Abstract
Uroporphyrinogen III synthase (U3S) catalyzes the asymmetrical cyclization of a linear tetrapyrrole to form the physiologically relevant uroporphyrinogen III (uro’gen III) isomer during heme biosynthesis. Here, we report four apoenzyme and one product complex crystal structures of the Thermus thermophilus (HB27) U3S protein. The overlay of eight crystallographically unique U3S molecules reveals a huge range of conformational flexibility, including a “closed” product complex. The product, uro’gen III, binds between the two domains and is held in place by a network of hydrogen bonds between the product’s side chain carboxylates and the protein’s main chain amides. Interactions of the product A and B ring carboxylate side chains with both structural domains of U3S appear to dictate the relative orientation of the domains in the closed enzyme conformation and likely remain intact during catalysis. The product C and D rings are less constrained in the structure, consistent with the conformational changes required for the catalytic cyclization with inversion of D ring orientation. A conserved tyrosine residue is potentially positioned to facilitate loss of a hydroxyl from the substrate to initiate the catalytic reaction.
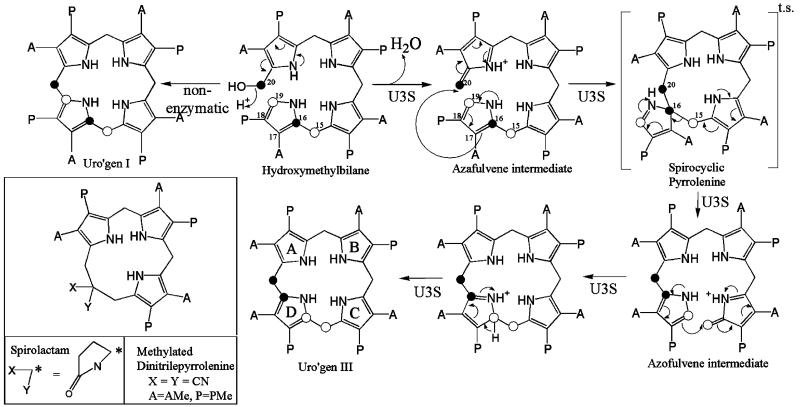
Tetrapyrrole cofactors such as heme, chlorophyll, cobalamin (vitamin B12), siroheme and coenzyme F430 are essential for oxygen transport, electron transport, photosynthesis, methionine synthesis in mammals, nitrite and sulfite assimilation, and methane production in methanogens. Tetrapyrrole cofactors share a multistep, branched biosynthetic pathway (1, 2). In mammals, the initial steps include the reaction of glycine and succinyl-CoA to produce 5-aminole-vulinic acid (ALA1), condensation of two ALA molecules to form the basic pyrrole structure (porphobilinogen, PBG), and assembly of four PBG pyrroles into a linear tetraypyrrole (hydroxymethylbilane, HMB). Subsequently, uroporphyrinogen III synthase (U3S; E.C. 4.2.1.75) cyclizes HMB to produce uroporphyinogen III (uro’gen III), the last common precursor of all tetrapyrrole cofactors. Remarkably, U3S catalyzed cyclization occurs with inversion of the fourth pyrrole ring (D ring) to form the III isomer of uroporphyrinogen. As illustrated in Figure 1, the U3S catalyzed reaction is believed to proceed via loss of substrate hydroxyl to form an initial azafulvene intermediate, which proceeds to a spiro-pyrrolenine transition state in which carbons 16 and 20 form a covalent bond. A second azafulvene intermediate is resolved by bond formation between substrate carbons 19 and 15 to produce the uro’gen III product (3-5). In the scheme, the D ring of the proposed spiro-pyrolenine transition state is excluded from the primary ring by connection of the A–D and C–D bridge carbon atoms through a single carbon atom of the D ring, C16. In support of this mechanism, a spirolactam derivative of the proposed transition state (Figure 1) has been shown to competitively inhibit the enzyme (6).
Figure 1.
The proposed mechanism of U3S catalyzed formation of uro’gen III (3). This figure resembles that of Roessner et al. (33). Spirolactam, a transition state mimic, has been shown to be a competitive inhibitor of U3S (36), and the structure of a synthetic precursor, methylated dinitrilepyrrolenine, has been determined (6). A = CH2CO2, P = CH2CH2CO2.
Defects in tetrapyrrole biosynthetic enzymes are associated with human disease, and mutations in U3S cause congenital erythropoietic porphyria (CEP), which is transmitted as an autosomal recessive trait (7, 8). In the absence of U3S activity, HMB autocyclizes to uroporphyrinogen I, which is released from cells and is fully oxidized to uroporphyrin I, where it reaches elevated levels in the urine and defines the biochemical manifestation of the disease. The severe phenotype associated with CEP was one of the first disorders of porphyrin biosynthesis described by Günther in 1911, and is often referred to as Günthers disease (9). The disorder has been described in multiple ethnic groups with approximately 200 cases reported worldwide. The developing erythroblast is the primary site for the overproduction of porphyrins and fluorescence of the marrow is a uniform finding. The defective erythroblasts are fragile and prone to peripheral hemolysis leading to varying degrees of anemia and splenomegaly. Homozygotes or compound heterozygotes with a phenotype of CEP generally have less than 10% of normal U3S activity; however, patients with mild signs of CEP may have up to 20% of normal activity (10, 11). Severe photosensitivity is common in patients, with skin fragility and cutaneous infections leading to photomutilation (12). The majority of known point mutations are expected to reduce enzyme activity by interfering with protein stability, such as by disrupting the packing of the hydrophobic core or by exposing hydrophobic residues to the solvent (13).
