Abstract
AGAMOUS clade genes encode MADS box transcription factors that have been shown to play critical roles in many aspects of flower and fruit development in angiosperms. Tomato possesses two representatives of this lineage, TOMATO AGAMOUS (TAG1) and TOMATO AGAMOUS-LIKE1 (TAGL1), allowing for an analysis of diversification of function after gene duplication. Using RNAi (RNA interference) silencing, transgenic tomato lines that specifically down-regulate either TAGL1 or TAG1 transcript accumulation have been produced. TAGL1 RNAi lines show no defects in stamen or carpel identity, but show defects in fruit ripening. In contrast TAG1 RNAi lines show defects in stamen and carpel development. In addition TAG1 RNAi lines produce red ripe fruit, although they are defective in determinacy and produce ectopic internal fruit structures. e2814, an EMS- (ethyl methane sulphonate) induced mutation that is temperature sensitive and produces fruit phenotypes similar to that of TAG1 RNAi lines, was also characterized. Neither TAG1 nor TAGL1 expression is disrupted in the e2814 mutant, suggesting that the gene corresponding to the e2814 mutant represents a distinct locus that is likely to be functionally downstream of TAG1 and TAGL1. Based on these analyses, possible modes by which these gene duplicates have diversified in terms of their functions and regulatory roles are discussed.
Keywords: AGAMOUS, flower development, fruit development, MADS box, tomato
Introduction
Numerous duplication events have occurred in the genomes of the lineages leading to present-day angiosperms, due in part to multiple occurrences of polyploidization (Wendel, 2000; Blanc and Wolfe, 2004). MADS box transcription factors have attracted particular interest due to their presence in high numbers in the core eudicots and their known roles in a great variety of plant developmental processes (Ng and Yanofsky, 2000). The duplication and subsequent diversification in function of MADS box genes may have played an important role in the origin and diversification of the angiosperms (Ng and Yanofsky, 2000; Irish, 2003; Soltis et al., 2007).
One sublineage of MADS box genes, the AG lineage, has been shown to have a diversity of roles in flower and fruit development across a number of angiosperm species. In gymnosperms, AG genes are expressed in microsporophylls, megasporophylls, and ovules (Tandre et al., 1995; Rutledge et al., 1998; Winter et al., 1999; Jager et al., 2003). Therefore, it has been suggested that the ancestral function of AG genes is in specifying male and female reproductive organs (Theissen et al., 2000; Kramer et al., 2004). In angiosperms, a duplication in the AG clade resulted in the euAG and PLE lineages within the core eudicots (Kramer et al., 2004). Functional analyses of AG clade genes in different core eudicot species have revealed interesting examples of functional conservation, diversification, and subfunctionalization (Bradley et al., 1993; Angenent et al., 1995; Davies et al., 1999; Kapoor et al., 2002; Nitasaka, 2003). In Antirrhinum majus, the euAG lineage gene is FARINELLI (FAR), and PLENA (PLE) corresponds to the PLE lineage gene (Causier et al., 2005). Loss-of-function analyses have shown that Antirrhinum PLE is necessary for stamen and carpel development, while FAR appears to be responsible only for aspects of pollen development in the stamens (Carpenter and Coen, 1990; Bradley et al., 1993; Davies et al., 1999). In Arabidopsis, a different parsing of these reproductive functions has occurred with the Arabidopsis euAG lineage gene, AGAMOUS (AG), specifying stamen and carpel identities, as well as floral determinacy (Yanofsky et al., 1990; Favaro et al., 2003). A recent duplication has resulted in two paralagous PLE lineage genes in Arabidopsis, SHATTERPROOF1 and SHATTERPROOF2 (SHP1 and SHP2), which are redundantly required for dehiscence zone formation in the silique, as well as aspects of ovule development (Liljegren et al., 2000; Pinyopich et al., 2003). These data indicate that the SHP genes have assumed novel roles in specifying development specific to a derived fruit type. Petunia, a euasterid like Antirrhinum, nonetheless has AG gene functions that are more similar to those of Arabidopsis. The Petunia euAG gene PMADS3 displays loss- and gain-of-function phenotypes similar to those of Arabidopsis AG (Tsuchimoto et al., 1993; Kater et al., 1998; Kapoor et al., 2002, 2005). Though no loss-of-function phenotype has been characterized for the Petunia PLE gene FBP6, overexpression studies have found that unlike PMADS3, FBP6 overexpression produces smaller petals, but does not produce homeotic transformations of sepals and petals (Kater et al., 1998).
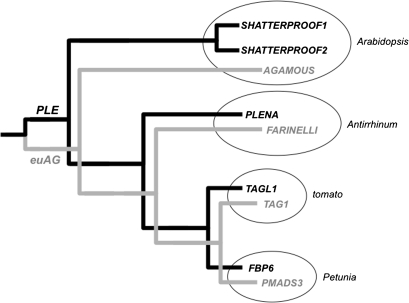
Currently, there are no available mutants of AG clade genes in tomato, although AG lineage genes have been identified and their expression patterns have been characterized to some extent (Busi et al., 2003; Hileman et al., 2006). TAG1 belongs to the euAG clade [Fig. 1 (Vrebalov et al., 2009)] and has been functionally characterized using antisense technology (Pnueli et al., 1994). These antisense analyses suggested that loss of TAG1 function was associated with homeotic transformations of stamens and carpels; however, these phenotypes could have reflected the coordinate loss of function of several AG-related genes in tomato. Functional analyses of TAGL1 have recently been carried out using RNA interference (RNAi), or by repressing its function using a dominant chimeric repressor construct (Itkin et al., 2009; Vrebalov et al., 2009). These analyses have demonstrated that TAGL1 has a unique role in regulating several aspects of ripening, particularly carotenoid accumulation, fleshy fruit expansion, and ethylene production. Furthermore, TAGL1 function cannot completely substitute for SHATTERPROOF function in Arabidopsis, indicating that these genes have diverged in their biochemical activities (Vrebalov et al., 2009).
Fig. 1.
AGAMOUS clade genes in four model species. A major duplication event occurred prior to the emergence of the core eudicots that gave rise to two clades of AGAMOUS-like MADS box genes. PLE and euAG lineages are shown in black and grey, respectively.
Because the functional analyses of TAGL1 and TAG1 have been carried out using transgenic lines that were generated in different backgrounds and using different strategies, it has been difficult to compare directly the phenotypes produced by loss of function of the AG lineage paralogues in tomato. In this study, loss-of-function-analyses of TAG1 and TAGL1 in the same genetic background (cv MicroTom) were carried out using RNAi. It was found that TAGL1 plays a novel role in regulating tomato fruit ripening and has a qualitatively distinct function from that of TAG1. As in earlier studies, it was found that TAG1 plays roles in tomato in specifying normal stamen and carpel development. In addition, though, it was also found that TAG1 RNAi lines produce fourth whorl fruits with defects in determinacy. A temperature-sensitive EMS (ethyl methane sulfonate) mutant has also been identified in tomato that has qualitatively distinct stamen defects and ‘fruit inside fruit’ development. This mutant, however, appears to correspond to a distinct locus that probably acts downstream of TAG1 and TAGL1. Together, these analyses demonstrate the extent of functional diversification between two closely related genes in a fleshy fruited species and the tremendous plasticity in the parsing of function in MADS box genes in the core eudicots, as well as illuminating aspects of the genetic pathway controlling fruit development in tomato.
