Abstract
Phosphorylation of the Kir3 channel by cAMP-dependent protein kinase (PKA) potentiates activity and strengthens channel-PIP2 interactions, whereas phosphorylation by protein kinase C (PKC) exerts the opposite effects (Keselman et al., Channels 1:113–123, 2007; Lopes et al., Channels 1:124–134, 2007). Unequivocal identification of phosphorylated residues in ion channel proteins has been difficult, but recent advances in mass spectrometry techniques have allowed precise identification of phosphorylation sites (Park et al., Science 313:976–979, 2006). In this study, we utilized mass spectrometry to identify phosphorylation sites within the Kir3.1 channel subunit. We focused on the Kir3.1 C-terminal cytosolic domain that has been reported to be regulated by several modulators. In vitro phosphorylation by PKA exhibited a convincing signal upon treatment with a phosphoprotein stain. The phosphorylated C terminus was subjected to mass spectrometric analysis using matrix-assisted lased desorption/ionization-time of flight mass spectroscopy (MS). Peptides whose mass underwent a shift corresponding to addition of a phosphate group were then subjected to tandem MS (MS/MS) in order to confirm the modification and determine its precise location. Using this approach, we identified S385 as an in vitro phosphorylation site. Mutation of this residue to alanine resulted in a reduced sensitivity of Kir3.1* currents to H89 and Forskolin, confirming an in vivo role for this novel site of the Kir3.1 channel subunit in its regulation by PKA.
Keywords: Phosphorylation, G-protein-gated inwardly rectifying potassium channels, Posttranslational modification, Ion channel regulation, Protein kinase A, Mass spectrometry, MALDI-TOF, Tandem MS
Introduction
Phosphorylation is a prevalent posttranslational modification mechanism regulating protein function. Several members of the inwardly rectifying potassium (Kir) channel family have been shown to be regulated by protein kinases. Protein kinase (PKA) and protein kinase C (PKC) have been shown to modulate Kir1 [16], Kir2 [6, 12, 14, 51, 56, 57], Kir4.2 [36], Kir4.1/Kir51 heterotetramers [39], and Kir6 [1].
GTP binding (G) protein-sensitive, inwardly rectifying potassium (GIRK or Kir3) channel subunits constitute the acetylcholine-activated K+ current (IK-ACh) responsible for slowing the heart rate [33, 43, 48]. Cloning of the atrial Kir3 channel revealed that it is a heterotetramer consisting of Kir3.1 and Kir3.4 subunits [15]. Acetylcholine-induced activation of Kir3 is mediated by direct interactions of the channel with the Gβγ subunits of the pertussis toxin- [55] sensitive Gi/o proteins [18]. Another Kir3 channel modulator is intracellular sodium (Na+), which has been shown to stimulate channel activity in a Gβγ-independent manner [38, 40, 45, 46, 54].
Regulation of ion channel activity by phosphatidylinositol 4,5-bisphosphate (PIP2) has been broadly demonstrated for a variety of membrane channels and transporters and has been extensively studied for the Kir channel family [19]. Moreover, several modulators of Kir channel activity have been shown to act through altering channel-PIP2 interactions. Interestingly, Gβγ and intracellular Na+ both enhance Kir3 channel-PIP2 interactions [9, 10, 40, 54].
PKA and PKC have also been shown to modulate Kir3 channel activity. PKA potentiates [21, 24, 27], while PKC inhibits [8, 13, 22] Kir3 currents. These kinases have been found to differentially affect Kir3 channel interactions with PIP2, namely PKA increasing and PKC reducing PIP2 sensitivity, thereby up- or down-regulating channel activity [13, 21]. However, identification of specific phosphorylation sites responsible for the functional effects of protein kinases has resorted to indirect approaches. In a study by Medina et al. [24], purified catalytic subunits of PKA and PKC were both shown to increase channel phosphorylation in vitro, as assessed by P32 incorporation into immunoprecipitated native atrial and heterologously expressed Kir3 channels. Native Kir3 channel currents were potentiated in the presence of ATP, but not in the presence of a nonhydrolysable ATP analog. Protein phosphorylation seemed to be involved rather than phosphorylation of inositides, since after PIP2-stimulated steady-state activity, hydrolyzable forms of ATP further potentiated Kir3 currents. Moreover, dephosphorylation by PP2A, a broad specificity serine/threonine phosphatase, reduced P32 incorporation and prevented Gβγ activation of Kir3 currents. Mutagenesis experiments pointed to a region between residues 373 and 419 that in the context of the Kir3.1, C terminus seemed to control channel phosphorylation. However, no specific residues could be identified within this region that accounted for this control. Direct PKA involvement was also suggested by studies showing that injection of cAMP and direct application of purified catalytic subunit of PKA potentiated Kir3 current and affected its single-channel properties [28]. In atrial cells, Kir3 has been shown to interact directly with PKA and two protein phosphatases, PP1 and PP2A [30]. Recently, serines 221 and 315, which form consensus PKA phosphorylation sites on the Kir3.1 C terminus, have been shown to play a role in PKA-mediated regulation of the Kir3.1/3.4 channel [21]. These results imply that Kir3 channel activity is regulated by direct PKA phosphorylation. However, the inability of these functional studies to conclusively attribute kinase-mediated effects to direct phosphorylation of the channel has prompted us to use a more direct approach towards identification of phosphorylation sites.
Mass spectrometry is a powerful tool towards direct identification of protein posttranslational modification sites. Recent advances in mass spectrometry techniques have allowed unambiguous identification of functionally relevant phosphorylation sites on several ion channel proteins [29, 35, 47, 52, 53]. In our experiments, we utilized the F137S mutation in Kir3.1 (Kir3.1*), which renders homomeric channels active and displays similar regulation characteristics as the Kir3.1/Kir3.4 heteromers [3, 50]. We analyzed both the full-length Kir3.1* and its purified cytosolic C-terminal domain using mass spectrometry. We examined the ability of the Kir3.1 C terminus to behave as a PKA substrate by subjecting it to in vitro phosphorylation. Using the matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) and tandem mass spectrometry (MS/MS) techniques, we unambiguously identified Kir3.1(S385) as a phosphorylation site. Interestingly, S385 is localized within the 373–419 region of Kir3.1, which was previously implicated as critical in mediating channel phosphorylation and enabling Gβγ gating [24]. We further tested the role of S385 in PKA-mediated effects on channel function and found its alanine mutant to exhibit a reduced response to PKA-mediated current potentiation.
Experimental procedures
Molecular biology
Human muscarinic receptor 2 (M2), Flag-Kir3.1, Kir3.1*, and Kir3.1* S385A DNA constructs were subcloned into pGEMHE and linearized by enzymatic digestion with Nhe1 (New England Biolabs, Beverly, MA, USA) and in vitro transcribed into cRNA using the mMessage mMachine (Ambion. Austin, TX, USA). cRNA concentration was estimated by subjecting the cRNA sample to formaldehyde gel electrophoresis and comparing the band intensity to an RNA molecular weight standard (Invitrogen Life Technologies, Carlsbad, CA, USA).
