Abstract
Nogo, MAG, and OMgp are myelin-associated proteins that bind to a neuronal Nogo-66 receptor (NgR/NgR1) to limit axonal regeneration after central nervous system (CNS) injury. Within Nogo-A two separate domains are known interact with NgR1. NgR1 is the founding member of three-member NgR family, whereas Nogo-A (RTN4-A) belongs to a four-member reticulon family. Here, we systematically map the interactions between these superfamilies, demonstrating novel nanomolar interactions of RTN2 and RTN3 with NgR1. Since RTN3 is expressed in the spinal cord white matter it may have a role in myelin inhibition of axonal growth. Further analysis of the Nogo-A and NgR1 interactions revealed a novel third interaction site between the proteins suggesting a trivalent Nogo-A interaction with NgR1. We also confirm here that MAG binds to NgR2, but not to NgR3. Unexpectedly, we found that OMgp interacts with MAG with a higher affinity than its affinity to NgR1. To better define how these multiple structurally distinct ligands bind to NgR1, we examined a series of Ala-substituted NgR1 mutants for ligand binding activity. We found that the core of the binding domain is centered in the middle of the concave surface of NgR1 leucine-rich repeat (LRR) domain and surrounded by differentially utilized residues. This detailed knowledge of the molecular interactions between NgR1 and its ligands is imperative when assessing the options to develop NgR1-based therapeutics for CNS injuries.
When nerve fibers of the brain and spinal cord in adult mammals are severed, little to no regrowth occurs. Astroglial scar and CNS myelin pose extrinsic barriers to regeneration (1,2). From CNS myelin, at least three proteins capable of inhibiting axonal growth in vitro are recognized: Nogo-A, MAG and OMgp (1,2). Nogo-A has several domains that participate in inhibiting axon growth. The hydrophilic Nogo-66 domain flanked by two hydrophobic segments is detectable on the oligodendrocyte surface (3,4). Together these three segments form a reticulon (RTN) homology domain (RHD) of about 200 amino acid (aa), characteristic of reticulon family members (5).
Nogo-66 binding provided the basis for the identification of a receptor for Nogo-A, termed NgR or NgR1 (6). Remarkably, MAG and OMgp also bind to NgR1 to inhibit axonal growth in vitro (7–9). NgR1 is a LRR-containing GPI-anchored neuronal protein; the structure of its LRR domain has been determined (10,11).
Perturbation of Nogo function by antibodies (12–14), by peptide, or by soluble NgR1 ectodomain (15–19) leads to enhanced axonal growth, plasticity and functional recovery after spinal injury or stroke. Genetic studies of Nogo-A (20–22) and NgR1 (23,24) have, however, found less clearcut evidence of their role in axonal regeneration. It is plausible that adaptive compensation for chronic genetic loss of NgR1 or Nogo-A may in part explain this observation.
Alternatively, the less pronounced genetic versus pharmacologic phenotype might relate to redundancy amongst the myelin inhibitory proteins and their signaling pathways. NgR1 is the founding member of three-member NgR family (11,25,26); Nogo-A belongs to a four member RTN family (5). MAG and OMgp have no known paralogs. A recent report demonstrates that in vitro MAG can bind and exert its inhibitory function via NgR2 as well NgR1 (27). The ability of OMgp to bind other NgR family members has not been assessed. The functions of other RTNs are largely enigmatic (28). As other RTNs are also present in the CNS and contain homologous RHDs we hypothesized earlier that they could interact with NgR family members (25). Here we map the NgR family receptor binding properties of all RTNs, MAG and OMgp. We also demonstrate that RTN3, which we found to bind to NgR1, is expressed in spinal cord white matter.
Several topologies of Nogo-A relative to the lipid bilayer have been supported experimentally. The extended length, 35 and 36 amino acids (3), of the hydrophobic segments flanking the Nogo-66-segment suggests that these segments might not be single-pass transmembrane regions. A recent report supports the existence of a conformation in which all three of the hydrophilic segments of the reticulons are on the same side of the lipid bilayer (29). This raises the possibility that carboxyl-terminal amino acids might contribute to NgR1 binding – a hypothesis we test here.
The molecular basis for NgR1 interaction with multiple ligands has not been defined. LRR domains are commonly involved in protein-protein interactions, presumably because the non-globular extended surface of LRR domain provides ample opportunities for high affinity interactions. Here, we show that NgR1 utilizes certain residues to interact with multiple ligands in a central binding region and other surrounding residues to interact with specific ligands. This data helps us to understand the ligand-specificity of different NgR family members and contributes to the elucidation of how CNS axonal plasticity and regeneration is limited by the interaction between multiple ligands and NgR family.
EXPERIMENTAL PROCEDURES
Recombinant DNA constructs
AP-Nogo-66, AP-Y4C (human Nogo-A aa 950–1018), AP-Nogo24 (human Nogo-A aa 995–1018) AP-MAG, AP-OMGP and AP-Lingo-1 constructs were described elsewhere (6,8,9,30,31). To generate additional AP fusion proteins, DNA fragments encoding 66 amino acid sequences of reticulon 1, 2 or 3 as shown in Fig. 1W, were cloned into pAPtag5 (kindly provided by Dr. J. Flanagan, (32)) with restriction enzyme XbaI. For the AP-carboxyl-terminal NogoA construct (AP-Nogo-C39), the DNA encoding last 39 amino acids of human Nogo-A was cloned into XbaI site of pAPtag5. An additional MAG-AP construct was generated by cloning sequence encoding MAG ectodomain devoid of signal peptide into HindIII and BglII restriction sites in pAPtag5. Mouse MAG, TLR4, NgR2 and NgR3 expression constructs contain Igκ signal peptide and myc and 6xHIS tags, followed by respective open reading frames devoid of signal peptides. These inserts were cloned into XbaI restriction site of a modified pSecTag2a in which stop-codon was replaced by the XbaI site. pSecTag2a was modified by using QuikChange II Site-Directed Mutagenesis Kit (Stratagene). Myc-NgR1 expression construct has been described earlier (6). Also, an additional AP-Nogo-66 construct based on pAPtag5 plasmid backbone was used in some experiments. This was created by subcloning insert from original pcAP-5-Nogo-66 plasmid (6) into pAPtag5 vector. All constructs were sequenced to confirm that no unwanted changes had occurred.
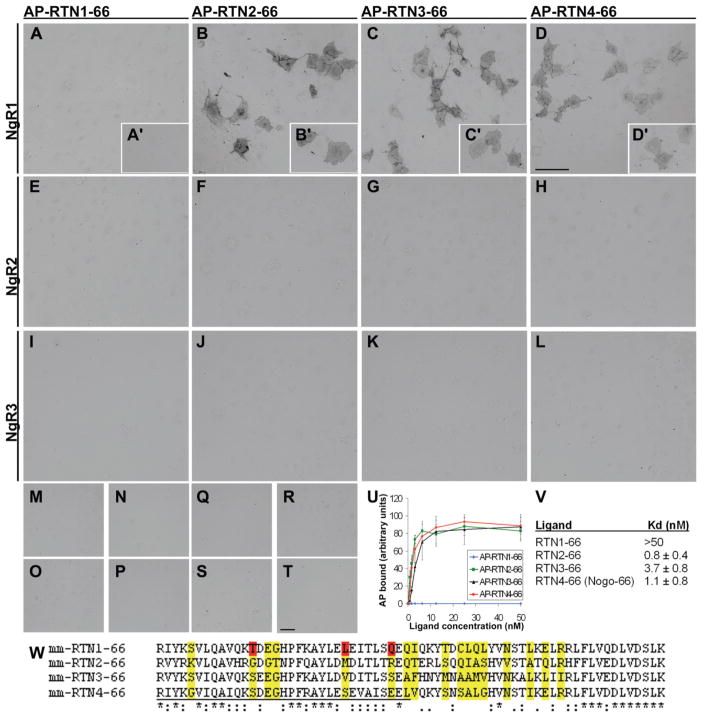
Figure 1. Analysis of interactions between reticulon family and leucine-rich repeat protein superfamily members shows that RTN2-66 and RTN3-66 interact specifically with NgR1.
