Summary
Background
The cohesin complex that mediates sister chromatid cohesion contains three core subunits: Smc1, Smc3, and Scc1. Heterotypic interactions between Smc1 and Smc3 dimerization domains create stable V-shaped Smc1/Smc3 heterodimers with a hinge at the center and nucleotide-binding domains (NBDs) at the ends of each arm. Interconnection of each NBD through their association with the N- and C-terminal domains of Scc1 creates a tripartite ring, within which sister DNAs are thought to be entrapped (the ring model). Crystal structures show that the Smc1/Smc3 hinge has a toroidal shape, with independent “north” and “south” interaction surfaces on an axis of pseudosymmetry. The ring model predicts that sister chromatid cohesion would be lost by transient hinge opening.
Results
We find that mutations within either interface weaken heterodimerization of isolated half hinges in vitro but do not greatly compromise formation of cohesin rings in vivo. They do, however, reduce the residence time of cohesin on chromosomes and cause lethal defects in sister chromatid cohesion. This demonstrates that mere formation of rings is insufficient for cohesin function. Stable cohesion requires cohesin rings that cannot easily open.
Conclusions
Either the north or south hinge interaction surface is sufficient for the assembly of V-shaped Smc1/Smc3 heterodimers in vivo. Any tendency of Smc proteins with weakened hinges to dissociate will be suppressed by interconnection of their NBDs by Scc1. We suggest that transient hinge dissociation caused by the mutations described here is incompatible with stable sister chromatid cohesion because it permits chromatin fibers to escape from cohesin rings.
Keywords: DNA
Highlights
► Unstable Smc1/3 hinge dimerization reduces the residence time of cohesin on chromatin
Introduction
Sister chromatid cohesion mediated by the cohesin complex is essential during mitosis for the attachment of sister kinetochores to microtubules in an amphitelic manner and thereby for the traction of sister chromatids to opposite cell poles during anaphase. Cohesin is composed of the Smc (structural maintenance of chromosome) proteins Smc1 and Smc3 and the non-Smc proteins Scc1 and Scc3. Smc1 and Smc3 possess Walker A and Walker B motifs within globular N- and C-terminal domains, respectively. Both proteins form 50 nm long intramolecular coiled coils with a central hinge/dimerization domain at one end and an ABC (ATP-binding cassette)-like ATPase head domain composed of N- and C-terminal domains at the other. Smc1 and Smc3 bind tightly to each other as a result of heterotypic interactions between their hinge/dimerization domains, forming stable V-shaped Smc1/Smc3 heterodimers. A remarkable aspect of cohesin is the manner in which its Scc1 (kleisin) subunit binds to the complex. Scc1's N- and C-terminal domains bind tightly to Smc1 and Smc3 ATPase heads, respectively, thereby forming a huge tripartite ring structure [1].
Cohesin is recruited to chromosomes by a separate Scc2/4 complex [2] in a process that requires hydrolysis of ATP bound to its Smc1 and Smc3 ATPase heads [3, 4]. Establishment of sister chromatid cohesion (but not mere association with chromatin) requires acetylation of Smc3's ATPase head by the Eco1 acetyltransferase [5–11]. Once established, cohesion is maintained, apparently without any cohesin subunit exchange [12, 13], from S phase until the onset of anaphase, whereupon cleavage of its kleisin subunit by a thiolprotease called separase opens the ring and triggers cohesin's dissociation from chromosomes [14, 15].
The finding that cohesin forms a ring raises the possibility that it associates with chromatin by entrapping DNAs inside its ring (the ring model). According to the simplest (“strongest”) version of this model, sister chromatid cohesion arises from the coentrapment of sister DNAs inside a single (monomeric) ring [16]. A weaker version of the model (the handcuff variant) postulates that cohesion arises instead from interactions between two different rings, each embracing separate sister chromatin fibers [17]. As predicted by the “strong” ring model, site-specific crosslinking of cohesin's Smc-Smc and Smc-kleisin interfaces traps circular sister minichromosome DNAs inside a covalently circularized cohesin ring [16]. If, as seems likely, entrapment is mediated by preassembled cohesin rings, then these must transiently open; cohesin must possess an entry gate, which has been proposed to be at the Smc1/Smc3 hinge interface [18].
Smc hinge domains are highly conserved from bacteria [19] to humans (A.K. et al., unpublished data). In both cases, shallow U-shaped hinge monomers interact to form a two-fold symmetric or pseudosymmetric torus with a shape not unlike that of DNA polymerase clamps, albeit with a much smaller hole (channel) in the middle. Crucially, dimerization arises from two potentially independent interactions surfaces at the “north” and “south” poles of the torus. The bipartite nature of the dimerization interface stems from pseudosymmetry of each hinge domain. A previous study concluded that one of the two interaction surfaces suffices for stable Smc1 and Smc3 hinge dimerization, at least when isolated half hinges are coexpressed in E.coli [20, 21]. If so, why is bipartite binding so conserved? According to both the strong and weak versions of the ring model, cohesin would not be able to associate stably with chromatin if hinges opened even transiently in postreplicative cells. Binding tight enough to prevent this might require both north and south interaction surfaces. To address this, we have investigated the effect both in vitro and in vivo of mutating key residues within the north and south interaction surfaces.
Results
Mutation of Either the North or South Hinge Interface Is Lethal
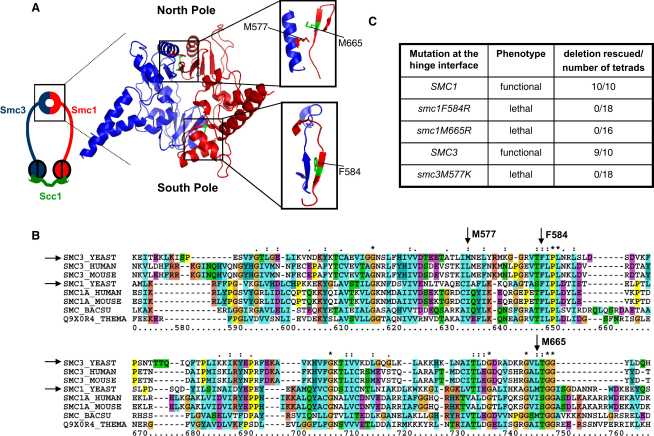
To investigate the importance of its two binding interfaces, we used sequence alignment to create a model of the yeast Smc1/Smc3 hinge based on the known homodimeric Thermotoga maritima hinge structure [19]. This identified residues, such as F584 and M665 in Smc1 and M577 in Smc3, whose substitution should affect binding (Figure 1A). All three residues (Figure 1B) are in highly conserved secondary structures [19]. Smc3 M577 and Smc1 M665 are situated in an α helix and a β sheet, respectively, that form extensive contacts at the north interface, whereas Smc1 F584 lies in a β sheet that interacts with an Smc3 β sheet at the south interface (Figure 1A inset). To maximize disruption, we mutated all three nonpolar residues to bulky charged residues, namely M577K, F584R, and M665R. To determine their phenotypes, we integrated either wild-type or mutant genes at ectopic sites (leu2 for SMC3 and ura3 for SMC1) within diploid strains heterozygous for the cognate smc deletion. Tetrad dissection revealed that wild-type but not the mutant genes complement the deletions. All three mutations are therefore lethal (Figure 1C). Other mutations within the hinge interface were viable (see Figure S1 available online), except smc3F590R, which had a phenotype similar to smc3M577K.