Materials and methods
Plant materials and growth conditions
Mutant line e2814 was provided by the Zamir Lab (http://zamir.sgn.cornell.edu/mutants/). Plants were grown in a greenhouse at Marsh Gardens (Yale University) under 16 h day and 8 h night conditions with auxiliary sodium lamps. Tomato (Solanum lycopersicum cv MicroTom) tissue culture experiments were carried out at 24 °C with a 16 h light and 8 h dark cycle. Transgenic tomato plants were grown at 22 °C with a 16 h light and 8 h dark cycle.
Transformation constructs and plant transformations
For tomato transformation experiments, RNAi constructs were generated using the Gateway System (Invitrogen, Carlsbad, CA, USA). A 222 bp region of the TAGL1 3′-untranslated region was amplified using the TAGL1FB1 primer containing an attB1 site (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCTGAAATTTGGGGTCAAGG-3′) and the TAGLIRB2 primer containing an attB2 site (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTGCATATTATTTATATAAGGC-3′). A longer 495 bp region of the TAGL1 C-terminal domain and 3′-untranslated region was amplified using the GL1FB1A primer containing an attB1 site (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTGAGCTGCAGAACGCCAACAT-3′) and the TAGL1RB2 primer.
A 262 bp region of the TAG1 3′-untranslated region was amplified using the TAG1FB1 primer containing an attB1 site (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTAATGTGCTTGAGAGATTGTC-3′) and the TAG1RB2 primer containing an attB2 site (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTGGACAGAAAAACAGTTGTGAA-3′).
A longer 430 bp region of the TAG1 C-terminal domain and 3′-untranslated region was amplified using the G1FB1A primer containing an attB1 site (5′-GGGGACAAGTTTGTACAAAAA-AGCAGGCTGAGCTGCAGAACGCCAACAT-3′) and the TAG1RB2 primer.
DNA products were amplified using the following program: 95 °C for 10 min, 40 cycles of 95 °C for 30 s, 50 °C for 45 s, 72 °C for 1 min, followed by 72 °C for 7 min.
These products were cloned into a pH7GWIWG2 (II) or pB7GWIWG2 (II) destination vector (Karimi et al., 2002; http://www.psb.ugent.be/gateway/index.php).
Transformation of tomato (S. lycopersicum cv MicroTom) wild-type cotyledon explants was performed as previously described (McCormick, 1991). The presence of the transgene was verified in the T0 generation by PCR using three sets of primers. HYG1-F (5′-GTTCGGTCGGCATCTACTCTATTC-3′) and HYG1-R (5′-TCGGCTCCAAC-AATGTCCTGAC-3′) amplified the hygromycin (HYG) resistance gene. BAR-F (5′-GCTGCCAG-AAACCCAGGTCA-3′) and BAR-R (5′-CGGACATGCCGGCGGTCTGC-3′) amplified the BAR resistance gene. The 35S2 primer (5′-CCTCTATATAAGGAAGTTCATTTCA-3′) and the gene-specific reverse primers with an attB2 site (TAG1RB2 or TAGL1RB2) amplified the region encompassing the end of the 35S promoter and the transgene. The PCR program for HYG was 95 °C for 10 min, 40 cycles of 95 °C for 30 s, 60 °C for 1 min, 72 °C for 1 min, followed by 72 °C for 7 min. The PCR program for BAR was 95 °C for 10 min, 40 cycles of 95 °C for 30 s, 60 °C for 1 min, 72 °C for 1 min, followed by 72 °C for 7 min. The PCR program for 35S2 and gene-specific product was 95 °C for 10 min, 40 cycles of 95 °C for 30 s, 50 °C for 45 s, 72 °C for 1 min, followed by 72 °C for 7 min.
Carotenoid extraction and HPLC
Carotenoid extraction and HPLC were performed according to a previously published protocol (Alba et al., 2005). Pooled green fruit [36 days post anthesis (dpa)] and red fruit (45 dpa) from transgenic and control lines were used. All samples were run with two technical and two biological replicates.
Quantitative RT-PCR
RNA from 5 g of green fruit (36 dpa) and red fruit (45 dpa) from control and transgenic lines was extracted using a previously published protocol (Griffiths et al., 1999). Quantitative real-time PCR was performed using three biological and three technical replicates for each sample using Taqman One-Step RT-PCR Master Mix Reagents (Applied Biosystems, Foster City, CA, USA) in a 20 μl total sample volume [10 μl of 2× Master Mix; 0.5 μl of 40× Multiscribe™, and RNase Inhibitor Mix; 900 nM of each primer; 250 nM Taqman MGB probe with VIC reporter dye; 3 μl of total RNA (150 ng total); and 4 μl diethylpyrocarbonate (DEPC)-H2O]. A standard curve was included on each plate for the specific gene being analysed using wild-type RNA. For each gene analysis, template-free and negative reverse transcriptase controls were included. The real-time PCR was performed on an ABI PRISM™ 7900HT Sequence Detection System using the following reaction conditions: reverse transcription at 48 °C for 30 min, 95 °C for 10 min, 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The ABI PRISM™ SDS version 2.1 software (Applied Biosystems) was used to calculate gene-specific threshold cycles (Cts) including the endogenous reference (18S) for every sample. Cts were calculated and relative quantitations using a standard curve method were used to calculate mRNA levels. Relative transcript levels were calculated using 18S controls. For each gene tested, the following primers and probes were used: IPP, 5′-TGATGGGAACAAGCCGATGT-3′ (forward), 5′-TGAACCTCCGCAAGAATTGTAA-3′ (reverse), 5′-CTACTGCTTCAGCTTC-3′ (probe); GGPS2, 5′-GTCCACTGGCATGGCTGCTG-3′ (forward), 5′-ATCAACAGCATTTGGTCCACCC-3′ (reverse), 5′-GTCAGTTCCTTGACCTTG-3′ (probe); PSY1, 5′-AACATATGCTAATGACTCCCGAGAGA-3′ (forward), 5′-ATGCGTTTGGGCCATCAA-3′ (reverse), 5′-TATGGTGCAGAAGAACA-3′ (probe); PSY2, 5′-GTCGCTGGTACAGTAGGATTGATG-3′ (forward), 5′-TCTCTGTCGTTGCCTTTGATTC-3′ (reverse), 5′-ATGGGCATTGCACC-3′ (probe); PDS, 5′-AGATTGTTATTGCTGGTGCAGG-3′ (forward), 5′-TGTGACCAGCATCTGCCAA-3′ (reverse), 5′-CTGTAGACAAACCACCCAA-3′ (probe); ZDS, 5′-ATCCTCTGATGGAAGCATGTATGTT-3′ (forward), 5′-GCATCAGCTTTTACAATTTTCTTCTG-3′ (reverse), 5′-TGGGCTTGCCATGTCAAAGGCC-3′ (probe); CRTISO, 5′-CAGGACAAGGTGTTATAGCTGTA-3′ (forward), 5′-GAGCACTGTCCAGCACATCTGAT-3′ (reverse), 5′-CTAAGTCAGCTGCAACAC-3′ (probe); LCY-B, 5′-TCGTCCTGGCTTGCGTATAGA-3′ (forward), 5′-TGTTCATCTTCTTCAATGCTCTTCA-3′ (reverse), 5′-CATGGTGGCTCGTTTAA-3′ (probe); CYC-B, 5′-GGCTCAATTCGACGTGATCA-3′ (forward), 5′-AGAGTGGTGAAGGGTCAACACA-3′ (reverse), 5′-CGGAGCTGGCCCTGCTGGG-3′ (probe); CRTR-B1, 5′-GAACGACGTTTTCGCCATAAC-3′ (forward), 5′-TGAGGCCTTTATGGAAGAAACC-3′ (reverse), 5′-AACGCTGTTCCAGCAATAGCCCTCCT-3′ (probe); and CRTR-B2, 5′-TGCCTTTTTCTGAAATCTTAGCTACA-3′ (forward), 5′-CTCGCCCAGTACTCCATTCC-3′ (reverse), 5′-TCTCTCGTTTGGCGCTGCCGT-3′ (probe).