RTN1-66 does not bind to NgR1 either at 6 nM (A) or at 3 nM (A′) concentration, whereas 6 nM RTN2-66 (B), RTN 3-66 (C), or RTN4-66/Nogo-66 (D) show strong binding. Similarly binding of 3 nM RTN 2-66, RTN 3-66, and RTN4-66 (B′, C′, and D′, respectively) is clearly detectable. No binding of RTNs 1-4 at 50 nM concentration to NgR2 (RTN1-4-66; Figs E-H, respectively) or to NgR3 (RTN1-4-66; Figs I-L, respectively) is detectable. None of the RTNs binds at 50 nM concentration to other tested members of the LRR superfamily Lingo-1 (RTN1-4-66: Figs M-P, respectively) or TLR4 (RTN1-4-66: Figs Q-T, respectively). (U) Determination of dissociation constant for RTN1-4 interactions with NgR1. (V) summary of obtained Kd values ±SEM for RTN1-4–NgR1 –interactions, as averaged from five experiments. (W) Sequence alignment of mouse RTN1-4 66 amino acid loop regions. Identical amino acids are indicated with asterisks; high physicochemical similarity of amino acids is indicated with colons, and similarity with dots. In RTN4, underlined amino acids 1–31 are indispensable for receptor binding. Non-conserved amino acids are highlighted in yellow, and putative residues in RTN1-66 responsible for the lost NgR1 affinity are highlighted in red. Scale bars in (D) and (T) are 100 μm.
NgR1 mutagenesis
NgR1 mutagenesis was accomplished using the Quick Change Multisite Directed Mutagenesis Kit (Stratagene). FLAG-tagged human NgR1 expression construct was used as a template. All NgR1 mutant constructs were analyzed by sequencing.
Recombinant proteins
Expression vectrors encoding AP-fusion proteins were transfected into HEK293T cells and conditioned media were collected after 5–7 days. In some cases conditioned media was concentrated using Amicon Ultra centrifugal filtration devices (Millipore). 6xHIS-tag containing AP-MAG, MAG-AP and AP-OMgp were purified from conditioned media using Ni-NTA affinity resin according to manufacturer’s guidelines (Qiagen).
COS-7 ligand binding assay
COS-7 binding assays were done as described earlier (6). Conditioned media containing AP-fused ligands or purified ligands were incubated with COS-7 cells transfected with indicated constructs for 1–2 hours at room temperature before washing and fixation. Bound AP was visualized by BCIP/NBT reaction.
Immunocytochemistry
Live-cell immuno-staining was performed by incubating cells in HBSS with 0.05% BSA and AP-conjugated anti-myc (9E10, Sigma) antibodies (dilution 1:200) or anti-NgR1 antibody (6) at room temperature or on ice for one hour followed by washing, fixing. Then cells were incubated for 1.5 hour in 65°C. Finally, bound antibodies were visualized by BCIP/NBT reaction.
In situ hybridization
In situ hybridization was performed as described (25). RTN-probes decribed earlier (25) were designed to recognize the splice-variants containing sequence that encodes 66 amino acid loop region.
RESULTS
NgR1 is a high-affinity receptor for several reticulon family members
We prepared amino-terminal AP-fusion proteins of all RTN 66-loop regions, and analyzed their binding properties to NgR family members (Fig. 1). We found that in addition to Nogo-66, RTN2-66 and RTN3-66 domains interact with NgR1, but not with other NgR family members or other related members of the type I transmembrane LRR protein superfamily tested (Fig. 1). The affinities of RTN2-66, RTN3-66, and Nogo-66 to NgR1 appear similar, with Kd-values for RTN2-66 and Nogo-66 around 1 nM and for RTN3-66 around 4 nM (Fig. 1V). RTN1-66 shows no affinity to NgR1 even at highest concentrations tested (50 nM).
Rtn3 and Nogo-A are expressed in the same glial cell population of spinal cord white matter
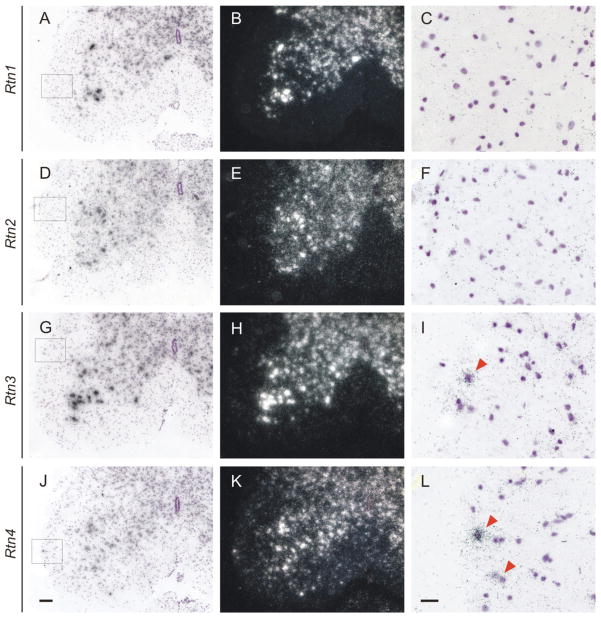
Earlier reports showed Nogo-A expression in CNS oligodendrocytes. However, the possible expression of other reticulons in spinal cord white matter has not been analyzed. We analyzed the expression of all reticulon mRNAs in adult mouse lumbar spinal cord by in situ hybridization. Consistent with earlier studies (25) we found that all reticulon mRNAs are expressed in neurons (Fig. 2). Interestingly, we found prominent expression of Rtn3 and Rtn4 mRNA transcripts in the white matter/lateral funiculus (Fig. 2; arrowheads). Nissl counterstaining of the sections enables large and weakly stained nuclei (neurons) to be distinguished from small and strongly stained nuclei (glia). We noted that glial cell nuclei of the stained cryosections display dichotomy in size distribution, and that Nogo-A and Rtn3 mRNAs are expressed by the same cell population characterized by larger and more weakly stained nuclei than other glial cells. The probes used in these experiments were designed to cover the conserved RHD regions and are relatively homologous: in the most homologous 203 nt sequence stretch Rtn3 and Nogo-A probes are 73% identical, but contain no identical nt stretches longer than 11 nt. Importantly, in the Purkinje cell layer of the cerebellum strong Nogo-A mRNA expression was observed, but no or low lever expression of Rtn3 mRNA was detected, thus confirming the specificity of the hybridization reaction (Supplementary Fig. 1). No signal was detected with control sense-probes (data not shown).
Figure 2. Rtn3, and Rtn4 (Nogo-A), but not Rtn1 or Rtn2, mRNAs are expressed in the white matter of the adult mouse spinal cord.
We analyzed by in situ hybridization the localization of the Rtn1 (A–C), Rtn2 (D–F), Rtn3 (G–I), and Rtn4 (J–L), mRNAs in adult mouse lumbal spinal cord. Prominent expression of all four transcripts was observed in neuronal cells in the gray matter. Furthermore Rtn3 and Rtn4 mRNAs were detected in non-neuronal cells (boxed areas). (Figs C, F, I, and L) Higher magnification images of boxed areas show prominent Rtn3 and Rtn4 signals, and that the signals overlap with non-neuronal cells with relatively larger nuclei (red arrowheads). Scale bar in (J) is 100 μm for lower and in (L) 20 μm for higher magnification images.