Electrophysiology in Xenopus oocytes
Oocytes obtained as previously described [17, 20] were injected with 2 ng each of Kir3.1*, Kir3.1* S385A, and M2 cRNAs. Electrophysiological recordings were performed 48 h after injection. The two-electrode voltage clamp technique was used to record whole-cell currents from oocytes, as previously described [7, 42, 44]. Briefly, a GeneClamp 500 amplifier (Axon Instruments, Foster City, CA, USA) was used, and data acquisition was carried out with Clampex pClamp8 software. The bath solution (high potassium, HK) contained (in mM) 91 KCl, 1 NaCl, 1 MgCl2, 5 KOH/HEPES (pH 7.4). Oocytes were perfused with HK to record control currents and HK containing 3 mM BaCl2 to block inwardly rectifying potassium currents. Currents were monitored in a series of 700 ms sweeps, during which the voltage was stepped from a 0-mV holding potential to 250 ms each +80 mV and −80 mV, repeated every 1 s. Currents were sampled at the end of 250 ms −80-mV pulse and were plotted in Clampfit (pClamp6.0, Axon Instruments) as the time course of current amplitudes. Both the HK and barium-insensitive currents were evaluated after reaching steady state. Kir3 current was calculated by subtracting the barium-insensitive current from the current elicited during the HK perfusion. Data were analyzed in Microcal Origin 6.0 software (Microcal Software, Northampton, MA, USA). For each injection group, five batches were tested (three to six oocytes per batch). Oocytes were incubated for 1–2h with either 50 μM H89 (Calbiochem, San Diego, CA, USA) or 50 μM Forskolin (Sigma-Aldrich, St. Louis, MI, USA). In each batch of oocytes expressing either Kir3.1* or Kir3.1*S385A, currents from H89- or Forskolin-treated oocytes were normalized to currents from untreated oocytes. Means of normalized basal and agonist-induced currents from each group are presented in bar graphs ±SEM.
Statistical analysis
The statistical significance of mean normalized currents was determined using two-tailed independent t test (Microcal Origin 6.0).
In vitro PKA phosphorylation and phospho-stain
We purified GST-tagged Kir3.1 (185–501; GST-Kir3.1C) as previously described [7]. Phosphorylation of 4 μg of GST-Kir3.1C with the catalytic subunit of PKA (cPKA) and dephosphorylation with alkaline phosphatase (AP; New England Biolabs, Ipswich, MA, USA) were carried out according to the manufacturer's protocols. In order to visualize phosphorylation of the GST-Kir3.1C, the sample was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and probed with Molecular Probes Pro-Q Diamond Phosphoprotein Gel stain (Molecular Probes, Invitrogen) that detects phospho-serine, threonine, and tyrosine residues. The gel was scanned on a Typhoon 9400 scanner (532ex, 560em; Proteomics Resource Center, Rockefeller University). The gel was then washed and stained for total protein with Novex Colloidal Blue stain (Invitrogen).
Matrix-assisted laser desorption/ionization–time of flight
GST-Kir3.1C was treated with AP or varying concentrations of cPKA as described above. GST-Kir3.1C was pulled down with glutathione-conjugated beads (GE Healthcare Bio-Sciences, Piscataway, NJ, USA) and washed with 50 mM ammonium bicarbonate. GST-Kir3.1C was eluted with three sequential incubations of glutathione sepharose in 20 μL of 10 mM reduced glutathione in 50 mM ammonium bicarbonate at room temperature for 10 min. The three elution fractions were pooled and reduced by incubation with 10 mM Tris (2-carboxyethyl) phosphine (Sigma-Aldrich) for 30 min at 37°C followed by alkylation for 1 h at room temperature with 50 mM iodoacetamide (Sigma-Aldrich). The pH of the sample was adjusted to 8 with 50 mM ammonium bicarbonate before digestion with 100 ng of sequencing grade Endoproteinase LysC (Roche, Indianapolis, IN, USA) for 4 h at 37°C, followed by digestion with 500 ng porcine, sequencing grade, modified, TPCK-treated trypsin (Promega, Madison, WI, USA). Matrix solution contained α-Cyano-4-hydroxycinnamic acid (4-HCCA) in 50% acetonitrile, 0.1% trifluoroacetic acid (TFA) in H2O. Peptides resulting form LysC and trypsin digestion of GST-Kir3.1C were mixed in a 1:1 ratio with the matrix solution. Of the peptide–matrix mix, 1.5 μL was deposited on the solid support. To desalt the sample, once matrix crystals started to form on the solid support but before the drying process was complete, we aspirated the remaining solution without disturbing the matrix/peptide crystals. Before MALDI-TOF MS, the crystals remaining on the solid support were briefly washed with cold 0.1% TFA. MALDI-TOF analysis was performed on a Voyager-DE STR BioSpectrometry Workstation (PerSeptive Biosystems, Ramsey, MN, USA). The spectrum was an average of 1,200 scans and ranged 800–4,000 m/z. All solvents were high-performance liquid chromatography (HPLC) grade.
Tandem MS
Prior to the analysis, the sample was desalted and purified using Poros 20 R2 beads (PerSeptive Biosystems) and ZipTipC18 (Millipore). Following cleanup, the sample was eluted with solution containing 50% acetonitrile, 0.1% trifluoroacetic acid in water, and mixed with 4-HCCA matrix in 50% acetonitrile and allowed to dry on the solid support. Comprehensive neutral loss scans [11, 23, 41] were performed in a linear ion trap mass spectrometer (LTQ, Thermo, San Jose) equipped with a vMALDI ion source. The instrument was programmed to fragment singly charged ions from a selected narrow window of 4 m/z units at a time, and fragment ions thus produced were then monitored in an MS2 window of 10 m/z units, which was centered at a value 98 m/z units lower than the center of the selected precursor ion window. These scans were performed successively, each time shifting the precursor ion window by four units from m/z 700 to m/z 4,000. Peptide (2,101.9 Da) from the AP-treated sample and 2,181.9 Da peptide from the cPKA-treated (625 U/μl) sample were subjected to MS2 analysis. Phosphorylation site identification was carried out by subjecting the neutral loss product (2,083.5 Da) detected in the MS2 scan of 2,181.9 Da peptide to an MS3 scan.
Flag-tagged Kir3 expression, pull-down, and in-gel digestion
Flag-Kir3.1* cRNA was injected into approximately 500 Xenopus oocytes. After 48–72 h of expression, the oocyte membrane fraction was collected, as previously described [25]. To extract membrane proteins, membrane pellets were resuspended in buffer containing 1% Triton-X100 and incubated at 4°C. Flag-Kir3.1* subunits were pulled down using EZView Red Anti-Flag Affinity Gel (Sigma-Aldrich) and eluted from beads with NuPAGE LDS (Invitrogen) gel loading buffer. Flag-Kir3.1* was subjected to SDS-PAGE and gel bands corresponding to glycosylated and core Flag-Kir3.1* were excised. In-gel digestion, peptide extraction, and sample desalting was carried out as described previously [2]. MALDI-TOF was carried out as described above.