RT-PCR
Total RNA from tomato tissue was isolated using Trizol (Invitrogen) according to the manufacturer's instructions. A 2.5 μg aliquot was used for cDNA synthesis using SuperScript III Reverse Transcriptase (Invitrogen) according to the manufacturer's instructions. The cDNA was diluted 1:10 and 1–2 ul was used for PCR. The following primers were used: TAG1-F (5′-AGCTCTTGCTGGAATGAAAC-3′), TAG1-R (5′-AAGCTCATGATAGTTTGATG-3′), TAGL1-F (5′-GCATTGGGCAGTTTAAGCCC-3′), TAGL1-R (5′-TCGCGACGAGAGTAATGAGG-3′), LeAP3-F (5′-GAGAAAATGCAAGAGCAGC-3′), LeAP3-R (5′-CAAAAGTAGTAATATCAGAGCC-3′), LePI-F (5′-CAATCAACTTACCCATAAAG-3′), LePI-R (5′-GATTAATTAGTGTTTCTAGC-3′), TM6-F (5′-CGAGAAAATGCAAGAAAACTTG-3′), TM6-R (5′-AGATCACGAGAACCAAATCC-3′), ACT1 (5′-GATGGATCCTCCAATCCAGACACTGTA-3′), and ACT2 (5′-GTATTGTGTTGGACTCTGGTGATGGTGT-3′).
The PCR program for TAGL1 and ACTIN was 94 °C for 5 min, 28 cycles of 94 °C for 30 s, 59 °C for 45 s, 72 °C for 1 min., followed by 72 °C for 10 min. The PCR program for TAG1 was 94 °C for 5 min, 29 cycles of 94 °C for 30 s, 59 °C for 45 s, 72 °C for 1 min, followed by 72 °C for 10 min. The PCR program for LeAP3, LePI, and TM6 was 95 °C for 10 min, 31 cycles of 95 °C for 30 s, 59 °C for 30 s, 72 °C for 1 min, followed by 72 °C for 10 min. Gel images were scanned and band intensities were normalized to ACTIN and quantified using NIH Image J (http://rsb.info.nih.gov/ij/).
Scanning electron microscopy
Plant tissue was fixed overnight in FAA (3.7% formaldehyde, 5% glacial acetic acid, 50% ethanol), then dehydrated to 100% ethanol. Samples were dried using a critical point dryer, sputter coated in gold, and analysed on a Zeiss ISI-SS40 scanning electron microscope.
Results
TAG1 and TAGL1 have overlapping but distinct expression patterns
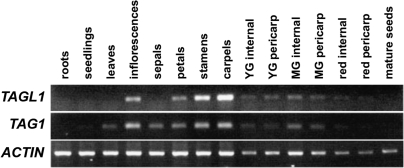
Previous studies have reported that TAG1 and TAGL1 are both expressed principally in flowers and in developing fruits (Busi et al., 2003; Hileman et al., 2006). To characterize this pattern of expression more explicitly, RT-PCR analyses were carried out using TAGL1 and TAG1 gene-specific primers (Fig. 2). TAGL1 expression was undetectable in roots, seedlings, leaves, and mature seeds. In flowers, TAGL1 transcripts were detected at low levels in petals, with stronger expression in stamens and carpels; expression was undetectable in sepals. TAGL1 was expressed in both pericarp and internal tissues of young green and mature green fruit in which the fruit are fully expanded. Expression in both the pericarp and internal tissues was diminished by the red fruit stage. Although TAGL1 expression was observed in developing ovules at earlier stages, mature seeds showed no expression. Expression patterns in flowers and fruit were similar for TAG1, with highest expression levels also seen in inflorescences, stamens, and carpels, with lower but detectable levels in later fruit stages. Unlike TAGL1, we observed detectable levels of TAG1 expression in leaves and sepals (Fig. 2).
Fig. 2.
TAGL1 and TAG1 expression levels in wild-type tissue. TAG1 and TAGL1 expression levels were detected by RT-PCR using dissected tissues from different stages as indicated. For fruit, pericarp was separated from the rest of the fruit tissue (internal) for analysis. YG, young green; MG, mature green fruit.
TAG1 RNAi lines show defects in stamen identity and floral determinacy
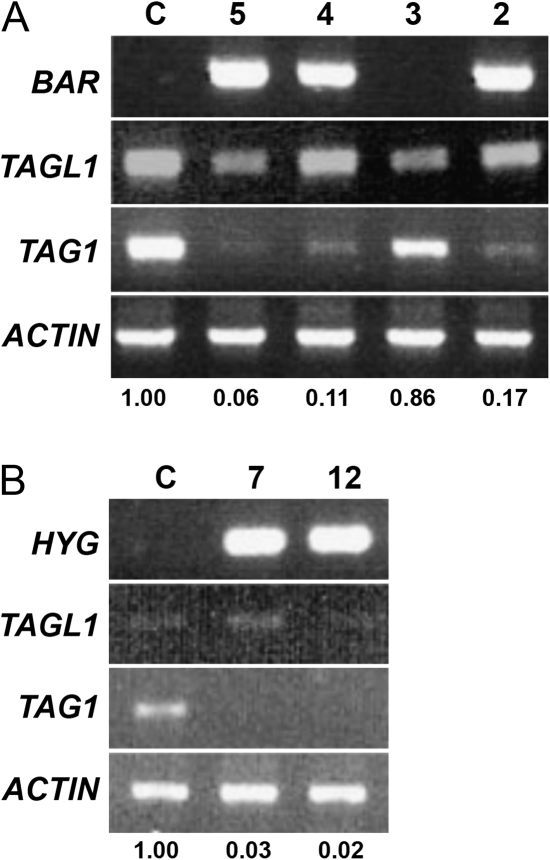
To explore TAG1 function in tomato cv MicroTom, an RNAi construct was generated utilizing a 262 bp region from the TAG1 3′-untranslated region, and this fragment was inserted into a Gateway RNAi vector. This construct was introduced into cv MicroTom using Agrobacterium-mediated T-DNA transfer. Seven independent TAG1 RNAi lines were recovered and verified for transgene integration by the presence of BAR or HYG resistance genes in the T0 generation. These lines showed a reduction in the levels of TAG1 transcripts, with three lines showing a significant reduction in TAG1 transcript abundance (Fig. 3A). These three lines showed a variety of floral defects (Fig. 4). It was found that these lines displayed slight defects in stamens, resulting in the production of smaller amounts of pollen compared with control lines. However, unlike previous reports (Pnueli et al., 1994), no transformation of fourth whorl carpels into sepalloid structures was observed. Instead, these RNAi lines still produced red fruit, but displayed varying degrees of a loss of determinacy (Fig. 4A, B). In some cases, a dramatic ‘fruit inside fruit’ phenotype was observed, in which ectopic fruit structures continued to develop in an indeterminate fashion (Fig. 4D). These lines did not produce seeds, and crosses with wild-type plants were also unsuccessful. Therefore, all characterizations of the phenotype were done in the T0 generation.
Fig. 3.