Nogo-A interaction with NgR1 involves three segments of Nogo-A
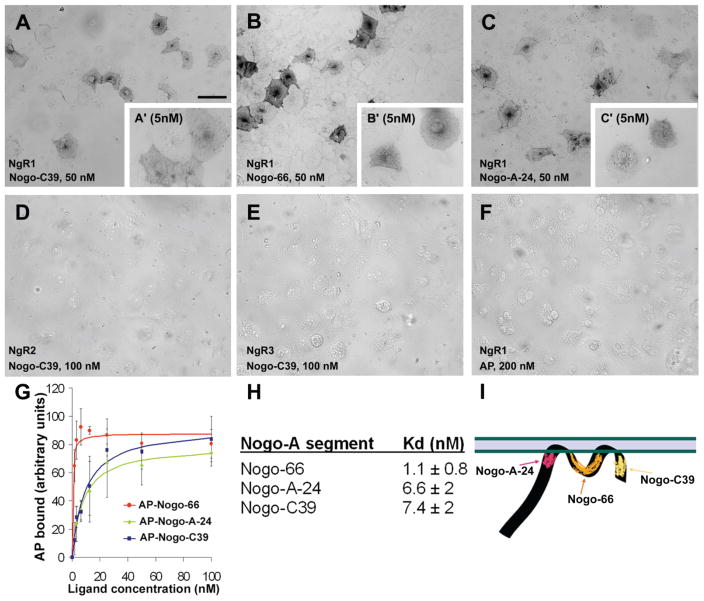
We produced an AP-fusion protein of the carboxy-terrminal 39 residues of Nogo-A (AP-Nogo-C39) and measured its affinity to NgR family members. We found that it interacts with high affinity and specificity only with NgR1 (Fig. 3). The NgR1 binding affinity (Kd) of AP-Nogo-C39 was determined to be nearly same as that of AP-Nogo-A-24 fragment; AP-Nogo-66, however, showed the highest affinity to NgR1. We also reconsidered the Kd value of Nogo-66–NgR1 interaction. The AP-Nogo-66 protein we used in earlier studies was found to have been proteolytically cleaved to a significant degree between the AP-moiety and Nogo66 region. Re-subcloning of the same insert into pAPtag5 plasmid serendipitously led to a formation of stable recombinant protein (Supplementary Fig. 2). This allowed us to determine Nogo-66 affinity for NgR1 more accurately than in previous studies and suggests that partial degradation of ligand is likely to have resulted in an underestimation of Nogo-66 affinity to NgR1. We conclude that Nogo-66 binds to NgR1 with Kd of 1 nM.
Figure 3. Carboxy-terminal segment of Nogo-A (Nogo-C39) binds to NgR1, but not to other NgR family members.
50 nM (A) and 5 nM (A′) Nogo-C39 (A); 50 nM (B) and 5 nM (B′) Nogo-66; and 50 nM (C) and 5 nM (C′) Nogo-A-24 fragments all bind to NgR1 with high affinity. No interaction of Nogo-C39 with NgR2 (D) or with NgR3 (E) is detected. AP-protein alone shows no affinity to NgR1 even at high (200 nM) concentration (F). (G) Determination of dissociation constant for interactions between different Nogo-A fragments and NgR1. Data is averaged from three experiments. (H) Summary of obtained Kd values ±SEM for interactions. (I) Schematic drawing (amino-terminus is not in scale) highlighting the different NgR1-binding segments of Nogo-A. Scale bar in (A) is 100 μm.
Carboxyl-terminal fragment of Nogo-A interacts exclusively with the LRR domain of NgR1
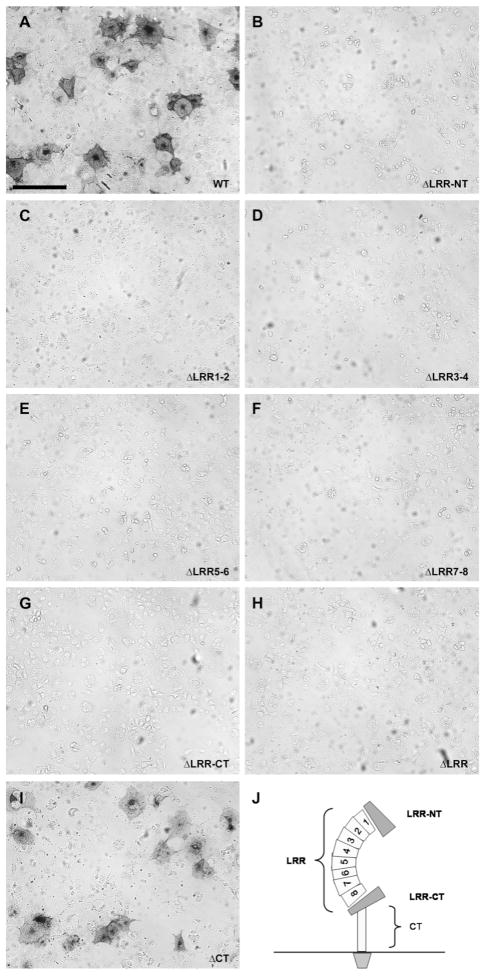
We mapped the interaction site of Nogo-C39 in NgR1 by using a series of deletion mutants lacking pairs of LRR or other structural elements in NgR1, as described in (33). The LRR domain of NgR1 is indispensable for Nogo-C39 binding (Fig. 4).
Figure 4. Leucine-rich repeat domain of NgR1 is necessary for the binding of carboxyl-terminal segment of Nogo-A.
Detected binding of Nogo-C39 fragment (at 50 nM concentration) to full-length NgR1 (A), deletion mutants lacking indicated LRR units (C–H), adjacent cysteine-rich flanking regions (B, G), or juxtamembrane stalk-region (“CT”) (I). Different structural units are indicated in (J). All NgR1 deletion constructs were detected on the plasma membrane by live-cell staining with anti-FLAG antibody (data not shown and (33)). Scale bar in (A) is 100 μm.
