Abstract
Seed storage proteins are of great importance in nutrition and in industrial transformation because of their functional properties. Brachypodium distachyon has been proposed as a new model plant to study temperate cereals. The protein composition of Brachypodium grain was investigated by separating the proteins on the basis of their solubility combined with a proteomic approach. Salt-soluble proteins as well as salt-insoluble proteins separated by two-dimensional gel electrophoresis revealed 284 and 120 spots, respectively. Proteins from the major spots were sequenced by mass spectrometry and identified by searching against a Brachypodium putative protein database. Our analysis detected globulins and prolamins but no albumins. Globulins were represented mainly by the 11S type and their solubility properties corresponded to the glutelin found in rice. An in silico search for storage proteins returned more translated genes than expressed products identified by mass spectrometry, particularly in the case of prolamin type proteins, reflecting a strong expression of globulins at the expense of prolamins. Microscopic examination of endosperm cells revealed scarce small-size starch granules surrounded by protein bodies containing 11S globulins. The presence of protein bodies containing glutelins makes B. distachyon closer to rice or oat than to wheat endosperm.
Keywords: 2D electrophoresis, Brachypodium, glutelin, grain, mass spectrometry, prolamin, protein bodies, seed storage protein
Introduction
Most people on earth rely on cereals for a large portion of their diet. World production of cereals reached 2525 million tonnes (Mt) in 2008 (FAOSTAT, 16 December 2009, http://faostat.fao.org/site/567/DesktopDefault.aspx?PageID=567). Among them, maize, rice, and wheat supply at least 70% of the total grain production. Cereal grains, despite their relatively low protein content, provide over 200 Mt of proteins for food and feed. The major grain proteins are storage proteins, accounting for about 60–80% of total proteins depending on species and varieties. Hence storage proteins have been extensively studied to increase the grain protein content and to modulate its protein composition for both nutrition and food processing. In wheat, storage proteins form the gluten network which plays a key role in the further processing of wheat flour into baked products such as pasta and noodles. When classified according to their solubility, grain storage proteins fall into three different fractions, albumins soluble in water, globulins soluble in dilute saline, and prolamins soluble in alcohol/water solutions in reducing conditions (Shewry and Thatam, 1990). Globulins are widely distributed in flowering plants, whereas prolamins which are characterized by a high content in proline and glutamine are restricted to grasses. Prolamins constitute the main endosperm storage proteins in wheat, barley, rye, and maize while globulins are the predominant storage proteins in oat and rice, accounting for about 70–80% of the total protein (Shewry and Halford, 2002). Cereal globulins, clearly related to the 11S type globulins, which accumulate in most dicot seeds (Derbyshire et al., 1976), are not salt-soluble and hence are referred to as glutelins (Shewry et al., 1995).
Numerous efforts have been made to improve the nutritional value and processing quality of cereal grains and also the resistance of cereals to biotic and abiotic stresses. Besides conventional breeding used for many years for crops improvement, transgenic technology developed in the past decade offers new opportunities to meet this goal (Bhalla, 2006; Dwivedi et al., 2008; Vasil, 2008). Such strategies require a sequenced genome or expressed sequence tag libraries and a better understanding of the biochemical mechanism underlying endosperm development and storage function. The use of model species has greatly accelerated our understanding of mechanisms and processes in plants kingdom. Brachypodium as a model species, not only for wheat, but also for other temperate grasses, offers many attractive attributes like a small genome (∼300 Mbp) recently sequenced (International Brachypodium Initiative, 2010), a short lifecycle, and easy to transform. Moreover, many resources are being developed by the scientific community (Doust, 2007; Huo et al., 2008; Vain et al., 2008; Vogel and Hill, 2008). Recent phylogenic analyses (Kellogg, 2001; Vogel et al., 2006) classified Brachypodium closer to wheat and barley than to rice, corn or sorghum. Despite the substantial knowledge accumulated on Brachypodium, little is known about its grain. Brachypodium grain is typical of the Poaceae family with a caryopsis size of 8 mm by 2 mm (Opanowicz et al., 2008). The presence of a crease, as in wheat or barley, is also interesting as this feature impacts the milling process (Laudencia-Chingcuanco and Vensel, 2008).
Up to now, the grain composition is poorly documented. A recent report on B. distachyon grain proteins revealed that the main storage proteins are of 12S and 7S globulin type (Laudencia-Chingcuanco and Vensel, 2008), although phylogenic analysis indicated that Brachypodium is ancestral but equally related to Avena and Triticum (Kellogg, 2001; Huo et al., 2009).
In this context, our work aimed at providing a deeper insight into the grain proteome of Brachypodium, with a special focus on storage proteins, their type and accumulation forms. Therefore, the seed storage proteins were separated by protein sequential extraction followed by 1-D and 2-D electrophoresis and identified by mass spectrometry. By investigating the subcellular localization of storage proteins in B. distachyon, it is also shown that they were deposited in small protein bodies.
Materials and methods
Plant materials and protein gel electrophoresis
In this study, B. distachyon grains were used from Bd21-3, a diploid inbred line isolated from the original accession Bd21 by Vogel and Hill (2008) and characterized for its ability to be transformed. Seed samples were gifts from Dr M Gonnaud from INRA Versailles.
All chemicals were of pure grade and purchased from Amersham Pharmacia Biotech (Uppsala, Sweden) and Sigma Chemical (St Louis, MO, USA).
Nitrogen content
The total amount of nitrogen (protein and non-protein) was evaluated according to the Kjeldahl method using a nitrogen-protein conversion factor of 5.7 adapted for cereals. (Mosse, 1990).
Amino acid analysis
The total amino acid composition of mature B. distachyon grains was determined after complete hydrolysis of finely ground dehulled grains in liquid nitrogen. For the quantification of sulphur amino acids, each sample was mixed with 10 μl of a solution of formic acid:hydrogen peroxide (9/11 v/v), incubated for 30 min at 20 °C before being dried under vacuum. After acid hydrolysis performed in 200 μl 6N HCl for 24 h at 110 °C, the amino acids were derivatized with phenyl isothiocyanate (PITC) and quantified by RP-HPLC on a Pico-Tag C18 column (3.9 mm i.d.×15 cm) according to Atanassova et al. (2003).
Protein extraction and quantification
Total extractable proteins were quantified after direct extraction of proteins from dry mature B. distachyon crushed dehulled grains in 0.062 M TRIS-HCl pH 6.8, and 2% SDS. Proteins were extracted using a sequential procedure. A hundred grains were first crushed into a fine powder in liquid nitrogen using a mortar and pestle. The first protein extract was obtained by adding 6 ml of buffer A (50 mM sodium phosphate buffer pH 8, and 0.5 M NaCl). After overnight stirring at 4 °C, the mixture was centrifuged at 10 000 g for 10 min and the supernatant corresponding to the albumins/globulins (A/G) fraction was recovered. The pellet was then washed with 10 ml of buffer A, centrifuged, and the supernatant pooled with the previous one. This fraction was dialysed against water and freeze-dried. The pellet was mixed with 6 ml of 50% propan-1-ol or ethanol and 1% DTT and stirred for 1 h before centrifuging at 10 000 g for 10 min. After recovering the supernatant, 3 ml of buffer UCT (8 M urea, 2% CHAPS, 2 M thiourea, and 18 mM DTT) was added to the pellet and stirred overnight. The protein fraction obtained in the recovered supernatant will be referred to as the UCT fraction. Protease inhibitors (Protease inhibitor complete, Roche, Mannheim, Germany) were added to all buffers.
Protein extracts concentration was determined by using the Non Interfering Protein Assay (Geno Technology, St Louis, MO), according to the supplier's recommendations.
Electrophoresis and Western blot analysis
E-1D: Protein samples were separated on 15% SDS-PAGE gels (13 cm length) according to Laemmli (1970) and stained with CBB (Coomassie Brilliant Blue R250 from Sigma, St Louis, MO, USA).