Results
Mass spectrometry analysis of full-length Kir3.1
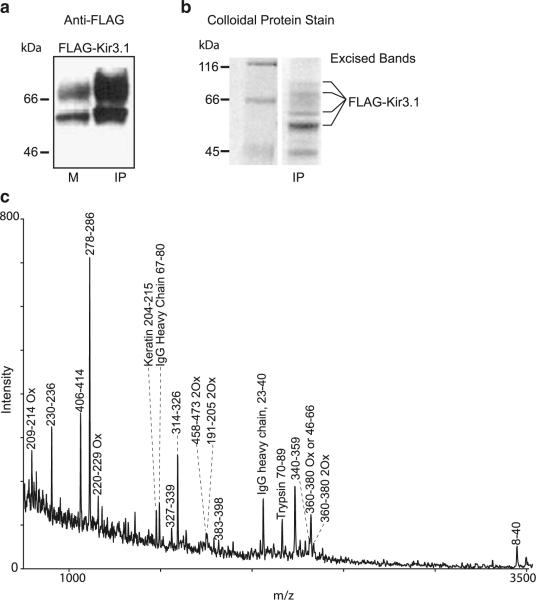
Flag-tagged Kir3.1* channel was expressed in Xenopus oocytes and immunoprecipitated from an oocyte membrane fraction with anti-Flag antibody conjugated beads (EZView affinity gel). Figure 1a shows the Western blot of oocyte membranes (M) and immunoprecipitated Flag-Kir3.1* (IP) probed with an anti-Flag antibody. The signal increase in the immunoprecipitated fraction is indicative of enrichment of Flag-Kir3.1*. Next, we proceeded with mass spectrometric analysis of the full-length Kir3.1*. The Flag-tagged Kir3.1* expressed in oocytes and isolated by immunoprecipitation was resolved on an SDS-PAGE gel (Fig. 1b). Several bands of molecular weight higher than 56 kDa associated with putative Kir3.1 were observed on both the Western blot (Fig. 1a) and the SDS-PAGE gel (Fig. 1b). These low-mobility bands are expected to be glycosylated forms of the Kir3.1 channel [15, 34]. Bands corresponding to the putative glycosylated forms and the non-glycosylated form of Kir3.1 (lowest, darkest band) were excised from the gel, pooled together, and in-gel-digested with modified TPCK-treated trypsin. Tryptic peptides were extracted from the gel pieces, desalted as described in the “Experimental procedures”, and then subjected to mass spectrometric analysis by MALDI-TOF (MALDI-TOF/MS). The most abundant peaks present in the mass spectrum in Fig. 1c correspond to tryptic peptides from Flag-Kir3.1*. Other peaks mapped to tryptic peptides derived from immunoglobulin, keratin, and auto-digested trypsin. The same mixture of tryptic peptides from Flag-Kir3.1* was also analyzed by HPLC-MS/MS using an electrospray ionization (ESI) ion trap mass spectrometer (LCQ, Thermo). ESI data included several peptides not detected by MALDI-TOF/MS (data not shown). The combined sequence coverage of Kir3.1 from both MALDI and ESI data was 38% of the entire sequence, or 47% of the cytosolic domain of the channel, a region that accounts for 49% of the serine and threonine amino acid residues in the cytosolic domain of Kir3.1. Peptides from the transmembrane region of Kir3.1* were not detected by either technique.
Fig. 1.
Mass spectrometry analysis of Kir3.1 using an in-gel trypsin digestion approach. a Anti-Flag Western blot of membrane (M) and EZView anti-Flag affinity gel precipitated (IP) fractions of Flag-tagged Kir3.1* expressed in Xenopus oocytes. b Colloidal stain of SDS-PAGE of precipitated Flag-Kir3.1*. Flag-Kir3.1* expressed in Xenopus oocytes was precipitated with EZView Anti-Flag affinity gel precipitated from membrane extracts of approximately 500 oocytes and subjected to SDS-PAGE. Protein gel bands marked on the right of the panel corresponding to Flag-Kir3.1* were excised for in-gel trypsin digestion. c MALDI-TOF mass spectrum of Flag-Kir3.1* after in-gel trypsin digestion taken in positive linear mode. Mass peaks are labeled according to the region of Kir3 subunit or other protein spanned by the corresponding peptide. Ox designates oxidized methionine. All mass values were within 0.5 Da of the predicted values from the protein sequence
Addition of the mass of a phosphate group, an 80-Da positive shift in a mass peak, would indicate phosphorylation of a peptide. We proceeded to test whether treatment with the PKA activator Forskolin or the inhibitor H-89 would result in the appropriate mass shifts in the Kir3.1 peptides compared to the non-phosphorylated untreated controls. Even though our results demonstrated that immunoprecipitation of Flag-Kir3.1* from Xenopus oocytes yielded sufficient amount of protein for mass spectrometry analysis, we were unable to observe a mass shift when comparing MALDI-TOF spectra from untreated, Forskolin-, or H89-treated groups in the full-length Kir3.1 channel protein.
In vitro phosphorylation screened by MALDI
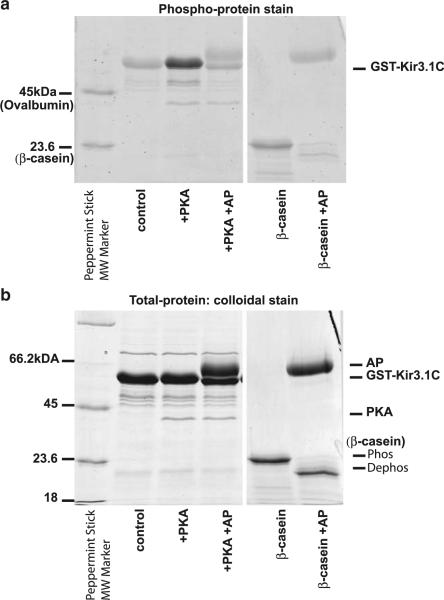
The C-terminal cytosolic domain of Kir3 harbors sites critical for interaction of the channel with PIP2, Na+, and Gβγ [19]. Interestingly, studies examining regulation of Kir3 activity by phosphorylation identified on the C terminus of the channel residues critical for physiological effects of phosphorylation [21, 22, 24]. In order to determine whether the Kir3.1 C terminus could behave as a substrate for PKA phosphorylation, we subjected the purified GST tagged C-terminal cytosolic domain of Kir3.1 (GST-Kir3.1C) to an in vitro phosphorylation assay. To phosphorylate GST-Kir3.1C, we incubated it with cPKA, and in order to dephosphorylate, we followed the cPKA treatment with AP. We also treated the constitutively phosphorylated β-casein with AP as a positive control of dephosphorylation (Fig. 2a). Next, we subjected the GST-Kir3.1C and β-casein samples to SDS-PAGE electrophoresis and stained the gel with phosphoprotein stain, which detects phosphorylated serines, threonines, and tyrosines. The intensity of the GST-Kir3.1C band under phospho-stain conditions was greater in the cPKA-treated sample compared to either the untreated (control) or the sample treated with cPKA followed by AP (Fig. 2a). When the same gel was re-stained for total protein with colloidal stain, the bands associated with GST-Kir3.1C showed the same intensity (Fig. 2b). These results indicate that greater intensity in the phospho-stain signal of the cPKA-treated GST-Kir3.1C was not due to greater amount of total protein but rather to PKA-mediated phosphorylation of the GST-Kir3.1 C terminus. In a similar assay, GST alone was not phosphorylated by PKA [21]; thus, our results suggest that the C terminus of Kir3.1 is a substrate for PKA.
Fig. 2.