The current level of structural understanding of the U3S mechanism is provided by crystal structures and mutagenic/biochemical analysis of the human enzyme (13), and by two Thermus thermophilus (strain HB8) structures that were recently deposited in the Protein Data Base (14) (PDB codes 1WCW and 1WD7) by the RIKEN Structural Genomics/Proteomics Initiative (15). The U3S structure comprises two α/β domains that each contain a central 5-stranded β-sheet flanked by α-helices; although the two domains possess similar folds, they differ in the position and angles of the helices. The two strands connecting the domains, which are presumably flexible in solution, adopt an extended β-sheet conformation in the human U3S crystal structure and display a range of less regular conformations in the T. thermophilus structures. Consistent with these structures, it has been proposed that the active site lies between the domains and that a significant conformational change might occur upon substrate binding (13).
In an effort to better understand the mechanism of U3S we set out to obtain a ligand protein complex. This effort failed using the human enzyme, but we did succeed in crystallizing and determining the structure of T. thermophilus (strain HB27) U3S (TtU3S) in complex with the reaction product, uro’gen III. In addition, we determined additional structures of apo T. thermophilus U3S, which collectively display an impressive (90°) range of interdomain angles. The uro’gen III product is seen to bind between the two domains, and specific interaction of the ligand carboxylate groups suggests that the defined closed conformation is formed in both substrate and product complexes. There is a notable lack of interactions between the enzyme and the product’s pyrrole rings, and the dominant use of enzyme main chain groups for hydrogen bonding interactions is consistent with the apparent lack of conserved active site residues (13). The precise coordination of A and B pyrrole carboxylate groups and solvent exposure of the D ring carboxylates is consistent with the model that the enzyme functions by restricting substrate conformations to productive orientations while allowing the D ring to rotate.
MATERIAL AND METHODS
The gene encoding the T. thermophilus (strain HB27) U3S (TtU3S) was obtained from a genomic DNA library (ATCC) and cloned into pET151/D-TOPO for expression with an N-terminal His-tag followed by a TEV protease cleavage site immediately preceding the full open reading frame. Recombinant protein expression in BL21(DE3)pLysS codon plus cells occurred by autoinduction (16). Protein was extracted from clarified whole cell lysate and purified over a Ni2+ affinity column (Qiagen). TEV cleavage was performed at room temperature overnight during dialysis into 20 mM Tris-HCl, 100 mM NaCl, pH 7.0 (all pH values were determined at room temperature, and all chromatography columns were run at 4 °C). Following size exclusion chromatography on a S200 column (Pharmacia) equilibrated in the same buffer supplemented with 1 mM DTT, the purified protein was concentrated to 8–12 mg · mL−1 for crystallization trials.
Four crystal structures were obtained: three apo (TtU3S1, TtU3S2, TtU3S3) and one product complex. The crystals typically grew in clusters that required manipulation to extract single crystals for data collection. All crystals were grown by vapor diffusion in sitting drop trays by mixing 2 μL of protein with 2 μL of precipitant solution and equilibrating the drop over 1 mL of precipitant solution. Small apo crystal forms grew at 4 °C within 1–7 days over the following reservoir solutions: TtU3S1, 1.2 M NaPO4, 0.8 M K2PO4 (~20 μm × 50 μm × 100 μm; deltoid shape); TtU3S2, 21–24% PEG MW 8000, 0.2 M Mg acetate, 0.1 M MES-HCl, pH 6.5, 0.2 M NaCl (100 μm × 200 μm; rectangular plate clusters, very thin in the third dimension); TtU3S3, 18–24% PEG MW 8000, 0.1 M Tris-HCl, pH 8.5, 0.2 mM MgCl2 (30 μm × 30 μm × 100 μm; rod clusters).
Uro’gen III was produced by chemical reduction of uroporphyrin III (Frontier Scientific, Logan, UT) using a method developed by H. Bergoniam and J.D.P. (in preparation). Briefly, 2 mg of uroporphyrin III was resuspended in 200 μL of H2O and 1,800 μL of methanol. The mixture was added to 5 mg of 10% palladium on activated carbon (PdC, Aldrich, St. Louis, MO) in a hydrogen atmosphere and stirred for 40–60 min. The PdC was removed using a glass fiber filter. The filtrate was dried under argon gas at 60 °C. The reduced uro’gen III was resuspended in 100 μL of 10 mg · mL−1 TtU3S in 20 mM Tris-HCl, 100 mM NaCl, 1 mM DTT, pH 7.0 prior to setting down crystal trays. Despite multiple attempts, only one drop containing the product complex crystals was obtained. The crystals (~30 μm 30 × μm × 30 μm; cube) grew anaerobically (using degassed solutions and incubated in an anaerobic chamber) over a reservoir of 28% PEG MW8000, 0.1 M MES-HCl, pH 6.5, 0.2 M Mg acetate at 16 °C in 4 days.
