Abstract
The use of the dominant negative mutant of Ras has been crucial in elucidating the cellular signaling of Ras in response to the activation of various membrane-bound receptors. Although several point mutants of Ras exhibit a dominant negative effect, the asparagine to serine mutation at position 17 (S17N) remains the most popular and the most effective at inhibiting the activation of endogenous Ras. It is now widely accepted that the dominant negative effect is due to the ability of the mutant to sequester upstream activators and its inability to activate downstream effectors. Here, we present the crystal structure of RasS17N in the GDP-bound form. In the three molecules that populate the asymmetric unit, the Mg2+ ion that normally coordinates the β-phosphate is absent because of steric hindrance from the Asn17 side chain. Instead, a Ca2+ ion is coordinating the α-phosphate. Also absent from one molecule is electron density for Phe28, a conserved residue that normally stabilizes the nucleotide’s guanine base. Except for Phe28, the nucleotide makes conserved interactions with Ras. Combined, the inability of Phe28 to stabilize the guanine base and the absence of a Mg2+ ion to neutralize the negative charges on the phosphates explain the weaker affinity of GDP for Ras. Our data suggest that the absence of the Mg2+ should also dramatically affect GTP binding to Ras and the proper positioning of Thr35 necessary for the activation of switch 1 and the binding to downstream effectors, a prerequisite for the triggering of signaling pathways.
Keywords: dominant negative, GDP, GTP-binding proteins, guanine nucleotide exchange, inhibition of signaling, inhibition, Mg2+ ion, Ras, S17N, site directed mutagenesis, X-ray crystallography
Ras is an essential component of signal transduction pathways that regulate cell growth, proliferation, differentiation, and apoptosis in response to the activation of membrane bound receptors (1, 2). The elucidation of the various Ras signaling pathways was primarily made possible by the wide use of two types of Ras mutants: dominant active and dominant negative. Mutants of Gly12 or Gln61 are typical dominant active mutants while the asparagine for serine mutant at position 17, RasS17N, is the most frequently used dominant negative form of Ras (3). The success of these mutants popularized the use of dominant active and dominant negative mutants to study the signaling of other Ras related GTP-binding proteins. While the biochemical properties of dominant active Ras mutants are well characterized, namely a slower rate of GTP hydrolysis combined with a faster intrinsic ability to exchange GTP for GDP resulting in a longer signaling state (4), the exact molecular details by which dominant negative Ras exerts their inhibitory function are a matter of debate in the literature. It is widely accepted that RasS17N blocks the ability of endogenous Ras to signal by sequestering and depleting the intracellular pool of available Ras activators or guanine nucleotide exchange factors (GEFs), thereby blocking the activation of endogenous Ras (5–7). The finding that the overexpression of a dominant active form of Ras (e.g. RasG12V) or an activator domain usually abolishes the inhibitory effect of dominant negative Ras supports the latter explanation (3, 8). However, other arguments have been also proposed (9, 10) including low affinity of RasS17N for GTP and the inability of GTP to induce an active RasS17N conformation necessary for the binding and activation of downstream effectors (5, 11). To complicate matters, the S17N mutant of Rap1A, a Ras-related protein, which a priori should behave like RasS17N, does not exhibit potent dominant negative properties since it is unable to inhibit the activation of Rap1A by its exchange factor, C3G, in vitro (12) and in cells (13) while various substitutions of Ser17 interact differently with the various Rap exchange factors (14).
In addition to its role as a tool to manipulate cellular signaling, the use of dominant negative Ras mutants showed promise in cancer gene therapy. For example, expression of RasS17N inhibited the growth of pancreatic cancer cells in a mouse model (15). Similarly, injection of adenovirus carrying the RasS17N mutation ameliorated the inflammatory reactions and suppressed bone destruction in arthritic joints of rats (16). Similarly, expression of another dominant negative mutant, RasN116Y, suppressed growth of metastatic human pancreatic cancer cells in the liver of nude mice (17).
Understanding the structure/function relationship of RasS17N at the molecular level should shed light on its cellular role and help design new tools to dissect Ras and other small G-proteins signaling. Ultimately, these new tools can be used in cancer gene therapy. In the present work, we describe the crystal structure of RasS17N bound to GDP and discuss the functional consequences of the mutation.