In vitro phosphorylation of the GST-tagged Kir3.1 C terminus. Purified GST-Kir3.1C was phosphorylated by treatment with cPKA or dephosphorylated following cPKA treatment with AP. The untreated (control), cPKA-, and the (cPKA + AP)-treated samples were loaded on SDS-PAGE. Phosphoprotein β-casein untreated and treated with AP served as control for AP activity and phosphoprotein detection by the stain. Peppermint Stick molecular weight standards included phosphoproteins β-casein and ovalbumin. a SDS-PAGE stained with Pro-Q Diamond Phosphoprotein gel stain. b SDS-PAGE gel in a was washed and stained for total protein with Novex Colloidal Blue stain (Invitrogen). Samples loaded in each lane are shown below the gel in each panel. Bands associated with AP, cPKA, GST-Kir3.1C as well as phosphorylated and AP dephosphorylated β-casein are marked on the right of the panel
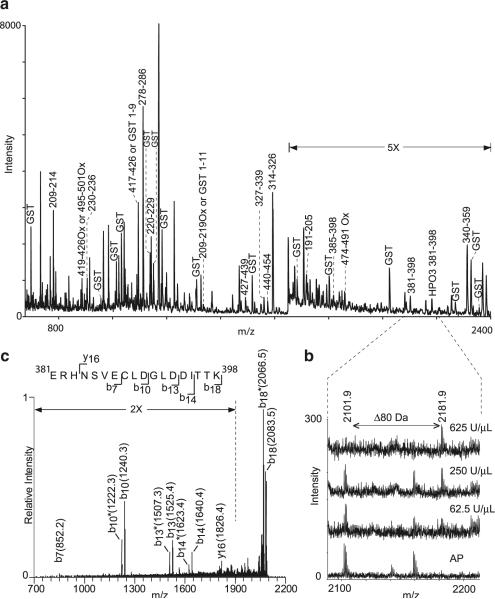
Next, we set out to determine the residues phosphorylated by an in vitro PKA reaction using mass spectrometry. GST-Kir3.1C was treated in separate experiments with AP or 62.5, 250, and 625 U/μl cPKA. In order to facilitate interpretation of the mass spectra, we aimed to minimize peptide contributions from cPKA, AP, and other contaminants present in either of these two enzyme preparations. Thus, after treatment with cPKA or AP, we affinity-purified GST-Kir3.1C using glutathione sepharose. Following elution, AP- or cPKA-treated GST-Kir3.1C was digested with trypsin and subjected to MALDI-TOF/MS. Mass peaks in the cPKA-treated samples were inspected for shifts of 80.0 Da relative to their counterparts in the AP-treated samples. The spectrum of 250 U/μl cPKA-treated GST-Kir3.1C is shown on Fig. 3a. A single peak with unmodified monoisotopic mass of 2,101.9 Da was observed to shift to 2,181.9 Da in the spectra of cPKA-treated samples (Fig. 3a, b). Figure 3b shows spectra from AP- and three cPKA-treated samples focusing on the region containing the two peaks of interest. The ratio between the 2,181.9- and the 2,101.9-Da peaks increased with increasing cPKA concentration, indicating that cPKA treatment results in the conversion of the lighter peptide into its 80 Da heavier counterpart. The mass peak at 2,101.9 Da corresponds to the theoretical mass of Kir3.1 peptide, which spans residues 381–398.
Fig. 3.
MALDI-TOF identification of possible phosphorylated peptides. Mass spectra, taken in positive reflectron mode, of cPKA- or AP-treated GST-Kir3.1C. a MALDI-TOF spectrum of GST-Kir3.1 treated with 250 U/μl cPKA is shown. Mass peaks corresponding to Kir3.1 C terminus tryptic peptides are labeled with the Kir3.1 subunit sequence residue numbers; tryptic peptides corresponding to GST are labeled. Ox oxidized methionine. All masses were within 0.05 Da of the predicted values from the protein sequence. b The selected region shown is between 2,075 and 2,225 m/z of the mass spectra from samples treated with alkaline phosphatase or 62.5, 250, and 625 U/μL cPKA The peptide peak at mass 2101.9 Da corresponding to residues 381–398 of Kir3.1 in the AP-treated sample was not observed in the 625 U/μl PKA treated sample. Treatment with 250 and 62.5 U/μl resulted in spectra containing both mass peaks. Monoisotopic m/z values are represented as M + H. c MS2 spectrum of 2,101.9-Da tryptic peptide. The singly charged species m/z 2101.9 from the AP-treated GST-Kir3.1C sample was fragmented with 35% relative collision energy (Thermo's nomenclature). Resulting yn+ and bn+ ions are noted on the sequence of the peptide corresponding to residues 381–398 of Kir3.1. Asterisk, neutral loss
We have confirmed the identity of the 2,101.9-Da peak as Kir3.1 peptide spanning residues 381–398 using tandem MS (MSn) analysis. The y and b series ions resulting from fragmentation of the 2,101.9-Da peptide are shown on the MS2 product ion scan (Fig. 3c). However, residues 381–398 contain one serine and two threonine residues either of which can be phosphorylated. Thus, we set out to determine whether the mass shift from 2,101.9 to 2,181.9, initially detected in the MALDI-TOF mass spectrum, was in fact due to the addition of a phosphate group and to determine which of the three candidate residues was modified.
Identification of Kir3.1 in vitro phosphorylation site using tandem MS
In order to address these questions, we subjected the GST-Kir3.1C samples treated with AP or varying cPKA amounts to MSn analysis. Phosphate groups on threonines and serines are more labile during fragmentation than the unmodified amino acids. The phosphate group is usually lost upon fragmentation as a neutral molecule of phosphoric acid (H3PO4) through a β elimination process [5, 23], which results in a mass decrease of 98 Da. Since the original amino acid was modified by a phosphate group, or a mass increase of 80 Da, after elimination of a phosphoric acid unit, the previously modified amino acid has its mass decreased by 18 Da. We subjected the AP- and cPKA-treated GST-Kir3.1C samples to a comprehensive neutral loss scan set to detect the loss of 98Da in search of potential phosphorylated peptides. Our results showed that there was no difference in the iongram between a blank run and AP-treated GST-Kir3.1C. However, cPKA-treated samples produced a few neutral loss peaks with intensities significantly above the noise threshold (data not shown). Since loss of the phosphoric acid unit is more prevalent than fragmentation along the peptide backbone, this peak dominates the MS2 spectrum and very little information can be gathered from MS2 experiments apart from the neutral loss itself. Therefore, we subsequently submitted the precursor ion of each putative candidate for phosphorylation to a further fragmentation stage or MS3.
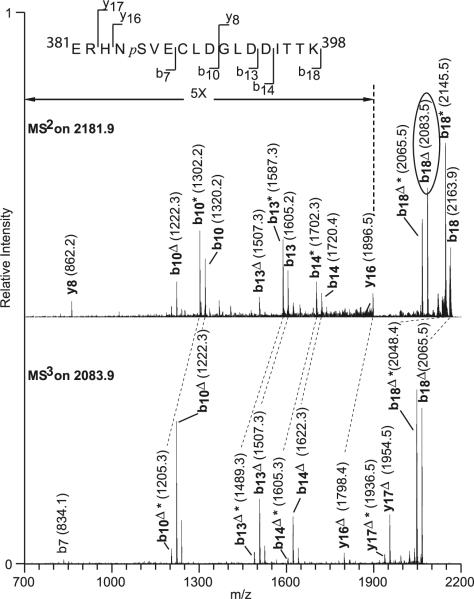
Figure 4 shows the sequence of the tryptic peptide 381–398 (top), the MS2 spectrum of the peptide before (middle), and the MS3 spectrum of the peptide after (bottom) the loss of a phosphoric acid unit from the peptide backbone, which produces a serine or threonine residue that is deficient in 18 mass units. The spectra show b and y ions mostly. In the MS2 spectra (middle), masses of b18+, y17+, and y16+ fragments that contained all three possible phosphorylation sites were all shifted by 80 Da compared to the theoretical mass of these peptides without any modifications. Interestingly, the peptides that showed the 80-Da shift, b7+, b10+, b13+, and b14+, contain only the serine 385 residue. In contrast, y8+ that contains only the threonine residues (Thr396 and Thr397) did not exhibit an 80-Da shift. Inspection of the fragments from the peptide after the neutral loss of phosphoric acid (2,083.5 Da) on the MS3 spectrum (bottom) shows that all observed b ions, b7+ through b18+, are mass-deficient in 18 units. This indicates that the elimination of the phosphoric acid unit occurred in a residue prior to the seventh amino acid, which eliminates the possibility that one of the threonine resides at positions 16 and 17 could have been modified. In addition, ions from the y series show a mass defect of 18 units for ions y16+ and y17+. This places the previously modified amino acid residue between positions 5 and 7, which includes a single serine residue. Thus, MS3 allowed us to unequivocally identify serine 385 as an in vitro PKA phosphorylation site. We also analyzed other peaks that appeared in the comprehensive neutral loss scan, but found that those peaks either produced signals below the detection levels in MS3 scans or were different enzyme cleavage products of the protein sequence around region 383–398.
