Abstract
Germin-like proteins (GLPs) have several proposed roles in plant development and defence. Two novel genes (Ps-GLP1 and 2) encoding germin-like protein were isolated from plum (Prunus salicina). Their regulation was studied throughout fruit development and during ripening of early and late cultivars. These two genes exhibited similar expression patterns throughout the various stages of fruit development excluding two important stages, pit hardening (S2) and fruit ripening (S4). During fruit development until the ripening phase, the accumulation of both Ps-GLPs is related to the evolution of auxin. However, during the S2 stage only Ps-GLP1 is induced and this could putatively be in a H2O2-dependent manner. On the other hand, the diversity in the Ps-GLPs accumulation profile during the ripening process seems to be putatively due to the variability of endogenous auxin levels among the two plum cultivars, which consequently change the levels of autocatalytic ethylene available for the fruit to co-ordinate ripening. The effect of auxin on stimulating ethylene production and in regulating Ps-GLPs transcripts was also investigated. These data, supported by their localization in the extracellular matrix, suggest that auxin is somehow involved in the regulation of both transcripts throughout fruit development and ripening.
Keywords: Auxin, ethylene, expression profile, flowering, germin-like protein, plum fruit development and ripening, subcellular localization
Introduction
Germins and germin-like proteins (GLPs) constitute a large and highly diverse family of ubiquitous plant proteins. Germin was first identified in germinating wheat embryos (Thomson and Lane, 1980) and was shown to play an important role in the plant defence response (Lane et al., 1986) as well as to possess oxalate oxidase activity (Lane et al., 1993). Proteins with sequence similarity to germins have been identified in various plant species and these are termed ‘germin-like proteins’ (GLPs). GLPs have a global low sequence identity with germins (Bernier and Berna, 2001). Despite the considerable sequence heterogeneity between different GLPs, they all contain localized amino acid sequence ‘signatures’ (Lane et al., 1991). Germins and the GLP gene family can be divided into two distinct groups. Members of the first group (the true germins) have relatively homogeneous sequences (Lane, 2000). However, the members of the second group (GLPs) are much more numerous and show high sequence divergence (Bernier and Berna, 2001). GLPs can be further divided into three subgroups based upon sequence conservation (Carter and Thornburg, 2000).
Plant GLPs have been found in various organs (leaves, roots, and floral tissues) and under different physiological conditions (seed germination, stress, and pathogen attack) (Membré et al., 1997; Wojtaszek et al., 1997). So far, extracellular manganese superoxide dismutase enzyme activity has been associated with many GLPs (Yamahara et al., 1999; Carter and Thornburg, 2000). Several putative roles have been suggested for GLPs. They were assumed to be structural proteins as a consequence of their localization in the extracellular matrix. On the other hand, other functions have also been proposed, including epidermal-specific protein (Wei et al., 1998), rhicadhesin receptor (Swart et al., 1994), low-affinity auxin-binding protein (Ohmiya et al., 1993), involved in circadian rhythms and floral induction (Heintzen et al., 1994; Ono et al., 1996; Vallelian-Bindschedler et al., 1998), and disease resistance (Park et al., 2004; Zimmermann et al., 2006; Godfrey et al., 2007).
In general, Japanese plums are climacteric fruits characterized by significant diversity for the date and rate of ripening (Abdi et al., 1997). It has been proposed that auxin can stimulate climacteric ethylene biosynthesis by activating ACS and ERF transcription (El-Sharkawy et al., 2008, 2009). Evaluation of the auxin content in peach fruits demonstrated that, concomitant with the climacteric ethylene production, a significant increase in IAA concentration could be detected in the mesocarp tissue (Miller et al., 1987). Thus, it seems that auxin is also a part of the mechanism that acts upstream of ethylene and controls the ripening of climacteric fruits by stimulating the onset and production of autocatalytic ethylene.
The isolation and characterization of two putative proteins encoding a germin-like protein in plum is reported here. Ps-GLPs gene expression in flowers, during fruit development as well as their protein localization was studied. The ability of auxin and auxin inhibitor to promote and inhibit, respectively, ethylene biosynthesis in fruit was investigated.
Materials and methods
Plant material and post-harvest treatments
Fruits from two Japanese plum (Prunus salicina L.) cultivars ‘Early Golden’ (EG) and ‘Shiro’ (SH) were harvested and treated as described previously (El-Sharkawy et al., 2007). Other tissues such as flowers and early developmental stages were collected from the ‘SH’ cultivar. All plant materials were frozen in liquid nitrogen and stored at –80 °C.
Fruit treatments
In order to evaluate IAA-induced ethylene production in plum fruit, mature ‘SH’ fruit were harvested as late as possible before fruit ethylene production had risen. The fruit were sterilized and treated with auxin-IAA (1, 10, and 100 μM) or with the auxin transport inhibitor, TIBA (50 μM). Fruit without any treatment were used as the control. After treatments, fruits were incubated at room temperature and ethylene production levels were measured daily. Mixed tissues of two fruit displaying a similar ethylene production at the same age were frozen for further analysis.
RNA isolation
Total RNA from fruit samples was extracted using the methods described by Boss et al. (1996). For vegetative tissues and flowers, total RNA was extracted using the Plant Total RNA purification kit (Norgen, Thorold, ON, Canada), as per the manufacturer's instructions. All RNA extracts were treated with DNase I (Promega, Madison, WI, USA) then cleaned up with RNeasy mini kit (Qiagen, Mississauga, ON, Canada).
Isolation and in silico analysis of plum cDNA sequences
Based on the sequence similarity of various GLPs, a pair of degenerate primers, GLP-F and GLP-R (primers from 1 and 2 in Table 1), was designed from the conserved regions to amplify the GLP orthologues from P. salicina. The isolated fragments were cloned in pGEM-T Easy vector (Promega), sequenced, and compared with database sequences using the BLAST program (Altschul et al., 1997). Extension of the partial cDNA clones was carried out using the 3′- and 5′- RACE kit (Invitrogen, Burlington, ON, Canada). Full-length amplification of cDNA sequences, designated as Ps-GLP1 and Ps-GLP2, was carried out using the Platinum Taq DNA Polymerase Alignment of Ps-GLP predicted protein sequences and the Neighbor–Joining tree construction was performed as described previously (El-Sharkawy et al., 2009).
Table 1.