Figure 1.
Model of the Cohesin Hinge from Yeast
(A) Model of the yeast cohesin hinge based on the bacterial Smc hinge from Thermotoga maritima. Amino acid residues used for the mutation at the dimerization surfaces of the Smc1 and Smc3 hinge domains are indicated.
(B) Sequence alignment of the hinge domain showing the conservation of F584 and M665 in Smc1 and M577 in Smc3.
(C) Mutations and their phenotype. The rightmost column represents the ability of ectopic copies of wild-type SMC1/SMC3 or mutant smc1/smc3, integrated into strains heterozygous for Δsmc1/Δsmc3, to rescue the deletion in tetrad dissection assays. Ideally, one rescue per tetrad is expected if the ectopic gene can complement the deletion of the endogenous gene. (See also Figure S1.)
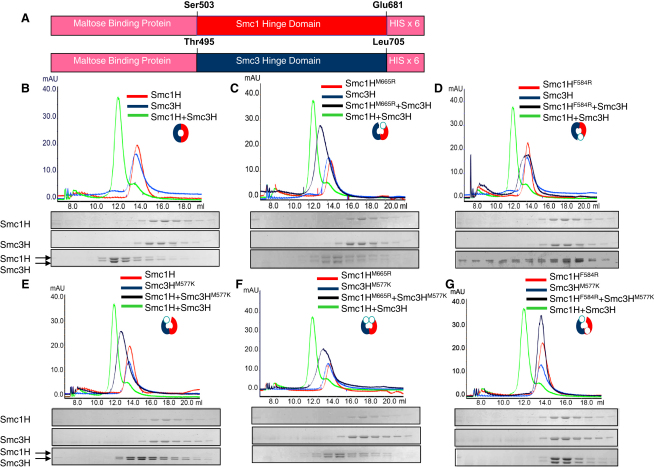
Effect of Mutations on Hinge Heterodimer Formation In Vitro
Wild-type and mutant hinge domains (Smc1 S503-E681 and Smc3 T495-L705) with maltose binding protein and 6× histidine at their N and C termini (Figure 2A) were expressed separately in E. coli and purified to homogeneity with Talon beads and size exclusion chromatography (Supplemental Experimental Procedures). All hinge domain fusion proteins, whether wild-type or mutant, eluted at volumes expected for monomers (Figures 2B–2E). An equimolar mixture (each at 10 μM) of wild-type hinges (Smc1H and Smc3H) migrated with an elution volume expected for heterodimers after prior incubation for 30 min at 25°C (Figure 2B), implying efficient dimerization in vitro. Mixtures of wild-type Smc3H and mutant Smc1H proteins [Smc1(F584R)H or Smc1(M665R)H] or wild-type Smc1H and Smc3(M577K)H eluted more rapidly than the individual proteins, but more slowly than fully wild-type mixtures (Figures 2C–2E). The elution volumes of Smc1(M665R)H/Smc3H and Smc1H/Smc3(M577K)H were intermediate between monomers and heterodimers, whereas that of Smc1(F584R)H/Smc3H was closer to that of monomers. These data imply that all three mutant hinges can still interact with wild-type partners but do so in an altered manner. The altered elution volumes imply either a major change in the conformation of heterodimers or, more likely, a reduction in binding affinities. If the mutations selectively disrupt either the north or south binding interface, then they might be expected to produce “half-opened” hinge dimers, which would be expected to migrate more rapidly and not, as observed, more slowly. The gel filtration data therefore indicate that the mutations reduce binding affinities, which was confirmed in the case of Smc1 M665R by a pull-down assay (Figures S2A–S2C). In contrast, the functional Smc1 I571K mutation had little or no effect on dimer formation as determined by gel filtration assay (Figure S2D).
Figure 2.
Mutant Hinge Domains of Cohesin Are Defective in Dimerization In Vitro
(A) Smc1 hinge domain (T509 to R676) and Smc3 hinge domains (A516 to R684) were fused with maltose binding protein domains at the N termini and with 6× His at the C termini to enable protein purification.
(B–G) Recombinant Smc hinge proteins were subjected to size exclusion chromatography either as monomers or as heterodimers with equimolar concentrations (10 mM). An overlay of the chromatograms for Smc1H, Smc3H, and Smc1H + Smc3H is presented. The chromatogram for the wild-type hinge dimer (B) was overlaid on all of the other chromatograms (C–G) to compare the shift of the wild-type dimer peak with mutant dimer peaks. (See also Figure S2.)
To test whether the residual binding of mutant hinges to their wild-type counterparts is due to the presence of an intact binding interface (either north or south) or due to incomplete inactivation of the mutated interface, we used gel filtration to measure the binding of Smc1(M665R)H with Smc3(M577K)H (hinges altered only at the north interface) or Smc1(F584R)H with Smc3(M577K)H (hinges altered at both interfaces). The former mixture had an elution profile similar to that of each single mutant mixed with cognate wild-type (Figure 2F), implying that complete inactivation of the north interface still permits heterodimer formation, albeit less efficiently than wild-type. In contrast, the elution volumes of Smc1(F584R)H and Smc3(M577K)H hinges were unaltered by their combination (Figure 2G), implying that mutation of both north and south interfaces abolishes heterodimer formation under these conditions. Our observations imply that both north and south hinge interfaces contribute to high-affinity binding but that each is sufficient for binding at lower affinity. Of the three mutations, Smc1 F584R has the most drastic binding defect. Interestingly, computational analysis suggests that F584R (but not M665R or the functional I571K) possibly causes a large reduction in the size of the small channel in cohesin's hinge (Figure S2E).