Fig. 4.
Tandem MS identifies S385 as a PKA phosphorylated residue. MS2 spectrum of 2,180.9-Da tryptic peptide fragments from the 625 U/μl PKA-treated GST-Kir3.1C. Fragmentation was carried out with 35% relative collision energy (Thermo's nomenclature). Subsequently, the phosphoric acid-depleted precursor ion b18Δ at m/z 2,083.9 (in oval) was selected for another fragmentation stage (MS3) using the same amount of collision energy. Ions from the b and y series are depicted indicating that the phosphorylated site is at serine 385. Dashed line indicates peak mass difference corresponding to loss of H3PO4; delta, loss of H3PO4 (98 Da); asterisk, neutral loss
Role of serine 385 on PKA regulation of Kir3.1 activity
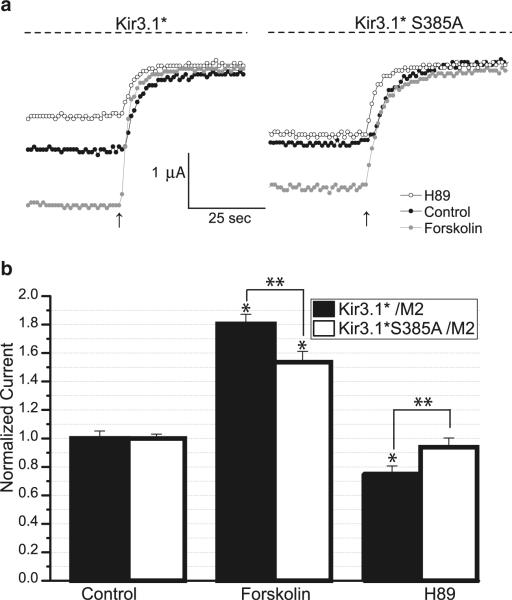
We have previously shown that treatment with the PKA inhibitor H89 reduced Kir3.1/3.4 heterotetrameric currents, while treatment with Forskolin, which stimulates adenylate cyclase, activated Kir3.1/3.4 currents [21]. Thus, we set out to determine whether serine 385 is involved in modulation of physiological function of Kir3.1 channel by PKA. We co-expressed Kir3.1* together with the M2 receptor [21] in Xenopus oocytes and recorded whole-cell currents using two-electrode voltage clamp from untreated oocytes (control) or oocytes pretreated with H89 or Forskolin. Figure 5a shows representative traces of whole-cell Kir3.1* currents as the membrane was held at −80 mV and perfused with a HK solution followed by 3 mM barium solution (arrow) to inhibit inwardly rectifying potassium current. The Kir3.1* current was calculated by subtracting the current remaining after barium inhibition from that in the presence of HK. The Kir3.1* channel in the untreated oocytes exhibited currents averaging 1.5 μA. In oocytes treated with H89, the average Kir3.1* current was 1.1 μA, while in the Forskolin-treated group, the current was 2.8 μA. The summary of several batches of oocytes, each normalized to Kir3.1* current from the untreated group, shows that H89 treatment reduced Kir3.1* current to 75% of control (Fig. 5b). In contrast, Forskolin treatment increased Kir3.1 current to 180% of control. Thus, as shown previously for the Kir3.1/Kir3.4 heterotetramer, H89 inhibited and Forskolin potentiated Kir3.1* currents, suggesting that PKA is involved in the regulation of the homomeric channel as well. Next, we co-expressed Kir3.1* S385A together with hM2 and recorded currents from untreated, H89-, and Forskolin-treated groups (Fig. 5a). In the untreated group, Kir3.1* S385A exhibited currents of 1.4 μA similar to that from Kir3.1*. In the H89-treated group, Kir3.1*S385A average current was 1.3 μA, which was not significantly different from the untreated group. Finally, in the Forskolin-treated group, the S385A mutant exhibited an average of 2.1 μA of current. After H89 treatment, Kir3.1* S385A current levels were not significantly different from control (Fig. 5b). Forskolin treatment increased the S385A mutant current to 140% of control, which, like Kir3.1*, was significantly different compared from control.
Fig. 5.
Effect of the S385A mutation on Kir3.1* currents. a Overlay of sample traces of two-electrode voltage clamp recording from untreated (black circles), 50 μM H89 (open circles), and 50 μM Forskolin (gray circles) oocytes expressing M2 and either Kir3.1* or Kir3.1*S385A. Membrane voltage was held at −80 mV. HK solution was applied to elicit and HK containing 3 mM BaCl2 was applied to inhibit the inwardly rectifying Kir3 currents. Arrow indicates start of BaCl2 perfusion. b Barium-sensitive currents from Forskolin and H89-treated groups expressing Kir3.1* (black bars) were normalized to the average of currents from the untreated group. Barium-sensitive currents from Forskolin and H89-treated groups expressing Kir3.1* S385A (open bars) were normalized to the average of currents from the untreated group. The bar graph represents summarized data from five batches, three to six oocytes for each group. Asterisk indicates statistically significant difference from currents in the untreated group as assessed by Student's t test (p<0.05), double asterisk indicates statistically significant difference between the indicated groups (p=0.05)
Thus, unlike Kir3.1*, the H89-mediated inhibition of PKA did not result in a significant current decrease of Kir3.1* S385A channel. Additionally, comparison of H89 current inhibition of Kir3.1* and Kir3.1*S385A showed a significant difference. Although Forskolin-induced current potentiation occurred in both Kir3.1* and Kir3.1* S385A, the Forskolin-mediated current of the wild type was significantly higher than that of the mutant.
Discussion
In this study, we employed mass spectrometric techniques aiming to identify and characterize phosphorylation sites for both the in vivo expressed full-length and for the purified C-terminal cytosolic domain of the Kir3.1 channel. We explicitly identified a novel PKA phosphorylation site at position Ser385 on the Kir3.1 channel C terminus and examined the effects of the S385A mutation on the channel's response to PKA. We report that S385 is an important PKA regulatory site that modulates channel activity.
We first aimed to establish that mass spectrometric analysis can be used to detect the full-length Kir3.1 channel expressed in a system in which its phosphorylation levels could be manipulated. Our previous study by Lopes et al. [21] described effects on the activity of the Kir3.1/3.4 heteromeric channel expressed in Xenopus oocytes as a function of PKA stimulation. Here, we focused on Kir3.1 homomers, since sites necessary for PKA-mediated effect on the heteromeric channel were found to reside on Kir3.1 [21] but not Kir3.4 [24]. We expressed and precipitated Flag-tagged Kir3.1* from Xenopus oocyte membranes for subsequent mass spectrometric analysis. Using MALDI-TOF mass spectrometry, we succeeded in detecting 47% of the sequence within the cytosolic domains of Kir3.1*. Absence of tryptic peptides from the transmembrane domains might be attributed to a reduced access to trypsin sites protected by tighter inter- and intra-subunit interactions or inefficient elution from the desalting media due to the hydrophobicity of this region. However, since protein kinase access may be limited to the cytosolic domains of Kir3.1, the poor coverage of the transmembrane domains was not viewed as critical for identification of PKA phosphorylation sites. Although we were able to detect approximately half of serines and threonines of the cytosolic domains of the channel, we could not detect phosphorylation sites, underscoring the need to optimize the coverage of these domains beyond 47%. Mass spectrometry has been successfully used to identify phosphorylated residues on channels phosphorylated at a high level, such as Kv2.1 and Kv1.2 channels [35, 53]. However, a lower degree of phosphorylation compared to Kv channels could influence detection of Kir3.1 phosphorylation using mass spectrometry. Phospho-stain comparing the in vitro PKA phosphorylated and AP dephosphorylated purified cytosolic domain of Kir3.1 shows a clear difference in signal. However, unlike Kv2.1 and Kv1.2 channels, which exhibited ~30-kDA mobility shift on SDS-PAGE as a result of AP treatment suggesting extensive degree of in vivo phosphorylation [26], we did not observe similar large mobility shifts between the cPKA- and the AP-treated Kir3.1 cytosolic domain. Thus, although metabolic labeling with P32 of the Kir3.1 channel expressed in HEK cells has demonstrated that this channel is in vivo phosphorylated [24], the phosphorylation levels could be low.