E-2D: Two-dimensional polyacrylamide gel electrophoresis (2-DE) was performed using the IPGphor Isoelectric Focusing System (Amersham Pharmacia Biotech, Uppsala, Sweden) for the first dimension, and the Hoefer SE 600 Ruby System (Amersham Pharmacia) for the second dimension. Prior to isoelectric focusing, the freeze-dried A/G fraction was solubilized in 250 μl of Destreak Rehydration Solution (Amersham Pharmacia) and 0.5% (v/v) IPG buffer (Amersham Pharmacia). The proteins obtained in buffer UCT were precipitated at –20 °C in acetone. Precipitated proteins were washed twice in acetone for 1 h at –20 °C and centrifuged at 10 000 g for 15 min at 4 °C. Dried protein pellets or freeze-dried proteins were dissolved in 250 μl of Destreak Rehydration Solution and 0.5% (v/v) IPG buffer. Isoelectric focusing of proteins was performed using 13 cm Immobiline dry strips NL pH 3–10 (Amersham Biosciences, Piscataway, NJ). The total voltage reached was 23 000 V. Prior to the second dimension, each strip was equilibrated, first with 6 ml of 50 mM TRIS-HCl, pH 8.8, 6 M urea, 30% v/v glycerol, and 2% SDS containing 64 mM DTT for 10 min and, subsequently, in the same buffer but containing 135 mM iodoacetamide. Proteins were then separated on the basis of their MW values in 15% SDS-PAGE gels using the Hoefer SE600 Electrophoresis unit (Amersham Biosciences). Gels were stained by CBB according to Devouge et al. (2007). Image acquisition was performed using a GS-710 Imaging Densitometer (BioRad) with a resolution of 300 micron pixel−1. Spot detection and image analysis were performed using ImageMaster 2D Platinum software (Amersham Pharmacia). Protein spots were detected and normalized based on the total spot volume. Quantification was performed from three replicates of gels loaded with 300 μg of proteins. The spotting was performed on gels with higher loadings, 500 μg for the salt extract and 700 μg for the UCT extract.
Protein identification by mass spectrometry
In-gel enzymatic digestion of proteins
Protein spots were removed manually and prepared for mass spectrometry analysis. The preparation of spots and their in-gel trypsin digestion was performed according to Devouge et al. (2007). The resulting peptide mixture was acidified by adding 1 μl of aqueous solution of formic acid (1%, v/v), and stored at –20 °C until analysis.
Mass spectrometry analysis
Protein identification was first attempted by the peptide-mass–fingerprint approach by means of MALDI-TOF mass spectrometry. Analyses were performed with a MALDI LR instrument equipped with a conventional 337 nm laser (Micromass/Waters). One μl of the sample was mixed with 1 μl of the matrix preparation (10 g l−1 α-cyano-4-hydroxycinnamic acid, 10 g l−1 2,5-dihydroxybenzoic acid, 70% (v/v) acetonitrile, and 0.1% (w/v) trifluoroacetic acid) and deposited onto the MALDI sample probe. Mass-data acquisitions were piloted by the Mass Lynx software (Micromass/Waters). Nanoscale capillary liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses of the digested proteins were further performed using a Switchos-Ultimate II capillary LC system (LC Packings/Dionex, Amsterdam, the Netherlands) coupled to a hybrid quadrupole orthogonal acceleration time-of-flight mass spectrometer (Q-TOF Global, Micromass/Waters, Manchester, UK). Chromatographic separation was conducted on a reverse-phase capillary column (Pepmap C18, 75 μm i.d., 15 cm length, LC Packings) at a flow rate of 200 nl min−1. The gradient consisted of a linear increase from 2% to 40% of acetonitrile in 50 min, followed by a rapid increase to 50% of acetonitrile within 10 min.
Mass-data acquisitions were piloted by the Mass Lynx software (Micromass/Waters) using the so-called ‘survey scan’ mode: MS data were recorded for 1 s on the mass-to-charge (m/z) range 400–1500, after which the three most intense ions (doubly or triply charged ions) were selected and fragmented in the collision cell (MS/MS measurements).
Protein identification: databank searching
Raw data collected during LC-MS/MS analyses were processed and further searched against databanks using Protein Lynx Global Server 2.1 (Micromass/Waters) and MASCOT Server 2.2 (Matrix Science). Protein identification was achieved by confronting mass data (MS and/or MS/MS spectra) against the putative Brachypodium protein database (release from 1 May 2009, http://www.brachypodium.org, International Brachypodium Initiative, 2010). One missed trypsin cleavage and one dynamic modification (methionine oxidation) were allowed per peptide. Mass accuracy was set to 150 ppm for parent ions and 0.3 Da for MS/MS fragments. Proteins were identified with a minimum of two MS/MS spectra matching the databank sequence, with MASCOT individual ion scores of more than 30 for the Brachypodium genome database (these thresholds correspond to P <0.05 in the MASCOT software). Protein identifications generated by single MS/MS spectra were manually inspected and validated only when the MS/MS spectra displayed a wide series of intense fragments which could be assigned to major predicted fragments (i.e. b or y ions) of the proposed peptide sequence.
In silico search of genes encoding storage proteins within the Brachypodium genome
In a first step, albumin, globulin, gliadin, glutelin, and glutenin sequences were selected from Uniref90 restricted to Viridiplantae (release-57.9, 13 October 2009) and manually curated. An exhaustive Basic Local Alignment Search Tool (BLAST) search for similar Brachypodium proteins was performed using the sequences retrieved from Uniref90, on the putative Brachypodium protein database (Bradi_1.0, release 1 May 2009) downloaded from http://www.brachypodium.org. The e-value cutoff for including sequences in the set of hypothetical Brachypodium storage proteins was set at 1 e-06. In order to confirm the finding of homologous proteins, this set was checked by performing a second BLAST against the UniRef100 database. Finally, for prolamins, features such as protein size and amino acid composition of the predicted proteins were used as a control.
Light microscopy
Transverse sections sampled from half grains were transferred into plastic moulds, fixed in formalin for 24 h at 20 °C, and processed for paraffin inclusion as described by Jamme et al. (2008). Microtome sections of 6–8 μm (Microm HM 340E, France) were deposited and mounted onto glass sides pretreated with VectaBond section adhesive (British BioCell International, UK) for light microscopy.
All the semi-thin sections (1 μm thick) were observed with a LEICA DMRD microscope. In order to visualize starch and intracellular proteins, sections were stained with 0.02 mol l−1 iodine for 2 min, then washed with water before being stained with fast green (1% in ethanol) for 5 min. They were extensively rinsed with water before examination.
Immunolabelling and transmission electron microscopy
Grains from which the lemma and palea had been removed were hydrated overnight at 4 °C in a wet chamber. Small pieces of hydrated grains were taken from the equatorial region of the grains. Samples for microscopy were prepared according to Philippe et al. (2007). Ultra-thin sections (80–100 nm) were cut on an ultramicrotome (MICROM MT-7000) equipped with a diamond knife and were collected on nickel grids. The polyclonal antibody anti-11S (Le Gall et al., 2005) was used as a molecular probe to detect 11S-like proteins in B. distachyon cross-sections. Sections were incubated in primary antibody diluted 200 times in phosphate-buffered saline (PBS) containing 1% (w/v) BSA and 0.05% (w/v) Tween 20. After washing with PBS buffer, plant material was incubated with a 20-fold dilution of anti-rabbit IgG (whole molecule) conjugated to 1 nm colloidal gold complexes, (Aurion, NL). The sections were washed with PBS and water. Labelling was intensified with the silver enhancement kits (Aurion, NL) according to the manufacturer's instructions.
Results
The type and relative abundance of seed storage components vary among species. In cereals, they accumulate in the endosperm and comprise proteins, carbohydrates, and neutral lipids.
The total nitrogen content of whole dehulled grains of B. distachyon reached 2.9±0.2% of the total dehulled grain. Using a conversion factor of 5.7, which is usual for cereals, the protein content was thus estimated to be around 16.6%. The starch content was found to be less than 10%, while cell wall polysaccharides reached 60% of the dehulled whole grain (F. Guillon, personal communication). The amino acid analysis of total proteins in the mature dehulled grain revealed a high content in Glx (20%) followed by Asx (10%), Pro (8.6%), and Gly (8.5%) (Table 1). The SDS-DTT extractable protein amount was estimated to be of 12.5±0.6%, indicating that proteins of this grain were difficult to extract completely.
Table 1.