We used isothermal titration calorimetry to estimate the binding constant of wild-type and mutant hinges. From the pattern of heat release (Figure S2F), we estimated the association constant of wild-type hinges to be 24 nM. No heat release could be detected when any of the three mutant hinge domains were titrated against the corresponding wild-type hinge domain (Figure S2G), precluding any estimate of their association constants via this method. We therefore used a competition assay to confirm that a mutated hinge binds less efficiently. Wild-type Smc1H or Smc1(M665R)H and Smc3-FLAG were mixed together with a wild-type Smc1-SNAP competitor in an equimolar ratio (1 μM each). Proteins bound to anti-FLAG beads were eluted with a 3× FLAG peptide and analyzed by Coomassie staining of SDS-PAGE. The M665R mutation greatly reduced the amount of Smc1 that binds the Smc3 hinge under these conditions (Figure S2A; A.K. et al., unpublished data). To estimate off rates, we coincubated Smc1 and Smc3-FLAG at high concentration (1 μM), diluted them to 50 nM, and incubated them in the presence of a large excess of competitor Smc1-SNAP (750 nM), and the amount of Smc1 associated with Smc3-FLAG was measured by western blotting after immunoprecipitation of Smc3-FLAG at various periods of time. This revealed that M665R greatly reduces both initial binding and stability of Smc1/Smc3 complexes (Figures S2B and S2C; A.K. et al., unpublished data).
Either North or South Interface Is Sufficient for Formation of Cohesin Complexes In Vivo
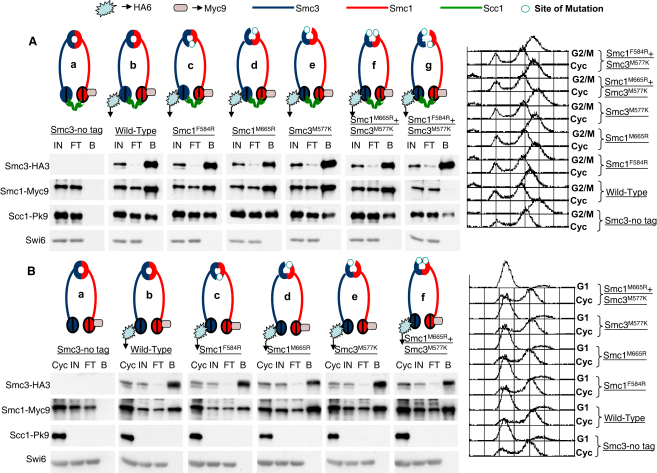
To investigate whether disrupting either the north or south interface affects cohesin assembly in vivo, we measured coimmunoprecipitation of Scc1 (tagged at its endogenous locus with nine copies of the pk epitope) and Smc1 (tagged with nine copies of the Myc epitope) with Smc3 proteins (tagged with three copies of the HA epitope). To avoid lethality caused by mutations, all strains expressed untagged wild-type Smc1 and Smc3 proteins from endogenous genes and Smc1-Myc9 and Smc3-HA3 proteins from single ectopic copies integrated at the ura3 and leu2 loci, respectively. To minimize cleavage of Scc1 by separase, we performed immunoprecipitations from soluble extracts prepared from yeast cells arrested in G2/M by nocodazole (Figure 3A). Wild-type or mutant Smc3 proteins were immunoprecipitated with HA-specific antibodies, and coprecipitation of Smc1-Myc9 (wild-type or mutant) and Scc1-pk9 was measured by western blot analysis. Importantly, coprecipitation of Scc1-pk9 and Smc1-Myc9 was dependent on Smc3's HA epitope (Figure 3Aa).
Figure 3.
Hinge Mutants of Smc1 and Smc3 Can Form Ring Complexes
(Aa–Ag) Coimmunoprecipitation assays analyzing the interaction of Smc3 with Smc1 and Scc1 in vivo. Soluble extracts were prepared from nocodazole-arrested yeast cells expressing Smc1-Myc9, Smc3-HA3, and Scc1-pk9 (Ab = K15106: SMC1-Myc9::URA3, SMC3-HA3::LEU2, SCC1-PK9::KanMX, MATa). Smc1 and Smc3 either were wild-type or carried hinge mutations (Ac = K15108: smc1(F584R)-Myc9; Ad = K16383: smc1(M665R)-Myc9; Ae = K15110: smc3(M577K)-HA3; Af = K16255: smc1(M665R)-Myc9,smc3(M577K)-HA3; Ag = K16345: smc1(F584R)-Myc9,smc3(M577K)-HA3). Smc3 was immunoprecipitated with an anti-HA-conjugated matrix, and bound fractions were probed with anti-HA, anti-Myc, and anti-Pk9 antibodies. The following abbreviations are used: IN, input 1×; FT, flowthrough 1×; B, bound fraction 10×. The right panel shows G2/M arrest in nocodazole for each strain used in the coimmunoprecipitation assays.
(Ba–Bf) Smc1 and Smc3 hinge dimerization is Scc1 independent. The experiment, as described in (A), was performed with soluble extract from the same yeast strains as in (A) but arrested in G1 by α factor. Fluorescence-activated cell sorting (FACS) profiles are shown at right; “Cyc” represents extract from cycling cells showing expression of Scc1-pk9. (See also Figure S3.)
Coprecipitation of Smc1-Myc9 and Scc1-pk9 with wild-type Smc3-HA3 protein (Figure 3Ab) was barely, if at all, affected by either F584R or M665R mutations in Smc1 (Figures 3Ac and 3Ad). Likewise, M577K in Smc3 had little or no effect on coprecipitation with wild-type Smc1-Myc9 or Scc1-pk9 (Figure 3Ae), or even with Smc1(M665R)-Myc9 (Figure 3Af). Interaction between Smc1 and Smc3 is only abolished when both north and south interfaces are disrupted. Thus, we detected little or no coprecipitation of Smc1(F584R)-Myc9 with Smc3(M577K)-HA3 (Figure 3Ag). These results suggest that interaction at either the north or south interface is sufficient for heterodimer formation in vivo. To test whether formation of dimers by mutant Smc proteins is facilitated by the interconnection of Smc1 and Smc3 ATPase head domains by Scc1 [22], we repeated these experiments with extracts prepared from yeast cells arrested in G1 by α factor in which Scc1 is not expressed as a result of low levels of transcription and persistent separase activity [15]. Even under these conditions, mutations affecting either the north or south interface permitted heterodimer formation, though Smc1 F584R appeared to reduce it (Figures 3Ba–3Bf).
We also measured dimer formation under conditions in which Myc-tagged wild-type or mutant Smc1 proteins (expressed from the ura3 locus) compete with untagged Smc1 protein (expressed from the endogenous gene) for binding to Smc3-HA6 (expressed from a single endogenous gene). Silver staining of proteins run on SDS-PAGE shows that, after stringent washes, equal amounts of Smc1 and Smc1-Myc9 copurify with Smc3-HA6 bound to anti-HA antibody-coupled agarose beads. F584R caused a reduction in the amount of the mutant Myc-tagged Smc1 protein and a corresponding increase in the untagged wild-type one (Figure S3). Surprisingly, Smc1(M665R)-Myc9 still competed effectively with endogenous Smc1 under these conditions, which contrasts with the inability of isolated mutant hinges to compete with wild-type in vitro (Figures S2A–S2C). We suggest that, in vivo, other mechanisms (for example, interconnection of nucleotide-binding domains [NBDs] by Scc1) stabilize Smc heterodimers containing Smc1 M665R.