Experimental Methods
Protein Preparation and Crystallization
RasS17N (residues 1–166) was cloned as a His-tagged protein in the pProEX-HTb vector and expressed in the Escherichia coli CodonPlus BL21(DE3) strain as described previously (18). Proteins were purified on Ni-NTA beads (Qiagen) followed by removal of the His-tag by the action of TEV protease and a gel filtration column (Superdex 200, GE Healthcare). The protein yield was high (> 50 mg per liter of bacterial culture) and the protein was stable showing little or no precipitation during the purification process. Initial crystals were grown at 20°C using the vapor diffusion, hanging drop method by mixing 3 µl of 20 mg/ml RasS17N (in 20 mM HEPES, 150 mM NaCl, 2 mM MgCl2, pH = 7.5) and 3 µl of a reservoir solution. The reservoir consisted of 15% (v/v) polyethylene glycol 400 (PEG400), 13% (w/v) PEG8000, 0.2 M calcium acetate, and 0.1 M Tris-HCl pH = 7.0. The crystals grew from heavy protein precipitates and were not suitable for X-ray diffraction experiments. Systematic search for amphiphilic molecules (Hampton Research Additives) showed that the addition of 5% (v/v) DMSO or 0.7% butanol to the reservoir resulted in single thin needle-like crystals, which were further seeded to yield larger (0.05 × 0.05 × 0.2 mm3) diffraction quality crystals. The GDP-bound form of RasS17N crystallized in space group I222 (a = 92.5 Å, b = 102.2 Å, c = 117.8 Å) with three molecules in the asymmetric unit corresponding to a Vm value (19) of 2.57 Å3/Da and an estimated solvent content of 52.2%.
Data Collection, Structure Determination, and Model Refinement
Diffraction intensities to 2.6 Å resolution were collected at 100 K at beamline X26C at the National Synchrotron Laboratory Source, Brookhaven National Laboratory, on a 2k × 2k CCD detector (ADSC), processed with the HKL2000 package, and scaled with SCALEPACK (20).
The structure of the RasS17N mutant was solved by molecular replacement in the program MOLREP (21) with the deposited coordinates of wild-type Ras as search model (PDB ID 4Q21) (22). The structure was refined in the program PHENIX (23) using TLS and tight non-crystallographic symmetry to final crystallographic residuals R/Rfree of 23.8/29.6%. The electron density of the GDP molecule was clear in all three molecules during refinement but no density was observed at the expected Mg2+ binding site in the final electron density maps. Stereochemistry was checked with the program PROCHECK (24). Data collection and refinement statistics are summarized in Table 1.
Table 1.
Data collection and refinement statistics.
| Data Collection |
|---|
| Resolution Range (Å) 44.7 – 2.60 |
| Rsym *, (%) overall (last resolution shell) 10.0 (57.9) |
| Completeness, (%) overall (last resolution shell) 99.8 (99.7) |
| Multiplicity, overall (last resolution shell) 5.6 (5.6) |
| <I>/<σ(I)> 17.5 (3.2) |
| N° of unique reflections 17,942 |
| Refinement Statistics |
| B factor, (A2) overall (from Wilson statistics+) 37.3 (56.6) |
| Rfree †, (%) overall (last resolution shell) 29.6 (29.2) |
| Rcryst ‡, (%) overall (last resolution shell) 23.8 (32.1) |
| rms deviation in bond length (Å) 0.012 |
| rms deviation in bond angle (°) 1.55 |
| Estimated coordinate error § (Å) 0.37/0.29 |
| Ramachandran plane ¶ (%) 94.7/5.3 |
Rsym = ∑i,hkl|<I(hkl)> - Ii(hkl)|/∑i,hklIi(hkl). Last resolution shell is 2.69 - 2.60 Å.
The Wilson statistics were calculated for the data 3.9 - 2.6 Å.
Rfree = ∑(hkl)∊T||Fobs| - |Fcalc||/∑(hkl)∊T|Fobs|, where T is the test set (37) obtained by randomly selecting 5 % of the data.
Rcryst = ∑(hkl)||Fobs| - |Fcalc||/∑(hkl)|Fobs|.
Estimated coordinate error based on Rfree/maximum likelihood.
Most favored/additional allowed regions.