MAG binds to NgR1 and NgR2 with moderate affinity and shows no affinity for NgR3
Venkatesh et al. (27) reported that MAG-Fc interacts with NgR2 and that NgR2 mediates MAG-dependent growth inhibitory signaling other than NgR1-mediated signalling. Previously we failed to observe AP-MAG fusion protein binding to human NgR2 (11). As Venkatesh et al. also failed to see AP-MAG–NgR2 -interaction it was proposed that AP tagging of MAG sterically interferes with the binding to NgR2, but not to NgR1 (27). Since then we have identified a non-conservative nucleotide variant in the PCR-derived NgR2 expression construct used in our earlier study. When using wild-type NgR2 expression plasmid, we found that AP-MAG binds with similar affinity to NgR2 and NgR1 (Fig. 5). Due to higher expression level of the transfected myc-NgR2 construct, we observed higher maximal binding of AP-MAG to NgR2 than to NgR1 expressing cells. However, as analyzed by Schatchard analysis, the dissociation constants for these interactions are essentially identical, using these constructs. MAG has no detectable affinity for NgR3. Binding experiments using MAG-AP resulted in similar results (data not shown)
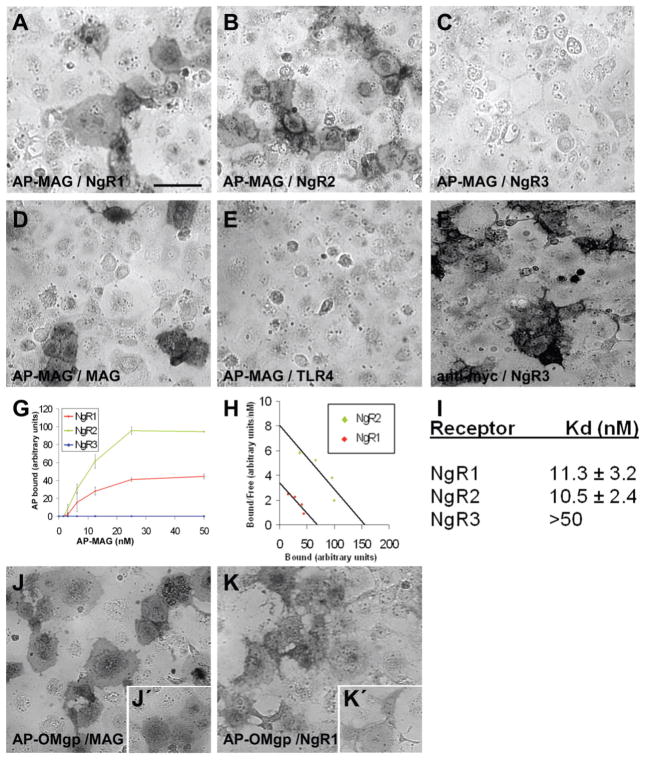
Figure 5. Interactions between MAG, OMgp and NgR family members.
15 nM MAG binds avidly to NgR1 (A) and NgR2 (B), but not to NgR3 (C), or to another member of LRR superfamily TLR4 (E). Homophilic binding of AP-MAG to MAG expressed on the cell surface was also detected (D). Cell-surface expression of myc-NgR3 was confirmed by live-cell staining with anti-myc antibody (F). (G) Analysis of MAG binding to NgRs as a function of ligand concentration. Data is averaged from four experiments. (H) Determination of Kd for MAG interaction with NgR1 and NgR2 by Scatchard analysis. (I) Summary of obtained Kd values ±SEM for MAG–NgRs -interactions. OMgp binds MAG ((J), 25 nM AP-OMgp and (J′), 12 nM AP-OMgp binding to MAG) with higher affinity than NgR1 ((K), 25 nM AP-OMgp and (K′), 12 nM AP-OMgp binding to NgR1). Scale bar in (A) is 100 μm.
OMgp binds MAG with very high affinity, NgR1 with moderate affinity, and does not interact with NgR2 or NgR3
Earlier OMgp was identified as a myelin inhibitory molecule interacting with NgR1 (9). To study if OMgp could also interact with NgR2 or NgR3, we prepared AP-OMgp recombinant protein and assessed its affinity to NgR family members. As reported earlier, we found that OMgp interacts with NgR1. Unexpectedly, among other conditions tested, we found that OMgp interacts with MAG and that this interaction has significantly higher affinity than the binding affinity of OMpg–NgR1 –interaction. The Kd for OMgp–MAG -interaction is 3–6 nM and 10–20 nM for OMgp–NgR1 -interaction (Fig. 5J-K and data not shown). We did not detect OMgp binding to NgR2 or NgR3 (data not shown).
A library of NgR1 mutants is expressed in a similar fashion than wild-type NgR1
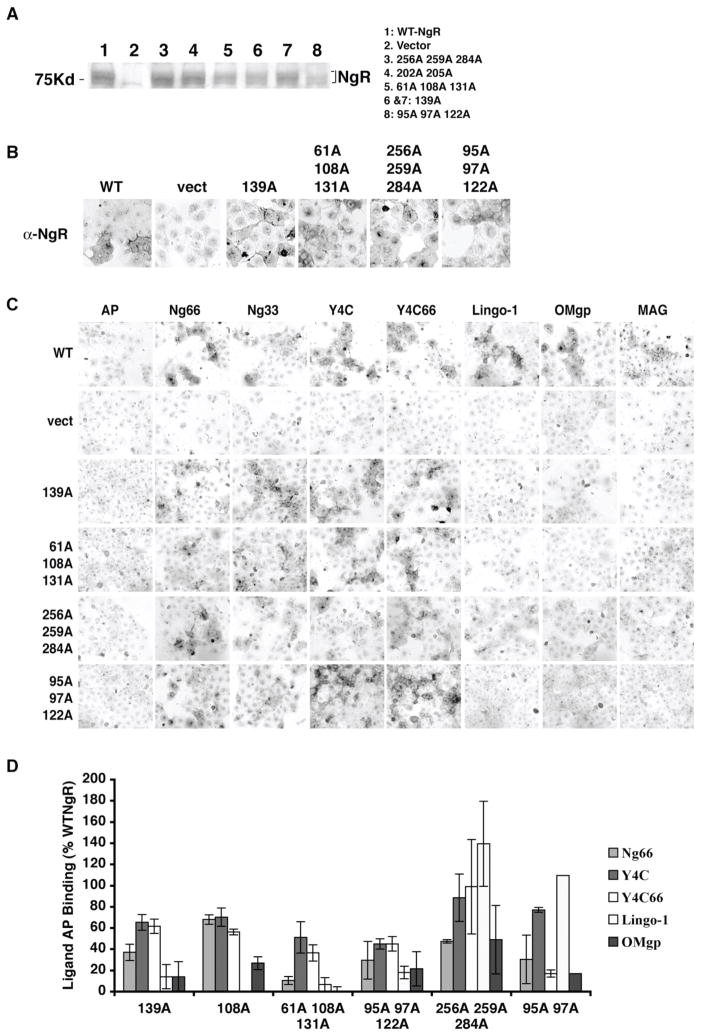
NgR1 has the capacity to bind Nogo-66, MAG, OMgp, and Lingo-1 plus Nogo-A-24 (6–9,31). To better define how multiple ligands with so wide structural diversity bind to the NgR1, we examined a series of Ala-substituted NgR1 mutants for ligand binding activity. Ala substitutions were generated for each of the charged residues predicted to be solvent accessible at the surface of the ligand binding LRR domain of NgR1 (10,11). We generated mutants in which 1–8 surface residues localized within 5 Å of one another were Ala-substituted. Because of the coiling nature of the LRR structure, residues juxtaposed on the protein surface are separated by approximately 25 residues in the primary structure. In addition to mutations in specific charged surface patches, other mutations were targeted to glycosylation sites (residues N82 and N179) and to regions predicted to be involved in ligand binding based on the NgR1 structure (10,11). A variant corresponding to a human polymorphism present in GenBank was also examined (D259N). None of the mutations alters the Leu residues critical for the tertiary structure of LRR structure or the Cys residues involved in disulfide bond formation in the amino and carboxyl terminal capping domains. The vast majority of such Ala surface substitution mutants are expressed as immunoreactive polypeptides with a molecular weight and an expression level indistinguishable from wild type NgR1 (Fig. 6A and data not shown). Those that where not expressed were excluded from further analysis. Moreover, all of the NgR1 mutants that were analyzed for ligand binding exhibited a cellular distribution in transfected COS-7 identical to that of the wild-type protein (Fig. 6B and data not shown). Notably, those mutations that remove both glycosylation sites in the LRR domain (aa 82 and 179), do not alter expression levels or surface localization, although the molecular weight is reduced by immunoblot analysis (data not shown).
Figure 6. Examples of NgR1 mutants that show differential binding to myelin ligands.