Association with Chromatin of Smc Proteins with Mutant Hinges
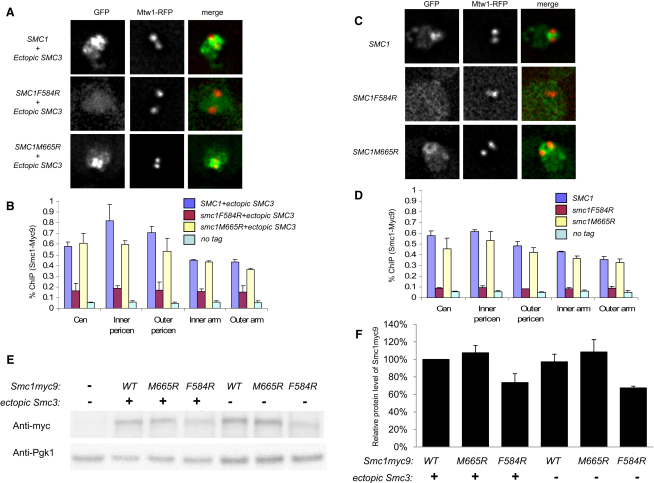
To better understand why Smc proteins with mutant hinges are dysfunctional despite forming cohesin complexes efficiently, we used two assays to measure their ability to associate with chromatin. We first used live-cell imaging. To do this, we created yeast strains in which GFP-tagged wild-type or hinge mutant Smc1 proteins were ectopically expressed. These strains also expressed HA3-tagged Smc3 from an ectopic locus as well as wild-type Smc1, Smc3, and RFP-tagged Mtw1 (a kinetochore component) from endogenous loci. Consistent with previous reports, we observed that pericentric Smc1-GFP forms a barrel-shaped structure along the axes of mitotic spindles (Figure 4A) [23]. F584R largely abolished this structure, whereas M665R had little adverse effect. Though Smc1(F584R)-GFP failed to form pericentric barrels, the protein was nevertheless concentrated within nuclei (Figure 4A). We next used quantitative PCR (qPCR) to measure DNA immunoprecipitation by Myc-specific antibodies following formaldehyde fixation and sonication-mediated DNA fragmentation (ChIP). This confirmed that Smc1-Myc9's association with centromeric, pericentromeric, and arm sequences from chromosome VI in cycling cells is greatly reduced by F584R but little affected by M665R (Figure 4B). Quantitative western blotting (Figures 4E and 4F; Figure S4A) showed that the level of Smc1(F584R)-Myc9 was about 70% of wild-type Smc1-Myc9. Given that only half of Smc1/Smc3 heterodimers associate with Scc1, which is therefore rate limiting for cohesin complex formation, the reduced protein levels might not contribute much to the reduced chromatin association. Irrespective of whether this is the case, the effect of F584R on chromatin association cannot be due merely to reduced protein levels, because the reduction in protein levels is far more modest than that of chromatin association.
Figure 4.
Chromatin Association of Hinge Mutants
(A) Live-cell imaging shows the localization of GFP-tagged Smc1 in yeast strains with ectopically expressed Smc3. Kinetochores are marked by RFP-tagged Mtw1 to detect the spindle axes. Wild-type Smc1-GFP (K16942: MATa, SMC1-GFP::URA3, SMC3-HA3::LEU2, MTW1-RFP::KanMX) and Smc1(M665R)-GFP (K16944: MATa, smc1(M665R)-GFP::URA3, SMC3-HA3::LEU2, MTW1-RFP::KanMX) are predominantly localized in the pericentromeric region of chromosomes and appear as barrel-shaped structures. Smc1(F584R)-GFP (K16891: MATa, smc1(F584R)-GFP::URA3, SMC3-HA3::LEU2, MTW1-RFP::KanMX) is enriched in the nuclear region but does not form any structure.
(B) ChIP-qPCR was performed for the quantitative measurement of chromatin association of 9× Myc-tagged Smc1 at known cohesin binding loci in the presence of an ectopic copy of Smc3. Crude extracts were prepared from asynchronous cultures K14133 (MATa, SMC1-Myc9::URA3, SMC3-HA3::LEU2), K14134 (MATa, smc1(F584R)-Myc9::URA3, SMC3-HA3::LEU2), K14137 (MATa, smc1(M665R)-Myc9::URA3, SMC3-HA3::LEU2), and K699 (no tag). Error bars represent standard deviation (SD); n = 2.
(C) Live-cell imaging for Smc1-GFP localization as in (A) without an ectopic copy of Smc3. Strains used were K16428: MATa, SMC1-GFP::URA3, MTW1-RFP::KanMX; K16642: MATa, smc1(M665R)-GFP::URA3, MTW1-RFP::KanMX; and K16611: MATa, smc1(F584R)-GFP::URA3, MTW1-RFP::KanMX. Wild-type Smc1 and Smc1 M665R still form the pericentromeric barrel structure, whereas Smc1 F584R does not enrich in the nucleus.
(D) Similar experiment as in (B), but with strains without an ectopic copy of Smc3. Crude extracts were prepared from the strains K11850 (MATa, SMC1-Myc9::URA3), K13858 (MATa, smc1(F584R)-Myc9::URA3), K13860 (MATa, smc1(M665R)-Myc9::URA3), and K699 (no tag). Error bars represent SD; n = 2. (See also Figure S4.)
(E) Yeast strains K699 (no tag), K14133 (MATa, SMC1-Myc9::URA3, SMC3-HA3::LEU2), K14134 (MATa, smc1(F584R)-Myc9::URA3, SMC3-HA3::LEU2), K14137 (MATa, smc1(M665R)-Myc9::URA3, SMC3-HA3::LEU2), K11850 (MATa, SMC1-Myc9::URA3), K13858 (MATa, smc1(F584R)-Myc9::URA3), and K13860 (MATa, smc1(M665R)-Myc9::URA3) were grown to exponential phase and lysed. To determine the protein levels of Smc1-Myc9 and endogenous Pgk1, 1 or 0.1 μg of total protein was loaded onto a SDS-PAGE gel. Smc1-Myc9 and endogenous Pgk1 were detected with anti-Myc antibody and anti-Pgk1 antibody. The chemiluminescence signal was detected by the ChemiDoc XRS+ system.
(F) The density of bands in (E) was quantitated by Quantity One 1-D Analysis Software. The protein level of Smc1-Myc9 was calculated as a ratio of the density of the Smc1-Myc9 band to the density of the corresponding Pgk1 band. The protein level of Smc1-Myc9 in strain K14133 was set as 100%. Error bars represent SD; n = 3.
