characteristics to Sl-ORT1-originated roots.
Fig. 5.
Morphological analysis of WT (Solanum lycopersicon cv. M82), Sl-ORT1 and interstock grafted plants. (A) An example of grafted plant morphology. (B) Lateral shoot number. (C) Lateral shoot fresh weight. (D) Total (main and lateral) shoot fresh weight. WT/WT/WT: WTscion-WTinterstock-WTstock; O/O/O: Sl-ORT1scion-Sl-ORT1interstock-Sl-ORT1stock; WT/O/WT: WTscion-Sl-ORT1interstock-WTstock; O/WT/O: Sl-ORT1scion-WTinterstock-Sl-ORT1stock. Number and weight of lateral shoots were measured for eight plants for each WT and Sl-ORT1 strain. The experiment was repeated three times. Means and standard deviations are shown; means of replicates were subjected to statistical analysis by multiple-range test. Different lowercase letters (a, b, c) above bars represent significantly different means (P ≤0.05). (This figure is available in colour at JXB online.)
Table 4.
Colonization rate by arbuscular mycorrhizal fungus (AMF) Glomus intraradices of WT, mutant (Sl-ORT1) and interstock grafted plants
| Planta | AMF colonization rateb |
| WT | 86.2±4.8 a |
| Sl-ORT1 | 1.1±0.6 c |
| WT/WT/WT | 84.8±2.4 a |
| O/O/O | 2.1±1.0 c |
| O/WT/O | 2.9±1.3 c |
| WT/O/WT | 61.9±5.9 b |
Six replicates were done for each treatment, in each replicate 10 plants were examined. Means and standard error were determined for all replicates; means of replicates were subjected to statistical analysis by multiple-range test.
WT/WT/WT: WTscion-WTinterstock-WTstock; O/O/O: Sl-ORT1scion-Sl-ORT1interstock-Sl-ORT1stock; O/WT/O: Sl-ORT1scion-WTinterstock-Sl-ORT1stock; WT/O/WT: WT scion-Sl-ORT1interstock-WT stock.
Different lowercase letters (a, b, c) represent significantly different means (P ≤0.05).
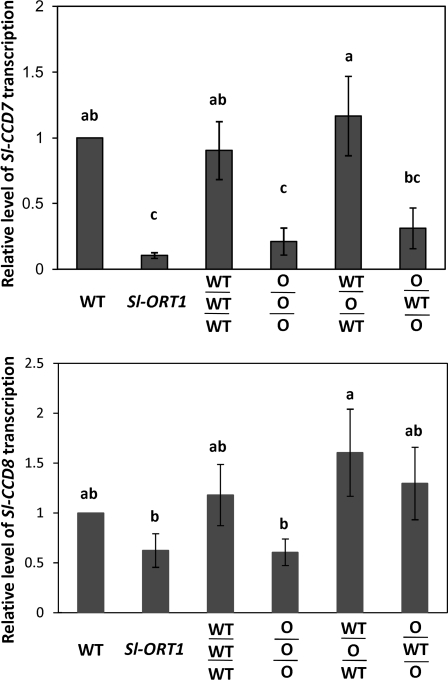
To characterize the effect of interstock grafting on roots further, the level of transcription of CCD8 and CCD7 was determined. CCD7/MAX3 and CCD8/MAX4 have been suggested to be key enzymes of carotenoid cleavage leading to strigolactone synthesis (Booker et al., 2004; Schwartz et al., 2004; Umehara et al., 2008), and to be expressed mainly in roots and lower stems (Booker et al., 2004; Bainbridge et al., 2005; reviewed by Dun et al., 2009).
To examine the expression pattern of the tomato (Sl) CCD7 and CCD8, a 900 bp fragment of the Sl-CCD8 gene was isolated (see Supplementary Fig. S3 at JXB online). A 400 bp fragment of the Sl-CCD7 gene was provided by Drs Klee and Vogel. Semi-quantitative PCR suggested a significantly reduced level of Sl-CCD7 in Sl-ORT1 mutant roots relative to the WT (Fig. 6). This reduction may suggest a reduced level of strigolactone synthesis in Sl-ORT1 roots. Sl-CCD8, which follows Sl-CCD7 in the biogenesis pathway of strigolatones (Schwartz et al., 2004; Umehara et al., 2008), was not significantly changed in Sl-ORT1 relative to the WT (Fig. 6).
Fig. 6.