The oligonucleotide primers used
| Name | Oligonucleotide sequence |
| 1- GLP-F | 5′-GCHGCAGTBACCCCTGCATTC-3′ |
| 2- GLP-R | 5′-RCCACCAAGAACACCCTTMAGCTT-3′ |
| 3- Ps-GLP1(F) | 5′-GCCCAATTTCCTGGTGTG-3′ |
| 4- Ps-GLP1(R) | 5′-CGGGGTGAGTGTGAAAGG-3′ |
| 5- Ps-GLP2(F) | 5′-CCCGGGTCTCCAAATTCT-3′ |
| 6- Ps-GLP2(R) | 5′-CCCCCAAGAACACCCTTC-3′ |
| 7- Ps-actin(F) | 5′-CTGGACCTTGCTGGTCGT-3′ |
| 8- Ps-actin(R) | 5′-ATTTCCCGCTCAGCAGTG-3′ |
| 9- Ps-GLP1(FG) | 5′-CGCGCGGATCCATGATTTTCCCTATCTTC-3′ |
| 10- Ps-GLP1(RG) | 5′-CGCGCGGATCCCATTAGTGCCACCAAGAA-3′ |
| 11- Ps-GLP2(FG) | 5′-CGCGCGGATCCATGCGCCAGGCAACGATG-3′ |
| 12- Ps-GLP2(RG) | 5′-CGCGCGGATCCCATTAGTACCCCCAAGAA-3′ |
Real-time quantitative RT-PCR
All RT-PCRs were performed as described previously (El-Sharkawy et al., 2007). Mx4000 v 4.20 software (Stratagene, La Jolla, CA, USA) was used to design gene-specific primers (primers from 3 to 8 in Table 1). To determine relative fold differences for each sample in each experiment, the Ct value for the two genes studied was normalized to the Ct value for β-actin and was calculated relative to a calibrator using the formula 2–ΔΔCt. The calibrator is the sample that exhibited the minimum level of transcripts in the whole experiment (‘SH’ fruits 105 DAB for Ps-GLP1 and 2).
Promoter isolation
Genomic DNA was extracted from immature leaves using the DNeasy Plant Maxi Kit (Qiagen). Promoters of Ps-GLP1 and Ps-GLP2 were isolated using the Universal Genome Walker Kit (Clontech, Palo Alto, CA, USA). Promoter sequence analysis was performed using PLACE Signal Scan Search program (Prestridge, 1991; Higo et al., 1999).
Protoplast isolation and transient expression of Ps-GLP::GFP fusion proteins
The coding sequences of Ps-GLP1 and 2 were cloned as a C-terminal fusion in frame with the GFP into the pGreen vector (Hellens et al., 2000) and expressed under the control of the 35S promoter. A high fidelity PCR system was used to amplify the ORFs using the following primers: Ps-GLP1(FG) and Ps-GLP1(RG) or Ps-GLP2(FG) and Ps-GLP2(RG) (primers from 9 to 12 in Table 1). The corresponding ORFs of Ps-GLP1 and 2 were cloned using the BamHI restriction site of the pGreen vector. Protoplasts used for transfection were obtained from suspension-cultured tobacco (Nicotiana tabacum) BY-2 cells according to the method described by Leclercq et al. (2005). Protoplasts were transfected by a modified polyethylene glycol method and analysed for GFP fluorescence by confocal microscopy as described by El-Sharkawy et al. (2009). All transient expression assays were repeated at least three times.
Results
Gene structure and organization
In the present work, two putative full-length germin-like protein cDNAs (Ps-GLP1 and 2) were isolated. Their deduced nucleotide and amino acid sequences exhibited strong identity to the peach GLPs Pp-ABP19 and Pp-ABP20, respectively (Ohmiya et al., 1998). The Ps-GLP1 sequence is 893 bp in length with a predicted open reading frame (ORF) encoding a protein of 209 amino acids. The 5′-, 3′-non-coding, and poly (A+) sequences were 42, 206, and 17 bp, respectively. The full-length Ps-GLP2 cDNA is 862 bp in length with a predicted ORF encoding a protein of 214 amino acids. The 5′-, 3′-non-coding, and poly (A+) sequences were 63, 132, and 24 bp, respectively. Multiple alignments of Ps-GLP sequences with other reported true germin/GLP genes highlighted a number of conserved motifs and structural similarities that are common to the plant GLP subfamily (Bernier and Berna, 2001). The deduced amino acid sequences of Ps-GLP1 and 2 comprise a conserved extracellular targeting peptide signal located at the N-terminus that is characteristic of the GLP gene family (see Supplementary Fig. S1 at JXB online). A signal peptide search (Nielsen et al., 1997) showed that a cleavage prediction of the hydrophobic signal targeting peptide would occur after Ala-18 and Ala-23 of the Ps-GLP1 and 2, respectively. In addition, the predicted protein of both sequences contains the three highly conserved germin/GLP oligopeptides characteristic of GLPs (Bernier and Berna, 2001). Box-A (QDFCVAD), includes a cysteine residue (Cys-24 and Cys-29 for Ps-GLP1 and 2, respectively), which is followed by a second cysteine (Cys-39 and Cys-44 for Ps-GLP1 and 2, respectively) believed to form an internal disulphide bridge of the extracellular domain (Woo et al., 2000). Box B is (G-P-H-HPGASEXXXXX-G) and box C is (GXXHFQXNG), in which X corresponds to any hydrophobic amino acid. Boxes B and C contain the three histidines and the glutamate residues involved in heavy metal ion-binding site (Dunwell and Gane, 1998; Woo et al., 1998). Consistent with GLP from other plant species (Carter et al., 1998), a single potential N-glycosylation site was identified in both cDNAs. The RGD-like tripeptides (KGD) motif sequence, thought to be involved in protein–protein interactions, was also present in both sequences (Labouré et al., 1999).
The promoter sequence from Ps-GLP1 (1194 bp, EU310511) and Ps-GLP2 (1031 bp, EU310510) contain several predictive cis-acting elements presumed to be involved in transcriptional regulation. Both Ps-GLP 5′-UTR regions hold ARF binding element (SAUR 15a, TGTCTC) located at positions –170 and –761 for Ps-GLP1, and –408 for Ps-GLP2 (Ulmasov et al., 1999). The ARF motif is found in the promoter of genes up-regulated by IAA (Goda et al., 2004). Also, a single binding element (LECPLEACS2, TAAAATAT) in Ps-GLP1 (at position –900) and two in Ps-GLP2 (at positions –627 and –898), proven to regulate ACC synthase gene expression in tomato (Matarasso et al., 2005), were identified. In addition, an identical sequence to tobacco NtBBF1 (ACTTA) was detected uniquely in the Ps-GLP1 promoter region at position –91. The NtBBF1 motif box is believed to be required for auxin induction (Baumann et al., 1999).