RESULTS
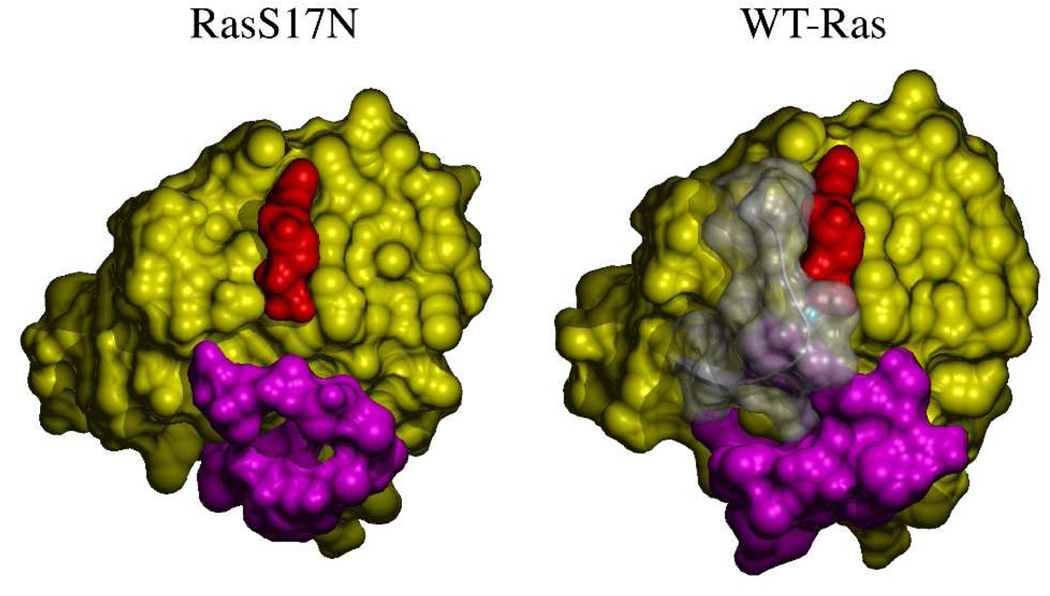
RasS17N•GDP crystallized in space group I222 with three molecules (hereafter A, C, and E) in the asymmetric unit. The structure of RasS17N was refined to crystallographic residuals R/Rfree of 23.8/29.6% and good stereochemistry (Table 1). Overall, the structure is characterized by high temperature factors for each of the three copies and weak electron density for the side chain of some surface exposed residues. This result is consistent with the high temperature factor (56.6 Å2) deduced from Wilson statistics (see Table 1). The electron density is clear for the main chain of all three molecules outside the switch 1 (residues 24 to 40) and switch 2 (residues 57 to 75) regions. The switch regions are the solvent exposed regions of Ras that change conformation following GTP hydrolysis and γ-phosphate release. They also constitute binding domains for effectors and regulators (25). The switch 1 electron density is clear for chain E but not for chain A or C. Interestingly, switch 1 is involved in crystal packing in chain E, weakly in chain A but not in chain C. The electron density of Phe28 in chain A is weak and absent in chain C (Figure 1A) suggesting that in chain C and contrary to what one would expect, the phenyl ring is not stabilizing the guanine of the GDP. As a result, the accessible surface area of GDP in chains A and C has increased (Figure 1B). The electron density for switch 2 residues 57 to 71 is absent in chain A and E. In both chains, switch 2 is not involved in crystal packing. In chain C, the electron density for residues 60 to 64 is absent and only the density of the main chain of this region’s remaining residues is clear. Combined with previous Ras structures where at least the N-terminal domain of switch 1 is involved in crystal packing, our data suggest that crystal packing might have contributed to the accepted conformation of Phe28 in stabilizing the guanine nucleotide.
Figure 1.
GDP-environment in RasS17N. A- Simulated annealing (Fobs – Fcalc) electron density omit map (blue mesh) shown at 3.0 σ cutoff and calculated with phases deduced from the final model after removal of the GDP and its coordinating Ca2+ ion from chain C. The final model is shown in pale green as a ribbon diagram. Asn17 and GDP are shown in ball-and-stick representation. The switch 1 region of WT-Ras with Phe28 in ball-and-stick representation is shown in transparent white for reference after superposition. Note the lack of density for Phe28. B- Comparison of the solvent accessibility of the GDP (red) in WT-Ras and in chain C of RasS17N shown in identical orientation and in yellow surface representation. The switch 1 and 2 are represented as white transparent and magenta surfaces, respectively. The Mg2+ ion is shown as a cyan sphere. Figures were prepared with POVScript (35), povray (www.povray.org), and MSMS (36).
As for all known Ras structures, the overall fold outside the switch regions is conserved in the RasS17N structure. The electron density of Asn17 is clear in all three copies and its side chain is not making any contact with the rest of the protein. The mutation has not disturbed the structure of the P-loop or interfered with the interactions made by this loop and the guanine nucleotide (Figure 2A). Comparison of the GDP-binding site in WT-Ras and RasS17N shows that with the exception of Phe28, the interactions made by the GDP and Ras including those made by the P-loop, the 116NKXD119 and 145SAK147 motifs are conserved in RasS17N.
