Abstract
Nerve growth factor (NGF) plays an important role in regulating mammalian neuronal/embryonic development, angiogenesis, and other physiological processes and has recently been investigated as a potential treatment for the neurodegenerative disorder, Alzheimer disease. In this study, we provide evidence that human NGF may also function as a metalloproteinase inhibitor, based on studies of NGF from snake venom. Originally, our aim was to isolate snake venom metalloproteinases targeting platelet receptors and/or ligands relevant to hemostasis and thrombosis, using Ni2+-agarose as a purification step based on the conserved metal ion-coordination motif in venom metalloproteinases. However, subsequent analysis of cobra (Naja kaouthia) venom led to the unexpected discovery that cobra venom NGF bound to Ni2+-agarose, eluting at ∼15 mm imidazole, enabling a one-step purification. The identity of the purified protein was confirmed by mass spectrometry and N-terminal sequence analysis. Partial co-purification of NGF within metalloproteinase-enriched venom fractions led us to test whether NGF affected metalloproteinase activity. Venom NGF potently inhibited metalloproteinases isolated from the same or different venom and specifically bound to purified Nk metalloproteinase immobilized on agarose beads. Human NGF also interacted with human metalloproteinases because it blocked metalloproteinase-mediated shedding of the platelet collagen receptor, glycoprotein (GP)VI, and associated with recombinant ADAM10 by surface plasmon resonance. Together, these results suggest that NGF can function as a metalloproteinase inhibitor.
Keywords: ADAM ADAMTS, Glycoprotein, Platelet, Proteolytic Enzymes, Receptor Structure-Function
Introduction
The identification and characterization of mammalian nerve growth factor (NGF)3 was aided by the serendipitous discovery of a functionally related snake venom NGF (1, 2). Venom NGF potently induces neural outgrowth, and its presence in reptilian venom glands pointed to rat salivary glands as a major source of mammalian NGF (2). Subsequently, NGF was found to play a role in mammalian neuronal/embryonic development and in human diseases including the neurodegenerative disorder, Alzheimer disease (2–5). In this regard, the use of NGF gene therapy (where autologous fibroblasts were genetically modified to express NGF) to treat Alzheimer disease in phase 1 trials has been reported (6). NGF may have other physiological functions. Mammalian NGF (designated as “β subunit”) regulates the esterase activity of a γ subunit of an NGF-containing oligomeric complex, where dissociation of NGF from the complex increases esterase activity (7, 8). Increased amounts of NGF or pro-NGF are also present at inflammatory sites in lung and other tissue, and NGF is implicated in promoting wound healing (9–12). However, the reason why NGF is a significant component in snake venom (1, 2) and its role in envenomation are not known.
Snake venom metalloproteinase-disintegrins have provided unique probes for analyzing platelet receptors, including glycoprotein (GP)Ibα (the major ligand-binding subunit of the GPIb-IX-V complex) or the collagen receptor, GPVI, which form an adhesion/signaling complex on platelets that initiates thrombus formation (13–17). For example, we previously showed that mocarhagin, a cobra venom metalloproteinase-disintegrin, selectively cleaves GPIbα within an anionic sequence of the extracellular domain at Glu282/Asp283 and abolishes binding of von Willebrand factor and other ligands (17, 18). In contrast, alborhagin, a viper venom metalloproteinase-disintegrin, potently activates platelets via an interaction with GPVI (19). Surface expression of platelet GPVI and GPIbα is also regulated by metalloproteinase-mediated shedding, involving activation of endogenous platelet metalloproteinases of the ADAM (a disintegrin and metalloproteinase) family (20–26).
Our original aim was to isolate metalloproteinase-disintegrins that target platelet receptors or their ligands by fractionating snake venoms on Ni2+-agarose, an approach based on the conserved metal ion coordination sequence (HEXXHXXGXXH) within the catalytic domain of venom metalloproteinase-disintegrins (13, 27). The use of this approach is supported by binding of a purified venom metalloproteinase to Ni2+-agarose, eluting at ∼10 mm imidazole (27). However, when crude cobra (Naja kaouthia) venom was applied to Ni2+-agarose, an ∼13-kDa protein bound to the resin, eluting at ∼15 mm imidazole. Characterization of this protein and its partial co-isolation with metalloproteinase-enriched venom fractions led us to test whether venom NGF regulated metalloproteinase activity. These results suggest that NGF acts as an inhibitor of metalloproteinase-disintegrins.