Similar results were obtained when wild-type and mutant Myc-tagged or GFP-tagged Smc1 proteins were expressed in cells containing only a single SMC3 gene, i.e., when tagged and untagged Smc1 subunits competed for a limiting amount of Smc3. Under these conditions, Smc1(M665R)-GFP still formed pericentric barrels in live cells, whereas Smc1(F584R)-GFP did not even accumulate within nuclei (Figure 4C). Under these conditions, F584R caused an even greater reduction in association of Smc1-Myc9 with chromosome VI sequences, whereas M665R still had little effect (Figure 4D). These results suggest that when Smc3 protein is limiting, Smc1 F584R protein fails to compete with wild-type Smc1 protein. It therefore fails to form Smc1 F584R/Smc3 heterodimers and no longer accumulates within nuclei. However, when Smc3 is no longer limiting, Smc1 F584R binds to Smc3 and accumulates within nuclei, presumably as Smc1 F584R/Smc3 heterodimers, but despite this largely fails to engage with chromosomes.
We also compared the distribution of wild-type and mutant Smc1 proteins (expressed as Myc-tagged proteins from ectopic gene copies) in chromosome spreads prepared from cycling cells. To compare mutant and wild-type chromosomes directly, we mixed mutant and wild-type cells before spheroplasting, and the two types of nuclei were distinguishable by the presence in wild-type cells of TetR-GFP foci associated with centromere-linked Tet operators. This revealed that M665R modestly reduces chromatin association whereas F584R abolishes it completely (Figure S4B).
We tested chromatin association of Smc3 M577K with a time-course ChIP-qPCR assay. Log-phase yeast cultures expressing either wild-type Smc3-HA3 or Smc3(M577K)-HA3 were arrested in G1 phase by α factor and then released into fresh medium at 25°C, after which they underwent DNA replication (by 40 min; Figure S4C). Quantitative ChIP revealed that M577K caused only a modest reduction in Smc3's association with centromeric, pericentromeric, or arm DNA sequences (Figure S4D). Finally, hybridization of ChIP samples to microarrays (ChIP-on-chip) revealed that the distribution along chromosome VI of Smc1 M665R and Smc3 M577K proteins did not differ greatly from their wild-type counterparts (Figures S4E and S4F; data not shown).
Smc3 Acetylation in Heterodimers with Mutant Hinges
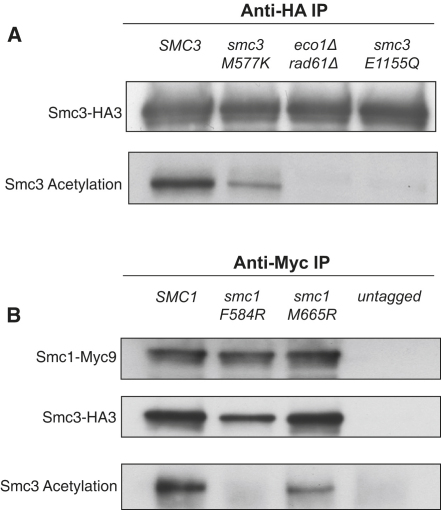
To test whether the lethality of hinge mutants could be due to a lack of Smc3 acetylation [5, 9–11], which also prevents cohesion establishment without greatly compromising chromatin association [8], we used an acetyl-lysine-specific antibody to measure the acetylation status of immunoprecipitated Smc3-HA3. Cells lacking Eco1 but kept alive by deletion of RAD61 lack Smc3 acetylation; Smc3 M577K merely reduced Smc3 acetylation, whereas Smc3 E1155K, which prevents hydrolysis of ATP bound to Smc3's NBD and largely eliminates cohesin's association with chromosomes, abolished it (Figure 5A). We also analyzed Smc1 hinge mutations with a variation of the assay, measuring the amounts of Smc3 and acetylated Smc3 in immunoprecipitates of wild-type or mutant Smc1 proteins. This revealed that M665R modestly reduced Smc3 acetylation, whereas F584R abolished it (Figure 5B). We conclude that mutant cohesin complexes that associate poorly (or not at all) with chromatin, such as Smc3 E1155Q and Smc1 F584R, are never stably acetylated, whereas those that can associate with chromatin, such as Smc1 M665R and Smc3 M577K, are acetylated with an efficiency comparable with their ability to associate with chromatin. The notion that chromatin association may be essential for acetylation is consistent with the finding that Smc3 is barely, if at all, acetylated in scc2-4 mutants [9]. Importantly, the degree of Smc3 acetylation within Smc1/Smc3 M577K or Smc1 M665R/Smc3 complexes is greater than that found in eco1-1 mutants growing at the permissive temperature [5]. The lethality caused by these two hinge mutants therefore cannot be attributed to reduced Smc3 acetylation per se.
Figure 5.
Acetylation of Smc3
Soluble extracts prepared from log-phase cells were subjected to coimmunoprecipitation by either anti-HA antibody (A) or anti-Myc antibody (B), and the acetylation of immunoprecipitated Smc3 in both panels was subsequently measured by an acetyl-lysine-specific antibody via western blotting. Strains used were K15106 (MATa, SMC1-Myc9::URA3, SMC3-HA3::LEU2, SCC1-PK9::KanMX), K15110 (MATa, smc3(M577K)-HA3::LEU2, SCC1-PK9::KanMX), K15794 (MATa, SMC3-HA3::LEU2, Δsmc3::HIS3, Δeco1::hphMX, Δrad61::NatMX), and K13561 (MATa, smc3(E1155Q)-HA3, MATa) in (A) and K15106, K15108 (MATa, smc1(F584R)-Myc9, SMC3-HA3::LEU2, SCC1-PK9::KanMX), K16383 (MATa, smc1(M665R)-Myc9, SMC3-HA3::LEU2, SCC1-PK9::KanMX), and K699 (u strain) in (B).
The Hinge's North Interface Is Necessary for Minichromosome Cohesion
Why, if Smc1 M665R and Smc3 M577K have little or no effect on cohesin complex formation and cause only modest decreases in chromatin association and Smc3 acetylation, are these mutations lethal? To test their ability to confer sister chromatid cohesion, we replaced the endogenous genes by heat-inducible degradation alleles, td-smc1 or td-smc3, expressed from tetracycline-repressible promoters. These strains also express the degron-specific E3 ubiquitin ligase Ubr1 from the GAL1-10 promoter and proliferate normally at 25°C in the absence of galactose or doxycycline. Wild-type SMC1 or SMC3, hinge mutant alleles, or empty vector (YIplac211) alone were integrated at the ura3 loci of appropriate td-smc1 or td-Smc3 strains. Wild-type but not mutant alleles enabled proliferation under restrictive conditions, namely at 37°C in the presence of doxycycline and galactose (data not shown).