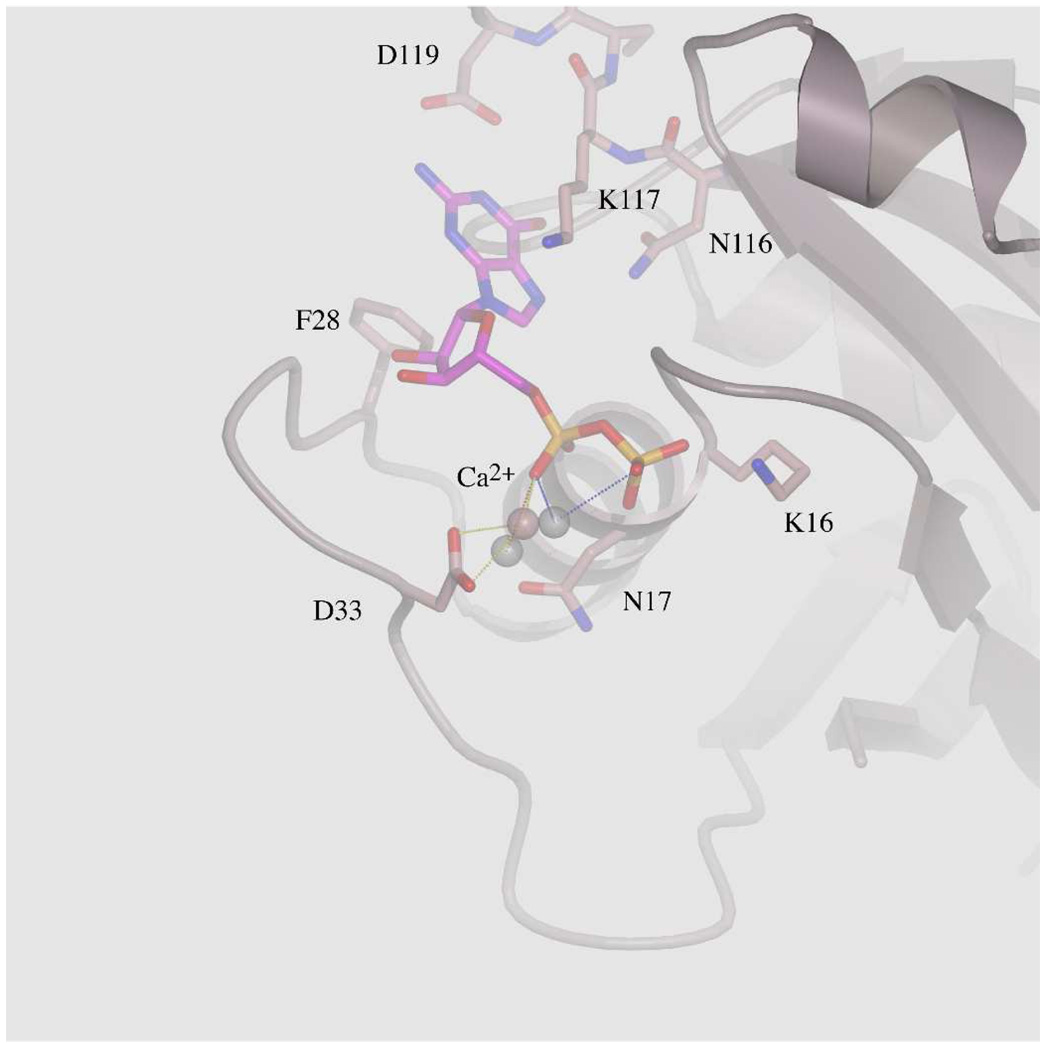
Figure 2.
A- The environment of the GDP-phosphates in chain E is represented with the corresponding Ca2+ (pink sphere). For simplicity, only important residues are shown in ball-and-stick representation. Asn17 is shown in surface and ball-and stick representations. Ser17, Asp57, the Mg2+ ion (green sphere) and its coordinating water molecules (red spheres) of WT-Ras are overlaid for comparison. Dotted lines represent electrostatic interactions. The P-loop of both WT-Ras and RasS17N are superposed showing that the GDP makes conserved interaction with RasS17N. B- Overlay of the GDP environment between the three molecules in the asymmetric unit showing how the Ca2+ ions differ in the way they coordinate α- and β-phosphates. For simplicity, only chain E is represented. Asp33 coordinates the Ca2+ ion only in this chain.