TAG1 RNAi lines show reduction in TAG1 expression levels. Semi-quantitative RT-PCR on buds from control and TAG1 RNAi lines. TAG1 levels were normalized to actin and are shown relative to the control sample. Expression of TAGL1 was also examined in all lines as indicated. (A) RNAi lines made using a shorter (262 bp) fragment for silencing. Line 3 was scored as negative for transformation. Lines 2, 4, and 5 show a significant reduction in TAG1 RNA levels. (B) RNAi lines made using a longer (430 bp) fragment for silencing. Lines 7 and 12 show almost complete lack of TAG1 transcripts. BAR, BASTA resistance marker; HYG, hygromycin resistance marker, indicating the presence of the transgene.
Fig. 4.
RNAi silencing of TAG1 produces defects in stamens and affects floral determinacy. (A) Control flower ∼3 dpa. (B) TAG1 RNAi line 4 flower made using a shorter (262 bp) fragment for silencing. (C) TAG1 RNAi line 7 flower made using a longer (430 bp) fragment for silencing. (D) TAG1 RNAi line 2 with the ‘fruit inside fruit’ phenotype. (E) Control (left) and TAG1 RNAi line 7 (right) carpels. (F) Control (left) and TAG1 RNAi line 7 (right) stamens showing petalloid tissue. (G) Control red, ripe fruit. (H) Range of phenotypes of TAG1 RNAi line 7 fruit: (top) ectopic exocarp tissue, (bottom left) ‘fruit inside fruit’ (bottom centre and right) internal flower structures.
To determine if the TAG1 RNAi (262 bp construct) phenotypes reflected only a partial loss of function, RNAi lines were also generated using a longer region (430 bp) encompassing both the 3’ coding region and 3′-untranslated region of the TAG1 gene, since longer double-stranded RNA transcripts may produce a stronger phenotype. Two transgenic lines were generated in the MicroTom cultivar and verified for the presence of the transgene in the T0 generation. These lines showed almost complete loss of endogenous TAG1 transcripts (Fig. 3B). Like the 262 bp construct lines, these TAG1 RNAi lines were also sterile, so all phenotypic characterizations were done in the T0 generation. In these 430 bp construct lines a severe phenotype was observed in flowers (Fig. 4). These flowers produced stamens with petalloid tissue (Fig. 4F). In addition, a strong loss of determinacy was observed at anthesis (Fig. 4E), resulting in a dramatic ‘fruit inside fruit’ phenotype (Fig. 4G, H). In common with the 262 bp TAG1 RNAi construct, the 430 bp TAG1 RNAi construct still resulted in the formation of red fruit for both transgenic lines, and did not display any transformation of carpels into sepalloid structures.
Studies were conducted to examine whether TAGL1 expression was affected in the TAG1 RNAi lines (Fig. 3). Little difference was seen in the levels of TAGL1 transcripts in the TAG1 RNAi lines, suggesting that TAG1 does not regulate the expression of TAGL1. These observations also indicated that TAGL1 is not targeted by the TAG1 RNAi construct. Since TAG1 and TAGL1 are the most closely related MADS box genes in the tomato genome (Vrebalov et al., 2009), these observations support the gene-specific targeting of TAG1 by the two RNAi constructs.
TAGL1 functions in regulating fruit ripening
To assess TAGL1 function in tomato cv MicroTom, two RNAi constructs designed to reduce TAGL1 transcript levels were generated. One construct was designed to target a 222 bp region encompassing the 3′-untranslated region of TAGL1; a second construct was also generated using a longer region (495 bp) that targeted both the C-terminal domain and the 3′-untranslated region of TAGL1. Both constructs were predicted to be gene specific in that they correspond to a variable region with low similarity to other MADS box genes. These constructs were introduced into wild-type tomato (cv MicroTom) plants via Agrobacterium-mediated transformation and independently derived lines were verified for the presence of the transgene. Ten lines were generated with the 222 bp RNAi construct and one line with the 495 bp RNAi construct that were characterized for their effects on down-regulation of endogenous TAGL1 expression in homozygous T1 plants (Fig. 5). There was wide variation in the extent of down-regulation of TAGL1, but all lines appeared to be gene specific in their effects in that the expression of TAG1 was not noticeably affected (Fig. 5).
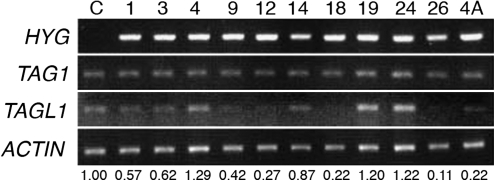
Fig. 5.
TAGL1 expression levels in TAGL1 RNAi lines. Semi-quantitative RT-PCR on green fruit from control (C) and TAGL1 RNAi lines. TAGL1 levels were normalized to actin and are shown relative to the control sample. Lines 1–26 were produced using the 222 bp fragment for silencing; line 4A was produced using the longer (495 bp) fragment for silencing. Expression of TAG1 was also examined by RT-PCR in all lines. HYG, hygromycin resistance marker indicating the presence of the transgene.
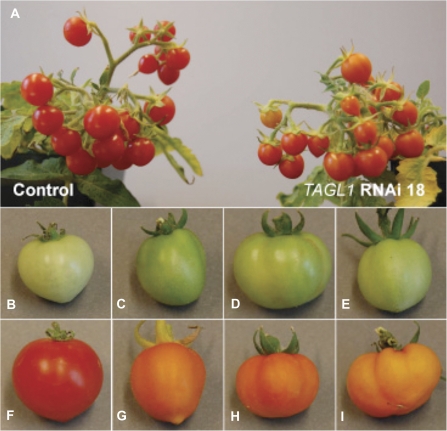
RNAi lines 1, 3, 9, 12, 18, 26 (all targeting the 222 bp region) and line 4A (targeting the 495 bp region) all showed an ∼50% reduction or greater in the expression of TAGL1 (Fig. 5). In these seven lines a striking fruit phenotype was observed where all fruits produced had a distinctive orange colour, compared with the normal bright red fruits produced in wild-type and control lines (Fig. 6F–I, and data not shown). Also, TAGL1 RNAi fruits at the mature green stage (∼36 dpa) were also darker green than control fruits (Fig. 6B–E). The RNAi lines showed no other defects in vegetative, flower, or fruit development, set seeds normally, and produced viable progeny. It was also observed that the differences in colour were not due to a delay in ripening, since experimental lines produced orange fruit at the same number of days post-anthesis as control lines and stopped changing colour at ∼45 dpa (data not shown). The fruits from TAGL1 RNAi lines never appeared bright red, but remained orange to orange-red. This phenotype is similar to that produced by RNAi-induced repression of TAGL1 in cv Ailsa Craig (Vrebalov et al., 2009), as well as in experiments in which TAGL1 expression was down-regulated using a chimeric repressor (Itkin et al., 2009). However, no noticeable change in pericarp thickness was observed as has been documented for down-regulation of TAGL1 in cv Ailsa Craig (Vrebalov et al., 2009). This could reflect the smaller fruit size and overall thinner pericarp that is characteristic of cv MicroTom which could potentially obscure a TAGL1 RNAi pericarp phenotype.
Fig. 6.
RNAi silencing of TAGL1 affects fruit colour. (A) Overview of control (left) and RNAi line TAGL1-18 (right) plants showing differences in fruit colour. (B–E) Fruit 36 dpa; (F–I) fruit 45dpa. (B) and (F) Control line mature green (36 dpa) and red (45 dpa) fruits. (C) and (G) RNAi line TAGL1-4A. Note the pointy fruit in (G). (D) and (H) RNAi line TAGL1-9. (E) and (I) RNAi line TAGL1-18.