(A) To analyze the expression of mutant NgR1 constructs COS-7 cells were transfected with plasmids encoding indicated constructs, lysates were subjected to SDS-PAGE, and Western blotting was performed with anti-NgR1 antibodies. (B) Immunocytochemical detection of indicated mutant NgR1 proteins at the surface of transfected COS-7 cells. Live COS-7 cells were incubated at 4°C with anti-NgR1 antibodies prior to fixation and incubation with labeled secondary antibodies. (C) Binding of AP or AP fused NgR1 ligands to COS-7 cells expressing different NgR1 mutants as indicated. AP-Nogo33: AP fusion protein of the amino-terminal 33 residues of Nogo-66. The concentrations of ligands applied were: AP, 30 nM; AP-Nogo-66 (Ng66), 5 nM; AP-Nogo33 (Ng33), 10 nM; AP-Y4C,10 nM; AP-Y4C66, 0.5 nM; AP-Lingo-1, 10 nM; AP-OMGP, 10 nM; AP-MAG, 30 nM. These concentrations are close to the binding Kd of these proteins to NgR1, so that any decrease in Kd is reflected in staining intensity. (D) Quantitation of AP ligand binding to NgR1 mutants expressed as a percentage of binding to wild-type NgR1.
Myelin ligand binding to NgR1 requires overlapping but separate residues
We used the library of 74 NgR1 mutants to test for their AP-Nogo-66, AP-Amino-Nogo-Y4C, AP-Nogo-C39, AP-MAG, AP-OMgp, and AP-Lingo-1 binding ability. The properties of the NgR1 mutants fall into one of three major categories (Table I and Fig. 7B). A number of Ala substituted NgR1 variants bind all of the ligands at wild-type levels. We conclude that the corresponding residues do not play an essential role in ligand interactions. Many of these residues are situated on the convex side of the NgR1 structure, indicating that this surface is not a primary site for these interactions. In addition, a significant extent of the concave surface is dispensable for ligand binding. Glycosylation at residue 179 is not essential for ligand binding. The D259N polymorphic variant exhibits normal ligand binding properties (Table I).
Table I.
Summary of NgR1 mutants’ ligand binding properties.
| No binding | Binding to all ligands | Differential binding |
|---|---|---|
| 163 | 61 | 82 |
| 82,179 | 92 | 108 |
| 133,136 | 122 | 139 |
| 158,160 | 127 | 210 |
| 182,186 | 131 | 78, 81 |
| 232, 234 | 138 | 87, 89 |
| 67, 68, 71 | 151 | 89, 90 |
| 111, 113, 114 | 176 | 95, 97 |
| 114, 117, 163 | 179 | 108, 131 |
| 182, 186, 210 | 227 | 256, 259 |
| 210, 232, 234 | 250 | 36, 38, 61 |
| 67, 68, 95, 97 | D259N | 61, 108, 131 |
| 87, 89, 133, 136 | 36, 38 | 95, 97, 122 |
| 182, 186, 158, 160 | 63, 65 | 114, 117, 139 |
| 111, 113, 114, 138 | 114, 117 | 117, 119, 120 |
| 117, 119, 120, 139 | 127, 151 | 216, 218, 220 |
| 95, 97, 188, 189, 191, 192 | 127, 176 | 220, 223, 224 |
| 202, 205, 227, 250, 277, 279 | 143, 144 | 237, 256, 259 |
| 95, 97, 117, 119, 120, 188, 189 | 189, 191 | 256, 259, 284 |
| 196, 199 | 237, 256, 284 | |
| 202, 205 | 63, 65, 87, 89 | |
| 267, 269 | 196, 199, 220, 223, 224 | |
| 277, 279 | 211, 213, 237, 256, 259, 284 | |
| 189, 191, 237 | 189, 191, 211, 213, 237, 256, 259, 284 | |
| 189, 191, 284 | ||
| 202, 205, 227 | ||
| 202, 205, 250 | ||
| 296, 297, 300 | ||
| 171, 172, 175, 176 | ||
| 292, 296, 297, 300 | ||
| 171, 172, 175, 176, 196, 199 |
Ala-substituted NgR1 mutants were tested for their binding to AP-Nogo-66, AP-Y4C, AP-Y4C66, AP-Nogo-C39, AP-Lingo-1, AP-OMgp and AP-MAG, and they fall into three categories: (1) Mutants that lose binding to all NgR1 ligands. (2) Mutants that still maintain binding to all NgR1 ligands. (3) Differential binding mutants that bind some ligands but lose binding to other ligands. The D259N mutant is an asparagine substitution to mimic a human polymorphism.
Figure 7. Mapping of ligand binding sites in NgR1 and summary of observed interactions.
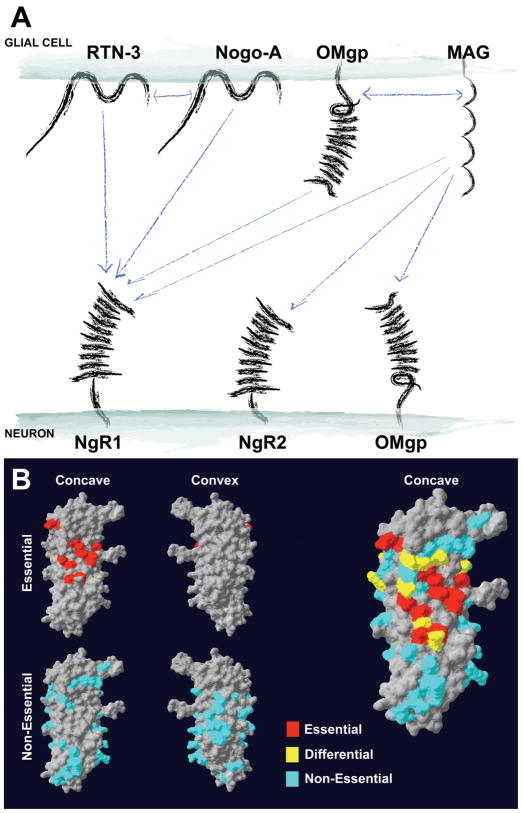
(A) Summary of observed interactions between ligands expressed in the oligodendrocytes and their neuronal receptors. Bidirectional arrows denote potential interactions in cis between proteins presented on the oligodendrocyte plasma membrane. (B) Summary of 74 Ala substitution human NgR1 mutants’ ability to bind Nogo-66, MAG and OMgp ligands. Residues required for the binding of all three ligands (red), some ligands but not others (yellow), and not required for ligand binding (blue) are highlighted. This illustration was made using SwissPdbViewer software.
A second group of mutants exhibits weak or no binding to each of the ligands (Table I and Fig. 7B). One interpretation is that these residues are required for NgR1 folding, so that their substitution with Ala results in misfolded protein with no ligand binding. However, there are several reasons to favor the alternative hypothesis that many of these residues contribute to the binding of multiple NgR1 ligands in a common binding pocket. Critically, the NgR1 expression levels and subcellular distribution are not altered for these mutants (data not shown). In contrast, misfolded proteins might be expected to be unstable and mislocalized. Notably, the majority of those residues that cannot be mutated to Ala without losing affinity to all ligands are clustered near one another. Thus, we conclude that the NgR1 surface created by residues 67/68, 111/113, 133/136, 158/160, 163, 182/186, and 232/234 constitutes a primary binding site for these ligands. Mouse and human NgR1 are identical at all 13 of these positions, supporting a conserved functional role for these residues. Human and mouse NgR1 differs from human and mouse NgR2 in four of these residues (68R>L; 133H>Q; 232Y>E), whereas human and mouse NgR1 differs from human and mouse NgR3 in three of these residues (113S>G; 136H>Y; 232Y>F). The non-conservative changes at these sites may account for the inability of NgR2 and NgR3 to bind several ligands specific for NgR1. Removal of both NgR1 N-linked glycosylation sites (82/179) abrogated binding to all ligands. Since the 82/179 mutant is expressed at the cell surface, the lack of binding indicates that glycosylation contributes to either protein folding or ligand binding directly.