Two features however, distinguish the RasS17N model from known GDP-bound structures of Ras (22, 26–30). First, there is no density attributable to a Mg2+ ion in the final electron density map. In all GDP-bound structures of Ras, a Mg2+ ion is found coordinated to the β-phosphate, the hydroxyl group of Ser17, and four water molecules. One of these water molecules is stabilized by the carboxylic group of Asp57 of the conserved 57DTAGQ61 motif. In the RasS17N structure, the side chain of Asn17 deprives the metal ion from a well-positioned coordinating oxygen atom, and consequently eliminates the only direct interaction between the Mg2+ ion and Ras (Figure 2A). Efforts to model a water molecule that would substitute the Oγ of Ser17 in coordinating the Mg2+ ion fail because of steric clashes with the Cβ of Asn17. Consequently, the metal ion cannot be octahedrally coordinated and therefore cannot bind to the GDP, consistent with previous data showing a ~30 fold lower affinity of the Mg2+ ion upon mutating Ser17 (11). In the final electron density map however, a strong electron density near the α-phosphate is clear in each of the three Ras chains (Figure 1A and Figure 2B). Effort to model a Mg2+ or Na+ ion or a water molecule gave unsatisfactory results. For example, refining a Mg2+ ion in chain E resulted in a residual peak at 3σ in a difference Fourier map and a relatively low temperature factor for the Mg2+ ion. The strong electron density map can be explained by a Mg2+ or Na+ ion in chains A and C but the distances to the α- and β-phosphates (2.7, 2.8, and 3.2 Å) were inconsistent with a Mg2+ ion. In addition, a Na+ ion or a water molecule does not fully neutralize the GDP negative charges. For these structural and chemical reasons, a Ca2+ ion was modeled and refined in each positive peak near the α-phosphate. These Ca2+ ions likely come from the crystallization medium. One could also argue that the Ca2+ ion has displaced the Mg2+ ion. However, Ras has been previously crystallized in calcium acetate yet the Mg2+ was clearly visible in the final electron density map (30). The Ca2+ ions sit in different sites in each chain. All three are positioned 2.6 to 2.8 Å from one Pα-oxygen. In chain C, the Ca2+ ion is also 3.0 Å from one Pβ-oxygen. In chain E, the side chain carboxylate group of Asp33 and one water molecule are also coordinating the Ca2+ ion. These ions are 3.6 to 3.9 Å away from the expected Mg2+ ion-binding site towards the α-phosphate. Their positive charges neutralize the negative charges of the phosphates. We thus conclude that in solution, the absence of the Mg2+ ion due to the S17N mutation prevents the shielding of the negative charges of the phosphates. Combined with the increase in flexibility of Phe28, this electrostatic effect explains the lower affinity of guanine nucleotides to RasS17N (11). Such an electrical shield should play an even more important role when the more negative GTP is bound to Ras rather than GDP, consistent with experimental data showing that GTP has weaker affinity for the Ser17 mutant of Ras than GDP (10, 11).
Second, in WT-Ras, the side chain of Asp57 of the conserved 57DTAGQ61 motif makes a hydrogen bond with the hydroxyl group of Ser17 and coordinates the Mg2+ ion through a water molecule (Figure 2A). The absence of the Mg2+ ion and of the hydroxyl group of Ser17 in RasS17N destabilizes Asp57 and increases its flexibility. Thus, the S17N mutation increases the flexibility of an already mobile switch 2 region.
DISCUSSION
The S17N mutant of Ras was initially isolated by screening Ras for mutants with preferential GDP over GTP binding using random mutagenesis (3) and was subsequently shown to inhibit the proliferation of NIH 3T3 cells when expressed alone or in the background of dominant active mutations such as A59T or Q61L. Co-expression of a transforming Ras mutant or of a Ras activator homology domain reverses the inhibitory effect of the S17N mutation suggesting that the dominant negative effect is due to the inhibition of the signaling of endogenous Ras by sequestering the available GEFs (3, 8). This in vivo property of RasS17N combined with the conservation of Ser17 in all GTP-binding proteins popularized the use of this mutant to study the signaling pathways of Ras and of other GTPases. At the biochemical level, the S17N mutation decreased the affinity for the GDP and GTP by a factor of 27 and 1,000 respectively, while the affinity for the GEF is increased by a factor of 5.4 when compared to WT-Ras; as for the downstream effectors, their binding to RasS17N is severely compromised (10).