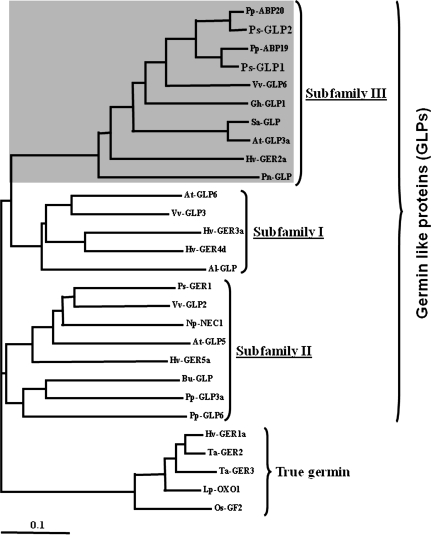
A phylogenetic tree, comprising 28 germin/GLP sequences from 16 species, was generated (Fig. 1). The dendrogram analysis defines that germins/GLPs could be divided into two main groups, true germins and GLPs. However, the members of the GLPs could be further divided into three main subfamilies (I, II, and III). Establishing a correlation between phylogenesis and functional properties of the encoded proteins based on published data allow a number of functional characteristics to be outlined. The true germin subfamily comprises most of the proteins that have oxalate oxidase enzyme activity (Lane et al., 1993; Le Deunff et al., 2004). GLP gene members corresponding to subfamilies I and II comprise GLP proteins exhibiting superoxide dismutase activity (Carter et al., 1999; Yamahara et al., 1999; Zimmermann et al., 2006; Godfrey et al., 2007). The Pisum sativum GLP, Ps-GER1, belongs to subfamily II and has been reported to function as a Rhicadhesin receptor (Swart et al., 1994). Ps-GLP1 and 2 are members of subfamily III that may have a common role as regulatory proteins involved directly or indirectly in auxin metabolism (Khuri et al., 2001). This group includes low-affinity auxin-binding proteins from peach (Ohmiya et al., 1993, 1998), cotton GLP1 important for cell wall expansion (Kim et al., 2004), as well as GLPs known to have a role linked to circadian rhythms and floral induction in A. thaliana (Staiger et al., 1999), Hordeum vulgare (Vallelian-Bindschedler et al., 1998), Sinapis alba (Heintzen et al., 1994), and Pharbitis nil (Ono et al., 1996). Despite strong sequence diversity between the different gene members of the four groups, many of them confer resistance against pathogen infection (Carter and Thornburg, 2000; Park et al., 2004; Zimmermann et al., 2006; Godfrey et al., 2007).
Fig. 1.
Phylogenetic relationships between P. salicina [Ps-GLP1 (EU310513), Ps-GLP2 (EU310512)]; P. persica [Pp-ABP19 (U79114), Pp-ABP20 (U81162)]; V. vinifera [Vv-GLP2 (ABH09468), Vv-GLP3 (AAQ63185), Vv-GLP6 (ABL60875)]; G. hirsutum [Gh-GLP1 (AAO92740)]; S. alba [Sa-GLP (P45854)]; A. thaliana [At-GLP3a (P94072), At-GLP5 (AAB51569), At-GLP6 (P92997)]; H. vulgare [Hv-GER1a (ABG46232), Hv-GER2a (ABG46233), Hv-GER3a (ABG46234), Hv-GER4d (ABG46236), Hv-GER5a (ABG46237)]; P. nil [Pn-GLP (P45853)]; A. lentiformis [Al-GLP (BAA78563)]; P. sativum [Ps-GER1 (CAB65369)]; N. plumbaginifolia [Np-NEC1 (Q9SPV5)]; B. unguiculata [Bu-GLP (BAC53790)]; P. patens [Pp-GLP3a (BAD86499), Pp-GLP6 (BAD86502)]; T. aestivum [Ta-GER2 (P15290), Ta-GER3 (P26759)]; L. perenne [Lp-OXO1 (CAC19429)]; and O. sativa [Os-GF2 (ABF98325)] based on full-length amino acid sequence. True germin, germin like protein (GLP) I, II, and III correspond to the different germin/GLP gene subfamilies.
Ps-GLP gene expression in flower and during fruit development
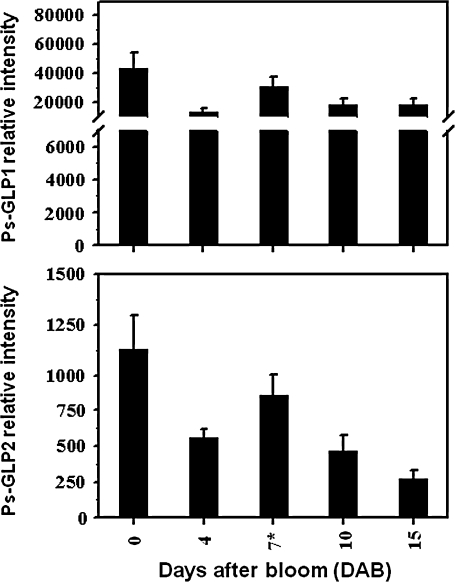
In flower and during early fruit development (0–15 DAB), both transcripts showed a similar pattern of accumulation. Ps-GLP mRNAs were highly expressed at bloom and decreased slightly afterward (∼4 DAB). The expression of the two Ps-GLPs was considerably re-stimulated after fertilization (∼7 DAB) and gradually declined thereafter, 10–15 DAB (Fig. 2).
Fig. 2.
Steady-state transcript levels of Ps-GLP1 and 2 assessed by QRT-PCR during flower and early fruit development of the ‘SH’ cultivar. The fertilized flowers stage is marked with an asterisk. The experiments were carried out in triplicate. The x-axis represents the developmental stages indicated by the number of days after bloom (DAB). Relative intensity in the y-axis refers to the fold difference in gene expression relative to ‘SH’ fruits 105 DAB for Ps-GLP1 and 2.
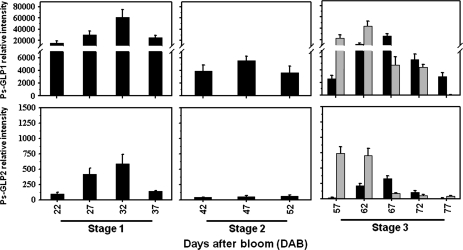
The stone fruit development period could be divided into three different stages (El-Sharkawy et al., 2007). The first stage (Stage 1, 22–37 DAB) illustrated by intense cell division and differentiation, and rapid growth caused by massive quantities of auxin (Miller et al., 1987). During this period of fruit development, both transcripts gradually increased to a peak (∼32 DAB) and declined thereafter (Fig. 3; Stage 1). In the second stage (Stage 2, 42–52 DAB); the endocarp hardens to form a solid stone, both genes clearly showed significant inhibition in their transcription levels (Fig. 3; Stage 2). However, the inhibition of Ps-GLP2 transcripts was more brutal than those of Ps-GLP1. A slight increase in Ps-GLP1 mRNA was detected throughout stage 2 (∼47 DAB) and declined thereafter, whereas that of Ps-GLP2 was weakly expressed. During Stage 3 (57–77 DAB), when the pulp (mesocarp) was separated from the seed (endocarp+embryo) both Ps-GLPs accumulated similarly in the pulp or seed (Fig. 3; Stage 3). However, their pattern of expression in the pulp was totally different from that in the seed. In the pulp, both transcripts peaked at ∼67 DAB, whereas, in the seed, both mRNAs were strongly detected at the beginning (∼57–62 DAB) and declined thereafter to reach their basal levels at the end of Stage 3, ∼77 DAB. Interestingly, the start of inhibition of both transcripts in the seed coincided with their accumulation peak in the pulp, ∼67 DAB.