Composition of Brachypodium (Brachy) grain in amino acids, expressed in g per 100 g AA compared with that of wheat (Shoup et al., 1966), barley (Lange et al., 2007), and rice calculated from Lasztity (1996)
| AA | Ala | Arg | Asx | Cys | Glx | Gly | His | Ile | Leu | Lys | Met | Phe | Pro | Ser | Thr | Trp | Tyr | Val |
| Brachy | 7 | 4.7 | 9 | 3 | 20 | 8.5 | 2.2 | 4 | 6.8 | 2 | 2.5 | 3.6 | 8.6 | 7.6 | 3.6 | nd | 0.8 | 5.9 |
| Wheat | 3.5 | 4.4 | 4.9 | 2.4 | 32.3 | 4.0 | 2.4 | 3.4 | 6.7 | 2.8 | 1.2 | 4.6 | 10.6 | 4.5 | 2.8 | nd | 1.73 | 4.2 |
| Barley | 3.8 | 5.0 | 5.6 | 2.2 | 28.7 | 3.9 | 2.3 | 4.0 | 7.4 | 3.4 | 1.7 | 6.3 | 16.5 | 4.6 | 3.4 | nd | nd | 5.4 |
| Rice | 4.9 | 9.1 | 7.8 | 2.1 | 17.6 | 4.1 | 2.5 | 4.0 | 8.0 | 3.7 | nd | 5.9 | 6.1 | 5.0 | 3.2 | nd | nd | 5.6 |
Further characterization of the grain proteome
Sequential fractionation of the proteome
In a first experiment, proteins were extracted with SDS-DTT buffer and analysed by SDS-PAGE; the profile revealed many bands with at least eight major bands (Fig. 1) with MW varying from 70 kDa to 10 kDa. Usually, grain storage proteins are classified based on their differential solubility in water, salt, and alcohol solutions (Osborne, 1924). Therefore, in a second experiment, these solubility properties were exploited to perform a sequential extraction of storage proteins in order to achieve a more detailed description of the B. distachyon grain proteome. Each of these fractions was then analysed by SDS-PAGE (Fig. 1). Lane 2 shows the result of the first extraction step, using 0.5 M NaCl. Analysis of this extract in reducing (lane 2) and non-reducing (not shown) conditions indicated that the three main bands (65, 57, and 40 kDa) observable on Fig. 1 lane 2 correspond to monomers. The remaining pellet was subsequently extracted with alcoholic solutions. Only traces of proteins were recovered and detected as fuzzy bands. This additional step extracting prolamins specifically indicates that this type of storage protein is present in very low amounts in B. distachyon grains. The pellet remaining after alcoholic extraction was finally extracted with urea buffer in reducing conditions. Its analysis (lane 3) revealed a large number of bands of MW varying from 70 kDa to 20 kDa with five major bands between 66 kDa and 25 kDa. By comparing the SDS extract profile with the other extracts, it seems that one band around 10 kDa was not found using the sequential extraction.
Fig. 1.
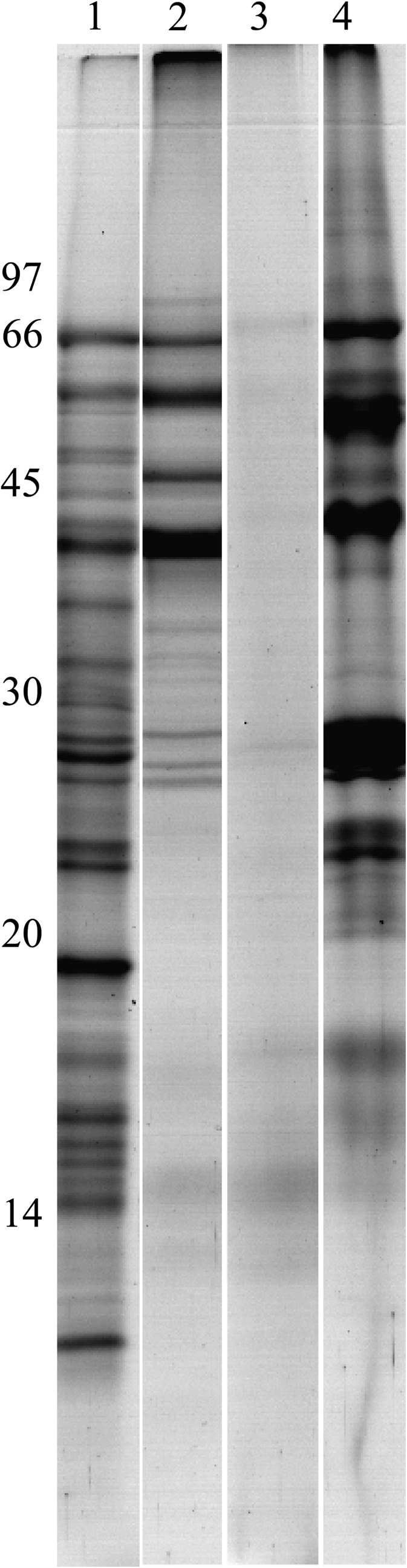
Polyacrylamide gel (15%) electrophoresis of fractions recovered during the sequential extraction. Lane 1, total SDS extract; 2, salt-soluble fraction; 3, alcohol-soluble fraction; 4, UCT-soluble fraction. All samples were reduced.
Towards more comprehensive 2D proteome maps
Given the small amount of extractable proteins in the alcoholic fraction obtained above, only two steps of protein extraction were performed to achieve a more comprehensive proteome map using 2D electrophoresis. Two proteome maps were obtained, the first after performing the extraction in the presence of salts (Fig. 2) and the second after extraction of the residual pellet in urea and reducing agents (Fig. 3). Proteins were separated in a pH range of 3–10 and a MW range varying from 10 kDa to 100 kDa. The major spots from the two-gel electrophoresis were excised and subjected to mass spectrometry analysis for tentative protein identification. Identification was done against the available putative Brachypodium protein database. Since these sequences are not yet annotated, tentative functional annotations were assessed by running a BLAST homology search of Brachypodium protein sequences against the NCBI nr database. The results are summarized in Table 2 (BLASTP 2.2.21; Altschul et al., 1997) and the complete peptide sequences for each protein are reported in supplementary data (see Supplementary Table S1 available at JXB online).
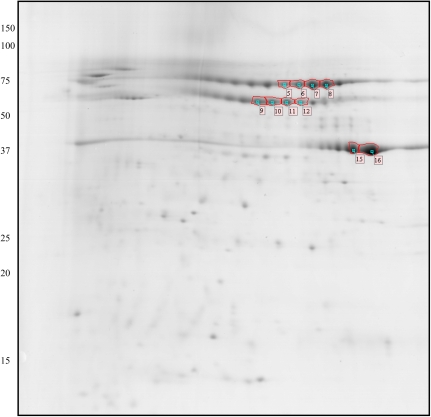
Fig. 2.
2D gel of Brachypodium proteins extracted at moderate ionic strength. Approximately 500 μg of protein sample was loaded onto an IPG 3–10 strip in the first dimension, and separated using a 15% SDS-PAGE gel in the second dimension. Proteins spots were visualized by Coomassie staining. The identified and labelled spots are annotated according to the spot number used in Table 3. (This figure is available in colour at JXB online.)
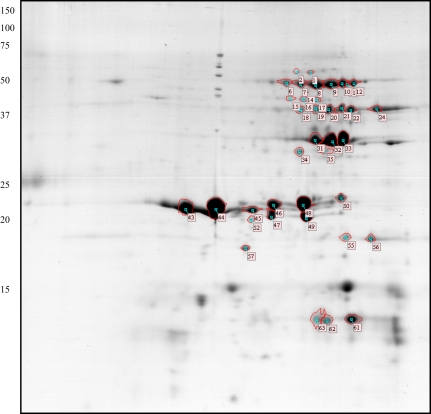
Fig. 3.
Preparative gel of Brachypodium UCT-extracted proteins. Approximately 700 μg of protein sample was loaded onto an IPG 3–10 strip in the first dimension, and separated using a 15% SDS-PAGE gel in the second dimension. Proteins spots were visualized by Coomassie staining. The identified and labelled spots are annotated according to the spot numbers used in Table 3. (This figure is available in colour at JXB online.)
Table 2.