Our structural data explain and confirm many of the previously described properties of Ser17 mutants (5, 10). The replacement of the conserved Ser17 by an Asn, an Ala or a Cys disrupts the only interaction the Mg2+ ion makes with Ras, at the same time the hydrophobic or longer side chains prevent a water molecule from substituting the hydroxyl group of Ser17. This combined effect destabilizes the octahedral coordination of the metal ion and disable it from binding the GDP, consistent with previous data showing a lower affinity of the Mg2+ ion when Ser17 is mutated (11). Although the affinity of the Mg2+ ion was measured using the S17A mutant, we believe that the Asn, Ala, and Cys mutants of Ser17 destabilize the Mg2+ ion for the same reasons resulting in the same biological outcome (5). Therefore, the affinity for the Mg2+ ion for these mutants should be similar. On the other hand, the absence of a side chain at position 17 in the S17G mutant should allow a water molecule to replace the hydroxyl group of Ser17 and to coordinate the Mg2+ ion so that it is present in both the GDP- and GTP-bound form of this mutant. This would allow a tight binding of the GTP, the adoption of an active conformation by Thr35 and switch 1, and therefore binding of this mutant to downstream effectors. This assumption is consistent with the finding that RasS17G has no inhibitory effect when transfected in cells (5).
The loss of the Mg2+ ion results in an increase in the flexibility of the switch regions of Ras. In part this is due to the increase in the flexibility of the conserved Phe28 and Asp57 of switch 1 and 2, respectively, which we were unable to model in the final electron density map unless they were involved in crystal packing. How the loss of the Mg2+ ion is transmitted to Phe28 is unclear especially since Phe28 and its immediate surrounding is not coordinating the metal ion in WT-Ras. One likely explanation is that the increase in the OFF-rate of the nucleotide in RasS17N destabilizes Phe28. How does the destabilization of Phe28 affect the OFF-rate of the nucleotide? Recently characterized mutants of Ras where Phe28 is not stabilizing the guanine base showed that the OFF-rate of the GDP is weakly affected by the removal of this residue (29, 31). Consequently, our data support the hypothesis that Mg2+ binding is essential for high nucleotide affinity. In regard to Asp57, whether the destabilization of this residue is a direct effect of the removal of the Mg2+ ion or, conversely, that the elimination of the hydroxyl group of Ser17 destabilizes Asp57, which in turn weakens binding of the Mg2+ ion, or both is a possibility. We favor the former hypothesis since the S17G mutant is not inhibitory.
We do not have the structure of the GTP-bound form of RasS17N but it is likely that this structure also lacks the Mg2+ ion for the same reasons discussed in this work. The absence of a Mg2+ ion makes it even harder for Ras to neutralize the negative charges of the triphosphates consistent with the high dissociation of the GTP for this mutant (10). The absence of the Mg2+ ion should also prevent Thr35 of the switch 1 region from adopting the conformation it has in active Ras (32). Since the Thr35/Mg2+ ion interaction is essential for the switch 1 region to adopt an effector binding conformation necessary for proper signaling, it is therefore likely that this mutant does not bind to effectors even when GTP-bound, consistent with its inability to signal (3, 10).
The Mg2+ free structure presented here contrasts with the structure of the homologous RhoA solved in the absence of magnesium (33). In the latter structure, the switch regions are well defined and pull away from the GDP, which is loosely bound to the protein. In addition, Lys18 interacts with Glu64 (Lys 16 and Glu62 in Ras) and Ala61 (Ala59 in Ras) is near the Mg2+ binding site. Although these residues are conserved between Ras and Rho, in RasS17N they do not adopt the same conformation as in Mg2+ free RhoA. Modeling Ala59 and switch 2 in all three copies as in the latter structure, which also mimics the conformation of switch 2 in nucleotide free Ras in complex with Sos (34), does not lead to steric clashes. Why the switch regions of RasS17N did not adopt the conformation they have in Mg2+ free RhoA is not clear.
ACKNOWLEDGMENT
We thank Annie Héroux for technical assistance at beamline X26C.
FUNDING: Research in NN’s laboratory is supported in part by grants from the NIH (CA-115611), National Science Foundation (MCB-0316600), and DOD (NF060060). The National Synchrotron Light Source is supported by the Department of Energy and NIH, and beamline X26C was supported in part by Stony Brook University and its Research Foundation.
ABBREVIATIONS
- DTT
dithiothreitol
- GEF
guanine nucleotide exchange factor
- Ni-NTA
nickel-nitrilotriacetic acid
- TEV
Tobacco Etch virus
- WT
wild-type
Footnotes
The coordinates of the GDP-bound form of RasS17N were submitted to the protein data bank (PDB ID 3LO5).