Fig. 3.
Steady-state transcript levels of Ps-GLP1 and 2 during Stage 1, Stage 2, and Stage 3 of ‘SH’ fruit development using the whole fruit (Stage 1 and Stage 2). However, during Stage 3 of fruit development the expression was determined in pulp (black bars) and in seeds (grey bars). The experiments were carried out in triplicate. The x-axis represents the developmental stage indicated by number of days after bloom (DAB) and by the name of stage. Other details are as described in Fig. 2.
The expression of Ps-GLPs during fruit ripening
To evaluate the possible role of the two Ps-GLP proteins in fruit ripening, QRT-PCR analysis was carried out to determine their accumulation profile throughout ripening of early ‘EG’ and late ‘SH’ plum cultivars.
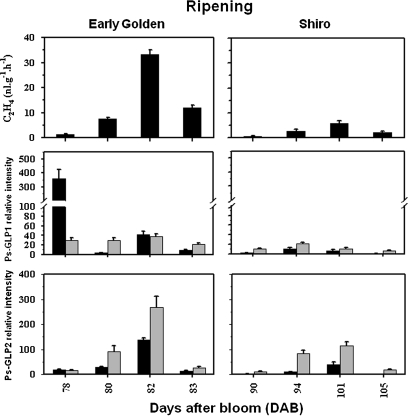
In both cultivars the fruits produced ethylene throughout ripening. ‘EG’ fruit displayed an early, rapid ripening, and a short and rapid (maximal 5 d) ethylene production profile. ‘SH’ fruit exhibited a suppressed climacteric pattern. The fruit ripened slower and later than ‘EG’. The ethylene production in ‘SH’ fruit reached a maximum at 11 d after the onset of ethylene emission (Fig. 4).
Fig. 4.
Ethylene evolution and steady-state transcript levels of Ps-GLP1 and 2 during early ‘EG’ (left panel) and late ‘SH’ (right panel) plum fruit ripening. The expression was quantified in pulp (black bars) and in seeds (grey bars). The experiments were carried out in triplicate. The x-axis represents the developmental stage indicated by number of days after bloom (DAB). Other details are as described in Fig. 2.
Both plum cultivars showed a dramatic decrease in Ps-GLPs transcript levels throughout ripening. In ‘EG’ pulp, relatively high Ps-GLP1 mRNA levels were detected in non-climacteric fruit (∼78 DAB) and strongly declined thereafter (Fig. 4); however, Ps-GLP2 was weakly detected. As ripening progressed and higher levels of autocatalytic ethylene produced, both transcripts increased relatively in the climacteric phase (∼82 DAB). In ‘EG’ seed, Ps-GLP1 mRNA was temporally quite constitutive during ripening, however, those of Ps-GLP2 increased to a peak ∼82 DAB as in the pulp but at much higher levels (Fig. 4). The peak of Ps-GLP2 mRNA, in the whole ‘EG’ fruit, coincided with the climacteric peak of ethylene production.
Throughout ripening of the late cultivar ‘SH’ (90–105 DAB), Ps-GLP1 mRNA remained at a minimal level or was almost undetectable in the whole ‘SH’ fruit. The accumulation pattern of Ps-GLP2 in the whole ‘SH’ fruit (pulp and seed), coincided well with the evolution of ethylene. However, Ps-GLP2 transcripts in the ‘SH’ pulp were at least three times lower than in ‘SH’ seeds (Fig. 4).
Auxin treatment stimulates ethylene biosynthesis and Ps-GLPs transcription in late plum fruit
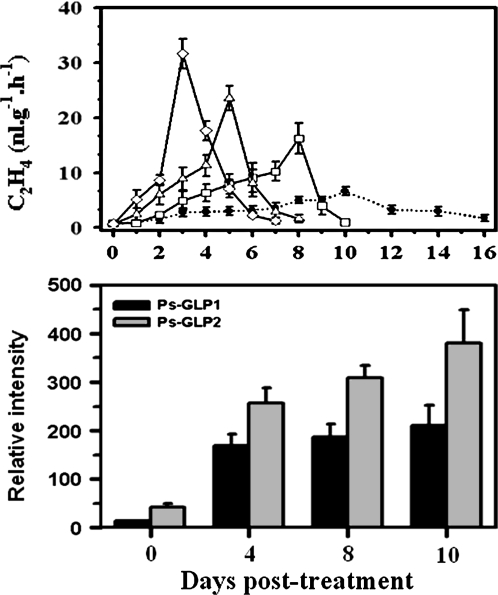
The previous results suggest that the suppressed climacteric phenotype in ‘SH’ fruits could partially be due to inadequate quantities of auxin to co-ordinate ripening. To test this hypothesis, ‘SH’ fruits were treated with different concentrations of IAA and 50 μM of the auxin inhibitor, TIBA; untreated ‘SH’ fruit were used as the control. TIBA-treated fruits were unable to ripen autonomously and their ethylene production remained low even after 35 d post-treatment (data not shown). Untreated, control fruits displayed a climacteric peak after 10 d, with a corresponding ethylene production at the peak of 6.6 nmol g−1 h−1 (Fig. 5A). Fruits that were treated with 1, 10, and 100 μM of IAA, exhibited a climacteric peak after 8, 5, and 3 d post-treatment, respectively; with a corresponding ethylene production at the peak of 16.2, 23.6, and 31.6 nmol g−1 h−1, respectively (Fig. 5A).
Fig. 5.
(A) Ethylene production of ‘SH’ plum fruit treated with auxin (filled circles) 0 μM, (open squares) 1 μM, (open triangles) 10 μM, and (open diamonds) 100 μM. Fruit treated with 0 μM auxin served as the control. (B) The steady-state mRNA levels for Ps-GLP1 and 2 in ‘SH’ fruit during ripening at room temperature after treatment with 10 μM IAA. The x-axis in each figure represents days after auxin treatment. Other details are as described in Fig. 2.
‘SH’ fruits treated with 10 μM of IAA and 50 μM TIBA were selected to study the accumulation pattern of Ps-GLPs transcripts. TIBA treatment completely inhibited Ps-GLPs transcription in the fruit (data not shown). However, auxin treatment triggered a dramatic increase in the accumulation level and/or pattern of the two Ps-GLP transcripts, suggesting a possible role for auxin (Fig. 5B). In IAA-treated fruits, Ps-GLPs transcription were steadily augmented along with the progression of fruit ripening and continued to increase past the climacteric peak, reaching their maximal levels in post-climacteric fruit, 10 d after treatment.