The third group of Ala substituted NgR1 mutants exhibits selective loss of binding for some ligands but not others (Tables I and II). The preservation of binding affinity for at least one ligand by each member of this class demonstrates the Ala replacements do not prevent NgR1 folding and surface expression. Most of the NgR1 residues responsible for differential ligand binding are situated at the perimeter of the primary binding site described above. Many of these substitutions reduce or eliminate MAG, OMgp and Lingo-1 binding without diminishing binding by Nogo-66, Nogo-Y4C or Nogo-C39. The simplest interpretation of this topographic relationship is that MAG, OMgp and Lingo-1 require not only a central ligand binding domain that is partially shared with multiple NgR1-interacting Nogo-A fragments, but also an adjacent group of residues for high affinity binding. This adjacent region includes amino acids 78/81, 87/89, 89/90, 95/97, 108, 119/120, 139, 210, and 256/259. Mouse and human NgR1 are identical at 11 of these 14 residues and similar at 13 of 14. Human NgR2 exhibits less conservation at these 14 positions with 8 identical and 6 non-identical amino acids (78R>G, 81R>S, 89H>F, 95R>T, 97D>Y, 259D>A) as compared to human NgR1. One of these changes is conservative (similar/nonidentical aa; 89H>F) and 5 are non-conservative (dissimilar aa). For human NgR3 there are 7 identical and 7 non-identical aa (78R>S, 81R>P, 89H>Y, 95R>Y, 97D>H, 120S>T, 259D>G) at these positions as compared to human NgR1. Two of these changes are conservative (89H>Y, 120S>T), two are moderately conservative (97D>H, 259D>G), and 3 are non-conservative. The lack of amino acid conservation at these sites may account for the inability of NgR2 to bind OMgp, and NgR3 to bind MAG and OMgp.
Table II.
List of NgR1 mutants that show differential binding to NgR1 ligands.
| Residues | Nogo-66 | Y4C | Nogo-C39 | Y4C-66 | Lingo-1 | OMgp | MAG | anti-NgR1 |
|---|---|---|---|---|---|---|---|---|
| WT | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| 82 | ++ | ++ | ++ | ++ | + | + | − | ++ |
| 108 | ++ | ++ | ++ | ++ | tr | + | − | ++ |
| 139 | + | + | + | + | − | tr | − | ++ |
| 210 | + | + | + | + | − | − | − | + |
| 78, 81 | ++ | ++ | ++ | ++ | tr | ++ | N/A | ++ |
| 87, 89 | ++ | ++ | ++ | ++ | − | + | + | ++ |
| 89, 90 | + | + | + | + | − | − | − | ++ |
| 95, 97 | + | + | + | + | + | tr | tr | ++ |
| 108, 131 | + | + | + | + | − | + | tr | ++ |
| 256, 259 | ++ | ++ | ++ | ++ | ++ | + | − | ++ |
| 36, 38, 61 | + | + | + | ++ | tr | + | tr | ++ |
| 61, 108, 131 | + | + | + | + | − | − | − | ++ |
| 95, 97, 122 | + | + | + | + | − | tr | tr | ++ |
| 114, 117, 139 | + | + | + | + | − | − | − | ++ |
| 117, 119, 120 | ++ | ++ | ++ | ++ | tr | tr | − | ++ |
| 216, 218, 220 | ++ | ++ | ++ | ++ | + | + | tr | ++ |
| 220, 223, 224 | + | + | + | + | − | − | tr | ++ |
| 237, 256, 259 | tr | tr | tr | + | + | tr | − | ++ |
| 256, 259, 284 | + | + | + | ++ | ++ | + | − | ++ |
| 63, 65, 87, 89 | ++ | ++ | ++ | ++ | − | + | − | ++ |
| 237, 256, 284 | ++ | ++ | ++ | ++ | ++ | + | − | ++ |
| 211, 213, 237, 256, 259, 284 | − | − | − | − | ++ | − | − | ++ |
| 189, 191, 211, 213, 237, 256, 259, 284 | − | − | − | − | ++ | − | N/A | ++ |
Binding of Ala-substituted NgR1 mutants to NgR1 ligands were compared to wild-type NgR1 and the levels of binding were categorized as ++ (WT level), + (weaker than wild type), tr (trace binding), − (no binding), N/A (not determined).
Of special interest is glycosylation site at residue 82. Mutating this glycosylation site to Ala reduces MAG, OMgp and Lingo-1 binding. Interestingly NgR2, which binds MAG, has a potential glycosylation site at Asn82, whereas in NgR3 this has been naturally replaced with Ala and concomitantly affinity to MAG has been lost. This suggests that sugar moieties attached to Asn82 contribute to NgR1 interaction with these particular ligands. Consistent with this model, MAG interaction with neurons expressing NgR1 and NgR2 has been shown to be at least partially sialic acid-dependent, and both receptors have been shown to likely be highly sialylated glycoproteins (27).
All three Nogo-A fragments were found to interact with residues located on the central portion of the concave side of LRR domain. We did not identify mutants that display differentially reduced affinity for a certain Nogo-A segment. However, it is possible that higher-resolution mapping of NgR1 residues involved in ligand-binding could reveal differences in their binding sizes.
DISCUSSION
The current study extends our understanding of how myelin inhibitors interact with the NgR family: NgR1 binds three linear segments of Nogo-A as well as MAG and OMgp; mutagenesis defined overlapping NgR1 binding sites for different ligands; RTN2 and RTN3 also bind NgR1 with high affinity; NgR2 binds MAG but not RTNs; and finally, NgR3 binds none of the known NgR family ligands
As all RTNs are also present in the CNS and as they all contain homologous 66 amino acid loop regions, we hypothesized earlier that that they could interact with NgR family members (25). Here we found that RTN2 and RTN3 interact with NgR1. Because of the high sequence similarity between RTN-66 regions, they very likely interact with the same site in NgR1. We localized this binding site to the center of the concave side of LRR domain by systematic mutagenesis. The several amino acid changes between NgR1 and NgR2 and NgR3 in this core binding region are likely to explain the specificity of this interaction. Earlier we showed that the first 31 aa ((15), underlined in Fig. 1W) in Nogo-66 region are critical for receptor binding. Analysis of different RTN-66 regions suggests that the amino acid changes in RTN1 (highlighted in red in Fig. 1W) could account for the loss of its affinity to NgR1. The amino-acids 36–41 in RTN-66 regions show considerable sequence diversity. This carboxyl-terminal part of Nogo-66 is instrumental for activating NgR1 downstream signaling (15). As our earlier results showed that GST-RTN1-66 and GST-RTN3-66 recombinant proteins do not cause growth cone collapse (4), other RTNs could thus function as NgR1 antagonists blocking Nogo-66-induced NgR1 activation. However, as ~95% of these GST proteins are misfolded and in inclusion bodies, it is plausible that the remaining protein fraction might be inactive as well. While no Rtn1 or Rtn3 mRNA expression has been detected in the optic nerve (4), expression of other reticulons in spinal cord white matter had not been previously analyzed. We found that RTN3 mRNA transcript containing the 66 aa loop region is expressed in spinal cord white matter at levels similar to that of Nogo-A. Earlier studies showed that different splice forms of endogenous RTN4 interact with each other (34), and that Nogo-B interacts with RTN3 (35). As the Nogo-B–RTN3 -interaction was mediated by the RHD (35), Nogo-A and RTN3 may also form complexes in glial cells. The stoichiometry of RTN3-RTN4 complexes might determine their function.