Relative transcription level of Sl-CCD7 and Sl-CCD8 in WT (Solanum lycopersicon cv. M82), Sl-ORT1 and interstock grafted plant whole-roots. WT/WT/WT: WTscion-WTinterstock-WTstock; O/O/O: Sl-ORT1scion-Sl-ORT1interstock-Sl-ORT1stock; WT/O/WT: WTscion-Sl-ORT1interstock-WTstock; O/WT/O: Sl-ORT1scion-WTinterstock-Sl-ORT1stock. The experiment was performed in eight replicates; in each replicate, the resulting amount-of-amplification product values were normalized to that of the WT. Means and standard error were determined from all replicates; means of replicates were subjected to statistical analysis by multiple-range test. Different lowercase letters (a, b) above bars represent significantly different means (P ≤0.05).
In O/WT/O plants, Sl-CCD7 transcription in roots was only slightly though not statistically significant induced in comparison to Sl-ORT1 or O/O/O plants (Fig. 6). Hence, in O/WT/O plants, WT interstock may also not restore the WT characteristics to Sl-ORT1-originated roots with respect to Sl-CCD7 transcription.
A significant reduction (albeit to a lesser extent than in O/WT/O plants) in mycorrhizal sensitivity was also recorded for WT/O/WT plants (Table 4). This reduced sensitivity was not accompanied by a reduction in Sl-CCD7 transcription (Fig. 6) and may reflect the minor reduction in AMF sensitivity that occurs in most grafted plants (see also Table 3).
Discussion
The first tomato strigolactone-deficient mutant (Sl-ORT1) is reported here. The mutant displays several phenotypes, such as resistance to the parasitic plant Orobanche (Dor et al., 2010), a lower level of mycorrhizal colonization and increased lateral shoot branching (this study). Also, WT interstock was found sufficient to rescue the hyper-branching phenotype of the tomato mutant. Similar properties of strigolactone-deficient mutants have been reported for other plant species (Napoli, 1996; Foo et al., 2001; Booker et al., 2004; Gomez-Roldan et al., 2008; Umehara et al., 2008), supporting the notion of a general mechanism of strigolactone activity in plants (Klee, 2008; Leyser, 2009; Santner and Estelle, 2009; Schachtschabel and Boland, 2009).
In root extracts of the Sl-ORT1 mutant, two of the strigolactones previously identified in WT tomato, solanacol and didehydro-orobanchol (López-Ráez et al., 2008), were present in minute amounts relative to the WT. This is in accordance with the reduced ability of Sl-ORT1 root extracts to induce AMF hyphal branching (Akiyama et al., 2005; Akiyama and Hayashi, 2006; Besserer et al., 2006, 2008; Gomez-Roldan et al., 2008; Yoneyama et al., 2008). Hyphal branching of AMF is considered to be an important step that enhances the fungi's ability to reach a plant host root (Nagahashi and Douds, 2000). This step is probably more important for infection by spore inoculum than that by whole inoculum; the latter contains vegetative hyphae and colonized root segments in addition to spores. Hence, whole inoculum, which requires a lesser extent of hyphal branching, is more infective to Sl-ORT1 than spore inoculum.
One key enzyme, CCD7, which has previously been identified as involved in strigolactone biosynthesis pathways in other plant species (Booker et al., 2004; Schwartz et al., 2004; Umehara et al., 2008; reviewed by Dun et al., 2009), exhibited reduced transcription levels in roots of the tomato mutant in comparison to the WT. Together, these results suggest a reduced level of strigolactone synthesis in the Sl-ORT1 mutant compared to the WT, and a correlation between Sl-CCD7 levels of transcription and strigolactone synthesis in tomato; this is also suggested for other plant species (Foo et al., 2005; Johnson et al., 2006; Dun et al., 2009; Hayward et al., 2009).
The Sl-ORT1 mutant had more side branches than the WT; this is in agreement with other studies that have described strigolactone-deficient mutants in Arabidopsis, pea, and rice, suggesting that strigolactones, or their derived substances, are important for shoot branching (Gomez-Roldan et al., 2008; Umehara et al., 2008; Brewer et al., 2009; Ferguson and Beveridge, 2009). It was found that, in Sl-ORT1, the increase in lateral shoot branching prevailed in the metamers only prior to the acquisition of a sympodial architecture; after that, regular plant architecture resumes. Hence, in tomato, strigolactones may be involved in the control of lateral shoot branching during vegetative growth, prior to inflorescence formation. Reduced lateral shoot branching was restored to that of WT in Sl-ORT1 following exogenous application of the synthetic strigolactone GR24, demonstrating that addition of strigolactone may complement the mutant phenotype, restoring it to that of the WT.