All crystals were cryoprotected in a solution of the precipitant made up with 20% glycerol and cryocooled by plunging into liquid nitrogen prior to data collection on beamlines X25 and X29 at the National Synchrotron Light Source, Brookhaven National Laboratory. Data were processed using the HKL suite of programs (17). Molecular replacement was performed with PHASER (18) using the T. thermophilus (strain HB8) U3S structure deposited to the PDB by the RIKEN Structural Genomics/Proteomics Initiative (pdb code 1WD7) (15). Solutions for all structures were only found when the two α/β domains of U3S were separated into independent search models. The β-strand linker was rebuilt in all structures using O (19), and final modeling and structure validation were performed using the program COOT (20). The uro’gen III ligand and corresponding refinement constraints were built using the SKETCHER module of CCP4 (21). All refinements, including 10 segment TLS(22) refinements, were performed using REFMAC (23,24). Crystallographic statistics are given in Tables 1 and 2.
Table 1.
Data Collection Statistics
| TtU3S1 | TtU3S2 | TtU3S3 | product complex |
|
|---|---|---|---|---|
| space group | P31 | P212121 | P21 | P21 |
| cell dimens |
a = b = 63.83 c = 58.58 |
a = 40.86 b = 63.24 c = 90.68 |
a = 40.75 b = 89.96 c = 74.98 β = 101.32 |
a = 35.74 b = 66.48 c = 55.46 β = 91.76 |
| no. of obs refs | 147186 | 75146 | 244799 | 73535 |
| no. of unique refs | 38923 | 14308 | 65660 | 19239 |
| high res (Å) | 2.07–2.0 | 2.12–2.0 | 1.66–1.60 | 1.97–1.90 |
| completeness (%) | 99.9 (100) | 99.(100) | 94.2(59.8) | 94.4(66.7) |
| Rsyma (%) | 8.6 (42.5) | 9.1(49.5) | 4.1 (35.1) | 7.5(54.8) |
| av I/σ(I) | 39.5 (7) | 18.4(3.3) | 30.3(2.2) | 31.7(2.3) |
| mosaicity (deg) | 0.2 | 0.47 | 0.35 | 0.73 |
Rsym = Σ|I − 〈I〉|/ΣI, where 〈I〉 is the average intensity from multiple observations of symmetry related reflections.
Table 2.
Refinement Statistics
| TtU3S1 | TtU3S2 | TtU3S3 | uro’gen III complex |
|
|---|---|---|---|---|
| resolution range (Å) | 30.0–2.0 | 30–2.0 | 30–1.6 | 30.0–1.9 |
| no. of protein atoms | 2005 | 1944 | 4055 | 1950 |
| no. of solvent molecules | 225 | 115 | 466 | 124 |
| Nn. of ligand molecules | 4 SO4 | 1 GOL | 0 | 1 UR3 |
| Rfactor (%)a | 18.0 | 20.4 | 18.9 | 19.3 |
| Rfree (%) | 23.2 | 25.9 | 23.6 | 24.0 |
| rmsd (bond lengths) (Å) | 0.011 | 0.01 | 0.014 | 0.011 |
| rmsd (bond angles) (deg) 〈B〉 (Å2) |
1.24 | 1.22 | 1.41 | 1.39 |
| main chain | 18.7 | 27.3 | 21.6 | 31.3 |
| side chain | 21.9 | 29.8 | 24.9 | 34.2 |
| water molecules | 35.7 | 34.3 | 31.4 | 41.7 |
| ligands | 50.2 | 44.9 | - | 48.3 |
| no. of Φ/Ψ angles (%) most favored allowed |
94.8 5.2 |
93.8 5.8 |
94.2 5.7 |
92.8 7.2 |
Rfactor = Σ∥Fo| − |Fc∥/Σ|Fo| × 100 over 95% of the data. Rfree = same calculation on 5% data not used in refinement.
RESULTS AND DISCUSSION
Structures of T. thermophilus U3S
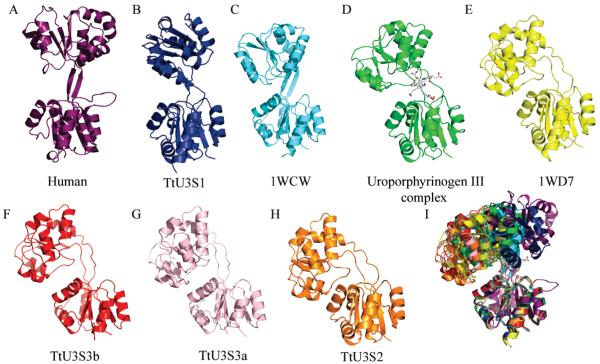
The structure of apo U3S from T. thermophilus (HB27) (TtU3S) was determined in three different crystal forms, two of which contained one molecule in the asymmetric unit and one of which contained two molecules in the asymmetric unit, to give a total four crystallographically independent views of the unliganded enzyme. A crystal structure of TtU3S in complex with uro’gen III (product) was also determined. Attempts to obtain a complex between spirolactam (a kind gift of Alan Spivey) and the human U3S failed as did attempts to form a complex between T. thermophilus U3S and PBG. Curiously, all of the T. thermophilus crystal forms grew from very similar conditions, and the drops of apo enzyme often contained more than one form. The different apo and product complex structures were refined against data collected to 1.6–2.0 Å with Rfree values of 18.0–20.4% and good geometry (Tables 1, 2).