List of identified proteins from 2D gels of Brachypodium, displayed both on Fig. 2(salt-soluble proteins) and on Fig. 3 (UCT extracted proteins)
| Salt soluble fraction (Fig. 2) | |||||||||
| Spot | Instr | Bradi_1.0 | MW | pI | Nb uni pep | % Cov | Score | e-value | Best homologue protein name |
| 5 | Q-TOF | Bradi1g13040.1 | 64 670 | 7.67 | 5 | 13 | 389 | globulin 3 [Triticum aestivum] | |
| 6 | Q-TOF | Bradi1g13040.1 | 64 670 | 7.67 | 10 | 23 | 634 | globulin 3 [Triticum aestivum] | |
| 7 | Q-TOF | Bradi1g13040.1 | 64 670 | 7.67 | 15 | 33 | 1300 | globulin 3 [Triticum aestivum] | |
| 8 | Q-TOF | Bradi1g13040.1 | 64 670 | 7.67 | 7 | 15 | 516 | globulin 3 [Triticum aestivum] | |
| 9 | Q-TOF | Bradi1g05910.1 | 56 024 | 6.65 | 12 | 27 | 1109 | globulin 2 [Zea mays] | |
| 10 | Q-TOF | Bradi1g05910.1 | 56 024 | 6.65 | 6 | 15 | 422 | globulin 2 [Zea mays] | |
| 11 | Q-TOF | Bradi1g05910.1 | 56 024 | 6.65 | 12 | 23 | 1149 | globulin 2 [Zea mays] | |
| 12 | Q-TOF | Bradi1g05910.1 | 56 024 | 6.65 | 3 | 8 | 171 | globulin 2 [Zea mays] | |
| 15 | Q-TOF | Bradi2g02140.1 | 39 841 | 8.44 | 13 | 40 | 804 | xylanase inhibitor 725OS [Triticum aestivum] | |
| 16 | Q-TOF | Bradi2g02140.1 | 39 841 | 8.44 | 20 | 51 | 1503 | xylanase inhibitor 725OS [Triticum aestivum] | |
| UCT fraction (Fig. 3) | ||||||||||
| Spot | Instr | Bradi_1.0 | Mw | pI | Nb uni pep | % | Score | e value | Best homologue protein name | 11S polypeptide |
| 1 | Q-TOF | Bradi2g38060.1 | 54 748 | 7.63 | 7 | 20 | 524 | 11S globulin [Avena sativa] | non-maturated | |
| 6 | Maldi | Bradi1g13040.1 | 64 670 | 7.67 | 15 | 34 | 236 | 8.10E-20 | globulin 3 [Triticum aestivum] | |
| 7 | Maldi | Bradi1g13040.1 | 64 670 | 7.67 | 41 | 283 | 1.60E-24 | globulin 3 [Triticum aestivum] | ||
| 8 | Q-TOF | Bradi1g13040.1 | 64 670 | 7.67 | 17 | 37 | 1563 | globulin 3 [Triticum aestivum] | ||
| 9 | Maldi | Bradi1g13040.1 | 64 670 | 7.67 | 41 | 253 | 1.60E-21 | globulin 3 [Triticum aestivum] | ||
| 10 | Q-TOF | Bradi1g13040.1 | 64 670 | 7.67 | 21 | 47 | 2017 | globulin 3 [Triticum aestivum] | ||
| 11 | Maldi | Bradi1g13040.1 | 64 670 | 7.67 | 41 | 269 | 4.10E-23 | globulin 3 [Triticum aestivum] | ||
| 12 | Q-TOF | Bradi1g13040.1 | 64 670 | 7.67 | 21 | 48 | 1981 | globulin 3 [Triticum aestivum] | ||
| 14 | Q-TOF | Bradi4g28220.1 | 54 322 | 6.68 | 7 | 15 | 407 | seed storage globulin [Avena sativa ] | ||
| 15 | Q-TOF | Bradi1g05910.1 | 56 024 | 6.56 | 10 | 24 | 928 | globulin 2 [Zea mays] | ||
| 16 | Q-TOF | Bradi1g05910.1 | 56 024 | 6.56 | 10 | 24 | 961 | globulin 2 [Zea mays] | ||
| 17 | Q-TOF | Bradi1g05910.1 | 56 024 | 6.56 | 14 | 36 | 1259 | globulin 2 [Zea mays] | ||
| 18 | Q-TOF | Bradi2g38060.1 | 54 748 | 7.63 | 13 | 35 | 1020 | 11S globulin [Avena sativa] | non maturated | |
| 19 | Maldi | Bradi2g38060.1 | 54 748 | 7.63 | 37 | 162 | 2.00E-12 | 11S globulin [Avena sativa] | non maturated | |
| 20 | Q-TOF | Bradi2g38060.1 | 54 748 | 7.63 | 19 | 52 | 1529 | 11S globulin [Avena sativa] | non maturated | |
| 21 | Maldi | Bradi4g28220.1 | 54 322 | 6.68 | 27 | 128 | 5.10E-09 | seed storage globulin [Avena sativa ] | non maturated | |
| 22 | Maldi | Bradi2g38060.1 | 54748 | 7,63 | 15 | 41 | 164 | 1.30E-12 | 11S globulin [Avena sativa] | non maturated |
| Bradi2g40840.1 | 55314 | 7,68 | 9 | 20 | 72 | 2.2E-3 | 11S globulin [Avena sativa] | |||
| 24 | Q-TOF | Bradi2g38060.1 | 54748 | 7.63 | 16 | 48 | 1255 | 11S globulin [Avena sativa] | non maturated | |
| 31 | Q-TOF | Bradi3g17070.1 | 37141 | 6.77 | 4 | 23 | 260 | prolamin [Brachypodium distachyon] | ||
| 32 | Q-TOF | Bradi3g17070.1 | 37141 | 6.77 | 4 | 19 | 364 | prolamin [Brachypodium distachyon] | ||
| 33 | Q-TOF | Bradi3g17070.1 | 37141 | 6.77 | 5 | 24 | 427 | prolamin [Brachypodium distachyon] | ||
| 34 | Q-TOF | Bradi3g17070.1 | 37141 | 6.77 | 1 | 4 | 82 | prolamin [Brachypodium distachyon] | ||
| 35 | Q-TOF | Bradi3g17070.1 | 37141 | 6.77 | 1 | 4 | 79 | prolamin [Brachypodium distachyon] | ||
| 43 | Q-TOF | Bradi2g38060.1 | 54748 | 7.63 | 15 | 32 | 1322 | 11S globulin [Avena sativa] | acidic polypeptide | |
| 44 | Maldi | Bradi2g38060.1 | 54748 | 7.63 | 25 | 121 | 2.60E-08 | 11S globulin [Avena sativa] | acidic polypeptide | |
| 45 | Q-TOF | Bradi2g38060.1 | 29618 | 5.85 | 5 | 13 | 555 | 11S globulin [Avena sativa] | acidic polypeptide | |
| 46 | Q-TOF | Bradi4g28220.1 | 54322 | 6.68 | 9 | 16 | 671 | seed storage globulin [Avena sativa ] | acidic polypeptide | |
| 47 | Q-TOF | Bradi4g29130.1 | 37213 | 9.15 | 4 | 12 | 397 | hypothetical protein | acidic polypeptide | |
| OsI_06680 [Oryza sativa] | ||||||||||
| On SwissProt Glutelin type-B 5 | ||||||||||
| 48 | Q-TOF | Bradi4g28220.1 | 54322 | 6.68 | 7 | 21 | 525 | seed storage globulin [Avena sativa ] | acidic polypeptide | |
| 49 | Q-TOF | Bradi2g40840.1 | 29817 | 6.58 | 8 | 19 | 535 | 11S globulin [Avena sativa] | acidic polypeptide | |
| 50 | Maldi | Bradi2g38060.1 | 54748 | 7.63 | 25 | 101 | 2.60E-06 | 11S globulin [Avena sativa] | acidic polypeptide | |
| 52 | Q-TOF | Bradi2g38060.1 | 29618 | 5.85 | 4 | 12 | 383 | 11S globulin [Avena sativa] | acidic polypeptide | |
| 55 | Q-TOF | Bradi4g28220.1 | 22137 | 7.2 | 9 | 21 | 818 | seed storage globulin [Avena sativa ] | basic polypeptide | |
| 56 | Q-TOF | Bradi4g28220.1 | 22137 | 7.2 | 8 | 18 | 832 | seed storage globulin [Avena sativa ] | basic polypeptide | |
| 57 | Q-TOF | Bradi2g38060.1 | 29618 | 5.85 | 6 | 13 | 381 | 11S globulin [Avena sativa] | acidic polypeptide | |
| 61 | Q-TOF | Bradi1g50300.1 | 18873 | 7.55 | 4 | 41 | 557 | putative avenin-like a precursor [Triticum aestivum] | ||
| 62 | Q-TOF | Bradi1g50300.1 | 18873 | 7.55 | 4 | 41 | 430 | putative avenin-like a precursor [Triticum aestivum] | ||
| 63 | Q-TOF | Bradi1g50300.1 | 18873 | 7.55 | 4 | 41 | 324 | putative avenin-like a precursor [Triticum aestivum] | ||
The columns correspond to an assigned protein spot number, the mass spectrometry technique used for identification, the protein identity as referred to in the Bradi_1.0 (release May 2009: http://www.brachypodium.org), its theoretical molecular weight (Mw), its isoelectric point (pI), the number of unique peptides matched, the per cent sequence coverage, the MASCOT individual ion score (LC-MS/MS on the QTOF instrument) or expected value for the protein hit (MALDI-MS), the NCBI database best homologue protein name and the type of 11S polypeptide obtained from mass spectrometry analysis of the peptides.