REFERENCES
- 1.Vojtek AB, Der CJ. Increasing complexity of the Ras signaling pathway. J Biol Chem. 1998;273:19925–19928. doi: 10.1074/jbc.273.32.19925. [DOI] [PubMed] [Google Scholar]
- 2.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 3.Feig LA, Cooper GM. Inhibition of NIH 3T3 cell proliferation by a mutant ras protein with preferential affinity for GDP. Mol Cell Biol. 1988;8:3235–3243. doi: 10.1128/mcb.8.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Der CJ, Finkel T, Cooper GM. Biological and biochemical properties of human rasH genes mutated at codon 61. Cell. 1986;44:167–176. doi: 10.1016/0092-8674(86)90495-2. [DOI] [PubMed] [Google Scholar]
- 5.Farnsworth CL, Feig LA. Dominant inhibitory mutations in the Mg(2+)-binding site of RasH prevent its activation by GTP. Mol Cell Biol. 1991;11:4822–4829. doi: 10.1128/mcb.11.10.4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Powers S, O'Neill K, Wigler M. Dominant yeast and mammalian RAS mutants that interfere with the CDC25-dependent activation of wild-type RAS in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:390–395. doi: 10.1128/mcb.9.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stacey DW, Feig LA, Gibbs JB. Dominant inhibitory Ras mutants selectively inhibit the activity of either cellular or oncogenic Ras. Mol Cell Biol. 1991;11:4053–4064. doi: 10.1128/mcb.11.8.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schweighoffer F, Cai H, Chevallier-Multon MC, Fath I, Cooper G, Tocque B. The Saccharomyces cerevisiae SDC25 C-domain gene product overcomes the dominant inhibitory activity of Ha-Ras Asn-17. Mol Cell Biol. 1993;13:39–43. doi: 10.1128/mcb.13.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jung V, Wei W, Ballester R, Camonis J, Mi S, Van Aelst L, Wigler M, Broek D. Two types of RAS mutants that dominantly interfere with activators of RAS. Mol Cell Biol. 1994;14:3707–3718. doi: 10.1128/mcb.14.6.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cool RH, Schmidt G, Lenzen CU, Prinz H, Vogt D, Wittinghofer A. The Ras mutant D119N is both dominant negative and activated. Mol Cell Biol. 1999;19:6297–6305. doi: 10.1128/mcb.19.9.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.John J, Rensland H, Schlichting I, Vetter I, Borasio GD, Goody RS, Wittinghofer A. Kinetic and structural analysis of the Mg(2+)-binding site of the guanine nucleotide-binding protein p21H-ras. J Biol Chem. 1993;268:923–929. [PubMed] [Google Scholar]
- 12.van den Berghe N, Cool RH, Horn G, Wittinghofer A. Biochemical characterization of C3G: an exchange factor that discriminates between Rap1 and Rap2 and is not inhibited by Rap1A(S17N) Oncogene. 1997;15:845–850. doi: 10.1038/sj.onc.1201407. [DOI] [PubMed] [Google Scholar]
- 13.Hogan C, Serpente N, Cogram P, Hosking CR, Bialucha CU, Feller SM, Braga VM, Birchmeier W, Fujita Y. Rap1 regulates the formation of E-cadherin-based cell-cell contacts. Mol Cell Biol. 2004;24:6690–6700. doi: 10.1128/MCB.24.15.6690-6700.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dupuy AG, L'Hoste S, Cherfils J, Camonis J, Gaudriault G, de Gunzburg J. Novel Rap1 dominant-negative mutants interfere selectively with C3G and Epac. Oncogene. 2005;24:4509–4520. doi: 10.1038/sj.onc.1208647. [DOI] [PubMed] [Google Scholar]
- 15.Stoll V, Calleja V, Vassaux G, Downward J, Lemoine NR. Dominant negative inhibitors of signalling through the phosphoinositol 3-kinase pathway for gene therapy of pancreatic cancer. Gut. 2005;54:109–116. doi: 10.1136/gut.2004.046706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamoto A, Fukuda A, Seto H, Miyazaki T, Kadono Y, Sawada Y, Nakamura I, Katagiri H, Asano T, Tanaka Y, Oda H, Nakamura K, Tanaka S. Suppression of arthritic bone destruction by adenovirus-mediated dominant-negative Ras gene transfer to synoviocytes and osteoclasts. Arthritis Rheum. 2003;48:2682–2692. doi: 10.1002/art.11214. [DOI] [PubMed] [Google Scholar]