Most of Nogo-A is localized to endoplasmic reticulum where it serves essential functions in a wide variety of cells (29). Accumulating evidence shows that at least a fraction of oligodendrocyte plasma membrane Nogo-A is in a conformation where amino-terminus of the protein faces extracellular milieu (3,34). Recently, we demonstrated that a second Nogo-A-specific domain, termed Nogo-A-24 (human Nogo-A aa 995–1018) immediately amino terminal to the first hydrophobic segment, has separate high affinity for NgR1 (31). Here we report that the carboxyl terminus following the second carboxyl-terminal hydrophobic region in RHD also interacts with NgR1 with high affinity. It is possible that Nogo-A, like several other proteins, could adapt multiple conformations to encompass different functions or to target the protein into different cellular compartments (for review, see (36)). The carboxyl-terminal end of Nogo-A possesses an ER-targeting sequence -K-X-K-X-X. This pentapeptide has to be cytosolic to be recognized. Thus it is possible that the population of Nogo-A protein molecules that present their carboxyl-termini extracellularly or on the luminal side of the ER might be enriched on the plasma membrane.
As the three Nogo-A segments that bind to NgR1 are closely connected to each other in a single polypeptide chain, it is plausible that they can not interact with several distant sites in a single NgR1 molecule. Interestingly, we noted that the binding-sites of Nogo-66, Amino-Nogo Y4C fragment, and Nogo-C39 fragment of Nogo-A on NgR1 overlap. It is feasible that these fragments might not interact simultaneously with one NgR1 monomer, but that tripartite NgR1 ligand would engage NgR1 clustering. The extracellular domain of NgR1 has significant affinity for surface-bound NgR1, and given the presumably high local concentration of GPI-anchored NgR1 in lipid rafts, receptor clustering could be facilitated by this basal level homophilic adhesion (33). Consistent with receptor clustering model, clustering of MAG or Nogo-66 has been shown to increase their potency to activate NgR1 signaling and downstream RhoA activation (37). Earlier we noticed that fusion of the Nogo-A-24, which is able to bind but not activate NgR1 to the NEP1-32 antagonist peptide creates a potent agonist peptide. This result also raised the possibility that bivalent or multivalent interactions of ligands with NgR1 are critical for its activation.
The observed very high affinity interaction between OMgp and MAG suggests a model where a ternary complex consisting of OMgp, MAG and NgR1 could regulate specific aspects of oligodendrocyte-neuron-interactions. At least in some cases, OMgp could also serve as a high-affinity neuronal ligand for MAG. This is supported by a report that OMgp is expressed at low levels in oligodendrocytes, whereas neuronal expression of OMgp, as detected by immunohistochemistry and in situ hybridization, is prominent (38). Interestingly, the reported neuronal OMgp expression pattern overlaps (e.g. in layer V of the cerebral cortex, pyramidal cells of the hippocampus) with that of NgR1. Thus OMgp could contribute to MAG binding in these cells and further downstream signaling might depend on the formation of a ternary complex of MAG, OMgp, and NgR1. Neuronal signaling triggered by MAG–OMgp interaction may also be independent of NgR1.
Since NgR1’s structure is now defined (10,11), we probed its surface for ligand binding sites by Ala substitutions. There appears to be a binding domain located in the central region on concave side of NgR1 LRR domain required by Amino-Nogo-A-24, Nogo-66, Nogo-C39, MAG and OMgp ligands. In addition, different ligands require particular residues surrounding this central site. Because all ligands require surface residues centered on the mid-portion of the concave face of NgR1, their mechanism for activating NgR1 signaling may be similar. Similarly to the case of internalin–E-cadherin (39) and glycoprotein Ibα–von Willebrand factor (40)-complexes, the concave side of the receptor serves as a ligand docking site. Previous work had been divided as to whether binding sites for Nogo-66 and MAG were separate or overlapping. Using NEP1–40 antagonist of Nogo-66, we did not observe inhibition of MAG interactions with NgR1 (8). With a sterically encumbered AP-Nogo-66 ligand, some competition with MAG-Fc binding to NgR1 was detected. Our findings are consistent with partial competition between ligands.
Because NgR1 is considered a target for the development of axonal regeneration therapeutics (41), the definition of this central binding domain shared by multiple ligands may facilitate the design and development of small molecule therapeutics blocking all NgR1 ligands. In contrast, if each ligand had been found to require completely separate residues for binding with high affinity, then the challenge of developing blockers of all myelin protein action at NgR1 would be significantly higher.
Lingo-1 has been reported as a component of a signal transducing NgR1 complex (30). It is notable that the residues required for Lingo-1 binding to NgR1 are very similar to those for the ligands MAG and OMgp. Because Lingo-1 in also expressed by oligodendrocytes, the binding analysis suggests that it might act as a ligand for NgR1.
The systematic mapping of all interactions between myelin inhibitory ligands and related molecules and NgR family members gives us better insight on the possible redundancy in signaling pathways and the specificity of earlier described interactions. Novel ligand-receptor interactions were elucidated so that future studies can characterize them in greater functional detail. The identification of a central ligand binding domain holds the promise that general NgR1 antagonists regeneration after CNS injury. may be created to possibly promote axonal
Supplementary Material
Acknowledgments
We thank Dr. S. Mi from Biogen Idec, Inc. for AP-Lingo-1 and AP-OMgp constructs, Dr. J. Flanagan (Harvard Medical School) for pAPtag5 plasmid, and Dr Eric Schmidt for kindly providing data presented in Supplementary Fig. 2. We thank Dr. Noam Harel for critical comments on the manuscript. This work was supported by grants to S.M.S. from the NIH and from Biogen Idec, Inc.