Sl-ORT1 shoots grafted to WT roots exhibited reduced lateral branching and restoration to a WT-like phenotype, indicating that, as in other plant species, WT roots may complement the shoot phenotype, probably via strigolactone supplementation (reviewed by Dun et al., 2009). WT shoots grafted to Sl-ORT1 roots exhibited a WT shoot phenotype, indicating that WT stems are sufficient to support lateral branching suppression, and suggesting that, as in other plant species, strigolactones are also synthesized to some extent in the shoots (reviewed by Dun et al., 2009).
Reduced lateral shoot branching was also evident in interstock-grafted plants which included WT interstock grafted between a mutant rootstock and mutant scion (O/WT/O grafted plants), a phenomenon which has also been reported for other plant species (Napoli, 1996; Foo et al., 2001; Booker et al., 2004).
Due to findings suggesting that strigolactones are produced in shoots and roots, but moves in the root-to-shoot direction (Napoli, 1996; Beveridge et al., 1997, 2000; reviewed by Dun et al., 2009) and, accordingly, that roots are the main site of CCD7 and CCD8 expression (Booker et al., 2004; Bainbridge et al., 2005), the phenomenon of WT interstock complementation of mutant scions may suggest that the WT interstock itself may produce sufficient amounts of the necessary components (strigolactones or their derivatives) to confer a reduction in shoot branching.
Lack of both induction in the roots of Sl-CCD7 transcription and sensitivity to AMF suggests that, in the O/WT/O plants, the ability of the WT interstock to restore the mutant shoot phenotype does not encompass the restoration of WT characteristics in mutant roots, such as the ability to synthesize strigolactones. The WT interstock probably produces a sufficient amount of components such as strigolactones, their metabolites, or other unknown secondary messengers that migrate towards the shoot apex, to confer a significant reduction in shoot branching; this is in accordance with the hypothesis raised by Foo et al. (2001), Brewer et al. (2009), and Ferguson and Beveridge (2009).
What might be the possible role of Sl-ORT1 gene? Due to the reduced transcription level of Sl-CCD7, it might be that Sl-ORT1 encodes Sl-CCD7. Another possibility is that Sl-ORT1 works as a regulator of Sl-CCD7 expression. Sl-CCD8 expression level is not significantly affected in the Sl-ORT1 mutant, whereas in other plant species (e.g. pea), CCD8 (RMS1) expression is stimulated by a mobile feedback signal (probably strigolactones; Foo et al., 2005; Beveridge et al., 2009). The fact that in Sl-ORT1 mutants, despite a reduction in CCD7 transcription, and reduced levels of strigolactones, CCD8 expression levels were not altered, may suggest an interaction between control of CCD7 expression and feedback regulation of CCD8 transcription.
To conclude, characterization of the first tomato strigolactone-deficient mutant supports the putative general role of strigolactones in a diversity of plant species as messengers of suppression of lateral shoot branching.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Morphological analysis of WT (Solanum lycopersicon cv. M82), Sl-ORT1, and hypocotyl grafted plants, at each of the shoot nodes.
Supplementary Fig. S2. Morphological analysis of WT (Solanum lycopersicon cv. M82), Sl-ORT1, and interstock grafted plants, at each of the shoot nodes.
Supplementary Fig. S3. The tomato CCD8 gene fragment sequence.
Supplementary Material
Acknowledgments
We would like to thank Yogev Rosianski, Doron Meir, and Bruria Ben-Dor for their technical help. We are grateful to Drs Harry Klee and Jonathan Vogel for providing us with a fragment of the tomato CCD7 sequence. The research was funded by the Chief Scientist Foundation, Ministry of Agriculture.