Subcellular localization of Ps-GLP1 and 2 proteins
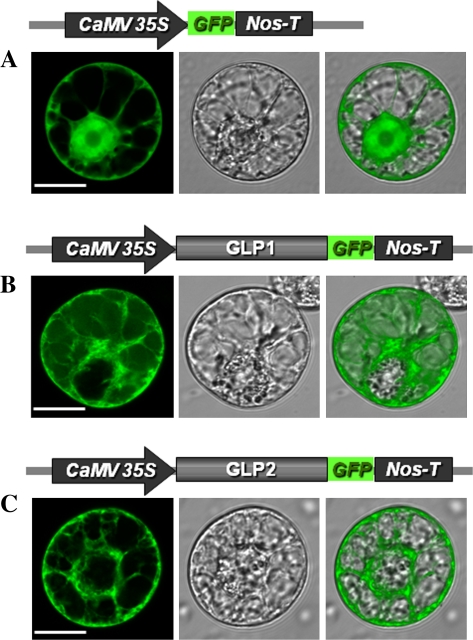
The presence of a putative signal peptide at the N-terminus of each Ps-GLP protein suggested their extracellular targeting localiztion. Ps-GLP1 and 2 coding regions were fused to the GFP tag and were transiently expressed in tobacco protoplasts. Fluorescence microscopy analysis demonstrated that control cells transformed with GFP alone displayed fluorescence ubiquitously as expected (Fig. 6A). By contrast, Ps-GLP1::GFP and Ps-GLP2::GFP fusion proteins were totally excluded from the nucleus and preferentially localized in the plasma membrane and the nuclear envelope (Fig. 6B, C).
Fig. 6.
Subcellular localization of Ps-GLP proteins fused to the GFP tag. Constructs consisting of either the control 35S::GFP, or 35S::GLP1-GFP or 35S::GLP2-GFP were used transiently to transform N. tabacum protoplasts. The subcellular localizations of the GFP protein under the control of 35S (A), the GLP1-GFP (B) or GLP2-GFP (C) fusion protein were analysed using confocal laser scanning microscopy. Light micrographs (centre panel) and fluorescence (left panel) images are merged (right panel) to illustrate the different locations of the two proteins. The length of the bar corresponds to 10 μm.
Discussion
In this study, the molecular characterization of two putative genes encoded for germin-like protein from plum is reported. GLP genes are members of a large multigene family exhibiting diverse patterns of expression and function (Bernier and Berna, 2001). Although sequence identity can be as low as 40% among the different germins/GLPs, there are highly conserved signature elements. One of the most important conserved elements is the presence of a signal peptide in the N-terminal region, which supported the hypothesis of an apoplastic and/or plasma membrane localization. Our results showed that both proteins are, as expected, exclusively localized in the plasma membrane outside the nucleus.
In the dendrogram, a number of well-defined branches have both Arabidopsis and other plant species genes but lack Prunus sequences, suggesting that there are likely to be as yet unidentified GLP genes within the Prunus genome. Twelve and seven GLPs that belong to the three different GLP subfamilies were characterized in Arabidopsis and grapes, respectively (Carter et al., 1998; Godfrey et al., 2007). Ps-GLP1 and 2 proteins belong to GLP subfamily III, which may consist of regulatory proteins with a possible involvement in auxin metabolism.
Assuming that the sequences isolated in this study encode functional GLP proteins, their expression profile was studied in different plant tissues and under various conditions in order to determine their role in fruit development. Bernier and Berna (2001) reported that at least one GLP mRNA has to be present in a specific organ and/or during a certain developmental stage throughout the entire plant life cycle. This simple analysis shows that GLPs are expressed in all plant parts and at all developmental stages.
Ps-GLP1 and 2 transcripts showed very close expression patterns in flowers and during early fruit development (0–37 DAB). Their strong accumulation during flowering reflects their important role in floral induction. Arabidopsis GLP3 mRNA belongs to the same GLP group (subfamily III) and was found to be most abundant in leaves and flowers (Membré et al., 1997). From the available data, it seems that both proteins are also required for regulating the abundant cell division during early embryo development, which occurs during this stage. Mathieu et al. (2006) demonstrated that hybrid larch GLP (LmGER1) played a central role in somatic embryo formation via the regulation of cell wall remodelling necessary for correct development.
Although both transcripts rapidly declined during S2, Ps-GLP1 mRNA was still present in considerable levels, while, that of Ps-GLP2 was totally absent. In order to determine the role of Ps-GLP1, it was essential to understand clearly the physiological aspects that characterize the S2 stage. As mentioned previously, during this stage there is hardly any increase in fruit size (no evidence of cell division and the expansion process), which coincided with a significant reduction in auxin content (Miller et al., 1987) suggesting a minor role for auxin. The absence of auxin during this stage probably explains the significant reduction in Ps-GLPs levels. However, the only fruit development process during this stage is the lignification of the endocarp in a H2O2-dependent manner to form a solid stone (seed). The role of H2O2 in the lignification of the cell wall was clearly determined by Ros Barceló (2005). These results suggest that the transcription of Ps-GLP1 could be altered due to the high levels of endogenous H2O2. The expression of both Ps-GLPs during this stage, along with their response to exogenous H2O2 treatment and the evolution of endogenous H2O2 content, suggest that only Ps-GLP1 protein can be involved in the lignification of the endocarp (see Supplementary Fig. S2 at JXB online).
It has been demonstrated that during the S3 stage the auxin content as well as various auxin-related genes starts to increase rapidly in the whole fruit (Miller et al., 1987; Trainotti et al., 2007). In our studies, a steady rise in the expression of both Ps-GLPs was observed during this stage suggesting that the up-regulation of the two transcripts is likely due to the increase in endogenous auxin content. Although, both transcripts have the same trend of accumulation in the pulp or in the seed, their expression seems to be tissue-specific since their accumulation profile in the pulp was totally different from those in the seed. To determine the role of Ps-GLPs during this stage, it is necessary to distinguish between the two tissues (pulp and seed). Accelerated cell division followed by cell expansion is a signature event of S3 that results in a significant increase in fruit size. Auxin regulates plant cell division, elongation, and differentiation through signal transduction (Taiz, 1984; Abel and Theologis, 1996; Christian et al., 2006). Plum GLP1 and 2 could be components of this signal network that mediates cell division and expansion. Kim and Triplett (2004) showed that the highest accumulation of cotton GLP1 transcripts is positively correlated with the stages of maximal cotton fibre expansion. The finding of auxin-responsive cis-elements in some plant GLP promoters associated with the increase of their expression in response to auxin treatment give further credence to the idea that GLPs may be involved in plant cell expansion (Berna and Bernier, 1997; this paper). Further, the findings that Ps-GLPs accumulation in the seed preceded that of pulp by at least 10 d indicate that seeds stimulate fruit growth and ripening by supplying auxin.