We proceeded by focusing on in vitro phosphorylation of the C terminus of Kir3.1. The C termini of Kir3 channels contain sites of interaction with several molecules that modulate channel activity [19]. Moreover, Kir3.1 is unique among other Kir3 subunits not only because it co-assembles with all other Kir3 subunit to augment their activity [3] but also in that it possesses a longer C terminus (~80 amino acids) that is in part responsible for the stimulating heteromeric activity by potentially serving as a target for regulation distinct from other Kir3 subunits [4]. Use of the purified GST-tagged Kir3.1 C terminus ensured that sample amounts would not be a limiting factor and enabled us to carry out trypsin digestion in solution. A GST protein tag allowed us to separate the Kir3.1 C terminus from cPKA, AP, or other contaminants following the in vitro reaction, simplifying interpretation of the mass spectra. Subjecting GST-Kir3.1 C terminus to MALDI-TOF, we succeeded in detecting 51% of Kir3.1 C terminus sequence. By comparing MALDI-TOF mass spectra of the Kir3.1 C terminus treated with cPKA and AP, we identified a potential phosphopeptide with an 80-Da shift upon cPKA treatment, characteristic of a single phosphate addition. Using tandem MS, we confirmed the sequence of the peptide and that the 80-Da shift was indeed due to an addition of a phosphate revealing its position on serine 385. We tested the functional significance of this in vitro phosphorylated residue in an in vivo system under conditions that either stimulated or inhibited PKA. Mutation of serine 385 to alanine significantly reduced the sensitivity of Kir3.1* channel currents to PKA activity. Our data strongly suggest that S385 phosphorylation contributes significantly to the PKA-dependent effects on Kir3.1 channel.
In a recent study, we showed that mutation of Ser 221 or 315 of Kir3.1 abolished the effect of PKA inhibitor H89 on Kir3.1/3.4 heterotetramer [21]. Serine 385, Serine 401 and Threonine 407 have been identified to be a crucial PKA phosphorylation site of GIRK1 by the Schreibmayer laboratory (in vitro and functionally) [49]. The corresponding residues in Kir1.1 had been previously shown to play an important role in the PKA effects in that channel [16]. Medina et al. [24] showed that in the heteromeric Kir3.1/3.4 channel, the Kir3.1 but not the Kir3.4 subunit was phosphorylated both in vivo and in vitro. Additionally, the region between 373 and 419 of Kir3.1 was found to be critical for channel phosphorylation, and the C-terminal region beyond residue 373 was critical for PP2A regulation of Kir3.1/Kir3.4 heteromers. Yet, identification of critical residues within the 373–419 region remained elusive. Our study identifies S385 as an important contributor to the PKA-dependent stimulation of Kir3.1 currents.
We have previously shown that PKA-dependent phosphorylation increased the apparent affinity of Kir3 channels to PIP2 [21]. Moreover, Liou et al. [16] showed that the Kir1.1 residues corresponding to Kir3.1(S221) and Kir3.1 (S315) controlled a similar sensitivity of Kir1.1 to PIP2. Several crystal structures capturing the cytosolic domains of Kir3.1 alone [32, 37] or as a part of a full-length chimeric channel with a bacterial Kir transmembrane domain [31] have been solved. Interestingly, the S221 and S315 residues of Kir3.1 are part of consensus sites involving Arg residues that are responsible for interactions of Kir3 with PIP2 [19]. Although Ser385 is not resolved in any of the available structures, in order to gain insight into the structural basis of the effect of phosphorylation at this site, it will be important to explore whether it is one of PKA phosphorylated sites that affects PIP2 sensitivity [13, 21] or other PIP2-independent inter- or intra-molecular interactions.
Acknowledgments
We thank members of the Logothetis Lab for helpful discussions. We thank Heikki Vaananen and Vasileios Petrou for the preparation of oocytes. This work was supported by a National Institute of Health grant (HL-54185) to D. E. L., National Institute of Health grant (CA88325) and resource grant (RR017802) to R.W., and by National Institutes of Health grant (RR00862) to Brian T. Chait, Head of the Laboratory for Mass Spectrometry and Gaseous Ion Chemistry at the Rockefeller University. D.E.L. is an Established Investigator of the American Heart Association.
References
- 1.Alekseev AE, Hodgson DM, Karger AB, Park S, Zingman LV, Terzic A. ATP-sensitive K+ channel channel/enzyme multimer: metabolic gating in the heart. J Mol Cell Cardiol. 2005;38:895–905. doi: 10.1016/j.yjmcc.2005.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brasaemle DL, Dolios G, Shapiro L, Wang R. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J Biol Chem. 2004;279:46835–46842. doi: 10.1074/jbc.M409340200. [DOI] [PubMed] [Google Scholar]
- 3.Chan KW, Sui JL, Vivaudou M, Logothetis DE. Control of channel activity through a unique amino acid residue of a G protein-gated inwardly rectifying K+ channel subunit. Proc Natl Acad Sci U S A. 1996;93:14193–14198. doi: 10.1073/pnas.93.24.14193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan KW, Sui JL, Vivaudou M, Logothetis DE. Specific regions of heteromeric subunits involved in enhancement of G protein-gated K+ channel activity. J Biol Chem. 1997;272:6548–6555. doi: 10.1074/jbc.272.10.6548. [DOI] [PubMed] [Google Scholar]
- 5.DeGnore JP, Qin J. Fragmentation of phosphopeptides in an ion trap mass spectrometer. J Am Soc Mass Spectrom. 1998;9:1175–1188. doi: 10.1016/S1044-0305(98)00088-9. [DOI] [PubMed] [Google Scholar]
- 6.Fakler B, Brandle U, Glowatzki E, Zenner HP, Ruppersberg JP. Kir2.1 inward rectifier K+ channels are regulated independently by protein kinases and ATP hydrolysis. Neuron. 1994;13:1413–1420. doi: 10.1016/0896-6273(94)90426-x. [DOI] [PubMed] [Google Scholar]
- 7.He C, Zhang H, Mirshahi T, Logothetis DE. Identification of a potassium channel site that interacts with G protein betagamma subunits to mediate agonist-induced signaling. J Biol Chem. 1999;274:12517–12524. doi: 10.1074/jbc.274.18.12517. [DOI] [PubMed] [Google Scholar]
- 8.Hill JJ, Peralta EG. Inhibition of a Gi-activated potassium channel (GIRK1/4) by the Gq-coupled m1 muscarinic acetylcholine receptor. J Biol Chem. 2001;276:5505–5510. doi: 10.1074/jbc.M008213200. [DOI] [PubMed] [Google Scholar]