As expected, TtU3S adopts the same overall fold as previously determined U3S structures (PDB codes 1JR2, 1WD7, and 1WCW) (13, 15). The first 8 and last 4 residues are generally disordered. Domain 1 comprises residues 8–39 and 168–260, domain 2 comprises residues 47–161, and the two domains are connected by two linker sequences comprising residues 40–46 and 162–167. The structures of domains 1 and 2 are each highly conserved between the different U3S structures. The overlap of multiple individual domains for the five unique TtU3S (HB27) molecules using the maximum likelihood approach and the program THE-SEUS (25) yields root-mean-square deviations (RMSDs) of 0.46 Å (domain 1; 123 Cα) and 0.77 Å (domain 2; 114 Cα), respectively. TtU3S (HB27) is 98% identical to TtU3S (HB8) with only 4 amino acid substitutions, all of which are within the first 10 residues. Pairwise comparisons typically display RMSDs of 0.4 Å and 0.6 Å for overlaps on domain 1 and domain 2, respectively. Despite a sequence identity between the T. thermophilus and human enzymes of only 14%, the structural alignment has a Z-score of 14.8 using the program DALI (26), and RMSDs on structurally equivalent residues of domain 1 (115/121 Cα) and domain 2 (113/127 Cα) are 2.7 Å and 2.5 Å, respectively.
The short linker segments between the two domains form an antiparallel β-ladder in the maximally extended human enzyme, but adopt less regular conformations in the TtU3S structures and are often poorly ordered in the apo structures. Linker flexibility facilitates an impressive array of relative domain orientations, and superposition of all structures on domain 1 shows a wide range of relative displacements of domain 2, including separations of up to 23.6 Å in centroid position and rotations of up to 90° (Figure 2) (LSQKAB) (27). The relative motions do not result from a single hinge point in the linker region, but reflect gradual shifts in phi/psi angles over the course of the linkers, and a wrapping of the first strand (residues 39–46) around the second (residues 162–167). The overall impression is that the linker will be highly mobile in solution with domains experiencing a wide range of relative orientations in the absence of a bound ligand.
Figure 2.
Crystal structures of U3S: (A) human (PDB code, 1JR2); (B, F, G, H) four apo T. thermophilus (HB27) U3S structures; (C, E) T. thermophilus (HB8) apo-U3S (PDB code 1WCW and 1WD7); (D) T. thermophilus (HB27) U3S bound to product, uro’gen III. (I) Overlap of all these U3S structures on domain 1 reveals substantial flexibility of the linker region and relative motion of the second domain. Structures have been positioned in the figure according to the angular displacement of the domain 2 relative to the most extended structure of human U3S.
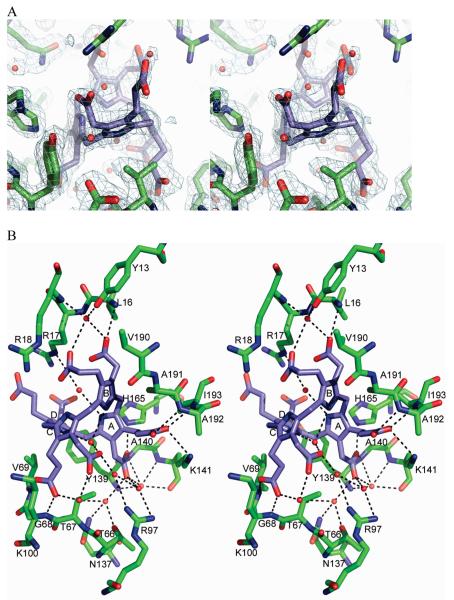
Structure of the Product Complex
The molecular replacement phased map of the cocrystallized product complex revealed strong density for the bound uro’gen III located between the two domains (Figure 3). This binding site is consistent with sequence conservation and NMR chemical shift changes observed in interface residues upon addition of HMB derivatives to U3S (13, 28). There are no major conformational changes observed within domains 1 or 2, although the two domains are much closer to each other than in any of the apo structures, consistent with the model that the flexible domains close around the bound substrate.
Figure 3.
U3S:uro’gen III product complex. (A) Final 2Fo – Fc electron density surrounding the uro’gen III ligand and nearby active site residues. (B) Stereo representation of U3S active site and bound uro’gen III. The acetate and propionate side chains of the four pyrrole rings (A, B, C and D) form hydrogen bonds to main chain amides, water molecules and two nonconserved side chains.