It is almost certain that the series of modifications that transform a mature green fruit into a ripe fruit occur during S3 (El-Sharkawy et al., 2008, 2009) and involve many different metabolic pathways. Therefore, the factors that control the transition of a fruit from the end of growth to the onset of ripening are of primary importance. In climacteric fruits most aspects of the ripening process are triggered and maintained by ethylene (Lelièvre et al., 1997). Previous studies showed that the endogenous auxin content significantly increased in the fruit during ripening, concomitant with the production of climacteric ethylene (Miller et al., 1987). In addition, treatment of fruit with synthetic auxin enhanced both fruit development and ripening (Augustí et al., 1999; Ohmiya, 2000). Furthermore, the evaluation of four different ethylene biosynthesis elements (ACC synthesis) and seven ethylene-responsive transcription factors (ERFs) in plum fruits (i.e. the same cultivars studied in the present work) revealed that auxin can affect ethylene production by increasing the transcription levels of different Ps-ACS and Ps-ERF mRNAs, which play an important role in determining the level of autocatalytic ethylene production and the capacity of the fruit to ripen (El-Sharkawy et al., 2008, 2009). Finally, Arteca and Arteca (2008) observed that different parts of Arabidopsis plants produced various levels of ethylene in response to IAA treatment. The level of IAA-induced ethylene depends on the concentration of auxin, age of the tissue (the youngest leaves showing the greatest stimulation), and the organ position (IAA-induced ethylene occurred in the root and inflorescence tips more than the regions below this).
To investigate further a possible autonomous role played by auxin, the expression of Ps-GLP1 and 2 was studied during ‘EG’ and ‘SH’ fruit ripening. Generally, the accumulation of the two Ps-GLP transcripts in the seed was higher or at least equal to their levels in the pulp during ripening of both plum cultivars. The Ps-GLP1 expression pattern follow the ethylene evolution profile in ‘EG’ mesocarp. On the other hand, its mRNA was hardly detected in ‘SH’ fruit. However, Ps-GLP2 accumulated in a similar pattern as that of ethylene production in the fruits of both cultivars. Although Ps-GLP2 transcript abundance in the seed of both cultivars seems to be ethylene-dependent, it is not likely to be the case as the seed at this stage is much lignified and dry resulting in the inhibition of ethylene biosynthesis (Rodrìguez-Gacio and Matilla, 2001).
The differences in the accumulation levels and/or pattern of both transcripts throughout ripening of early and late fruits might be partially due to the variation in auxin contents and/or ethylene produced among the two plum cultivars. Such variations affect the capacity of the fruit to produce and respond to ethylene, which results in the differentiation in ripening behaviour thereafter (Trainotti et al., 2007; El-Sharkawy et al., 2008, 2009). The scarcity of auxin in the late cultivar can consequently affect the levels of autocatalytic ethylene available for the fruit to maintain ripening. Treatment of late ‘SH’ fruit with auxin significantly accelerated auxin-induced ethylene production and sensitivity. The fruits restored the typical climacteric pattern to a comparable level with those of early ‘EG’ fruits. Moreover, it seems that the concentration of auxin correlated positively with the precocity of the climacteric phase and the rate of ethylene production at the peak.
Taken together, auxin seems to be accumulated rapidly and in higher levels in the early cultivar, which leads to the up-regulation of different transcripts associated with auxin, including auxin and ethylene elements (Trainotti et al., 2007; El-Sharkawy et al., 2008, 2009). Such high levels of ripening-related proteins (auxin- and ethylene-related proteins) resulted in the early transition of the mature green fruit into the ripening stage. Once the fruit initiates autocatalytic ethylene, the ripening process will be enhanced in both auxin- and ethylene-dependent manners. Therefore, the possibility of regulating many ethylene-related genes by auxin is the best alternative that explains the significant accumulation of such transcripts during ripening in an ethylene-independent manner (El-Sharkawy et al., 2008, 2009; Ziliotto et al., 2008). However, the expression profiles of Ps-GLP1 and 2 in response to auxin and auxin inhibitor application suggests that the two transcripts are putatively accumulated in the fruit during ripening in an auxin-dependent manner.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Amino acid sequence alignment of the P. salicina genes, Ps-GLP1 and Ps-GLP2 with closely related sequences from other species.
Supplementary Fig. S2. Hydrogen peroxide (H2O2) levels in developing plum fruits and the effect of externally applied H2O2 on the expression of Ps-GLP1 and Ps-GLP2 during the early stages of fruit growth.
Supplementary Material
Acknowledgments
We thank the Early Researchers Award of the Ontario Ministry of Innovation, the Canadian Foundation for Innovation, the Ontario Innovation Trust, the Ontario Ministry of Agriculture, Food and Rural Affairs, and the Niagara Tender Fruit Marketing Board for financial assistance to SJ. We also thank the MITACS-Accelerate program for partial assistance to IE-S during the study period.
Glossary
Abbreviations
- DAB
days after bloom
- EG
‘Early Golden’
- GLP
germin-like protein
- SH
‘Shiro’
- TIBA
2,3,5-triiodobenzoic acid
References
- Abdi N, Holford P, McGlasson WB, Mizrahi Y. Ripening behavior and responses to propylene in four cultivars of Japanese type plums. Postharvest Biology and Technology. 1997;12:21–34. [Google Scholar]
- Abel S, Theologis A. Early genes and auxin action. Plant Physiology. 1996;111:9–17. doi: 10.1104/pp.111.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arteca RN, Arteca JM. Effects of brassinosteroid, auxin, and cytokinin on ethylene production in Arabidopsis thaliana plants. Journal of Experimental Botany. 2008;59:3019–3026. doi: 10.1093/jxb/ern159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustí M, Almela V, Andreu I, Juan M, Zacarias L. Synthetic auxin 3,5,6-TPA promotes fruit development and climacteric in Prunus persica L. Batsch. The Journal of Horticultural Science and Biotechnology. 1999;74:556–560. [Google Scholar]
- Baumann K, De Paolis A, Costantino P, Gualberti G. The DNA binding site of the Dof protein NtBBF1 is essential for tissue-specific and auxin-regulated expression of the rolB oncogene in plants. The Plant Cell. 1999;11:323–333. doi: 10.1105/tpc.11.3.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berna A, Bernier F. Regulated expression of a wheat germin gene in tobacco: oxalate oxidase activity and apoplastic localization of the heterologous protein. Plant Molecular Biology. 1997;33:417–429. doi: 10.1023/a:1005745015962. [DOI] [PubMed] [Google Scholar]