- 17.Takeuchi M, Shichinohe T, Senmaru N, Miyamoto M, Fujita H, Takimoto M, Kondo S, Katoh H, Kuzumaki N. The dominant negative H-ras mutant, N116Y, suppresses growth of metastatic human pancreatic cancer cells in the liver of nude mice. Gene Ther. 2000;7:518–526. doi: 10.1038/sj.gt.3301125. [DOI] [PubMed] [Google Scholar]
- 18.Hall BE, Yang SS, Boriack-Sjodin PA, Kuriyan J, Bar-Sagi D. Structure-based mutagenesis reveals distinct functions for Ras switch 1 and switch 2 in Sos-catalyzed guanine nucleotide exchange. J Biol Chem. 2001;276:27629–27637. doi: 10.1074/jbc.M101727200. [DOI] [PubMed] [Google Scholar]
- 19.Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 20.Otwinowski Z, Borek D, Majewski W, Minor W. Multiparametric scaling of diffraction intensities. Acta Crystallogr A. 2003;59:228–234. doi: 10.1107/s0108767303005488. [DOI] [PubMed] [Google Scholar]
- 21.Vagin A, Teplyakov A. MOLREP: An Automated Program for Molecular Replacement. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- 22.Milburn MV, Tong L, deVos AM, Brunger A, Yamaizumi Z, Nishimura S, Kim SH. Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic ras proteins. Science. 1990;247:939–945. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- 23.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 24.Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 25.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 26.Kraulis PJ, Domaille PJ, Campbell-Burk SL, Van Aken T, Laue ED. Solution structure and dynamics of ras p21.GDP determined by heteronuclear three- and four- dimensional NMR spectroscopy. Biochemistry. 1994;33:3515–3531. doi: 10.1021/bi00178a008. [DOI] [PubMed] [Google Scholar]
- 27.Ito Y, Yamasaki K, Iwahara J, Terada T, Kamiya A, Shirouzu M, Muto Y, Kawai G, Yokoyama S, Laue ED, Walchli M, Shibata T, Nishimura S, Miyazawa T. Regional polysterism in the GTP-bound form of the human c-Ha-Ras protein. Biochemistry. 1997;36:9109–9119. doi: 10.1021/bi970296u. [DOI] [PubMed] [Google Scholar]
- 28.Hall BE, Bar-Sagi D, Nassar N. The structural basis for the transition from Ras-GTP to Ras-GDP. Proc Natl Acad Sci U S A. 2002;99:12138–12142. doi: 10.1073/pnas.192453199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ford B, Skowronek K, Boykevisch S, Bar-Sagi D, Nassar N. Structure of the G60A mutant of Ras: implications for the dominant negative effect. J Biol Chem. 2005;280:25697–25705. doi: 10.1074/jbc.M502240200. [DOI] [PubMed] [Google Scholar]
- 30.Ford B, Hornak V, Kleinman H, Nassar N. Structure of a transient intermediate for GTP hydrolysis by ras. Structure. 2006;14:427–436. doi: 10.1016/j.str.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 31.Ford BA, Boykevisch S, Zhao C, Kunzelmann S, Bar-Sagi D, Herrmann C, Nassar N. Characterization of a Ras mutant with identical GDP- and GTP-bound structures. Biochemistry. 2009 doi: 10.1021/bi901479b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pai EF, Kabsch W, Krengel U, Holmes KC, John J, Wittinghofer A. Structure of the guanine-nucleotide-binding domain of the Ha-ras oncogene product p21 in the triphosphate conformation. Nature. 1989;341:209–214. doi: 10.1038/341209a0. [DOI] [PubMed] [Google Scholar]
- 33.Shimizu T, Ihara K, Maesaki R, Kuroda S, Kaibuchi K, Hakoshima T. An open conformation of switch I revealed by the crystal structure of a Mg2+-free form of RHOA complexed with GDP. Implications for the GDP/GTP exchange mechanism. J Biol Chem. 2000;275:18311–18317. doi: 10.1074/jbc.M910274199. [DOI] [PubMed] [Google Scholar]
- 34.Boriack-Sjodin PA, Margarit SM, Bar-Sagi D, Kuriyan J. The structural basis of the activation of Ras by Sos. Nature. 1998;394:337–343. doi: 10.1038/28548. [DOI] [PubMed] [Google Scholar]
- 35.Fenn TD, Ringe D, Petsko GA. POVScript+: A Program for Model and Data Visualization Using Persistence of Vision Ray-Tracing. J. Appl. Cryst. 2003;36:4. [Google Scholar]
- 36.Sanner MF, Olson AJ, Spehner JC. Reduced surface: an efficient way to compute molecular surfaces. Biopolymers. 1996;38:305–320. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 37.Brünger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]