To measure sister chromatid cohesion, we first arrested strains containing a 7.5 kb centromere-containing minichromosome in G1 with α factor at 25°C and depleted functional Smc proteins by transferring the cells to medium containing doxycycline and galactose at 37°C. Cells were subsequently transferred to medium containing galactose, doxycycline, and nocodazole (instead of α factor) at 37°C. Fluorescence-activated cell sorting (FACS) analysis showed that DNA replication was complete by 100 min after release from α factor (Figure 6A), and western blots documented complete degradation of endogenous Smc1 or Smc3 proteins (Figure 6B) and expression of either wild-type or mutant proteins expressed from the ura3 locus (Figure 6B).
Figure 6.
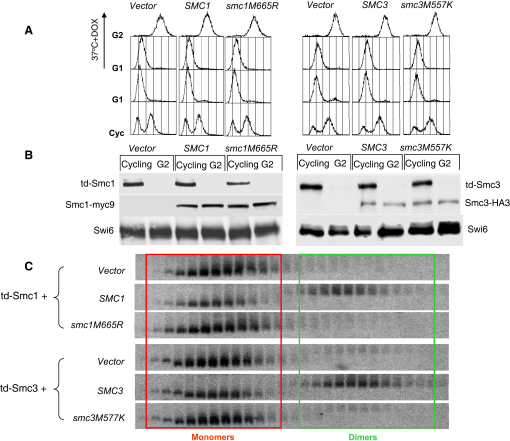
Mutations in the Hinge Domain of Smc1/Smc3 Destroy Cohesin's Ability to Coentrap Sister Minichromosomes
Exponential-phase cells of strains K16338 (MATa, td-SMC1, YIplac211::URA3), K16335 (td-SMC1, SMC1-Myc9::URA3), and K16339 (MATa, td-SMC1, smc1(M665R)-Myc9::URA3) or K16341 (MATa, td-SMC3, YIplac211::URA3), K16342 (MATa, td-SMC3, SMC3-HA3::URA3), and K16343 (MATa, td-SMC3, smc3(M577K)-HA3::URA3) harboring a 7.5 kb minichromosome were arrested with α factor at 25°C. Endogenous Smc1 or Smc3 was degraded by shifting cultures to degron-mediated proteolysis conditions (YEP gal, 5 μg/ml doxycycline [DOX], 37°C) for 1 hr. Subsequently, the cells were released from G1 arrest to YEP gal medium containing 5 μg/ml doxycycline and 10 μg/ml nocodazole. After 100 min at 37°C, the cells were lysed and extracts were fractionated by sucrose gradient centrifugation followed by gel electrophoresis. Minichromosome DNA was detected by Southern blotting.
(A) FACS analysis shows that all strains completed DNA replication in the above conditions.
(B) Western blot (right) showing that endogenous HA-tagged td-Smc1 proteins are completely degraded and hence that ectopically expressed Myc-tagged Smc1 is the only version of Smc1 in the minichromosome cohesion assay (left). In the td-Smc3 strains, endogenous HA-tagged td-Smc3 proteins are also completely degraded, and ectopically expressed HA-tagged Smc3 proteins are the only version of Smc3.
(C) Minichromosome DNA was detected by Southern blotting. Monomers and dimers are marked inside the red and green boxes, respectively.
Crude extracts prepared from these cells were fractionated by centrifugation through sucrose gradients followed by electrophoresis in agarose gels, and the 7.5 kb minichromosome was detected by Southern blotting (Figure 6C). This revealed that about half of the minichromosomes were present as dimers in cells expressing wild-type Smc proteins but that few if any were found as dimers in cells transformed with an empty vector. Crucially, very few dimers were detected in cells expressing Smc1 M665R or Smc3 M577K in td-smc1 or td-smc3 cells, respectively. Though it does not prevent chromatin association or Smc3 acetylation, disruption of the hinge's north interface prevents establishment of stable sister chromatid cohesion.
The Hinge's North Interface Is Necessary for Cohesin's Stable Association with Pericentromeric Chromatin
Why do Smc1 M665R and Smc3 M577K mutant proteins fail to confer sister chromatid cohesion? According to the ring model, both sister chromatid cohesion and stable association with chromatin depend on retention of chromatin fibers inside tripartite cohesin rings. If, as is likely, Smc1 M665R (Figures S2A–S2C) and Smc3 M577K increase hinge dissociation, even if only transiently, then the mutations would permit escape of chromatin fibers that had previously been entrapped by mutant complexes. According to this hypothesis, the mutations should reduce cohesin's residence time at specific chromosomal loci. To test this, we compared recovery of fluorescence after photobleaching (FRAP) of wild-type or mutant GFP tagged Smc proteins associated with pericentromeric chromatin [23]. Either Smc1-GFP or Smc1(M665R)-GFP was expressed from genes integrated at the ura3 locus of diploid cells with wild-type endogenous genes. Smc1-GFP fluorescence forms a barrel-like structure situated between spindle pole bodies in metaphase cells in which sister kinetochores have bioriented and therefore partially separated [23]. A portion of the Smc1-GFP barrel was photobleached by a 200 ms laser exposure, and fluorescence of the bleached area was measured at 15 s intervals for 8 min. As reported previously, Smc1-GFP fluorescence recovered little, if at all, during this period (Figure 7A). In contrast, fluorescence recovered with a half-life of about 70 s when a similar experiment was performed with HTB2-GFP (data not shown) [5, 23]. These data suggest that there is little or no de novo recruitment of wild-type cohesin complexes to the vicinity of centromeres at this stage of the cell cycle, and because there is little change in level of fluorescence without bleaching, there can be little dissociation (see Supplemental Experimental Procedures).
Figure 7.