- Bernier F, Berna A. Germins and germin-like proteins: plant do-all proteins. But what do they do exactly? Plant Physiology and Biochemistry. 2001;39:545–554. [Google Scholar]
- Boss PK, Davies C, Robinson SP. Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera cv. Shiraz grape berries and the implications for pathway regulation. Plant Physiology. 1996;111:1059–1066. doi: 10.1104/pp.111.4.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C, Thornburg RW. Tobacco Nectarin I. Purification and characterization as a germin-like, manganese superoxide dismutase implicated in the defence of floral reproductive tissues. Journal of Biological Chemistry. 2000;275:36726–36733. doi: 10.1074/jbc.M006461200. [DOI] [PubMed] [Google Scholar]
- Carter C, Graham RA, Thornburg RW. Arabidopsis thaliana contains a large family of germin-like proteins: characterization of cDNA and genomic sequences encoding 12 unique family members. Plant Molecular Biology. 1998;38:929–943. doi: 10.1023/a:1006038117130. [DOI] [PubMed] [Google Scholar]
- Carter C, Graham RA, Thornburg RW. Nectarin I is a novel, soluble germin-like protein expressed in the nectar of Nicotiana sp. Plant Molecular Biology. 1999;41:207–216. doi: 10.1023/a:1006363508648. [DOI] [PubMed] [Google Scholar]
- Christian M, Steffens B, Schenck D, Burmester S, Böttger M, Lüthen H. How does auxin enhance cell elongation? Roles of auxin-binding proteins and potassium channels in growth control. Plant Biology. 2006;8:346–352. doi: 10.1055/s-2006-923965. [DOI] [PubMed] [Google Scholar]
- Dunwell JM, Gane PJ. Microbial relatives of seed storage proteins: conservation of motifs in a functionally diverse superfamily of enzymes. Journal of Molecular Evolution. 1998;46:147–154. doi: 10.1007/pl00006289. [DOI] [PubMed] [Google Scholar]
- El-Sharkawy I, Kim WS, El-Kereamy A, Jayasankar S, Svircev AM, Brown DCW. Isolation and characterization of four ethylene signal transduction elements in plums (Prunus salicina L.) Journal of Experimental Botany. 2007;58:3631–3643. doi: 10.1093/jxb/erm213. [DOI] [PubMed] [Google Scholar]
- El-Sharkawy I, Kim WS, Jayasankar S, Svircev AM, Brown DCW. Differential regulation of four members of ACC synthase gene family in plum. Journal of Experimental Botany. 2008;59:2009–3027. doi: 10.1093/jxb/ern056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sharkawy I, Sherif S, Mila I, Bouzayen M, Jayasankar S. Molecular characterization of seven genes encoding ethylene-responsive transcriptional factors during plum fruit development and ripening. Journal of Experimental Botany. 2009;60:907–922. doi: 10.1093/jxb/ern354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S. Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiology. 2004;134:1555–1573. doi: 10.1104/pp.103.034736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey D, Able AJ, Dry IB. Induction of a grapevine germin-like protein (VvGLP3) gene is closely linked to the site of Erysiphe necator infection: a possible role in defence? Molecular Plant–Microbe Interactions. 2007;20:1112–1125. doi: 10.1094/MPMI-20-9-1112. [DOI] [PubMed] [Google Scholar]
- Heintzen C, Fischer R, Melzer S, Kappeler S, Apel K, Staiger D. Circadian oscillations of a transcript encoding a germin-like protein that is associated with cell walls in young leaves of the long-day plant Sinapis alba L. Plant Physiology. 1994;106:905–915. doi: 10.1104/pp.106.3.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Edwards AE, Leyland NR, Bean S, Mullineaux P. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Molecular Biology. 2000;42:819–832. doi: 10.1023/a:1006496308160. [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Research. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Pesacreta TC, Triplett BA. Cotton-fibre germin-like protein. II. Immunolocalization, purification, and functional analysis. Planta. 2004;218:525–535. doi: 10.1007/s00425-003-1134-0. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Triplett BA. Cotton-fibre germin-like protein. I: Molecular cloning and gene expression. Planta. 2004;218:516–524. doi: 10.1007/s00425-003-1133-1. [DOI] [PubMed] [Google Scholar]
- Khuri S, Bakker FT, Dunwell JM. Phylogeny, function, and evolution of the cupins, a structurally conserved, functionally diverse superfamily of proteins. Molecular Biology and Evolution. 2001;18:593–605. doi: 10.1093/oxfordjournals.molbev.a003840. [DOI] [PubMed] [Google Scholar]
- Labouré AM, Faik A, Mandaron P, Falconet D. RGD-dependent growth of maize calluses and immunodetection of an integrin-like protein. FEBS Letters. 1999;442:123–128. doi: 10.1016/s0014-5793(98)01634-2. [DOI] [PubMed] [Google Scholar]
- Lane BG. Oxalate oxidase and differentiating surface structure in wheat: germins. The Biochemical Journal. 2000;49:309–321. doi: 10.1042/0264-6021:3490309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane BG, Dunwell JM, Ray JA, Schmitt MR, Cuming AC. Germin, a protein marker of early plant development, is an oxalate oxidase. Journal of Biological Chemistry. 1993;268:12239–12242. [PubMed] [Google Scholar]
- Lane BG, Grzelczak ZF, Kennedy TD, Kajioka R, Orr J, DíAgostino S, Jaikaran A. Germin: compartmentation of two forms of the protein by washing growing wheat embryos. Biochemistry and Cell Biology. 1986;64:1025–1037. [Google Scholar]
- Lane BG, Bernie F, Dratewka-Kos E, Shafai R, Kennedy TD, Pyne C, Munro JR, Vaughan T, Walters D, Altomare F. Homologies between members of the germin gene family in hexaploid wheat and similarities between these wheat germins and certain Physarum spherulins. Journal of Biological Chemistry. 1991;266:10461–10469. [PubMed] [Google Scholar]
- Leclercq J, Ranty B, Sanchez-Ballesta MT, Li Z, Jones B, Jauneau A, Pech JC, Latche A, Ranjeva R, Bouzayen M. Molecular and biochemical characterization of LeCRK1, a ripening-associated tomato CDPK-related kinase. Journal of Experimental Botany. 2005;56:25–35. doi: 10.1093/jxb/eri003. [DOI] [PubMed] [Google Scholar]
- Le Deunff E, Davoine C, Le Dantec C, Billard JP, Huault C. Oxidative burst and expression of germin/oxo genes during wounding of ryegrass leaf blades: comparison with senescence of leaf sheaths. The Plant Journal. 2004;38:421–431. doi: 10.1111/j.1365-313X.2004.02056.x. [DOI] [PubMed] [Google Scholar]
- Lelièvre JM, Latché A, Jones B, Bouzayen M, Pech JC. Ethylene and fruit ripening. Physiologia Plantarum. 1997;101:727–739. [Google Scholar]
- Matarasso N, Schuster S, Avni A. A novel plant cysteine protease has a dual function as a regulator of 1-aminocyclopropane-1-carboxylic acid synthase gene expression. The Plant Cell. 2005;17:1205–1216. doi: 10.1105/tpc.105.030775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu M, Lelu-Walter MA, Blervacq AS, David H, Hawkins S, Neutelings G. Germin-like genes are expressed during somatic embryogenesis and early development of conifers. Plant Molecular Biology. 2006;61:615–627. doi: 10.1007/s11103-006-0036-5. [DOI] [PubMed] [Google Scholar]