TAGL1 regulates carotenoid biosynthetic gene expression and carotenoid accumulation
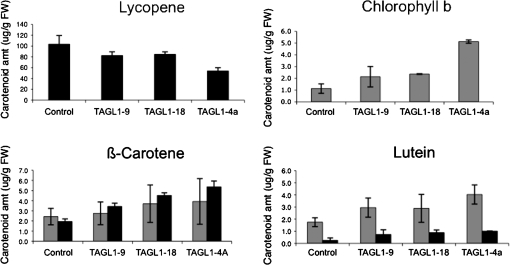
The dramatic alteration in fruit pigmentation in the RNAi lines suggested that TAGL1 controls the expression of the carotenoid biosynthetic gene pathway. Using HPLC, the amounts of phytoene, phytofluene, lycopene, lutein, α-carotene, β-carotene, chlorophyll a, and chlorophyll b were measured in mature green (∼36 dpa) and ripe fruit (∼45 dpa) from three RNAi lines and one control line. Specific alterations were found in the levels of chlorophyll b, as well as in the levels of β-carotene, lutein, and lycopene in the RNAi lines as compared with controls (Fig. 7). In all RNAi lines, increased levels of β-carotene and of lutein were found in both mature green and ripe fruit. Beta-carotene is orange-red while lutein is yellow-orange, and alteration in the relative levels of these carotenoids presumably is responsible for the orange colour phenotype observed in the RNAi lines. Lycopene levels appeared to be somewhat lower in the RNAi lines. In a parallel set of analyses, Vrebalov et al. (2009) also observed an increase in levels of β-carotene and lutein, and a reduction in the levels of lycopene in RNAi induced down-regulation of TAGL1 in tomato cv Ailsa Craig, supporting the observations of specific alterations in the levels of these carotenoids in TAGL1-repressed fruit. It was also observed that chlorophyll b levels were higher in green fruit from all three RNAi lines as well (Fig. 7), which could explain the darker green phenotype of mature green stage TAGL1 RNAi fruit.
Fig. 7.
Chlorophyll and carotenoid levels in TAGL1 RNAi lines. Chlorophyll b and carotenoid levels were determined using mature green (36 dpa; grey bars) and ripe (45 dpa; black bars) fruit from three different TAGL1 RNAi lines as well as controls, by HPLC using mean HPLC peak areas (n=2). Standard errors are shown.
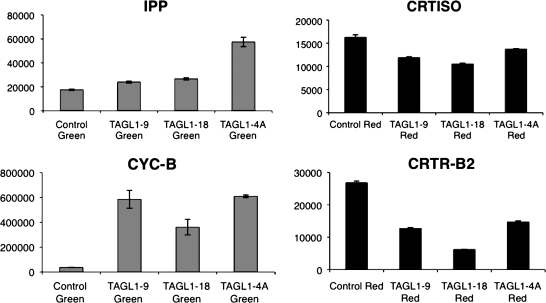
Using quantitative RT-PCR, gene expression levels of a number of carotenoid biosynthetic genes were also analysed, including IPP, encoding isopentenyl diphosphate; PSY1, phytoene synthase 1; PSY2, phytoene synthase 2; PDS, phytoene desaturase; CrtISO, carotenoid isomerase; ZDS, ζ-carotene desaturase; LCY-B, lycopene β-cyclase; CRTR-B1, β-ring carotene hydroxylase; and CrtR-b2, β-ring carotene hydroxylase (chromoplast specific). Significant changes were found in the expression of several of these genes in the RNAi lines, as well as developmental differences in some cases in terms of how such genes were expressed. In particular, it was found that the chromoplast-specific lycopene β-cyclase (CYC-B) gene was expressed at significantly higher levels in green fruits in TAGL1 RNAi lines compared with control lines (Fig. 8). This would account for the orange colour and higher levels of β-carotene seen in mature TAGL1 RNAi fruits, as has been observed in mutants that overexpress CYC-B (Ronen et al., 2000). These observations are also consistent with those reported in Vrebalov et al. (2009) in which levels of β-carotene as well as lutein were higher in TAGL1-suppressed tomato cv Ailsa Craig fruit. Levels of IPP expression were also somewhat higher in green fruit from RNAi lines. Carotenoid isomerase (CrtISO) gene expression levels were reduced in red fruit of TAGL1 RNAi lines, as were the levels of CRTR-b2 transcripts (Fig. 8). As CrtlISO and CRTR-b2 are expressed predominantly in chromoplasts as opposed to chloroplasts (Liu et al., 2003; Galpaz et al., 2006), their down-regulation in TAGL1 RNAi lines also probably contributes to the orange phenotype of the resulting fruit.
Fig. 8.
Transcript levels of carotenoid biosynthesis genes in TAGL1 RNAi lines. Relative transcript levels were determined by quantitative RT-PCR using gene-specific primers on green (grey bars) and red (black bars) fruit. 18S RNA was used as an internal control to normalize the relative level of each transcript. Standard errors of three replicates are shown. IPP, encoding isopentenyl diphosphate; CrtISO, carotenoid isomerase; CYC-B, lycopene β-cyclase (chromoplast specific); and CrtR-b2, β-ring carotene hydroxylase (chromoplast specific).
The e2814 mutant resembles the TAG1 RNAi fruit phenotype and shows stamen defects
Currently, no stable mutants of TAG1 or TAGL1 have been identified in tomato. In an effort to try to identify such a mutant, the available tomato mutant collection generated through EMS and fast-neutron mutagenesis (http://zamir.sgn.cornell.edu/mutants) was utilized. There are currently 3417 phenotypically characterized mutants available in the inbred M82 background. One mutant, e2814, was listed in the database as a recessive mutant showing partial sterility and a ‘fruit inside fruit’ phenotype. Homozygous e2814 mutant plants were examined and it was noted that the ‘fruit inside fruit’ phenotype was only observed when plants were exposed to higher temperatures (approximately 30-37 degrees C) in the spring and summer. Plants grown in cooler temperatures in the autumn produced wild-type flowers and fruits and produced viable seeds, indicating that e2814 is temperature sensitive.
It was found that the ‘fruit inside fruit’ phenotype closely resembled the TAG1 RNAi phenotype observed in more severely affected fruits (Fig. 9). In some cases, a more dramatic ‘fruit inside fruit’ phenotype in the e2814 mutants was observed, where an almost intact second fruit was growing within a larger fruit (Fig. 9L). In all fruits examined, the fourth whorl always produced red fruit and in no cases was transformation of carpels into green sepal-like structures seen. There were also no seeds found in fruits with this severe phenotype.
Fig. 9.
Phenotypes of the e2814 mutant. (A) Wild-type cv M82 flower. (B) e2814 mutant flower showing green, narrow stamens. (C) e2814 mutant flower shows conversion of all stamens to green, carpelloid structures. (D) Wild-type M82 carpel (left) and e2814 carpel with fused stamens (right). (E) Scanning electron microscopy image of an e2814 carpel and a fused, carpelloid stamen. (F) Epidermal cells of a wild-type M82 carpel. (G) Epidermal cells of a wild-type M82 stamen. (H) Epidermal cells of carpelloid, fused stamens from (E). (I) Wild-type M82 ripe fruit. (J) TAG1-9 RNAi fruit. (K) e2814 mutant fruit resembles the TAG1 RNAi fruit phenotype. (L) e2814 mutant fruit with severe ‘fruit inside fruit’ phenotype. Bars=1040 mm (E), 13 mm (F and H), 35 mm (G).