The product tetrapyrrole ring is largely surrounded by the enzyme (Figure 4), with a total of 688 Å2 of U3S surface area buried against the ligand. Solvent channels run through the active site, and ten water molecules form 14 interactions with the uro’gen III. Few hydrophobic contacts are made with the ligand. Val69 from domain 2 sits adjacent to the D ring pyrrole while Val190, Ala191 and Ile193 (in domain 1) pack behind the B ring. The cyclic tetrapyrrole adopts a highly puckered “two-up, two-down” conformation, where rings A and C are pointing in one direction and rings B and D are pointing in the opposite direction (Figure 3B). The pyrrole NH groups do not interact with the enzyme but instead form hydrogen bonds with ordered water molecules. This is in contrast to the structure of the following enzyme in the heme biosynthetic pathway, uroporphyrinogen decarboxylase, which binds its product with all four pyrrole NH groups pointing in one direction and coordinated by a single active site aspartate side chain (29). In the conformation seen here for TtU3S, direct interaction between the enzyme and uro’gen III NH groups is prevented by the disposition of the ligand’s carboxylate side chains on both sides of the tetrapyrrole.
Figure 4.
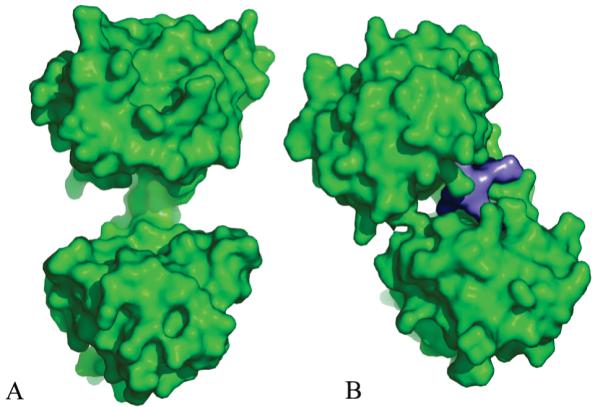
Domain closure upon ligand binding. (A) Surface representation of human apo U3S, which adopts the most open conformation observed in the various crystal structures. (B) Product bound T. thermophilus U3S showing how the domains close around the uro’gen ligand (purple). Both panels are in the same orientation as Figure 2.
Most of the direct hydrogen bonds between uro’gen III and TtU3S are between the product’s carboxylate side chains and main chain NH groups of the enzyme (Figure 3B). The protein side chains that form hydrogen bonds with the product are Lys141, His165, and Gln194, which coordinate the A ring propionate, A ring acetate, and B ring acetate, respectively. None of these residues is highly conserved, and all of their coordinated tetrapyrrole carboxylates also form hydrogen bonds with main chain amides that appear sufficient to define the uro’gen III conformation. Consistent with the failure of an earlier effort to identify critically important residues for U3S catalysis (13), these observations explain why the U3S active site does not contain many invariant amino acid side chains (Supporting Information).
The uro’gen III A ring acetate forms hydrogen bonding interactions with the main chain amides of Gly140 from domain 2 and the NE2 of linker residue His165. The A ring propionate hydrogen bonds with the main chain amides of Ala192 and Ile193, and the Lys141 NZ atom from domain 1 (Figure 3B). Because these A ring interactions involve both domains, they appear to be especially important for the induction of domain closure upon ligand binding. Binding of the B ring acetate with the Leu16 NH further constrains the uro’gen III and may explain the observation that substrate binding is abolished when the acetate and propionate groups of rings A or B are swapped (30, 31). These hydrogen bonds and the constricted environment of the A and B ring pyrroles also indicate that the first two rings of the substrate become conformationally constrained upon binding the enzyme.
The C ring propionate forms hydrogen bonds with the NH groups of Gly28 and Val69, although the C ring pyrrole binding site appears to be relatively open and the acetate does not directly contact protein. This is consistent with a model in which the C ring undergoes some limited motion during catalysis and the observation that swapping the two side chains at this site does not significantly affect catalytic activity (30, 31). The D ring carboxylates extend into solvent and do not make any interaction with the enzyme, consistent with the finding that swapping the D ring acetate and propionate side chains does not significantly alter catalytic efficiency (30, 31). This also suggests that the motions of the D ring, which has to flip during the reaction, are relatively unconstrained.
Comparison with Related U3S Enzymes
U3S is one of the most highly diverged of the heme biosynthetic enzymes, with a sequence identity of only 14% between the human and T. thermophilus enzymes. We therefore performed a structure-based sequence alignment of the human and T. thermophilus sequences and further aligned the Arabidopsis (32) and Anacystis nidulans sequences (33) on the basis of threading calculations using the program PSIPRED (34) (Supporting Information). This analysis indicates that seven residues are invariant. Three of the invariant residues are glycines, Gly22, Gly90 and Gly214 that lie on tight turns and adopt phi/psi angles disfavored for other residues. Two others, Pro29 and Ala94, lie on a strand (domain 1) and helix (domain 2), respectively, and point into hydrophobic pockets of the enzyme. Thus, five of the seven invariant residues perform structural roles. Thr217 lies on a surface loop and points its hydroxyl toward the bound product at a distance of 5.1 Å. Thr228 appears to be conserved because it contributes to catalysis; mutation of the corresponding human Thr228 to Ala resulted in the greatest activity loss of residues studied, although this residue is not essential for catalysis because the mutant protein still retained 32% of wild-type activity. The one other invariant residue, Tyr155, is discussed below.