References
- Akiyama K, Hayashi H. Strigolactones: chemical signals in fungal symbionts and parasitic weeds in plant roots. Annals of Botany. 2006;97:925–931. doi: 10.1093/aob/mcl063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama K, Matsuzaki K, Hayashi H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature. 2005;435:824–827. doi: 10.1038/nature03608. [DOI] [PubMed] [Google Scholar]
- Bainbridge K, Sorefan K, Ward S, Leyser O. Hormonally controlled expression of the Arabidopsis MAX4 shoot branching regulatory gene. The Plant Journal. 2005;44:569–580. doi: 10.1111/j.1365-313X.2005.02548.x. [DOI] [PubMed] [Google Scholar]
- Bennett T, Sieberer T, Willett B, Booker J, Luschnig C, Leyser O. The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Current Biology. 2006;16:553–563. doi: 10.1016/j.cub.2006.01.058. [DOI] [PubMed] [Google Scholar]
- Besserer A, Bécard G, Jauneau A, Roux C, Séjalon-Delmas N. GR24, a synthetic analog of strigolactones, stimulates the mitosis and growth of the arbuscular mycorrhizal fungus Gigaspora rosea by boosting its energy metabolism. Plant Physiology. 2008;148:402–413. doi: 10.1104/pp.108.121400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besserer A, Puech-Pagès V, Kiefer P, Gomez-Roldan V, Jauneau A, Roy S, Portais J, Roux C, Bécard G, Séjalon-Delmas N. Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biology. 2006;4:1239–1247. doi: 10.1371/journal.pbio.0040226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge CA, Dun EA, Rameau C. Pea has its tendrils in branching discoveries spanning a century from auxin to strigolactones. Plant Physiology. 2009;151:985–990. doi: 10.1104/pp.109.143909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge CA, Symons GM, Murfet IC, Ross JJ, Rameau C. The rms1 mutant of pea has elevated indole-3-acetic acid levels and reduced root sap zeatin riboside content but increased branching controlled by graft transmissible signal(s) Plant Physiology. 1997;15:1251–1258. [Google Scholar]
- Beveridge CA, Symons GM, Turnbull CG. Auxin inhibition of decapitation-induced branching is dependent on graft-transmissible signals regulated by genes Rms1 and Rms2. Plant Physiology. 2000;123:689–698. doi: 10.1104/pp.123.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker J, Auldridge M, Wills S, McCarty D, Klee H, Leyser O. MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Current Biology. 2004;14:1232–1238. doi: 10.1016/j.cub.2004.06.061. [DOI] [PubMed] [Google Scholar]
- Bouwmeester HJ, Roux C, Lopez-Raez JA, Bécard G. Rhizosphere communication of plants, parasitic plants and AM fungi. Trends in Plant Science. 2007;12:224–230. doi: 10.1016/j.tplants.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Brewer PB, Dun EA, Ferguson BJ, Rameau C, Beveridge CA. Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant Physiology. 2009;150:482–493. doi: 10.1104/pp.108.134783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David-Schwartz R, Badani H, Wininger S, Levy AA, Galili G, Kapulnik Y. Identification of a novel genetically controlled step in mycorrhizal colonization: plant resistance to infection by fungal spores but not extra-radical hyphae. The Plant Journal. 2001;27:561–569. doi: 10.1046/j.1365-313x.2001.01113.x. [DOI] [PubMed] [Google Scholar]
- Dor E, Alperin B, Wininger S, Ben-Dor B, Somvanshi VS, Koltai H, Kapulnik Y, Hershenhorn J. Characterization of a novel tomato mutant resistant to Orobanche and Phelipanche spp. weedy parasites. Euphytica. 2010;171:371–380. [Google Scholar]
- Dun EA, Brewer PB, Beveridge CA. Strigolactones: discovery of the elusive shoot branching hormone. Trends in Plant Science. 2009;14:364–372. doi: 10.1016/j.tplants.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Eshed Y, Abu-Abied M, Saranga Y, Zamir D. Lycopersicon esculentum lines containing small overlappping introgressions from L. pennellii. Theoretical and Applied Genetics. 1992;83:1027–1034. doi: 10.1007/BF00232968. [DOI] [PubMed] [Google Scholar]