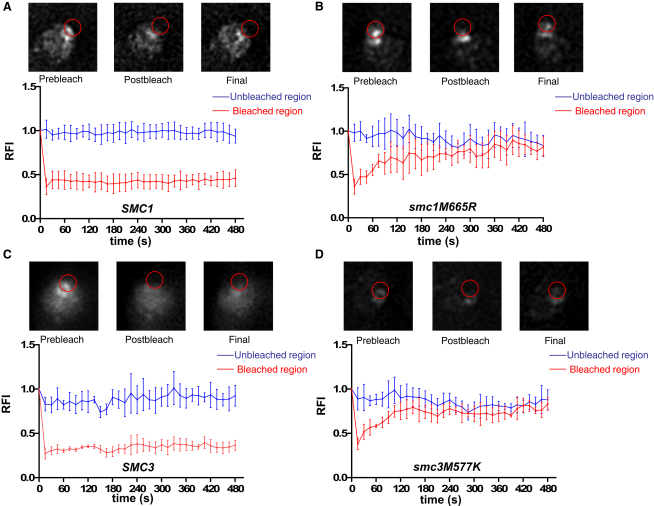
Mutations in the Smc Hinge Domain Cause Unstable Association of Cohesin with Chromosomes
Stability of cohesin's association with chromosomes was measured mainly in pericentromeric regions by means of fluorescence recovery after photobleaching (FRAP) in diploid yeast cells. A portion of the barrel-shaped fluorescent signal was bleached by exposure to an argon laser for 200 ms. Images were captured every 15 s for 8 min postbleaching to measure the recovery rate. Red circles in the upper panels of (A)–(D) depict the region of interest that was bleached and analyzed for recovery. Relative fluorescence intensities (RFI) of unbleached (blue) and bleached (red) signals are plotted over time in the lower panels. Wild-type Smc1-GFP (A; K16252: SMC1-GFP::URA3) and wild-type Smc3-GFP (C; K16113: SMC3-GFP) had no recovery in any of the cells analyzed. Smc1(M665R)-GFP (B; K16253) recovered with t1/2 = 45.2 ± 12.84 s (n = 6). Smc3(M577K)-GFP (D; K16114) recovered with t1/2 = 28.57 ± 7.2 s (n = 6). Error bars represent SD.
The pattern of Smc1(M665R)-GFP fluorescence resembles that of wild-type; that is, it has a similar barrel-like structure, confirming that the mutant complexes do indeed associate with centromeric chromatin. Unlike Smc1-GFP, fluorescence associated with Smc1(M665R)-GFP barrels recovers after photobleaching, indicating rapid de novo loading of mutant complexes at this location. Because there is no change in the amount of fluorescence associated with the mutant barrels in the absence of photobleaching, the increased loading must be balanced by increased dissociation, and we estimate from the rate of fluorescence recovery that Smc1(M665R)-GFP has a half life of 45.2 ± 12.84 s (Figure 7B).
We used a similar method to compare the stability of wild-type Smc3-GFP and Smc3(M577K)-GFP. In this case, we created heterozygous diploid strains in which one copy of SMC3 was substituted via homologous recombination by a version expressing either Smc3-GFP or Smc3(M557K)-GFP. Whereas Smc3-GFP fluorescence did not recover within 8 min following photobleaching, Smc3(M557K)-GFP recovered rapidly, leading us to estimate a half life of ∼28.57 ± 7.2 s (Figures 7C and 7D). We conclude that disruption of the hinge's north interface prevents the stable association of cohesin with centromeric chromatin.
Discussion
The Smc1/Smc3 hinge dimerization interface is exceptional in that interaction takes place via two potentially independent surfaces that together form the torus-shaped hinge. Mutations affecting the north interface (Smc1 M665R or Smc3 M577K) or south interface (Smc1 F584R) reduce hinge binding affinity but do not eliminate interaction either in vivo or in vitro. Smc1 F584R has the most severe effect on binding. Under conditions where the mutant protein has to compete with wild-type protein, it barely binds its partner in vivo. However, even this mutant binds its partner and forms cohesin complexes when there is no competition with wild-type protein.
Our finding that combining Smc3 M577K and Smc1 F584R abolishes binding both in vitro and in vivo suggests that these two mutations have crippling effects on north and south interactions, respectively. In the absence of either interaction, Smc1 and Smc3 no longer interact stably at physiological concentrations. Conversely, our finding that hinges still interact in vitro and cohesin complexes still form in vivo when Smc3 M577K and Smc1 M665R are combined, which together should abolish any interaction at the north interface, implies that the south interface is sufficient to drive Smc1/Smc3 dimerization in vivo. Though not as clearly demonstrated, the same may be true for the north interface. Our data do not exclude the possibility that engagement of Smc1 and Smc3 NBDs when ATP is bound to both heads is also capable of bringing the two proteins together. However, unlike association mediated by their hinge domains, this interaction is presumably not a stable one, because it would be destroyed by ATP hydrolysis.
Though just one hinge interaction surface is sufficient for cohesin complex formation, both are essential for sister chromatid cohesion. What goes wrong when individual interfaces are disrupted? There is overwhelming evidence, from chromosome spreads, ChIP-qPCR, ChIP-on-chip, and live-cell imaging of GFP-tagged proteins that mutations like Smc3 M577K and Smc1 M665R, which disrupt the north interface, do not abolish cohesin's association with chromatin. Moreover, cohesin complexes containing either of these two mutations are acetylated by Eco1 (albeit less efficiently than wild-type), a process that normally only occurs when cohesin associates with chromosomes (this work and [18]). Crucially, the mutations have less severe effects on chromatin association than the scc2-4 allele or on acetylation than the eco1-1 allele when the latter are grown at the permissive temperature. A defect in chromatin association or Smc3 acetylation per se cannot therefore explain the lethality caused by Smc3 M577K or Smc1 M665R.
Chromatin association requires hydrolysis of ATP bound to both Smc heads and is thought to involve entrapment of chromatin fibers by cohesin rings [3]. There is little or no evidence that either Smc3 M577K or Smc1 M665R prevents this process, or even the (initial) establishment of sister chromatid cohesion during DNA replication. However, it is clear that the mutations have a dramatic effect on the fate of cohesin complexes once they have associated with chromatin. Whereas fluorescence attributable to wild-type GFP-tagged Smc proteins associated with centromeric chromatin during mitosis fails to recover after photobleaching, consistent with little or no turnover, fluorescence associated with Smc1 M665R or Smc3 M577K recovers after one or two minutes, implying rapid turnover.
The more rapid turnover of mutant cohesin complexes is not merely an indirect consequence of the fact that they do not form stable sister chromatid cohesion, because GFP-tagged cohesin complexes that cannot be acetylated by Eco1, namely Smc3 K112R K113R mutants, also do not form sister chromatid cohesion but nevertheless form centromeric barrels whose fluorescence does not recover after photobleaching [5]. We therefore suggest that the primary defect caused by Smc1 M665R or Smc3 M577K mutations is a failure of cohesin to remain stably associated with chromatin. If, as seems likely, stable association requires entrapment of chromatin fibers by cohesin rings, then instability caused by Smc1 M665R or Smc3 M577K mutations can be explained by a greater tendency of mutant hinges to open transiently, permitting escape of previously entrapped chromatin fibers. The phenotype of these mutant hinges is therefore fully consistent with the ring model. Indeed, had the mutations not reduced the stability of cohesin's association with chromatin, this would have constituted a grave challenge to the model.