Prediction of ligand binding by the human enzyme from the known interactions of the T. thermophilus U3S product complex is complicated by the low sequence similarity and the large-scale domain motions that must occur upon binding. In order to build a plausible model of the human enzyme–product complex, we performed independent alignments of each domain and of the linker segment of the unliganded human U3S structure onto the TtU3S complex. In the resulting model, substitution of two residues by proline eliminates main chain amides that hydrogen bond with product in the T. thermophilus U3S complex. Additionally, two residues in the human model would need to be repositioned, for example by adopting an alternative rotamer conformation, in order to avoid steric clash. Overall, the human U3S product complex model seems to be reasonable if approximate.
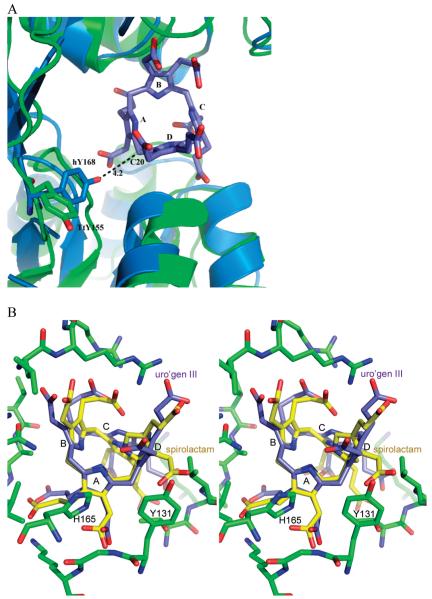
A study of the U3S protein from A. nidulans reported that mutation of Tyr166 to Phe resulted in an inactive enzyme (33). In contrast, our earlier study concluded that the equivalent Tyr168Phe mutation in human U3S only caused 50% reduction of activity (13). Concern that our assays may have been performed under conditions of limiting substrate (33) is not valid because data were taken entirely from the range in which the reaction was linear, which is only expected to occur when substrate is saturating (13). Inspection of the different U3S structures reveals that this conserved tyrosine is located on the linker connecting the two domains and adopts a variety of conformations. In the TtU3S product complex, the equivalent Tyr155 residue adopts a solvent exposed conformation and is separated from the A and D rings of the uro’gen III product by Tyr131, with the closest interatomic distance between Tyr155 and the product B ring being 7 Å (Figure 5A). Our modeling suggests that, in the context of the human U3S structure, Tyr168 might approach the uro’gen III bridge carbon, C20, and be well positioned to hydrogen bond with the tyrosine to the HMB hydroxyl (Figure 5B). In this speculative model, the putative catalytic tyrosine would contribute to loss of the substrate hydroxyl and formation of the azafulvene intermediate, as suggested for the A. nidulans enzyme (33), with the different tyrosine contexts possibly contributing to differences in enzymatic activity for U3S enzymes from the different species.
Figure 5.
Comparative modeling. (A) Overlap of the individual domains and linker region of the human U3S (cyan) on the TtU3S structure (green) reveals the alternate side chain position of a conserved tyrosine residue (TtU3S Tyr155, human Tyr168) that may function in removal of the C20 hydroxyl of the substrate HMB. (B) Stereo representation of uro’gen III and a modeled spiropyrollenine transition state in the active site of U3S. Rings A and B pack against the U3S protein, and rings C and especially D are more open to solvent. The modeled spiro-intermediate macrocycle is smaller in diameter and easily fits into the active site with minimal changes. Close approach between the D ring side chains and Val69 may determine the pucker of the intermediate.
Implications for Catalytic Mechanism
The HMB substrate autocyclizes in the absence of U3S to form the symmetrical uro’gen I isomer with a half-life of only 4 min (Figure 1) (35). U3S therefore likely functions in part by restricting HMB to conformations that avoid attack of carbon 19 on carbon 20 and formation of the I isomer, but allow attack of carbon 16 on carbon 20 and formation of the III isomer product. Unfortunately, the product complex does not provide a clear indication of how this is achieved in detail. To further investigate the mechanism we have modeled a plausible transition state based on both the uro’gen III-bound crystal structure and a crystal structure of a methylated dinitrilepyrrolenine (6) (Figure 1). This molecule is a synthetic precursor to the known competitive inhibitor spirolactam, and its crystallization demonstrated that the contracted ring structure is energetically viable (6) (Figures 1, 5B). To generate the transition state model, the dinitrilepyrrolenine was positioned in the active site by overlap of the A and B pyrroles on the product complex structure, which as discussed above are the most tightly coordinated and constrained components of the product complex. Our model depicts the methoxy side chains of the dinitrile derivative as carboxylates, and rotates the flexible side chains into a position most matching the bound uro’gen III structure (Figure 5B). The nitrile groups were replaced with a full pyrrole ring, complete with side chains, and the macro-ring pucker and orientation of the D ring carboxylates were positioned to minimize conflicts with the enzyme.