Floral phenotypes were also observed in plants grown at higher temperatures. Unlike the conversion of stamens to petals or petalloid structures as seen in TAG1 RNAi-induced loss-of-function phenotypes, e2814 flowers showed conversion of stamens into carpel-like structures (Fig. 9B). In some cases, stamens were converted into green, twisted style-like tissue (Fig. 9C). Closer examination of the mutant third and fourth whorl structures revealed green or yellow style-like structures of varying thickness in place of third whorl stamens. Also the fourth whorl style of e2814 mutant flowers appeared twisted and larger than in the wild type (Fig. 9D). There was also fusion of third whorl structures to the fourth whorl (Fig. 9E). Scanning electron microscopy of the fused third whorl structures showed that the epidermal cells somewhat resemble wild-type fourth whorl epidermal cells (Fig. 9F–H).
e2814 probably represents a mutation in a novel gene
To test the hypothesis that the lesion in e2814 may correspond to a mutation in TAG1, RNA was isolated from wild-type and e2814 floral buds, the RNA was reverse transcribed to cDNA, and the full-length coding region of TAG1 from these cDNAs was sequenced. No differences between wild-type and the e2814 cDNA were found at the nucleotide level (data not shown). Sequencing of the full-length coding region of cDNA corresponding to the TAGL1 gene also revealed no differences at the nucleotide level (data not shown).
e2814 does not affect expression of MADS box genes involved in floral development
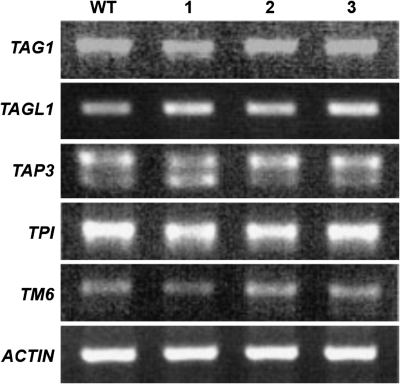
Although the sequence of the full-length coding region of TAG1 was unchanged in the e2814 mutant line, changes in transcript levels of TAG1 were also investigated. Since the phenotype appears to be temperature sensitive, flowers were collected from mutant lines that showed a distinct floral phenotype and from the same lines that did not show a phenotype. Using cDNA made from these tissue samples and wild-type tissue samples grown in the same conditions, the levels of TAG1 transcripts were checked using RT-PCR. No changes in transcript levels were observed in any of the mutant samples compared with the wild type (Fig. 10). TAGL1 expression levels were also unchanged (Fig. 10). Since e2814 flowers have defects in stamen development, transcript levels of B class genes in tomato were also examined (de Martino et al., 2006). Semi-quantitative RT-PCR of TAP3, TPI, and TM6 also showed no difference in expression levels in the e2814 mutant (Fig. 10).
Fig. 10.
e2814 lines show no change in the expression of several MADS box genes. RT-PCR on flowers from M82 wild-type (WT); e2814 flowers showing a wild-type phenotype (1), and stamen and carpel phenotypes (2, 3).
Discussion
TAG1 and TAGL1 have distinct roles in fruit development
A duplication event early in the diversification of the core eudicots has led to two AGAMOUS clades, the euAG and the PLE lineages (Kramer et al., 2004). Functional analyses of euAG and PLE genes in Arabidopsis and Antirrhinum have demonstrated that these paralogues have diversified in a distinct manner in each species (Yanofsky et al., 1990; Davies et al., 1999; Liljegren et al., 2000; Causier et al., 2005). The roles of the tomato members of these lineages, TAG1 and TAGL1, were examined and it was shown that they have unique and distinct functions in fruit development.
The RNAi studies of TAG1 demonstrate that loss of function of this gene in tomato cv MicroTom results in a loss of determinacy in the fourth whorl, resulting in a ‘fruit inside fruit’ phenotype, as well as defects in stamen identity. This phenotype differs from previously published results in which TAG1 expression was down-regulated using antisense technology and produced transformation of fourth whorl organs into sepal-like organs (Pnueli et al., 1994). There are two possible explanations for this discrepancy. One possibility is that the phenotype from the TAG1 RNAi lines represents a weak phenotype and low levels of TAG1 expression are still present and can still act to specify fourth whorl carpel identity. In Arabidopsis, studies of AG partial loss-of-function situations have shown that carpel identity, stamen identity, and determinacy can be separated genetically (Mizukami and Ma, 1992; Sieburth et al., 1995). These studies support the idea that floral meristem determinacy requires the highest level of AG expression, and even slight reduction in AG levels through antisense, RNAi, or other disruptive mutations can cause defects in fourth whorl determinacy. However, in the present analyses, overt homeotic changes in carpel identity were never observed, even in the strongest loss-of-function TAG1 RNAi lines. The TAG1 RNAi lines produced in this study showed a range of phenotypes, even within the different fruits of the same transgenic line. In general, stronger phenotypes (particularly more severe ‘fruit inside fruit’ phenotypes) were observed in fruits derived from older flowers. However, in all cases, even in the fruits with dramatic ‘fruit inside fruit’ phenotypes, the fruits remained red, unlike the earlier antisense study. In contrast, Pnueli et al. (1994) produced a total of 14 antisense lines with aberrant flowers. Two lines exhibited the most extreme phenotypes, but all 14 lines displayed a consistent fourth whorl phenotype with carpels transformed to sepals, accompanied by loss of determinacy.
An alternative explanation to resolve the differences between these studies is that the homeotic transformations observed in the antisense analysis reflect a synthetic phenotype in which multiple AG lineage genes were coordinately down-regulated due to the strategy being used. In the present study, the RNAi construct was designed for silencing to only target TAG1, taking care to avoid stretches of 20–22 bp that matched TAGL1, the most closely related MADS box gene. As such, presumably this analysis of TAG1 RNAi-induced gene-specific silencing more accurately reflects the loss of function of TAG1 alone.
Although it is clear that, upon duplication, there has been subsequent diversification of function in many of the AG lineage genes, several studies have shown that, despite assuming new functions, some AG lineage genes may still retain some aspects of a presumed ancestral carpel identity function (Favaro et al., 2003; Pinyopich et al., 2003). In tomato, it was found that reduction of expression of TAGL1 by RNAi produces defects in fruit ripening. Down-regulation of TAGL1 has been associated with defects in carpel fleshiness as well (Vrebalov et al., 2009), supporting the idea of a conserved role in fourth whorl development. The TAGL1 RNAi-induced defects in tomato fruit ripening are associated with a reduction in chlorophyll b degradation, and alterations in carotenoid levels. This is consistent with other analyses that have demonstrated that TAGL1 function is required for normal accumulation of carotenoids, particularly lycopene, chlorophyll breakdown, and gene expression changes associated with ripening (Itkin et al., 2009; Vrebalov et al., 2009). Although TAGL1 is clearly playing specialized roles in controlling aspects of fruit ripening, it may still retain cryptic carpel identity function. In the case of TAG1 RNAi-induced repression carried out in this study, TAGL1 expression is unaffected and may still be specifying carpel identity. Coordinate down-regulation of TAG1, TAGL1, and potentially other closely related MADS box genes would be needed to test whether redundancy can explain the specification of carpel identity in tomato.
A new mutant in fruit development distinct from TAG1 and TAGL1
One of the difficulties in elucidating the function of TAG1 or TAGL1 in tomato is the lack of a stable mutant. Though antisense strategies, RNAi, and virus-induced gene silencing (VIGS) have provided many insights, these techniques have many drawbacks. Most notably, VIGS produces transient phenotypes and RNAi lines often produce weaker phenotypes after a few generations (Burch-Smith et al., 2004). Both VIGS and RNAi have the added problem of variable silencing, even within the same transgenic line. To that end, a stable mutant affecting carpel development, e2814, was identified in the hope that it would correspond to a mutation in an AG clade gene.