It is striking from Figs 2 and 3 that the two protein patterns from the two extracts are different. In both cases, only the main spots were considered for identification, as the choice was made to focus our work on storage proteins.
For the salt extract (Fig. 2), the 2-D gel displayed 284 spots. The six most intense spots are part of three groups of spots which appeared as clear strings on the gel. As commonly observed in proteomics, spots within a string of regularly separated spots frequently belong to the same protein family. The most intense spots from each of these three strings were thus excised and identified by mass spectrometry. One of the strings comprised 11 spots located around 72 kDa and with pI ranging from 6 to 8.2. Four of these spots were identified (spots 5, 6, 7, 8), as being the expression product of the gene Bradi1g13040.1, which was found to be homologous to globulin 3 [Triticum aestivum] (53% identity). A second string constituted by nine spots around 57 kDa and with pI between 6.1 and 8 was analysed. Four of these spots (spots 9, 10, 11, 12) were found to be the expression product of the gene Bradi1g05910.1, which was annotated as homologous to globulin-2 [Zea mays] (60% identity). Both identifications can be categorized as 7S globulins, and the combined intensities of the corresponding spots showed that this family represents 31% of the total protein content of the salt-soluble fraction.
The last string consisted of several major spots found at 37 kDa and with pI between 7.8 and 8.8. The two most abundant spots of this string (15, 16) were identified as expression products of Bradi2g02140.1. This gene product was found to be homologous to xylanase inhibitors, but with a significant homology covering only the N terminal sequence (from the N terminus to AA269). Similar proteins have been reported in wheat seed by Croes et al. (2008), but as they are not storage proteins they will not be considered further.
The electrophoresis of the urea extract (Fig. 3) revealed 120 spots among which 65 major spots were excised for identification and 52 were identified. Twenty-three well-resolved spots were identified as members of the 11S storage protein family. They correspond to expression products of five genes (Table 2). The well-known post-translational cleavage of 11S globulins was taken into consideration for the in silico databank search to identify the mature protein polypeptides. 11S globulins are synthesized during seed-filling as a precursor (preproprotein) form consisting of a single protein chain of about 60 kDa. Maturation of 11S globulin proproteins occurs leading to two polypeptides, one acidic polypeptide of about 35–40 kDa and one basic polypeptide of about 20 kDa (Dickinson et al., 1989). Accordingly, the 11S globulins were found at molecular weights distributed between 70 kDa and 18 kDa and various pIs ranging from 5 to 8. Six spots corresponding to non-maturated forms were detected around 52 kDa (proproteins), ten to the constitutive acidic polypeptides (around 30 kDa or less), and two to the basic polypeptides as reported in Table 2. When all spot volumes were considered, the polypeptides allocated to this 11S family accounted for 72% of the total spot intensities of this fraction. Besides the 11S protein spots, two sets of minor spots were identified as being 7S globulins. 7S globulins were already identified as major spots in the salt extract, and their presence in the urea extract might be due to an incomplete extraction with our saline extraction buffer, since the presence of this family is overwhelming in the salt extract and only accounts for 6% of the total protein amount in the urea extract. Seven spots were identified as prolamin type proteins: four corresponding to the ACF22781.1 prolamin [Brachypodium], and three spots of MW 19 kDa were identified as products of Bradi1g50300.1, which shares 47% identity with a putative avenin-like from Triticum aestivum (CAJ32654). It is interesting to note that the combined volumes of the seven prolamin spots represent only 5% of the expressed proteins found in this fraction, consistent with our previous finding that this family of proteins was especially poorly represented in B. distachyon grains (very few proteins were found to be extractable in alcoholic medium, see above).
In silico search of storage proteins
The putative Brachypodium protein database was searched to obtain a list of potential storage proteins, albumins (2S type), globulins, and prolamins. A first BLAST search ran against the putative Brachypodium protein database by taking, as inputs, the representative proteins of these families as extracted from the Uniref90 databank restricted to the taxonomy Viridiplantae (see Materials and methods). This yielded in a set of hypothetical storage proteins only for globulins and prolamins whereas no hits were found for 2S storage proteins. Three homologous sequences of 7S globulins were found in the putative Brachypodium protein database, 12 of 11S globulins and 9 of prolamins. A second BLAST has identified their best, and clearly annotated, homologues in UniRef100 (Table 3). According to the classification of the prolamin superfamily defined by Shewry and Thatam (1990), based on the annotation of their homologues, two of the sequences fall in the HMW group although their total residue number is really lower than that of classical HMW subunit prolamins as well as their calculated glutamine content. The others, because their cysteine content varies between 3% and 6.6% of their total amino acids, fall in the S-rich prolamin group (Table 3).
Table 3.
List of the in silico predicted storage proteins found among Bradi_1.0 sequences
| Brachypodium sequences | e-value | Homologue | Protein name | Exp | % Exp |
| (UniRef100) | |||||
| Type 11S | |||||
| Bradi2g38050.1 | 0 | Q38780 | 11S globulin OS=Avena sativa | yes | 40–50 |
| Bradi2g38060.1 | 0 | Q38780 | 11S globulin OS=Avena sativa | yes | |
| Bradi2g40840.1 | 0 | Q38780 | 11S globulin OS=Avena sativa | yes | |
| Bradi4g28220.1 | 0 | P12615 | 12S seed storage globulin 1 OS=Avena sativa | yes | |
| Bradi4g29130.1 | 3E-080 | O49258 | 12S globulin OS=Avena sativa | yes | |
| Bradi2g20860.1 | 5E-034 | Q0Q5E3 | Globulin 1 Tax=Triticum aestivum | ||
| Bradi2g37470.1 | 1E-178 | B6SLE7 | Legumin-like protein OS=Zea mays | ||
| Bradi2g37470.2 | 4E-141 | B6SLE7 | Legumin-like protein OS=Zea mays | ||
| Bradi2g37860.1 | 2E-158 | O49258 | 12Sglobulin OS=Avena sativa | ||
| Bradi2g38070.1 | 4E-147 | Q38780 | 11S globulin OS=Avena sativa | ||
| Bradi2g62590.1 | 8E-142 | B6TDD3 | Legumin-like protein OS=Zea mays | ||
| Bradi2g62590.2 | 6E-171 | B6TDD3 | Legumin-like protein OS=Zea mays | ||
| Type 7S | |||||
| Bradi1g05910.1 | E-138 | Q7M1Z8 | Globulin-2 OS=Zea mays | yes | 9–10 |
| Bradi1g13040.1 | 2E-159 | B7U6L4 | Globulin 3 OS=Triticum aestivum | yes | |
| Bradi1g63180.1 | 2E-164 | B6SK46 | Cupin family protein OS=Zea mays | ||
| Prolamin | |||||
| Type S-rich | |||||
| Bradi1g50300.1 | 2E-024 | Q2A784 | Putative avenin-like a OS=Triticum aestivum | yes | 3–4 |
| Bradi3g17070.1 | 2E-155 | C3SAE7 | Prolamine OS=Brachypodium distachyon | yes | |
| Bradi1g50200.1 | 3E-024 | Q2A784 | Putative avenin-like a OS=Triticum aestivum | ||
| Bradi1g50290.1 | 7E-024 | Q2A781 | Putative avenin-like a OS=Aegilops tauschii | ||
| Bradi2g33280.1 | 3E-025 | C3SAE7 | Prolamine OS=Brachypodium distachyon | ||
| Bradi2g38530.1 | 3E-11 | C3SAE7 | Prolamine OS=Brachypodium distachyon | ||
| Bradi2g39940.1 | 2E-028 | C3SAE7 | Prolamine OS=Brachypodium distachyon | ||
| HMW | |||||
| Bradi2g20870.1 | 7E-14 | Q8LKI8 | HMW glutenin subunit y | ||
| OS=Aegilops speltoides | |||||
| Bradi2g20910.1 | 2E-015 | Q670Q5 | High molecular weight glutenin subunit | ||
| 1Dy10.1 OS=Triticum aestivum |
The columns correspond to putative Brachypodium storage protein sequences (Bradi_1.0), e-value, entry number of the best homologue sequence as found in the Uniref100 databank and corresponding protein name, evidence at proteomic level (‘yes’ means that the gene was found to be expressed based on our 2D-E experiments), and approximate percentage of the total extractable proteins and estimated from 2D-E analysis. Three protein families are indicated: 11S globulin, 7S globulins, prolamins (divided into S-rich prolamins and HMW prolamins).