The transition state model is more compact than the product, with the distance between A and C pyrrole nitrogens reduced from 4.8 Å to 3.4 Å, and the diameter of the ring contracted from 7.0 Å to 6.2 Å when measured diagonally between bridge carbons. Despite these differences, it is trivial to accommodate all of the hydrogen bonds seen in the product complex crystal structure in the transition state complex model and the transition state model is structurally quite similar to the observed product complex structure (Figure 5B). Progression from the transition state to the second azofulvene intermediate and on to the product is expected to require easily accessible conformational shifts in the relatively unconstrained C and D pyrroles and only minor changes in the enzyme conformation.
Remaining questions of U3S mechanism include, how does the linear tetrapyrrole substrate bind, to what extent is the loss of hydroxyl facilitated by the conserved tyrosine residue in the enzyme from different species, and what conformational changes are required upon progression from substrate to the first azafulvene intermediate and on to the spiro-pyrrolenine transition state? Based upon the interactions seen in the product complex crystal structure and the known biochemistry (30, 31), we favor the model that the substrate A and B rings bind in the same manner as seen in the product complex. This suggests that the relative domain orientation is defined to a large extent by the A ring, whose side chains contact both U3S domains, and whose binding is likely to be a major trigger for domain closure and formation of the catalytic environment. Because the initial catalytic step appears to be loss of a hydroxyl from an A ring subsituent and that A ring binding likely defines the main features of the closed conformation, we speculate that large scale changes in domain orientation are not deployed during the catalytic cycle. Nevertheless, the chance of some functionally important enzyme conformational change remains a clear possibility. More complete understanding of U3S mechanism will require additional information, such as structures of substrate or intermediate complexes, perhaps reinforced by a computational analysis of possible reaction coordinates.
Supplementary Material
Footnotes
This work was supported by NIH RO1 GM56775 and P30 DK072437. Operations at the National Synchrotron Light Source (NSLS) are supported by the Department of Energy, Office of Basic Energy Sciences, and by the National Institute of Health (NIH). Data collection at the NSLS was funded by the National Center for Research Resources.
Data deposition: Model coordinates and structure factors were deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 3D8N, 3D8R, 3D8S, 3D8T).
- U3S
- uroporphyrinogen III synthase
- uro’gen III
- uroporphyrinogen III synthase
- ALA
- 5-aminolevulinic acid
- PBG
- porphobilinogen
- HMB
- hydroxymethylbilane
- RMSD
- root-mean-square deviation
- PDB
- Protein Data Bank
REFERENCES
- 1.Chadwick D, Ackrill K, editors. The Biosynthesis of the Tetrapyrrole Pigments. Vol. 180. John Wiley & Sons; New York: 1994. [Google Scholar]
- 2.Dailey H, editor. Biosynthesis of Heme and Chlorophylls. McGraw-Hill; 1990. [Google Scholar]
- 3.Mathewson J, Corwin A. Biosynthesis of pyrrole pigments: a mechanism for porphybilinogen polymerization. J. Am. Chem. Comm. 1961:1313–1315. [Google Scholar]
- 4.Battersby AR, McDonald E. Biosynthesis of porphyrins and corrins. Philos. Trans. R. Soc. London B. 1976;273:161–180. doi: 10.1098/rstb.1976.0007. [DOI] [PubMed] [Google Scholar]
- 5.Battersby AR, Fookes CJR, Matcham GWJ, McDonald E. Biosynthesis of the pigments of life: formation of the macrocycle. Nature. 1980;285:17–21. doi: 10.1038/285017a0. [DOI] [PubMed] [Google Scholar]
- 6.Stark WM, Baker MG, Raithby PR, Leeper FJ, Battersby AR. The spiro intermediate proposed for the biosynthesis of the natural porphyrins: Synthesis and properties of its macrocycle. J. Chem. Soc., Chem. Commun. 1985:1294–1296. [Google Scholar]
- 7.Anderson PC, Battersby AR, Broadbent HA, Fookes CJR, Hart GJ. Biosynthesis of porphyrins and related macrocycles. Part 17. Synthesis of modified hydroxymeth-ylbilanes and studies of their chemical and biological properties. Tetrahedron. 1986;42:3123–3135. [Google Scholar]
- 8.Romeo G, Levin EY. Uroporphyrinogen 3 cosyn-thetase in human congenital erythropoietic porphyria. Proc. Natl. Acad. Sci. U.S.A. 1969;63:856–863. doi: 10.1073/pnas.63.3.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Günther H. Die Haematoporphyria. Dtsch. Arch. Klin. Med. 1911;105:189–146. [Google Scholar]
- 10.Deybach JC, de Verneuil H, Phung N, Nordmann Y, Puissant A, Boffety B. Congenital erythropoietic porphyria (Günther’s disease): enzymatic studies on two cases of late onset. J. Lab. Clin. Med. 1981;97:551–558. [PubMed] [Google Scholar]