- 9.Ho IH, Murrell-Lagnado RD. Molecular mechanism for sodium-dependent activation of G protein-gated K+ channels. J Physiol 520 Pt. 1999;3:645–651. doi: 10.1111/j.1469-7793.1999.00645.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang CL, Feng S, Hilgemann DW. Direct activation of inward rectifier potassium channels by PIP2 and its stabilization by Gbetagamma. Nature. 1998;391:803–806. doi: 10.1038/35882. [DOI] [PubMed] [Google Scholar]
- 11.Hunter AP, Games DE. Chromatographic and mass spectrometric methods for the identification of phosphorylation sites in phosphoproteins. Rapid Commun Mass Spectrom. 1994;8:559–570. doi: 10.1002/rcm.1290080713. [DOI] [PubMed] [Google Scholar]
- 12.Karle CA, Zitron E, Zhang W, Wendt-Nordahl G, Kathofer S, Thomas D, Gut B, Scholz E, Vahl CF, Katus HA, Kiehn J. Human cardiac inwardly-rectifying K+ channel Kir(2.1b) is inhibited by direct protein kinase C-dependent regulation in human isolated cardiomyocytes and in an expression system. Circulation. 2002;106:1493–1499. doi: 10.1161/01.cir.0000029747.53262.5c. [DOI] [PubMed] [Google Scholar]
- 13.Keselman I, Fribourg M, Felsenfeld DP, Logothetis DE. Mechanism of PLC-mediated Kir3 current inhibition. Channels. 2007;1:113–123. doi: 10.4161/chan.4321. [DOI] [PubMed] [Google Scholar]
- 14.Kiesecker C, Zitron E, Scherer D, Lueck S, Bloehs R, Scholz EP, Pirot M, Kathofer S, Thomas D, Kreye VA, Kiehn J, Borst MM, Katus HA, Schoels W, Karle CA. Regulation of cardiac inwardly rectifying potassium current IK1 and Kir2.× channels by endothelin-1. J Mol Med. 2006;84:46–56. doi: 10.1007/s00109-005-0707-8. [DOI] [PubMed] [Google Scholar]
- 15.Krapivinsky G, Gordon EA, Wickman K, Velimirovic B, Krapivinsky L, Clapham DE. The G-protein-gated atrial K+ channel IKACh is a heteromultimer of two inwardly rectifying K(+)-channel proteins. Nature. 1995;374:135–141. doi: 10.1038/374135a0. [DOI] [PubMed] [Google Scholar]
- 16.Liou HH, Zhou SS, Huang CL. Regulation of ROMK1 channel by protein kinase A via a phosphatidylinositol 4,5-bisphosphate-dependent mechanism. Proc Natl Acad Sci U S A. 1999;96:5820–5825. doi: 10.1073/pnas.96.10.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Logothetis DE, Kammen BF, Lindpaintner K, Bisbas D, Nadal Ginard B. Gating charge differences between two voltage-gated K+ channels are due to the specific charge content of their respective S4 regions. Neuron. 1993;10:1121–1129. doi: 10.1016/0896-6273(93)90060-5. [DOI] [PubMed] [Google Scholar]
- 18.Logothetis DE, Kurachi Y, Galper J, Neer EJ, Clapham DE. The beta gamma subunits of GTP-binding proteins activate the muscarinic K+ channel in heart. Nature. 1987;325:321–326. doi: 10.1038/325321a0. [DOI] [PubMed] [Google Scholar]
- 19.Logothetis DE, Lupyan D, Rosenhouse-Dantsker A. Diverse Kir modulators act in close proximity to residues implicated in phosphoinositide binding. J Physiol. 2007;582:953–965. doi: 10.1113/jphysiol.2007.133157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Logothetis DE, Movahedi S, Satler C, Lindpaintner K, Nadal-Ginard B. Incremental reductions of positive charge within the S4 region of a voltage-gated K+ channel result in corresponding decreases in gating charge. Neuron. 1992;8:531–540. doi: 10.1016/0896-6273(92)90281-h. [DOI] [PubMed] [Google Scholar]
- 21.Lopes CMB, Remon JI, Matavel A, Sui JL, Keselman I, Medei E, Shen Y, Rosenhouse-Dantsker A, Logothetis DE. Protein kinase A modulates PLC-dependent regulation and PIP2-sensitivity of K+ channels. Channels. 2007;1:124–134. doi: 10.4161/chan.4322. [DOI] [PubMed] [Google Scholar]
- 22.Mao J, Wang X, Chen F, Wang R, Rojas A, Shi Y, Piao H, Jiang C. Molecular basis for the inhibition of G protein-coupled inward rectifier K(+) channels by protein kinase C. Proc Natl Acad Sci U S A. 2004;101:1087–1092. doi: 10.1073/pnas.0304827101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLachlin DT, Chait BT. Analysis of phosphorylated proteins and peptides by mass spectrometry. Curr Opin Chem Biol. 2001;5:591–602. doi: 10.1016/s1367-5931(00)00250-7. [DOI] [PubMed] [Google Scholar]
- 24.Medina I, Krapivinsky G, Arnold S, Kovoor P, Krapivinsky L, Clapham DE. A switch mechanism for G beta gamma activation of I(KACh) J Biol Chem. 2000;275:29709–29716. doi: 10.1074/jbc.M004989200. [DOI] [PubMed] [Google Scholar]
- 25.Mirshahi T, Robillard L, Zhang HL, Hebert TE, Logothetis DE. G beta residues that do not interact with G alpha underlie agonist-independent activity of K+ channels. J Biol Chem. 2002;277:7348–7355. doi: 10.1074/jbc.M109999200. [DOI] [PubMed] [Google Scholar]
- 26.Misonou H, Mohapatra DP, Park EW, Leung V, Zhen D, Misonou K, Anderson AE, Trimmer JS. Regulation of ion channel localization and phosphorylation by neuronal activity. Nat Neurosci. 2004;7:711–718. doi: 10.1038/nn1260. [DOI] [PubMed] [Google Scholar]
- 27.Mullner C, Vorobiov D, Bera AK, Uezono Y, Yakubovich D, Frohnwieser-Steinecker B, Dascal N, Schreibmayer W. Heterologous facilitation of G protein-activated K+ channels by beta-adrenergic stimulation via cAMP-dependent protein kinase. Journal of General Physiology. 2000;115:547–557. doi: 10.1085/jgp.115.5.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mullner C, Yakubovich D, Dessauer CW, Platzer D, Schreibmayer W. Single channel analysis of the regulation of GIRK1/GIRK4 channels by protein phosphorylation. Biophys J. 2003;84:1399–1409. doi: 10.1016/S0006-3495(03)74954-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neville DCA, Rozanas CR, Price EM, Gruis DB, Verkman AS, Townsend RR. Evidence for phosphorylation of serine 753 in CFTR using a novel metal-ion affinity resin and matrix-assisted laser desorption mass spectrometry. Protein Sci. 1997;6:2436–2445. doi: 10.1002/pro.5560061117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikolov EN, Ivanova-Nikolova TT. Coordination of membrane excitability through a GIRK1 signaling complex in the atria. J Biol Chem. 2004;279:23630–23636. doi: 10.1074/jbc.M312861200. [DOI] [PubMed] [Google Scholar]
- 31.Nishida M, Cadene M, Chait BT, MacKinnon R. Crystal structure of a Kir3.1-prokaryotic Kir channel chimera. EMBO J. 2007;26:4005–4015. doi: 10.1038/sj.emboj.7601828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishida M, MacKinnon R. Structural basis of inward rectification: cytoplasmic pore of the G protein-gated inward rectifier GIRK1 at 1.8 A resolution. Cell. 2002;111:957–965. doi: 10.1016/s0092-8674(02)01227-8. [DOI] [PubMed] [Google Scholar]