- Ferguson BJ, Beveridge CA. Roles for auxin, cytokinin and strigolactone in regulating shoot branching. Plant Physiology. 2009;149:1929–1944. doi: 10.1104/pp.109.135475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floss DS, Walter MH. Role of carotenoid cleavage dioxygenase 1 (CCD1) in apocarotenoid biogenesis revisited. Plant Signaling and Behavior. 2009;4:172–175. doi: 10.4161/psb.4.3.7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E, Bullier E, Goussot M, Foucher F, Rameau C, Beveridge CA. The branching gene RAMOSUS1 mediates interactions among two novel signals and auxin in pea. The Plant Cell. 2005;17:464–474. doi: 10.1105/tpc.104.026716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E, Turnbull CG, Beveridge CA. Long-distance signaling and the control of branching in the rms1 mutant of pea. Plant Physiology. 2001;126:203–209. doi: 10.1104/pp.126.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadkar V, David-Schwartz R, Nagahashi G, Douds DD, Jr, Wininger S, Kapulnik Y. Root exudate of pmi tomato mutant M161 reduces AM fungal proliferation in vitro. FEMS Microbiology Letters. 2003;223:193–198. doi: 10.1016/S0378-1097(03)00357-4. [DOI] [PubMed] [Google Scholar]
- Gal TZ, Aussenberg ER, Burdman S, Kapulnik Y, Koltai H. Expression of a plant expansin is involved in the establishment of root knot nematode parasitism in tomato. Planta. 2006;224:155–162. doi: 10.1007/s00425-005-0204-x. [DOI] [PubMed] [Google Scholar]
- Giovannetti M, Mosse B. An evaluation of techniques for measuring vesicular arbuscular mycorrhizal infection in roots. New Phytologist. 1980;84:489–500. [Google Scholar]
- Goldwasser Y, Yoneyama K, Xie X, Yoneyama K. Production of strigolactones by Arabidopsis thaliana responsible for Orobanche aegyptiaca seed germination. Plant Growth Regulation. 2008;55:21–28. [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, et al. Strigolactone inhibition of shoot branching. Nature. 2008;455:189–194. doi: 10.1038/nature07271. [DOI] [PubMed] [Google Scholar]
- Haas JH, Krikun J. Efficacy of endomycorrhizal-fungus isolates and inoculum quantities required for growth response. New Phytologist. 1985;100:613–621. [Google Scholar]
- Hayward A, Stirnberg P, Beveridge C, Leyser O. Interactions between auxin and strigolactone in shoot branching control. Plant Physiology. 2009;151:400–412. doi: 10.1104/pp.109.137646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AW, Gowda G, Wassanali A, Knox J, Monaco S, Razawi Z, Roseberry G. The preparation of synthetic analogues of strigol. Journal of the Chemical Society (Perkin transactions 1) 1981;1:1734–1743. [Google Scholar]
- Johnson X, Brcich T, Dun EA, Goussot M, Haurogné K, Beveridge CA, Rameau C. Branching genes are conserved across species. Genes controlling a novel signal in pea are coregulated by other long-distance signals. Plant Physiology. 2006;142:1014–1026. doi: 10.1104/pp.106.087676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joel DM, Hershenhorn H, Eizenberg H, Aly R, Ejeta G, Rich P, Ransom J, Sauerborn J, Rubiales D. Biology and management of weedy root parasites. Horticultural Reviews. 2007;33:267–349. [Google Scholar]
- Joel DM, Steffens JC, Matthews DE. Germination of weedy root parasites. In: Kigel J, Galili G, editors. Seed development and germination. New York: Marcel Dekker; 1995. pp. 567–597. [Google Scholar]
- Klee H. Plant biology: hormones branch out. Nature. 2008;455:176–177. doi: 10.1038/455176a. [DOI] [PubMed] [Google Scholar]
- Koltai H, Gadkar V, Kapulnik Y. Biochemical and practical views of arbuscular mycorrhizal fungus–host association in horticultural crops. In: Janick J, editor. Horticultural reviews. Hoboken: John Wiley & Sons, Inc; 2009. (in press) [Google Scholar]
- Leyser O. The control of shoot branching: an example of plant information processing. Plant, Cell and Environment. 2009;32:694–703. doi: 10.1111/j.1365-3040.2009.01930.x. [DOI] [PubMed] [Google Scholar]
- López-Ráez JA, Charnikhova T, Gómez-Roldán V, et al. Tomato strigolactones are derived from carotenoids and their biosynthesis is promoted by phosphate starvation. New Phytologist. 2008;178:863–874. doi: 10.1111/j.1469-8137.2008.02406.x. [DOI] [PubMed] [Google Scholar]