References
- 1.Yiu G, He Z. Nat Rev Neurosci. 2006;7(8):617–627. doi: 10.1038/nrn1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu BP, Cafferty WB, Budel SO, Strittmatter SM. Philos Trans R Soc Lond B Biol Sci. 2006;361(1473):1593–1610. doi: 10.1098/rstb.2006.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oertle T, van der Haar ME, Bandtlow CE, Robeva A, Burfeind P, Buss A, Huber AB, Simonen M, Schnell L, Brosamle C, Kaupmann K, Vallon R, Schwab ME. J Neurosci. 2003;23(13):5393–5406. doi: 10.1523/JNEUROSCI.23-13-05393.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.GrandPre T, Nakamura F, Vartanian T, Strittmatter SM. Nature. 2000;403(6768):439–444. doi: 10.1038/35000226. [DOI] [PubMed] [Google Scholar]
- 5.Oertle T, Klinger M, Stuermer CA, Schwab ME. Faseb J. 2003;17(10):1238–1247. doi: 10.1096/fj.02-1166hyp. [DOI] [PubMed] [Google Scholar]
- 6.Fournier AE, GrandPre T, Strittmatter SM. Nature. 2001;409(6818):341–346. doi: 10.1038/35053072. [DOI] [PubMed] [Google Scholar]
- 7.Domeniconi M, Cao Z, Spencer T, Sivasankaran R, Wang K, Nikulina E, Kimura N, Cai H, Deng K, Gao Y, He Z, Filbin M. Neuron. 2002;35(2):283–290. doi: 10.1016/s0896-6273(02)00770-5. [DOI] [PubMed] [Google Scholar]
- 8.Liu BP, Fournier A, GrandPre T, Strittmatter SM. Science. 2002;297(5584):1190–1193. doi: 10.1126/science.1073031. [DOI] [PubMed] [Google Scholar]
- 9.Wang KC, Koprivica V, Kim JA, Sivasankaran R, Guo Y, Neve RL, He Z. Nature. 2002;417(6892):941–944. doi: 10.1038/nature00867. [DOI] [PubMed] [Google Scholar]
- 10.He XL, Bazan JF, McDermott G, Park JB, Wang K, Tessier-Lavigne M, He Z, Garcia KC. Neuron. 2003;38(2):177–185. doi: 10.1016/s0896-6273(03)00232-0. [DOI] [PubMed] [Google Scholar]
- 11.Barton WA, Liu BP, Tzvetkova D, Jeffrey PD, Fournier AE, Sah D, Cate R, Strittmatter SM, Nikolov DB. Embo J. 2003;22(13):3291–3302. doi: 10.1093/emboj/cdg325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schnell L, Schwab ME. Nature. 1990;343(6255):269–272. doi: 10.1038/343269a0. [DOI] [PubMed] [Google Scholar]
- 13.Bregman BS, Kunkel-Bagden E, Schnell L, Dai HN, Gao D, Schwab ME. Nature. 1995;378(6556):498–501. doi: 10.1038/378498a0. [DOI] [PubMed] [Google Scholar]
- 14.Wiessner C, Bareyre FM, Allegrini PR, Mir AK, Frentzel S, Zurini M, Schnell L, Oertle T, Schwab ME. J Cereb Blood Flow Metab. 2003;23(2):154–165. doi: 10.1097/01.WCB.0000040400.30600.AF. [DOI] [PubMed] [Google Scholar]
- 15.GrandPre T, Li S, Strittmatter SM. Nature. 2002;417(6888):547–551. doi: 10.1038/417547a. [DOI] [PubMed] [Google Scholar]
- 16.Lee JK, Kim JE, Sivula M, Strittmatter SM. J Neurosci. 2004;24(27):6209–6217. doi: 10.1523/JNEUROSCI.1643-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li S, Strittmatter SM. J Neurosci. 2003;23(10):4219–4227. doi: 10.1523/JNEUROSCI.23-10-04219.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Baughman KW, Basso DM, Strittmatter SM. Ann Neurol. 2006 doi: 10.1002/ana.20953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li S, Liu BP, Budel S, Li M, Ji B, Walus L, Li W, Jirik A, Rabacchi S, Choi E, Worley D, Sah DW, Pepinsky B, Lee D, Relton J, Strittmatter SM. J Neurosci. 2004;24(46):10511–10520. doi: 10.1523/JNEUROSCI.2828-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simonen M, Pedersen V, Weinmann O, Schnell L, Buss A, Ledermann B, Christ F, Sansig G, van der Putten H, Schwab ME. Neuron. 2003;38(2):201–211. doi: 10.1016/s0896-6273(03)00226-5. [DOI] [PubMed] [Google Scholar]
- 21.Zheng B, Ho C, Li S, Keirstead H, Steward O, Tessier-Lavigne M. Neuron. 2003;38(2):213–224. doi: 10.1016/s0896-6273(03)00225-3. [DOI] [PubMed] [Google Scholar]
- 22.Kim JE, Li S, GrandPre T, Qiu D, Strittmatter SM. Neuron. 2003;38(2):187–199. doi: 10.1016/s0896-6273(03)00147-8. [DOI] [PubMed] [Google Scholar]
- 23.Zheng B, Atwal J, Ho C, Case L, He XL, Garcia KC, Steward O, Tessier-Lavigne M. Proc Natl Acad Sci U S A. 2005;102(4):1205–1210. doi: 10.1073/pnas.0409026102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim JE, Liu BP, Park JH, Strittmatter SM. Neuron. 2004;44(3):439–451. doi: 10.1016/j.neuron.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 25.Lauren J, Airaksinen MS, Saarma M, Timmusk T. Mol Cell Neurosci. 2003;24 (3):581–594. doi: 10.1016/s1044-7431(03)00199-4. [DOI] [PubMed] [Google Scholar]
- 26.Pignot V, Hein AE, Barske C, Wiessner C, Walmsley AR, Kaupmann K, Mayeur H, Sommer B, Mir AK, Frentzel S. J Neurochem. 2003;85(3):717–728. doi: 10.1046/j.1471-4159.2003.01710.x. [DOI] [PubMed] [Google Scholar]
- 27.Venkatesh K, Chivatakarn O, Lee H, Joshi PS, Kantor DB, Newman BA, Mage R, Rader C, Giger RJ. J Neurosci. 2005;25(4):808–822. doi: 10.1523/JNEUROSCI.4464-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan R, Shi Q, Hu X, Zhou X. Cell Mol Life Sci. 2006;63(7–8):877–889. doi: 10.1007/s00018-005-5338-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. Cell. 2006;124(3):573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 30.Mi S, Lee X, Shao Z, Thill G, Ji B, Relton J, Levesque M, Allaire N, Perrin S, Sands B, Crowell T, Cate RL, McCoy JM, Pepinsky RB. Nat Neurosci. 2004;7(3):221–228. doi: 10.1038/nn1188. [DOI] [PubMed] [Google Scholar]
- 31.Hu F, Liu BP, Budel S, Liao J, Chin J, Fournier A, Strittmatter SM. J Neurosci. 2005;25(22):5298–5304. doi: 10.1523/JNEUROSCI.5235-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flanagan JG, Cheng HJ, Feldheim DA, Hattori M, Lu Q, Vanderhaeghen P. Methods Enzymol. 2000;327:19–35. doi: 10.1016/s0076-6879(00)27264-9. [DOI] [PubMed] [Google Scholar]
- 33.Fournier AE, Gould GC, Liu BP, Strittmatter SM. J Neurosci. 2002;22 (20):8876–8883. doi: 10.1523/JNEUROSCI.22-20-08876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dodd DA, Niederoest B, Bloechlinger S, Dupuis L, Loeffler JP, Schwab ME. J Biol Chem. 2005;280(13):12494–12502. doi: 10.1074/jbc.M411827200. [DOI] [PubMed] [Google Scholar]
- 35.Qi B, Qi Y, Watari A, Yoshioka N, Inoue H, Minemoto Y, Yamashita K, Sasagawa T, Yutsudo M. J Cell Physiol. 2003;196(2):312–318. doi: 10.1002/jcp.10297. [DOI] [PubMed] [Google Scholar]
- 36.Levy D. Essays Biochem. 1996;31:49–60. [PubMed] [Google Scholar]
- 37.Niederost B, Oertle T, Fritsche J, McKinney RA, Bandtlow CE. J Neurosci. 2002;22(23):10368–10376. doi: 10.1523/JNEUROSCI.22-23-10368.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Habib AA, Marton LS, Allwardt B, Gulcher JR, Mikol DD, Hognason T, Chattopadhyay N, Stefansson K. J Neurochem. 1998;70(4):1704–1711. doi: 10.1046/j.1471-4159.1998.70041704.x. [DOI] [PubMed] [Google Scholar]
- 39.Schubert WD, Urbanke C, Ziehm T, Beier V, Machner MP, Domann E, Wehland J, Chakraborty T, Heinz DW. Cell. 2002;111(6):825–836. doi: 10.1016/s0092-8674(02)01136-4. [DOI] [PubMed] [Google Scholar]
- 40.Huizinga EG, Tsuji S, Romijn RA, Schiphorst ME, de Groot PG, Sixma JJ, Gros P. Science. 2002;297(5584):1176–1179. doi: 10.1126/science.107355. [DOI] [PubMed] [Google Scholar]
- 41.Lee DH, Strittmatter SM, Sah DW. Nat Rev Drug Discov. 2003;2(11):872–878. doi: 10.1038/nrd1228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.