- 33.Noma A, Difrancesco D, Trautwein W. Dose–response curve for acetylcholine (Ach) and carbamylcholine (Cch) in rabbit Sa-node. Pflugers Archiv–Eur J Physiol. 1978;377:R4. [Google Scholar]
- 34.Pabon A, Chan KW, Sui JL, Wu X, Logothetis DE, Thornhill WB. Glycosylation of GIRK1 at Asn119 and ROMK1 at Asn117 has different consequences in potassium channel function. J Biol Chem. 2000;275:30677–30682. doi: 10.1074/jbc.M005338200. [DOI] [PubMed] [Google Scholar]
- 35.Park KS, Mohapatra DP, Misonou H, Trimmer JS. Graded regulation of the Kv2.1 potassium channel by variable phosphorylation. Science. 2006;313:976–979. doi: 10.1126/science.1124254. [DOI] [PubMed] [Google Scholar]
- 36.Pearson WL, Dourado M, Schreiber M, Salkoff L, Nichols CG. Expression of a functional Kir4 family inward rectifier K+ channel from a gene cloned from mouse liver. J Physiol. 1999;514(Pt 3):639–653. doi: 10.1111/j.1469-7793.1999.639ad.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pegan S, Arrabit C, Zhou W, Kwiatkowski W, Collins A, Slesinger PA, Choe S. Cytoplasmic domain structures of Kir2.1 and Kir3.1 show sites for modulating gating and rectification. Nat Neurosci. 2005;8:279–287. doi: 10.1038/nn1411. [DOI] [PubMed] [Google Scholar]
- 38.Petit-Jacques J, Sui JL, Logothetis DE. Synergistic activation of G protein-gated inwardly rectifying potassium channels by the betagamma subunits of G proteins and Na(+) and Mg(2+) ions. J Gen Physiol. 1999;114:673–684. doi: 10.1085/jgp.114.5.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rojas A, Cui N, Su J, Yang L, Muhumuza JP, Jiang C. Protein kinase C dependent inhibition of the heteromeric Kir4.1–Kir5.1 channel. Biochim Biophys Acta. 2007;1768:2030–2042. doi: 10.1016/j.bbamem.2007.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenhouse-Dantsker A, Sui JL, Zhao Q, Rusinova R, Rodriguez-Menchaca AA, Zhang Z, Logothetis DE. A sodium-mediated structural switch that controls the sensitivity of Kir channels to PtdIns(4,5)P(2) Nat Chem Biol. 2008;4:624–631. doi: 10.1038/nchembio.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schlosser A, Pipkorn R, Bossemeyer D, Lehmann WD. Analysis of protein phosphorylation by a combination of elastase digestion and neutral loss tandem mass spectrometry. Anal Chem. 2001;73:170–176. doi: 10.1021/ac000826j. [DOI] [PubMed] [Google Scholar]
- 42.Schreibmayer W, Lester HA, Dascal N. Voltage clamping of Xenopus laevis oocytes utilizing agarose-cushion electrodes. Pflugers Archiv–Eur J Physiol. 1994;426:453–458. doi: 10.1007/BF00388310. [DOI] [PubMed] [Google Scholar]
- 43.Soejima M, Noma A. Mode of regulation of the ACh-sensitive K-channel by the muscarinic receptor in rabbit atrial cells. Pflugers Arch. 1984;400:424–431. doi: 10.1007/BF00587544. [DOI] [PubMed] [Google Scholar]
- 44.Stuhmer W. Electrophysiological recording from Xenopus oocytes. Methods Enzymol. 1992;207:319–339. doi: 10.1016/0076-6879(92)07021-f. [DOI] [PubMed] [Google Scholar]
- 45.Sui JL, Chan KW, Logothetis DE. Na+ activation of the muscarinic K+ channel by a G-protein-independent mechanism. J Gen Physiol. 1996;108:381–391. doi: 10.1085/jgp.108.5.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sui JL, Petit-Jacques J, Logothetis DE. Activation of the atrial KACh channel by the betagamma subunits of G proteins or intracellular Na+ ions depends on the presence of phosphatidylinositol phosphates. Proc Natl Acad Sci U S A. 1998;95:1307–1312. doi: 10.1073/pnas.95.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Surti TS, Huang L, Jan YN, Jan LY, Cooper EC. Identification by mass spectrometry and functional characterization of two phosphorylation sites of KCNQ2/KCNQ3 channels. Proc Natl Acad Sci U S A. 2005;102:17828–17833. doi: 10.1073/pnas.0509122102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trautwein W, Dudel J. Zum Mechanismus der Membranwirkung des Acetylcholin An der Herzmuskelfaser. Pflugers Archiv fur Die Gesamte Physiologie des Menschen und der Tiere. 1958;266:324–334. doi: 10.1007/BF00416781. [DOI] [PubMed] [Google Scholar]
- 49.Treiber F, Rosker C, Fritz R, Mullner C, Steinecker B, Schreibmayer WF. 2148-Pos on the role of PKA phosphorylation of GIRK1 and GIRK4 subunits in G-protein activation as well as in trafficking of the channel complex. Biophys J. 2008;94:2148. [Google Scholar]
- 50.Vivaudou M, Chan KW, Sui JL, Jan LY, Reuveny E, Logothetis DE. Probing the G-protein regulation of GIRK1 and GIRK4, the two subunits of the KACh channel, using functional homomeric mutants. J Biol Chem. 1997;272:31553–31560. doi: 10.1074/jbc.272.50.31553. [DOI] [PubMed] [Google Scholar]
- 51.Wischmeyer E, Karschin A. Receptor stimulation causes slow inhibition of IRK1 inwardly rectifying K+ channels by direct protein kinase A-mediated phosphorylation. Proc Natl Acad Sci U S A. 1996;93:5819–5823. doi: 10.1073/pnas.93.12.5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yan J, Olsen JV, Park KS, Li W, Bildl W, Schulte U, Aldrich RW, Fakler B, Trimmer JS. Profiling the phospho-status of the BKCa channel {alpha} subunit in rat brain reveals unexpected patterns and complexity. Mol Cell Proteomics. 2008;7:2188–2198. doi: 10.1074/mcp.M800063-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang JW, Vacher H, Park KS, Clark E, Trimmer JS. Trafficking-dependent phosphorylation of Kv1.2 regulates voltage-gated potassium channel cell surface expression. Proc Natl Acad Sci U S A. 2007;104:20055–20060. doi: 10.1073/pnas.0708574104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang H, He C, Yan X, Mirshahi T, Logothetis DE. Activation of inwardly rectifying K+ channels by distinct PtdIns (4,5)P2 interactions. Nat Cell Biol. 1999;1:183–188. doi: 10.1038/11103. [DOI] [PubMed] [Google Scholar]
- 55.Zhang QL, Doupnik CA. Gating properties of GIRK channels activated by receptors coupled selectively to PTX-insensitive G alpha (I/O) subunits and RGS proteins. Biophys J. 2002;82:200A. [Google Scholar]
- 56.Zhu G, Qu Z, Cui N, Jiang C. Suppression of Kir2.3 activity by protein kinase C phosphorylation of the channel protein at threonine 53. J Biol Chem. 1999;274:11643–11646. doi: 10.1074/jbc.274.17.11643. [DOI] [PubMed] [Google Scholar]
- 57.Zitron E, Kiesecker C, Luck S, Kathofer S, Thomas D, Kreye VA, Kiehn J, Katus HA, Schoels W, Karle CA. Human cardiac inwardly rectifying current IKir2.2 is upregulated by activation of protein kinase A. Cardiovasc Res. 2004;63:520–527. doi: 10.1016/j.cardiores.2004.02.015. [DOI] [PubMed] [Google Scholar]