Thus, although a similar number of genes encoding 11S globulins and prolamins was found, our experimental observations clearly showed that the genes encoding for globulins are much more highly expressed than those encoding for prolamins.
Several distinct spots were identified as the expression products of a single gene. This is quite common in proteomics, and mostly arises from post-translational modifications. Indeed, glycosylation was previously reported in pea for the 7S family (Casey and Domoney, 1999), phosphorylation was shown for the 11S in Arabidopsis (Wan et al., 2007), while non-specific proteolytic cleavages of 11S globulins in Arabidopsis seeds were observed by Gallardo et al. (2001). These modifications could explain the discrepancy between the number of detected spots and the number of genes identified in this study, although the extensive characterization of these modifications goes beyond the scope of this paper and would represent a dedicated work.
Subcellular localization of the main storage proteins
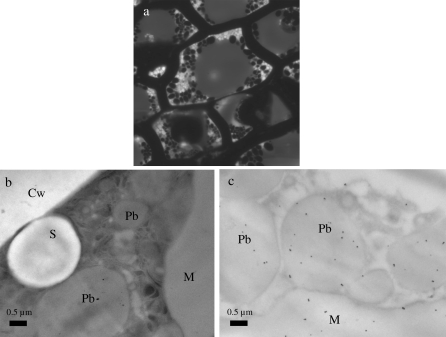
The endosperm is the main tissue of cereal grain and contributes as much as 75% of the total dehulled grain weight. The storage endosperm of Brachypodium consists of vacuolated thick cell walls, making it quite distinct from the storage endosperm of other cereals. A small amount of starch granules ranging in size from 2–4 μm was observed. They appear to be concentrated at the cell periphery and are surrounded by protein bodies with apparent sizes ranging from 0.5 μm to 2 μm (Fig. 4a). Figure 4b obtained by transmission electron microscopy shows an endosperm cell with, on the upper left side, part of a cell wall and within the cell, small starch granules together with almost spherical protein bodies surrounded by a membrane. A larger structure resembling larger protein bodies can be observed on the right side of the image. Since 11S globulin type proteins appears to be the major form of storage proteins in mature B. distachyon grain, it was necessary to localize them more precisely within the endosperm by means of immunocytochemical labelling studies (Fig. 4c). After labelling with anti-11S globulin serum, protein bodies as well as the larger irregular shaped structures were labelled, indicating the accumulation of 11S type globulin within these structures.
Fig. 4.
(a) Fluorescence micrograph of a cell in resin-embedded starchy endosperm showing the localization of proteins bodies around starch granules. (b, c) Transmission electron micrographs of a mature endosperm cell. (b) Immunolabelling control in which primary antibody anti-11S was substituted with preimmune serum. (c) Immunolabelling of protein bodies and proteic matrix with antibody to 11S proteins. S, starch granule; Pb, protein bodies; M, matrix; Cw, cell wall.
Discussion
The protein content of B. distachyon grain approaches the highest level measured in cereal grains, in fact it is close to that of oats. The amino acid composition characterized by a high content in amides reflects the predominance of storage proteins in the global grain proteome. Among these proteins only 75–80% were extractable probably because of the high content of cell wall polysaccharides resulting in a high viscosity of the extracts. The starchy endosperm cells of B. distachyon grains have thick cell walls compared with other cereals and appeared to be vacuolated. At this stage, the content of the vacuole is not known.
Storage proteins identification
The genome sequencing of Brachypodium is complete, but the genomic databank has not been functionally annotated until now. Therefore, the proteins cited in our study were annotated by running the BLAST alignment algorithm against the NCBI nr database. Identification of B. distachyon storage proteins showed that their closest homologues (best BLAST hit) mainly belong to Avena sativa (oat) and Triticum aestivum (wheat) with 38% and 35% of identified spots, respectively. Both species are members of the Pooideae subfamily which also includes B. distachyon.
Cereal grain protein composition has long been defined on the basis of their solubility in different media (Osborne, 1924). It was found that sequential extraction relying on these solubility properties enabled the whole grain proteome to be simplified and a deeper and more comprehensive description of the protein composition to be obtained. Among B. distachyon grain proteins, 10% were salt-soluble proteins, very few were soluble in alcoholic medium, whilst the overwhelming majority (90%) was soluble in urea buffer in reducing conditions. Most of these proteins were identified as globulins, and noticeably, only a very low amount of prolamins was found.
11S globulins
Within the globulins, the 11S family is the most represented in Brachypodium grain. Their identification revealed that they are the expression products of five genes among which Bradi2g38060 and Bradi2g38050 are identical and give indiscernible expression products (see Supplementary data Fig. S1 at JXB online). The 11S storage globulins are synthesized as a single polypeptide precursor or preproprotein, the signal sequence of which is removed cotranslationally. The resultant proprotein, consisting of a single protein chain of about 52 kDa, is further cleaved to form an acidic α-polypeptide of Mr ≈30 000 and a basic β-polypeptide of Mr ≈22 000 linked by a disulphide bond. All 11S proteins identified in this study displayed the specific cleavage site between the α- and β-polypeptides which lie between an asparagine and a glycine residue and has been highly conserved during the evolution of the 11S globulin (Dickinson et al., 1989; Sjödahl et al., 1991). Our results revealed the presence of three 11S globulin forms: the residual proprotein, and the α- and β-polypeptides. The presence of the proprotein might indicate that the maturation process giving rise to the formation of the α- and β-chains was not fully completed when developing seeds entered into quiescence. Similar results were reported in Arabidopsis seeds (Gallardo et al., 2001). Many α-polypeptides were identified, their presence at various pI probably results from post-translational modifications (Wan et al., 2007). Although they should be present, only β-polypeptides (theoretical pI of 7.2) coming from Bradi4g28220.1 were identified. The β-polypeptides arising from the other 11S globulins identified had theoretical pIs over 8.6 and are probably not visible in the 2D gel. The presence of fragments of α-polypeptides confirms that some proteolytic enzymes may be active during seed-filling and process storage proteins into chains capable of stable accumulation in mature seeds (Gruis et al., 2002).
Five other genes of the 11S type were identified by in silico homology search. Among these, three also contain the specific ‘NG’ cleavage site and are highly similar to the expressed genes that were detected. The alignment of these amino acid sequences (i.e. detected and predicted by in silico search) revealed that the β-polypeptides are more conserved than the α-polypeptides, the lengths of which show great variations (see Supplementary Fig. S1 at JXB online). This is a common feature of the amino acid sequences of 11S globulins (Sjödahl et al., 1991). The two other in silico predicted sequences, although belonging to the 11S globulins, are quite different. For these five genes, no expression products were found in our analysis.