The e2814 mutation is temperature sensitive, and at the restrictive temperature (∼30–37 °C), displays a ‘fruit inside fruit’ phenotype and partial sterility. The temperature sensitivity of a mutant is often associated with alterations in protein conformation that disrupt biological activity. Several temperature-sensitive mutations in MADS box genes have been identified (Bowman et al., 1989; Schwarz-Sommer et al., 1992; Zachgo et al., 1995); one of these, a mutation in the Antirrhinum DEFICIENS gene, has a deletion in a lysine residue in the K domain that probably disrupts protein–protein interactions at the restrictive temperature (Zachgo et al., 1995). However, sequencing of the TAG1 and TAGL1 coding regions from the e2814 mutant did not identify any such point mutations, suggesting that e2814 corresponds to a different locus.
Analyses of Arabidopsis AG have shown that it functions in part to regulate SPOROCYTELESS/NOZZLE and DAD1 directly, two genes that play roles in microsporogenesis and late stamen maturation (Ito et al., 2004, 2007). Along with regulation of jasmonic acid biosynthesis through control of DAD1, AG has also been shown to regulate the expression of GA4, an enzyme that catalyses the biosynthesis of gibberellin (Gomez-Mena et al., 2005). TAGL1, in addition to regulating carotenoid biosynthetic gene expression, also controls ethylene evolution during tomato fruit ripening (Itkin et al., 2009; Vrebalov et al., 2009). Based on the present expression analyses, the locus corresponding to e2814 is probably downstream of both TAG1 and TAGL1 in fruit development, but also appears to have a TAG1- and TAGL1-independent role in regulating stamen differentiation. This may be occurring in part through feedback controls through various hormonal response pathways. One possibility is that e2814 corresponds to one of the large number of ripening-associated transcripts that have recently been identified as being regulated by TAGL1 (Itkin et al., 2009). Identification of the gene corresponding to the e2814 mutant should be valuable in elucidating the network of genes involved in stamen and fruit development in tomato and facilitate better comparisons between how these regulatory networks may have evolved in different plant species.
Acknowledgments
We acknowledge the Tomato Genetics Resource Center at UC Davis and Dani Zamir (Hebrew University) for kindly providing tomato seed. We thank David Weiss (Hebrew University) for help during the early phases of this work, and the staff at Marsh Botanic Gardens for help with plant husbandry. This work was supported by the United States Department of Agriculture–Agricultural Research Service, National Science Foundation Plant Genome grants 05-01778 and 06-06595, BARD grant IS-3803-05, USDA-NRI grants 2007-02773 and 2006-35304-17323, and National Science Foundation grant DBI-0411960.
References
- Alba R, Payton P, Fei Z, McQuinn R, Debbie P, Martin GB, Tanksley SD, Giovannoni JJ. Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. The Plant Cell. 2005;17:2954–2965. doi: 10.1105/tpc.105.036053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angenent GC, Franken J, Busscher M, Van Dijken A, Van Went JL, Dons HJM, Van Tunen AJ. A novel class of MADS box genes is involved in ovule development in Petunia. The Plant Cell. 1995;7:1569–1582. doi: 10.1105/tpc.7.10.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. The Plant Cell. 2004;16:1667–1678. doi: 10.1105/tpc.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. Genes directing flower development in Arabidopsis. The Plant Cell. 1989;1:37–52. doi: 10.1105/tpc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley D, Carpenter R, Sommer H, Hartley N, Coen E. Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of Antirrhinum. Cell. 1993;72:85–95. doi: 10.1016/0092-8674(93)90052-r. [DOI] [PubMed] [Google Scholar]
- Burch-Smith TM, Anderson JC, Martin GB, Dinesh-Kumar SP. Applications and advantages of virus-induced gene silencing for gene function studies in plants. The Plant Journal. 2004;39:734–746. doi: 10.1111/j.1365-313X.2004.02158.x. [DOI] [PubMed] [Google Scholar]
- Busi MV, Bustamante C, D'Angelo C, Hidalgo-Cuevas M, Boggio SB, Valle EM, Zabaleta E. MADS-box genes expressed during tomato seed and fruit development. Plant Molecular Biology. 2003;52:801–815. doi: 10.1023/a:1025001402838. [DOI] [PubMed] [Google Scholar]
- Carpenter R, Coen ES. Floral homeotic mutations produced by transposon-mutagenesis in Antirrhinum majus. Genes and Development. 1990;4:1483–1493. doi: 10.1101/gad.4.9.1483. [DOI] [PubMed] [Google Scholar]
- Causier B, Castillo R, Zhou J, Ingram R, Xue Y, Schwarz-Sommer Z, Davies B. Evolution in action: following function in duplicated floral homeotic genes. Current Biology. 2005;15:1508–1512. doi: 10.1016/j.cub.2005.07.063. [DOI] [PubMed] [Google Scholar]
- Davies B, Motte P, Keck E, Saedler H, Sommer H, Schwarz-Sommer Z. PLENA and FARINELLI: redundancy and regulatory interactions between two Antirrhinum MADS-box factors controlling flower development. EMBO Journal. 1999;18:4023–4034. doi: 10.1093/emboj/18.14.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Martino G, Pan I, Emmanuel E, Levy A, Irish VF. Functional analyses of two tomato APETALA3 genes demonstrate diversification in their roles in regulating floral development. The Plant Cell. 2006;18:1833–1845. doi: 10.1105/tpc.106.042978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro R, Pinyopich A, Battaglia R, Kooiker M, Borghi L, Ditta G, Yanofsky MF, Kater MM, Colombo L. MADS-box protein complexes control carpel and ovule development in Arabidopsis. The Plant Cell. 2003;15:2603–2611. doi: 10.1105/tpc.015123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galpaz N, Ronen G, Khalfa Z, Zamir D, Hirschberg J. A chromoplast-specific carotenoid biosynthesis pathway is revealed by cloning of the tomato white-flower locus. The Plant Cell. 2006;18:1947–1960. doi: 10.1105/tpc.105.039966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Mena C, de Folter S, Costa MM, Angenent GC, Sablowski R. Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development. 2005;132:429–438. doi: 10.1242/dev.01600. [DOI] [PubMed] [Google Scholar]
- Griffiths A, Barry C, Alpuche-Solis A, Grierson D. Ethylene and developmental signals regulate expression of lipoxygenase genes during tomato fruit ripening. Journal of Experimental Botany. 1999;50:793–798. [Google Scholar]
- Hileman LC, Sundstrom JF, Litt A, Chen M, Shumba T, Irish VF. Molecular and phylogenetic analyses of the MADS-box gene family in tomato. Molecular Biology and Evolution. 2006;23:2245–2258. doi: 10.1093/molbev/msl095. [DOI] [PubMed] [Google Scholar]
- Irish VF. The evolution of floral homeotic gene function. Bioessays. 2003;25:637–646. doi: 10.1002/bies.10292. [DOI] [PubMed] [Google Scholar]
- Itkin M, Seybold H, Breitel D, Rogachev I, Meir S, Aharoni A. The TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. The Plant Journal. 2009;60:1081–1095. doi: 10.1111/j.1365-313X.2009.04064.x. [DOI] [PubMed] [Google Scholar]