The phenotype caused by Smc1 F584R is more surprising. This mutation disrupts the south interface, possibly destroying it completely, but residual binding at the north interface nevertheless permits formation of cohesin complexes in vivo. However, unlike Smc1 M665R or Smc3 M577K, cohesin complexes formed by Smc1 F584R associate poorly, if at all, with chromatin. Smc1 F584R is poorly crosslinked by formaldehyde with chromosome VI DNA sequences and fails to form barrel-shaped structures around mitotic centromeres. Consistent with this, Smc3 associated with Smc1 F584R is not acetylated by Eco1. It is conceivable that F584R merely has a more dramatic effect on the stability of cohesin's association with chromatin, causing it to be so unstable that it can never be detected on chromosomes. However, it is also possible that the mutation alters the structure of the hinge in a manner that is incompatible with the process that entraps chromatin fibers in the first place. It has been suggested that entrapment requires the transient opening of cohesin rings at the hinge interface [18], and if so, hinges may interact during the entrapment process with chromosomal proteins such as the Scc2/4 complex, a process that could be disrupted by Smc1 F584R.
Our observations are consistent with the previous finding that replacing three conserved glycine residues by alanine in either the Smc1 or the Smc3 hinge does not abolish dimer formation [21]. These more complex mutations are predicted to affect the north and south interfaces, respectively, and their combination abolishes dimer formation [21]. Our work implies that although “single” triple glycine mutations may not greatly affect dimer formation, they should cause lethality, a prediction that we have confirmed by genetic studies in yeast (data not shown). Crucially, our characterization of the dimerization process provides an essential basis for exploring more sophisticated aspects of hinge function, namely hinges' possible involvement in DNA entrapment.
Acknowledgments
We are grateful to Nasmyth laboratory members for useful discussions, R. Parton for assistance with microscopy, K. Bloom for providing useful plasmids, and C.H. Haering for critical reading of the manuscript. A.M. is supported by a Cancer Research UK grant. This research was supported by Cancer Research UK and the Wellcome Trust.
Published online: February 11, 2010
Footnotes
Supplemental Information includes four figures, one table, and Supplemental Experimental Procedures and can be found with this article online at doi:10.1016/j.cub.2009.12.059.
Supplemental Information
References
- 1.Nasmyth K., Haering C.H. The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- 2.Ciosk R., Shirayama M., Shevchenko A., Tanaka T., Toth A., Shevchenko A., Nasmyth K. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- 3.Arumugam P., Gruber S., Tanaka K., Haering C.H., Mechtler K., Nasmyth K. ATP hydrolysis is required for cohesin's association with chromosomes. Curr. Biol. 2003;13:1941–1953. doi: 10.1016/j.cub.2003.10.036. [DOI] [PubMed] [Google Scholar]
- 4.Weitzer S., Lehane C., Uhlmann F. A model for ATP hydrolysis-dependent binding of cohesin to DNA. Curr. Biol. 2003;13:1930–1940. doi: 10.1016/j.cub.2003.10.030. [DOI] [PubMed] [Google Scholar]
- 5.Rowland B.D., Roig M.B., Nishino T., Kurze A., Uluocak P., Mishra A., Beckouët F., Underwood P., Metson J., Imre R. Building sister chromatid cohesion: smc3 acetylation counteracts an antiestablishment activity. Mol. Cell. 2009;33:763–774. doi: 10.1016/j.molcel.2009.02.028. [DOI] [PubMed] [Google Scholar]
- 6.Skibbens R.V., Corson L.B., Koshland D., Hieter P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 1999;13:307–319. doi: 10.1101/gad.13.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivanov D., Schleiffer A., Eisenhaber F., Mechtler K., Haering C.H., Nasmyth K. Eco1 is a novel acetyltransferase that can acetylate proteins involved in cohesion. Curr. Biol. 2002;12:323–328. doi: 10.1016/s0960-9822(02)00681-4. [DOI] [PubMed] [Google Scholar]
- 8.Tóth A., Ciosk R., Uhlmann F., Galova M., Schleiffer A., Nasmyth K. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Unal E., Heidinger-Pauli J.M., Kim W., Guacci V., Onn I., Gygi S.P., Koshland D.E. A molecular determinant for the establishment of sister chromatid cohesion. Science. 2008;321:566–569. doi: 10.1126/science.1157880. [DOI] [PubMed] [Google Scholar]
- 10.Ben-Shahar T.R., Heeger S., Lehane C., East P., Flynn H., Skehel M., Uhlmann F. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–566. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J., Shi X., Li Y., Kim B.J., Jia J., Huang Z., Yang T., Fu X., Jung S.Y., Wang Y. Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol. Cell. 2008;31:143–151. doi: 10.1016/j.molcel.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Haering C.H., Schoffnegger D., Nishino T., Helmhart W., Nasmyth K., Löwe J. Structure and stability of cohesin's Smc1-kleisin interaction. Mol. Cell. 2004;15:951–964. doi: 10.1016/j.molcel.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 13.Uhlmann F., Nasmyth K. Cohesion between sister chromatids must be established during DNA replication. Curr. Biol. 1998;8:1095–1101. doi: 10.1016/s0960-9822(98)70463-4. [DOI] [PubMed] [Google Scholar]
- 14.Uhlmann F., Wernic D., Poupart M.A., Koonin E.V., Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- 15.Uhlmann F., Lottspeich F., Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- 16.Haering C.H., Farcas A.M., Arumugam P., Metson J., Nasmyth K. The cohesin ring concatenates sister DNA molecules. Nature. 2008;454:297–301. doi: 10.1038/nature07098. [DOI] [PubMed] [Google Scholar]
- 17.Zhang N., Pati D. Handcuff for sisters: A new model for sister chromatid cohesion. Cell Cycle. 2009;8:399–402. doi: 10.4161/cc.8.3.7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gruber S., Arumugam P., Katou Y., Kuglitsch D., Helmhart W., Shirahige K., Nasmyth K. Evidence that loading of cohesin onto chromosomes involves opening of its SMC hinge. Cell. 2006;127:523–537. doi: 10.1016/j.cell.2006.08.048. [DOI] [PubMed] [Google Scholar]
- 19.Haering C.H., Löwe J., Hochwagen A., Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell. 2002;9:773–788. doi: 10.1016/s1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- 20.Hirano M., Hirano T. Hinge-mediated dimerization of SMC protein is essential for its dynamic interaction with DNA. EMBO J. 2002;21:5733–5744. doi: 10.1093/emboj/cdf575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiu A., Revenkova E., Jessberger R. DNA interaction and dimerization of eukaryotic SMC hinge domains. J. Biol. Chem. 2004;279:26233–26242. doi: 10.1074/jbc.M402439200. [DOI] [PubMed] [Google Scholar]
- 22.Gruber S., Haering C.H., Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–777. doi: 10.1016/s0092-8674(03)00162-4. [DOI] [PubMed] [Google Scholar]
- 23.Yeh E., Haase J., Paliulis L.V., Joglekar A., Bond L., Bouck D., Salmon E.D., Bloom K.S. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr. Biol. 2008;18:81–90. doi: 10.1016/j.cub.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.