7S globulins
One-third of the identified globulins are 7S type globulins and may be related to the ‘vicilin-like’ globulins found in dicotyledonous seeds. Such proteins were also observed in maize, barley, and wheat seeds (Wallace and Kriz, 1991; Heck et al., 1993; Loit et al., 2009). They are often called ‘embryo-globulins’ and, in the case of maize, they exist as two saline-soluble forms (GLB1 and GLB2) (Kriz, 1989; Wallace and Kriz, 1991) located in the embryo and in the aleurone layer (Kriz and Schwartz, 1986). The two proteins identified in B. distachyon resemble these proteins, with a high sequence similarity and a same solubility behaviour. Therefore, it can be assumed that they are located both in the embryo and in the aleurone layer as in maize, as also suggested by Laudencia-Chingcuanco and Vensel (2008).
Other storage proteins
Besides the globulins, only two types of prolamins were identified in Brachypodium grain, making them minor storage proteins as in rice and oat. One is homologous to γ-gliadins and the other to an avenin-like. These two proteins are characterized by a high content in glutamine (22.2% and 20.0%) in cysteine (3.0% and 4.7%), and – for the one homologous to γ-gliadin, in proline (17.4%). Insight into the conserved domains showed that both belong to the AAI-LTSS superfamily, which encompasses seed storage proteins presenting a common pattern of eight cysteines that form four disulphide bridges. A recent study by Xu and Messing (2009) used syntenic alignment of chromosomal regions of multiple genomes from the Poaceae family, including rice and wheat, and identified three brachypodin loci that were prolamin-like in Brachypodium, named Bra1, Bra2, and Bra3. The phylogenetic analysis revealed that Bra1 and Bra2 belong to a same group,that the authors named ‘group II’, which include γ-, α-gliadins and LMW glutenins, while Bra3 belongs to ‘group III’ which includes only HMW glutenins. Following the classification of Xu and Messing (2009), our in silico study performed on a putative Brachypodium proteins database gave seven translated genes belonging to group II, and two to group III. In addition, our experimental data showed that the two expressed prolamins found in this work are of the S-rich type of prolamins and belong to group II. These results confirmed that group II prolamins are actually expressed in Brachypodium, while genes from group III might not be expressed as our data did not reveal any protein belonging to this group. It is noteworthy that the absence of expression of prolamin genes was already reported in wheat (Forde et al., 1985).
It is striking that, no protein belonging to the ‘group I’ has been found here, neither in our in silico search nor in our proteomics analyses. Group I encompasses the youngest genes among prolamins, which arose long after the split of the Poaceae family into three subfamilies (Xu and Messing, 2009). Therefore, our results strengthen the close relationships of Brachypodium with wheat, barley, and oat, all three belonging to the Pooideae subfamily.
The main storage proteins in cereals are prolamins and globulins which may occur as salt-soluble forms but also as salt-insoluble forms, in this case referred to as glutelins. The proportion of these two storage families varies among cereals, prolamins accounting for 50–60% of total endosperm proteins in Triticum (wheat), Zea (maize), and Hordeum (barley), but representing only 5–10% of endosperm proteins in Oryza (rice) and Avena (oat). As in other cereals, our results show that B. distachyon grains store these two main types of storage protein. Close to what was observed for rice and oat, 11S globulins are the predominant storage proteins in B. distachyon (their amount represented 50–60% of the total protein amount loaded on the gels, while the prolamins represented less than 12%) and they are encoded by a multigenic family of genes. Again, similar to what was observed in oat or rice grains, they were not soluble in salt buffers and may therefore be related to the glutelins which were found in oat and rice (Shotwell et al., 1988; Shewry and Halford, 2002).
Subcellular localization
Our study revealed the cellular location of glutelins in small-sized protein bodies, but also in large irregular shaped structures. As in rice, maize, oat, and barley, individual protein bodies are found in the central starchy endosperm cells in mature grains of Brachypodium. The larger structures will need to be characterized further but our observations suggest the presence of a protein matrix. Both cases can be found in cereals: in barley, the deposition of storage proteins was observed in protein bodies and the protein matrix, while in rice, the packaging of storage proteins occurs in two types of protein bodies (Lasztitky, 1996; Tanaka et al., 1980).
Conclusion
To the best of our knowledge, this study is the first to provide an insight into the grain biochemistry of one member of the Brachypodieae tribe which belongs to the same Pooideae subfamily as the Triticeae. By means of proteomic approaches on the one hand, and of immunocytolocalization on the other hand, our work brought two notable results about B. distachyon grain composition: first, its overall protein content is rather high, and approaches the level described in oat kernels; secondly, the accumulation of storage proteins occurs in protein bodies. As B. distachyon is the unique sequenced genome of the Pooideae family, these results provide the basis for further functional comparisons to other cereal grains, among which are those of agronomic interest such as wheat, rice, barley, and maize. The in silico homology search revealed the presence of genes characteristic of the two main storage protein families in seeds: the prolamins and the globulins. Although the number of these genes was equivalent for both types, the expression products identified were mostly glutelins (i.e. salt-insoluble globulins). Such a large amount of glutelins makes B. distachyon closer to Avena (oat) and Oryza (rice) than to wheat, in which prolamins represent the major form of storage proteins.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Table S1. Detailed results of the MASCOT search from LC-MS data.
Supplementary Fig. S1. Alignment of deduced amino acid sequences of the 11S type globulins from distachyon: the arrow indicates the position of the specific ‘NG’ cleavage site between the acidic and basic polypeptides.
Supplementary Material
Acknowledgments
This work was performed thanks to the database produced by the US Department of Energy Joint Genome Institute http://www.jgi.doe.gov/.
We would like to acknowledge G Deshayes for technical assistance in the proteomic analysis. We thank M Dalgalarrondo for amino acid analyses and Dr Y Popineau for comments on the manuscript.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanassova M, Choiset Y, Dalgalarrondo M, Chobert JM, Dousset X, Ivanova I, Haertlé T. Isolation and partial biochemical characterization of a proteinaceous anti-bacteria and anti-yeast compound produced by Lactobacillus paracasei subsp. paracasei strain M3. International Journal of Food Microbiology. 2003;87:63–73. doi: 10.1016/s0168-1605(03)00054-0. [DOI] [PubMed] [Google Scholar]
- Bhalla PL. Genetic engineering of wheat: current challenges and opportunities. Trends in Biotechnology. 2006;24:305–311. doi: 10.1016/j.tibtech.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Casey R, Domoney C. Pea globulins. In: Shewry PR, Casey R, editors. Seed proteins. Kluwer Academic Publishers; 1999. pp. 171–208. [Google Scholar]
- Croes E, Gebruers K, Luyten N, Delcour JA, Courtin CM. Variability of polymorphic families of three types of xylanase inhibitors in the wheat grain proteome. Proteomics. 2008;8:1692–1705. doi: 10.1002/pmic.200700813. [DOI] [PubMed] [Google Scholar]
- Derbyshire E, Wright DJ, Boulter D. Legumin and vicilin, storage proteins of legume seeds. Phytochemistry. 1976;15:3–24. [Google Scholar]
- Devouge V, Rogniaux H, Nési N, Tessier D, Guéguen J, Larré C. Differential proteomic analysis of four near-isogenic Brassica napus varieties bred for their erucic acid and glucosinolate contents. Journal of Proteome Research. 2007;4:342–353. doi: 10.1021/pr060450b. [DOI] [PubMed] [Google Scholar]