- Matusova R, Rani K, Verstappen FW, Franssen MC, Beale MH, Bouwmeester HJ. The strigolactone germination stimulants of the plant-parasitic Striga and Orobanche spp. are derived from the carotenoid pathway. Plant Physiology. 2005;139:920–934. doi: 10.1104/pp.105.061382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSteen P, Leyser O. Shoot branching. Annual Review of Plant Biology. 2005;56:353–374. doi: 10.1146/annurev.arplant.56.032604.144122. [DOI] [PubMed] [Google Scholar]
- Mouchel CF, Leyser O. Novel phytohormones involved in long-range signaling. Current Opinion in Plant Biology. 2007;10:473–476. doi: 10.1016/j.pbi.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Nagahashi G, Douds DD. Partial separation of root exudate components and their effects upon the growth of germinated spores of AM fungi. Mycological Research. 2000;104:1453–1464. [Google Scholar]
- Naik SP, Chanemougasoundharam A, Khurana PMS, Kalloo G. Genetic manipulation of carotenoid pathway in higher plants. Current Science. 2003;85:1423–1430. [Google Scholar]
- Napoli CA. Highly branched phenotype of the petunia dad1-1 mutant is reversed by grafting. Plant Physiology. 1996;111:27–37. doi: 10.1104/pp.111.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongaro V, Leyser O. Hormonal control of shoot branching. Journal of Experimental Botany. 2008;59:67–74. doi: 10.1093/jxb/erm134. [DOI] [PubMed] [Google Scholar]
- Phillips JM, Hayman DS. Improved procedures for clearing and staining parasitic and vesicular arbuscular mycorrhizal fungi for rapid assessment of infection. Transactions of the British Mycological Society. 1970;55:158–161. [Google Scholar]
- Pnueli L, Carmel-Goren L, Hareven D, Gutfinger T, Alvarez J, Ganal M, Zamir D, Lifschitz E. The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development. 1998;125:1979–1989. doi: 10.1242/dev.125.11.1979. [DOI] [PubMed] [Google Scholar]
- Santner A, Estelle M. Recent advances and emerging trends in plant hormone signaling. Nature. 2009;459:1071–1078. doi: 10.1038/nature08122. [DOI] [PubMed] [Google Scholar]
- Schachtschabel D, Boland W. Strigolactones: the first members of a new family of ‘shoot branching hormones’ in plants? ChemBioChem. 2009;10:221–223. doi: 10.1002/cbic.200800727. [DOI] [PubMed] [Google Scholar]
- Schmitz G, Theres K. Shoot and inflorescence branching. Current Opinion in Plant Biology. 2005;8:506–511. doi: 10.1016/j.pbi.2005.07.010. [DOI] [PubMed] [Google Scholar]
- Schwartz SH, Qin X, Loewen MC. The biochemical characterization of two carotenoid cleavage enzymes from Arabidopsis indicates that a carotenoid-derived compound inhibits lateral branching. Journal of Biological Chemistry. 2004;279:46940–46945. doi: 10.1074/jbc.M409004200. [DOI] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature. 2008;455:195–200. doi: 10.1038/nature07272. [DOI] [PubMed] [Google Scholar]
- Xie X, Kusumoto D, Takeuchi Y, Yoneyama K, Yamada Y, Yoneyama K. 2′-Epi-orobanchol and solanacol, two unique strigolactones, germination stimulants for root parasitic weeds, produced by tobacco. Journal of Agricultural and Food Chemistry. 2007;55:8067–8072. doi: 10.1021/jf0715121. [DOI] [PubMed] [Google Scholar]
- Xie X, Yoneyama K, Kusumoto D, Yamada Y, Yokota T, Takeuchi Y, Yoneyama K. Isolation and identification of alectrol as (+)-orobanchyl acetate, a germination stimulant for root parasitic plants. Phytochemistry. 2008;69:427–431. doi: 10.1016/j.phytochem.2007.07.017. [DOI] [PubMed] [Google Scholar]
- Yokota T, Sakai H, Okuno K, Yoneyama K, Takeuchi Y. Alectrol and orobanchol, germination stimulants for Orobanche minor, from its host red clover. Phytochemistry. 1998;49:1967–1973. [Google Scholar]
- Yoneyama K, Takeuchi Y, Sekimoto H. Phosphorus deficiency in red clover promotes exudation of orobanchol, the signal for mycorrhizal symbionts and germination stimulant for root parasites. Planta. 2007;225:1031–1038. doi: 10.1007/s00425-006-0410-1. [DOI] [PubMed] [Google Scholar]
- Yoneyama K, Xie X, Sekimoto H, Takeuchi Y, Ogasawara S, Akiyama K, Hayashi H, Yoneyama K. Strigolactones, host recognition signals for root parasitic plants and arbuscular mycorrhizal fungi, from Fabaceae plants. New Phytologist. 2008;179:484–494. doi: 10.1111/j.1469-8137.2008.02462.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.