- 11.Warner C, Poh-Fitzpatrick M, Zaider E, Tsai S, Desnick R. Congenital erythropoietic porphyria; a mild variant with low uroporphyrin I levels due to a missense mutation (A66V) encoding residual uroporphyrinogen III synthase activity. Arch. Dermatol. 1992;128:1243–1248. doi: 10.1001/archderm.128.9.1243. [DOI] [PubMed] [Google Scholar]
- 12.Anderson KE, Sassa S, Bishop D, Desnick RJ. The metabolic basis of inherited disease. McGraw-Hill; New York: 2001. Disorders of heme biosynthesis: X-linked sideroblastic anemia and the porphyrias; pp. 2991–3062. [Google Scholar]
- 13.Mathews MAA, Schubert HL, Whitby FG, Alexander KJ, Schadick K, Bergonia HA, Phillips JD, Hill CP. Crystal structure of human uroporphyrinogen III synthase. EMBO J. 2001;20:5832–5839. doi: 10.1093/emboj/20.21.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mizohata E, Matsuura T, Sakai H, Murayama K, Terada T, Shirouzu M, Kuramitsu S, Yokoyama S. Crystal structure of uroporphyrinogen III synthase from an extremely thermophilic bacterium Thermus thermophilus HB8. RIKEN Structural Genomics/Proteomics Initiative (RSGI).; Unpublished. [Google Scholar]
- 16.Studier F. Protein production by auto-induction in high-density shaking cultures. Protein Expression Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 17.Otwinowski Z. Oscillation data reduction program. In: Sawyer L, Isaacs N, Bailey S, editors. Data Collection and Processing. SERC Daresbury Laboratory; Warrington, U.K.: 1993. pp. 56–62. [Google Scholar]
- 18.McCoy A, Grosse-Kunstleve R, C S, Read R. Likelihood-enhanced fast translation functions. Acta Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 19.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 20.Emsley P, Cowtan K. Coot: Model-Building Tools for Molecular Graphics. Acta Crystallogr., D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 21.CCP4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr., D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 22.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr., D. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 23.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr., A. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 24.Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macro-molecular structures using FFT. Acta Crystallogr., A. 1999;55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- 25.Theobald D, Wuttke D. THESEUS: Maximum likelihood superpositioning and analysis of macromolecular structures. Bioinformatics. 2006;22:2171–2172. doi: 10.1093/bioinformatics/btl332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holm L, Sander C. Dali/FSSP classification of three-dimensional protein folds. Nucleic Acids Res. 1997;25:231–234. doi: 10.1093/nar/25.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kabsch W. A solution for the best rotation to relate two sets of vectors. Acta Crystallogr., A. 1976;32:922–923. [Google Scholar]
- 28.Cunha L, Kuti M, Bishop D, Mezei M, Zeng L, Zhou M, Desnick R. Human uroporphyrinogen III synthase: NMR-based mapping of the active site. Proteins. 2007;71:855–873. doi: 10.1002/prot.21755. [DOI] [PubMed] [Google Scholar]
- 29.Phillips J, Whitby F, Kushner J, Hill C. Structural basis for tetrapyrrole coordination by uroporphyrinogen decar-doxylase. EMBO J. 2003;22:6225–6233. doi: 10.1093/emboj/cdg606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Battersby AR, Fookes CJR, Pandey PS. Linear tetrapyrrolic intermediates for biosynthesis of the natural porphyrins. Experiments with modified substrates. Tetrahedron. 1983;39:1919–1926. [Google Scholar]
- 31.Pichon C, Atshaves BP, Xue T, Stolowich NJ, Scott AI. Studies on uro’gen III synthase with modified bilanes. Bioorg. Med. Chem. 1994;4:1105–1110. [Google Scholar]
- 32.Tan F-C, Cheng Q, Saha K, Heinemann IU, Jahn M, D J, Smith AG. Identification and characterization of the Arabidopsis gene encoding the tetrapyrrole biosynthesis enzyme uroporphyrinogen III synthase. Biochem. J. 2007;410:291–299. doi: 10.1042/BJ20070770. [DOI] [PubMed] [Google Scholar]
- 33.Roessner CA, Ponnamperuma K, Scott IA. Mutagenesis identified a conserved tyrosine residue important for the activity of uroporphyrinogen III synthase from Anacystis nidulans. FEBS Lett. 2002;525:25–28. doi: 10.1016/s0014-5793(02)03056-9. [DOI] [PubMed] [Google Scholar]
- 34.Bryson K, McGuffin L, Marsden R, Ward J, Sodhi J, Jones D. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33:W36–38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Battersby AR, Fookes CJ, Gustafson-Potter KE, Matcham GWJ, McDonald E. Proof by synthesis that unrearranged hydroxymethylbilane is the product from deaminase and the substrate for cosynthase in the biosynthesis of uro’gen-III. J. Chem. Soc., Chem. Commun. 1979:1155–1158. [Google Scholar]
- 36.Stark WM, Hart GJ, Battersby AR. Synthetic studies on the proposed spiro intermediate for biosynthesis of the natural porphyrins: Inhibition of cosynthase. J. Chem. Soc., Chem. Commun. 1986:465–467. [Google Scholar]
- 37.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins D, Thompson J. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.