- Dickinson CD, Hussein EHA, Nielsen CN. Role of post-translational cleavage in glycinin assembly. The Plant Cell. 1989;1:459–469. doi: 10.1105/tpc.1.4.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doust A. Architectural evolution and its implications for domestication in grasses. Annals of Botany. 2007;100:941–950. doi: 10.1093/aob/mcm040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi S, Perotti E, Ortiz R. Towards molecular breeding of reproductive traits in cereal crops. Plant Biotechnology Journal. 2008;6:529–559. doi: 10.1111/j.1467-7652.2008.00343.x. [DOI] [PubMed] [Google Scholar]
- Forde BG, Heyworth A, Pywell J, Kreis M. Nucleotide sequence of a B1 hordein gene and the identification of possible upstream regulatory elements in endosperm storage protein genes from barley, wheat and maize. Nucleic Acids Research. 1985;13:7327–7339. doi: 10.1093/nar/13.20.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SP, Puype M, Demol H, Vandekerckhove J, Job D. Proteomic analysis of arabidopsis seed germination and priming. Plant Physiology. 2001;126:835–848. doi: 10.1104/pp.126.2.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruis DF, Selinger DA, Curran JM, Jung R. Redundant proteolytic mechanisms process seed storage proteins in the absence of seed-type members of the vacuolar processing enzyme family of cysteine proteases. The Plant Cell. 2002;14:2863–2882. doi: 10.1105/tpc.005009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck GR, Chamberlain AK, Ho TH. Barley embryo globulin 1 gene, Beg1: characterization of cDNA, chromosome mapping and regulation of expression. Molecular and General Genetics. 1993;239:209–218. doi: 10.1007/BF00281620. [DOI] [PubMed] [Google Scholar]
- Huo N, Lazo G, Vogel J, et al. The nuclear genome of Brachpodium distachyon: analysis of BAC and sequences. Functional and Integrative Genomics. 2008;8:135–147. doi: 10.1007/s10142-007-0062-7. [DOI] [PubMed] [Google Scholar]
- Huo N, Vogel JP, Lazo GR, You FM, Ma Y, McMahon S, Dvorak J, Anderson OD, Luo MC, Gu YQ. Structural characterization of Brachypodium genome and its syntenic relationship with rice and wheat. Plant Molecular Biology. 2009;70:47–61. doi: 10.1007/s11103-009-9456-3. [DOI] [PubMed] [Google Scholar]
- International Brachypodium Initiative. Genome sequence analysis of the model grass. Brachypodium distachyon. Nature. 2010;463:763–768. doi: 10.1038/nature08747. [DOI] [PubMed] [Google Scholar]
- Jamme F, Robert R, Bouchet B, Saulnier L, Dumas P, Guillon F. Aleurone cell walls of wheat grain: high spatial resolution investigation using synchrotron infrared microspectroscopy. Applied Spectroscopy. 2008;62:895–900. doi: 10.1366/000370208785284448. [DOI] [PubMed] [Google Scholar]
- Kellogg EA. Evolutionary history of the grasses. Plant Physiology. 2001;125:1198–1205. doi: 10.1104/pp.125.3.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriz AL. Characterization of embryo globulins encoded by the maize Glb genes. Biochemical Genetics. 1989;27:239–251. doi: 10.1007/BF02401804. [DOI] [PubMed] [Google Scholar]
- Kriz AL, Schwartz D. Synthesis of globulins in maize embryos. Plant Physiology. 1986;82:1069–1075. doi: 10.1104/pp.82.4.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lange M, Vincze E, Wieser H, Schjoerring JK, Holm PB. Suppression of C-hordein synthesis in barley by antisense constructs results in a more balanced amino acid composition. Journal of Agricultural and Food Chemistry. 2007;55:6074–6081. doi: 10.1021/jf0709505. [DOI] [PubMed] [Google Scholar]
- Lasztity R. The chemistry of cereal proteins. 2nd edn. CRC Press; 1996. [Google Scholar]
- Laudencia-Chingcuanco DL, Vensel WH. Globulins are the main seed storage proteins in Brachypodium distachyon. Theoretical and Applied Genetics. 2008;117:555–563. doi: 10.1007/s00122-008-0799-y. [DOI] [PubMed] [Google Scholar]
- Le Gall M, Quillien L, Guéguen J, Rogniaux H, Sève B. Identification of dietary and endogenous ileal protein losses in pigs by immunoblotting and mass spectrometry. Journal of Nutrition. 2005;135:1215–1222. doi: 10.1093/jn/135.5.1215. [DOI] [PubMed] [Google Scholar]
- Loit E, Melnyk CW, MacFarlane AJ, Scott FW, Altosaar I. Identification of three wheat globulin genes by screening a Triticum aestivum BAC genomic library with cDNA from a diabetes-associated globulin. BMC Plant Biolology. 2009;9:93. doi: 10.1186/1471-2229-9-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosse J. Nitrogen-to-protein conversion factor for ten cereals and six legumes or oilseeds. A reappraisal of its definition and determination. Variation according to species and to seed protein content. Journal Agricultural and Food Chemistry. 1990;38:18–24. [Google Scholar]
- Opanowicz M, Vain P, Draper J, Parker D, Doonan JH. Brachypodium distachyon: making hay with a wild grass. Trends in Plant Science. 2008;13:172–177. doi: 10.1016/j.tplants.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Osborne TB. The vegetable proteins. 2nd edn. London: Longmans Green and Co; 1924. [Google Scholar]
- Philippe S, Tranquet O, Utille JP, Saulnier L, Guillon F. Investigation of ferulate deposition in endosperm cell walls of mature and developing wheat grains by using a polyclonal antibody. Planta. 2007;225:1287–1299. doi: 10.1007/s00425-006-0422-x. [DOI] [PubMed] [Google Scholar]
- Shewry PR, Halford NG. Cereal seed storage proteins: structures, properties and role in grain utilization. Journal of Experimental Botany. 2002;53:947–958. doi: 10.1093/jexbot/53.370.947. [DOI] [PubMed] [Google Scholar]
- Shewry PR, Tatham AS. The prolamin storage proteins of cereal seeds: structure and evolution. Biochemical Journal. 1990;267:1–12. doi: 10.1042/bj2670001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewry PR, Napier JA, Tatham AS. Seed storage proteins: structure and biosynthesis. The Plant Cell. 1995;7:945–956. doi: 10.1105/tpc.7.7.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shotwell MA, Afonso C, Davies E, Chesnut RS, Larkins BA. Molecular characterization of oat seed globulins. Plant Physiology. 1988;87:698–704. doi: 10.1104/pp.87.3.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoup FK, Pomeranz Y, Deyoe CW. Amino acid composition of wheat varieties and flours varying widely in bread-making potentialities. Journal of Food Science. 1966;31:94–101. [Google Scholar]
- Sjödahl S, Rödin J, Rask L. Characterization of the 12S globulin complex of Brassica napus. Evolutionary relationship to other 11–12S storage globulins. European Journal of Biochemistry. 1991;196:617–621. doi: 10.1111/j.1432-1033.1991.tb15857.x. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Sugimoto T, Ogawa M, Kasai Z. Isolation and characterization of two types of protein bodies in rice endosperm. Agricultural Biology and Chemistry. 1980;44:1633–1639. [Google Scholar]
- Vain P, Worland B, Thole V, Mckenzie N, Alves SC, Opanowicz M, Fish L, Bevan M, Snape JWW. Agrobacterium-mediated transformation of the temperate grass Brachypodium distachyon (genotype Bd 21) for T-DNA insertional mutagenesis. Plant Biotechnology Journal. 2008;6:236–245. doi: 10.1111/j.1467-7652.2007.00308.x. [DOI] [PubMed] [Google Scholar]
- Vasil IK. A short history of plant biotechnology. Phytochemistry Reviews. 2008;7:387–394. [Google Scholar]
- Vogel J, Hill T. High-efficiency Agrobacterium-mediated transformation of Brachypodium distachyon inbred line Bd21-3. Plant Cell Reports. 2008;27:471–478. doi: 10.1007/s00299-007-0472-y. [DOI] [PubMed] [Google Scholar]
- Vogel JP, Gu YQ, Twigg P, Lazo GR, Laudencia-Chingcuanco D, Hayden DM, Donze TJ, Vivian LA, Stamova B, Coleman-Derr D. EST sequencing and phylogenetic analysis of the model grass Brachypodium distachyon. Theoretical and Applied Genetics. 2006;113:186–195. doi: 10.1007/s00122-006-0285-3. [DOI] [PubMed] [Google Scholar]
- Wallace NH, Kriz AL. Nucleotide sequence of a cDNA clone corresponding to the maize globulin-2 gene. Plant Physiology. 1991;95:973–975. doi: 10.1104/pp.95.3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Ross AR, Yang J, Hegedus DD, Kermode AR. Phosphorylation of the 12 S globulin cruciferin in wild-type and abi1-1 mutant Arabidopsis thaliana (thale cress) seeds. Biochemical Journal. 2007;404:247–256. doi: 10.1042/BJ20061569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu JH, Messing J. Amplification of prolamin storage protein genes in different subfamilies of the Poaceae. Theoretical and Applied Genetics. 2009;119:1397–1412. doi: 10.1007/s00122-009-1143-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.