- Ito T, Ng KH, Lim TS, Yu H, Meyerowitz EM. The homeotic protein AGAMOUS controls late stamen development by regulating a jasmonate biosynthetic gene in Arabidopsis. The Plant Cell. 2007;19:3516–3529. doi: 10.1105/tpc.107.055467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Wellmer F, Yu H, Das P, Ito N, Alves-Ferreira M, Riechmann JL, Meyerowitz EM. The homeotic protein AGAMOUS controls microsporogenesis by regulation of SPOROCYTELESS. Nature. 2004;430:356–360. doi: 10.1038/nature02733. [DOI] [PubMed] [Google Scholar]
- Jager M, Hassanin A, Manuel M, Le Guyader H, Deutsch J. MADS-box genes in Ginkgo biloba and the evolution of the AGAMOUS family. Molecular Biology and Evolution. 2003;20:842–854. doi: 10.1093/molbev/msg089. [DOI] [PubMed] [Google Scholar]
- Kapoor M, Baba A, Kubo K, Shibuya K, Matsui K, Tanaka Y, Takatsuji H. Transgene-triggered, epigenetically regulated ectopic expression of a flower homeotic gene pMADS3 in Petunia. The Plant Journal. 2005;43:649–661. doi: 10.1111/j.1365-313X.2005.02481.x. [DOI] [PubMed] [Google Scholar]
- Kapoor M, Tsuda S, Tanaka Y, Mayama T, Okuyama Y, Tsuchimoto S, Takatsuji H. Role of petunia pMADS3 in determination of floral organ and meristem identity, as revealed by its loss of function. The Plant Journal. 2002;32:115–127. doi: 10.1046/j.1365-313x.2002.01402.x. [DOI] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- Kater MM, Colombo L, Franken J, Busscher M, Masiero S, Van Lookeren MM, Angenent GC. Multiple AGAMOUS homologs from cucumber and petunia differ in their ability to induce reproductive organ fate. The Plant Cell. 1998;10:171–182. doi: 10.1105/tpc.10.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EM, Jaramillo MA, Di Stilio VS. Patterns of gene duplication and functional evolution during the diversification of the AGAMOUS subfamily of MADS box genes in angiosperms. Genetics. 2004;166:1011–1023. doi: 10.1093/genetics/166.2.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren SJ, Ditta GS, Eshed Y, Savidge B, Bowman JL, Yanofsky MF. SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature. 2000;404:766–770. doi: 10.1038/35008089. [DOI] [PubMed] [Google Scholar]
- Liu YS, Gur A, Ronen G, Causse M, Damidaux R, Buret M, Hirschberg J, Zamir D. There is more to tomato fruit colour than candidate carotenoid genes. Plant Biotechnology Journal. 2003;1:195–207. doi: 10.1046/j.1467-7652.2003.00018.x. [DOI] [PubMed] [Google Scholar]
- McCormick S. Transformation of tomato with Agrobacterium tumefaciens. In: Lindsey K, editor. Plant tissue culture manual. Dordrecht: Kluwer; 1991. pp. 1–9. [Google Scholar]
- Mizukami Y, Ma H. Ectopic expression of the floral homeotic gene agamous in transgenic arabidopsis plants alters floral organ identity. Cell. 1992;71:119–131. doi: 10.1016/0092-8674(92)90271-d. [DOI] [PubMed] [Google Scholar]
- Ng M, Yanofsky MF. Three ways to learn the ABCs. Current Opinion in Plant Biology. 2000;3:47–52. doi: 10.1016/s1369-5266(99)00036-9. [DOI] [PubMed] [Google Scholar]
- Nitasaka E. Insertion of an En/Spm-related transposable element into a floral homeotic gene DUPLICATED causes a double flower phenotype in the Japanese morning glory. The Plant Journal. 2003;36:522–531. doi: 10.1046/j.1365-313x.2003.01896.x. [DOI] [PubMed] [Google Scholar]
- Pinyopich A, Ditta GS, Savidge B, Liljegren SJ, Baumann E, Wisman E, Yanofsky MF. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature. 2003;424:85–88. doi: 10.1038/nature01741. [DOI] [PubMed] [Google Scholar]
- Pnueli L, Hareven D, Rounsley SD, Yanofsky MF, Lifschitz E. Isolation of the tomato AGAMOUS gene TAG1 and analysis of its homeotic role in transgenic plants. The Plant Cell. 1994;6:163–173. doi: 10.1105/tpc.6.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronen G, Carmel-Goren L, Zamir D, Hirschberg J. An alternative pathway to beta-carotene formation in plant chromoplasts discovered by map-based cloning of beta and old-gold color mutations in tomato. Proceedings of the National Academy of Sciences, USA. 2000;97:11102–11107. doi: 10.1073/pnas.190177497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge R, Regan S, Nicolas O, Fobert P, Cote C, Bosnich W, Kauffeldt C, Sunohara G, Seguin A, Stewart D. Characterization of an AGAMOUS homologue from the conifer black spruce (Picea mariana) that produces floral homeotic conversions when expressed in Arabidopsis. The Plant Journal. 1998;15:625–634. doi: 10.1046/j.1365-313x.1998.00250.x. [DOI] [PubMed] [Google Scholar]
- Schwarz-Sommer Z, Hue I, Huijser P, Flor PJ, Hansen R, Tetens F, Lonnig W-E, Saedler H, Sommer H. Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO Journal. 1992;11:251–263. doi: 10.1002/j.1460-2075.1992.tb05048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth LE, Running MP, Meyerowitz EM. Genetic separation of third and fourth whorl functions of AGAMOUS. The Plant Cell. 1995;7:1249–1258. doi: 10.1105/tpc.7.8.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Ma H, Frohlich MW, Soltis PS, Albert VA, Oppenheimer DG, Altman NS, dePamphilis C, Leebens-Mack J. The floral genome: an evolutionary history of gene duplication and shifting patterns of gene expression. Trends in Plant Science. 2007;12:358–367. doi: 10.1016/j.tplants.2007.06.012. [DOI] [PubMed] [Google Scholar]
- Tandre K, Albert VA, Sundas A, Engstrom P. Conifer homologues to genes that control floral development in angiosperms. Plant Molecular Biology. 1995;27:69–78. doi: 10.1007/BF00019179. [DOI] [PubMed] [Google Scholar]
- Theissen G, Becker A, Di Rosa A, Kanno A, Kim JT, Munster T, Winter K-U, Saedler H. A short history of MADS box genes in plants. Plant Molecular Biology. 2000;42:115–149. [PubMed] [Google Scholar]
- Tsuchimoto S, van der Krol AR, Chua N-H. Ectopic expression of PMADS3 in transgenic petunia phenocopies the petunia blind mutant. The Plant Cell. 1993;5:843–853. doi: 10.1105/tpc.5.8.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J, Pan IL, Arroyo AJ, et al. Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1. The Plant Cell. 2009;21:3041–3062. doi: 10.1105/tpc.109.066936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel JF. Genome evolution in polyploids. Plant Molecular Biology. 2000;42:225–249. [PubMed] [Google Scholar]
- Winter K-U, Becker A, Munster T, Kim JT, Saedler H, Theissen G. MADS box genes reveal that gnetophytes are more closely related to conifers than to flowering plants. Proceedings of the National Academy of Sciences, USA. 1999;96:7342–7347. doi: 10.1073/pnas.96.13.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature. 1990;346:35–39. doi: 10.1038/346035a0. [DOI] [PubMed] [Google Scholar]
- Zachgo S, de Andrade Silva E, Motte P, Trobner W, Saedler H, Schwarz-Sommer Z. Functional analysis of the Antirrhinum floral homeotic Deficiens gene in vivo and in vitro by using a temperature-sensitive mutant. Development. 1995;121:2861–2875. doi: 10.1242/dev.121.9.2861. [DOI] [PubMed] [Google Scholar]