- Membré N, Berna A, Neutelings G, David A, David H, Staiger D, Vasquez JS, Raynal M, Delseny M, Bernier F. cDNA sequence, genomic organization and differential expression of three Arabidopsis genes for germin/oxalate oxidase-like proteins. Plant Molecular Biology. 1997;35:459–469. doi: 10.1023/a:1005833028582. [DOI] [PubMed] [Google Scholar]
- Miller AN, Walsh CS, Cohen JD. Measurement of indole-3-acetic acid in peach fruits (Prunus persica L. Batsch cv. Redhaven) during development. Plant Physiology. 1987;84:491–494. doi: 10.1104/pp.84.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, Heijne GV. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Engineering. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Ohmiya A. Effects of auxin on growth and ripening of mesocarp discs of peach fruit. Scientia Horticulturae. 2000;84:309–319. [Google Scholar]
- Ohmiya A, Motoyuki K, Sakai S, Hayashi T. Purification and properties of an auxin-binding protein from the shoot apex of peach tree. Plant and Cell Physiology. 1993;34:177–183. [Google Scholar]
- Ohmiya A, Tanaka Y, Kadowaki K, Hayashi T. Cloning of genes encoding auxin-binding proteins (ABP19/20) from peach: significant peptide sequence similarity with germin-like proteins. Plant and Cell Physiology. 1998;39:492–499. doi: 10.1093/oxfordjournals.pcp.a029396. [DOI] [PubMed] [Google Scholar]
- Ono M, Sage-Ono K, Inoue M, Kamada H, Harada H. Transient increase in the level of mRNA for a germin-like protein in leaves of the short-day plant Pharbitis nil during the photoperiodic induction of flowering. Plant and Cell Physiology. 1996;37:855–861. doi: 10.1093/oxfordjournals.pcp.a029022. [DOI] [PubMed] [Google Scholar]
- Park CJ, An JM, Shin YC, Kim KJ, Lee BJ, Paek KH. Molecular characterization of pepper germin-like protein as the novel PR-16 family of pathogenesis-related proteins isolated during the resistance response to viral and bacterial infection. Planta. 2004;219:797–806. doi: 10.1007/s00425-004-1290-x. [DOI] [PubMed] [Google Scholar]
- Prestridge DS. SIGNAL SCAN: a computer program that scans DNA sequences for eukaryotic transcriptional elements. Computer Applications in the Biosciences. 1991;7:203–206. doi: 10.1093/bioinformatics/7.2.203. [DOI] [PubMed] [Google Scholar]
- Rodrìguez-Gacio MC, Matilla AJ. The last step of the ethylene-biosynthesis pathway in turnip tops (Brassica rapa) seeds: alterations related to development and germination and its inhibition during desiccation. Physiologia Plantarum. 2001;111:273–279. doi: 10.1034/j.1399-3054.2001.1120216.x. [DOI] [PubMed] [Google Scholar]
- Ros Barceló A. Xylem parenchyma cells deliver the H2O2 necessary for lignification in differentiating xylem vessels. Planta. 2005;220:747–756. doi: 10.1007/s00425-004-1394-3. [DOI] [PubMed] [Google Scholar]
- Staiger D, Appel K, Trepp G. The Atger3 promoter confers circadian clock-regulated transcription with peak expression at the beginning of the night. Plant Molecular Biology. 1999;40:873–882. doi: 10.1023/a:1006278030024. [DOI] [PubMed] [Google Scholar]
- Swart S, Logman TJ, Smit G, Lugtenberg BJ, Kijne JW. Purification and partial characterization of a glycoprotein from pea (Pisum sativum) with receptor activity for rhicadhesin, an attachment protein of Rhizobiaceae. Plant Molecular Biology. 1994;24:171–183. doi: 10.1007/BF00040583. [DOI] [PubMed] [Google Scholar]
- Taiz L. Plant cell expansion: regulation of cell wall mechanical properties. Annual Review of Plant Physiology. 1984;35:585–657. [Google Scholar]
- Thomson EW, Lane BG. Relation of protein synthesis in imbibing wheat embryos to the cell-free translating capacities of bulk mRNA from dry and imbibing embryos. Journal of Biological Chemistry. 1980;255:5965–5970. [PubMed] [Google Scholar]
- Trainotti L, Tadiello A, Casadoro G. The involvement of auxin in the ripening of climacteric fruits comes of age: the hormone plays a role of its own and has an intense interplay with ethylene in ripening peaches. Journal of Experimental Botany. 2007;58:3299–3308. doi: 10.1093/jxb/erm178. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. Dimerization and DNA binding of auxin response factors. The Plant Journal. 1999;19:309–319. doi: 10.1046/j.1365-313x.1999.00538.x. [DOI] [PubMed] [Google Scholar]
- Vallelian-Bindschedler L, Mosinger E, Metraux JP, Schweizer P. Structure, expression and localization of a germin-like protein in barley (Hordeum vulgare L.) that is insolubilized in stressed leaves. Plant Molecular Biology. 1998;37:297–308. doi: 10.1023/a:1005982715972. [DOI] [PubMed] [Google Scholar]
- Wei Y, Zhang Z, Anderson CH, Schmelzer E, Gregersen PL, Collinge DB, Smedegaard-Petersen V, Thordal-Christensen H. An epidermis/papilla-specific oxalate oxidase-like protein in the defence response of barley attacked by the powdery mildew fungus. Plant Molecular Biology. 1998;36:101–112. doi: 10.1023/a:1005955119326. [DOI] [PubMed] [Google Scholar]
- Wojtaszek P, Stobiecki M, Bolwell GP. Changes in the composition of exocellular proteins of suspension-cultured Lupinus albus cells in response to fungal elicitors or CuCl2. Journal of Experimental Botany. 1997;48:2015–2021. [Google Scholar]
- Woo EJ, Dunwell JM, Goodenough PW, Pickersgill RW. Barley oxalate oxidase is a hexameric protein related to seed storage proteins: evidence from X-ray crystallography. FEBS Letters. 1998;437:87–90. doi: 10.1016/s0014-5793(98)01203-4. [DOI] [PubMed] [Google Scholar]
- Woo EJ, Dunwell JM, Goodenough PW, Marvier AC, Pickersgill RW. Germin is a manganese containing homohexamer with oxalate oxidase and superoxide dismutase activities. Nature Structure and Molecular Biology. 2000;7:1036–1040. doi: 10.1038/80954. [DOI] [PubMed] [Google Scholar]
- Yamahara T, Shiono T, Suzuki T, Tanaka K, Takio S, Sato K, Yamazaki S, Satoh T. Isolation of a germin-like protein with manganese superoxide dismutase activity from cells of a moss, Barbula unguiculata. Journal of Biological Chemistry. 1999;274:33274–33278. doi: 10.1074/jbc.274.47.33274. [DOI] [PubMed] [Google Scholar]
- Ziliotto F, Begheldo M, Rasori A, Bonghi C, Tonutti B. Transcriptome profiling of ripening nectarine (Prunus persica L. Batsch) fruit treated with 1-MCP. Journal of Experimental Botany. 2008;59:2781–2791. doi: 10.1093/jxb/ern136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann G, Bäumlein H, Mock HP, Himmelbach A, Schweizer P. The multigene family encoding germin-like proteins of barley: regulation and function in basal host resistance. Plant Physiology. 2006;142:181–192. doi: 10.1104/pp.106.083824. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.