EXPERIMENTAL PROCEDURES
Materials
Snake venom from N. kaouthia was obtained from Sigma. Murine anti-GPIbα monoclonal antibodies, AK2 directed against residues Leu36–Gln59, and WM23, against the mucin-like extracellular domain, have been previously described (14, 15, 18, 24). Murine monoclonal antibodies, 6B12, against an ectodomain epitope of human GPVI, and 14A2, against the platelet receptor CD151 that activates FcγRIIa, have also been described elsewhere (14, 20, 28). Glycocalicin, the soluble ectodomain fragment of human platelet GPIbα, was purified as described previously (14, 18, 29). The GPVI agonist, collagen-related peptide (CRP), with amino acid sequence GCO(GPO)10GCOG-NH2 where O represents hydroxyproline, was prepared as described elsewhere (14, 20). Recombinant human NGF and ADAM10 were purchased from R&D Systems, Minneapolis, MN.
Snake Venom Metalloproteinases
A cobra venom metalloproteinase-disintegrin, mocarhagin, was purified from Naja mocambique mocambique venom as described previously (18, 30). Mocarhagin cleaves platelet GPIbα at Glu282/Asp283 and abolishes binding of von Willebrand factor and anti-GPIbα antibody AK2 (14, 15, 18). Nk metalloproteinase from cobra (N. kaouthia) venom was purified using the same methods (27).
Ni2+-Agarose Chromatography
Crude lyophilized venom (0.1 g) from N. kaouthia was dissolved in 10 ml of TS buffer and loaded at ∼30 ml/h onto a 10 × 1-cm column of Ni2+-agarose equilibrated with TS buffer at room temperature (27). After washing with the same buffer until the A280 of the eluate returned to base line, bound protein was eluted by a linear 100-ml 0–30 mm imidazole gradient in TS buffer, and 5-ml fractions were collected. Aliquots of fractions were analyzed by electrophoresis on SDS 5–20% exponential gradient polyacrylamide gels and either stained with Coomassie Blue or Western-blotted with rabbit anti-mocarhagin antibody, as described previously (19). Fractions containing an ∼13-kDa protein were pooled, concentrated using an Amicon ultrafiltration device (YM10 membrane), and dialyzed into TS buffer.
Mass Spectrometry and N-terminal Sequence Analysis
Matrix-assisted laser desorption/ionization-time of flight mass spectrometry of purified venom NGF was carried out by Dr. Simon Harris, Monash University, Melbourne, Australia. N-terminal sequence analysis was performed using previously described methods (18, 19).
Snake Venom Metalloproteinase Assays
Snake venom metalloproteinase activity was assayed in two ways. First, a purified soluble extracellular portion of GPIbα, glycocalicin (∼30 μg/ml, final concentration) in TS buffer containing either 1 mm ZnCl2, 1 mm CaCl2 or 10 mm EDTA was treated with mocarhagin (5 μg/ml, final concentration) for 60 min at 22 °C in the absence or presence of venom NGF (0.1–60 μg/ml). Samples were made 10 mm in EDTA to stop the reaction and analyzed by SDS 5–20% polyacrylamide gel electrophoresis and immunoblotting with rabbit anti-glycocalicin IgG as described elsewhere (14).
Second, washed platelets were isolated from venous blood and resuspended at 1 × 108 platelets/ml in Tyrode's buffer as described previously (14, 19, 20). After treatment with either TS buffer alone or Nk metalloproteinase (5 μg/ml, final concentration) for 60 min at 22 °C in the presence of 1 mm ZnCl2, 1 mm CaCl2 and in the absence or presence of venom NGF (0.1–60 μg/ml), samples were made 0.1% (w/v) in BSA by adding 0.1 volume of a 1% (w/v) stock solution, and 40-μl samples were added in triplicate to microtiter wells coated with either 1% (w/v) BSA in TS buffer (BSA/TS) or anti-FLAG (negative control), WM23, or AK2 (50 μl of 1 μg/ml solution in TS buffer, for 3 h at 22 °C) as described previously (27). After aspiration of the coating solution, wells were blocked for 1 h with 100 μl BSA/TS and washed four times with 100-μl aliquots of BSA/TS before the addition of platelets. After 1 h, wells were aspirated and washed four times with BSA/TS, and platelet adhesion was determined using anti-glycocalicin IgG (1 μg/ml in TS buffer), washing four times with BSA/TS, and visualized using an horseradish peroxidase-conjugated anti-rabbit secondary IgG and the chromogenic substrate, tetramethylbenzidine (Sigma). The A450 was measured in a microtiter plate reader 3550 (Bio-Rad). Specific binding was calculated from total binding by subtracting the nonspecific binding to control (anti-FLAG) IgG-coated wells.
Binding of Venom NGF to Nk-coated Beads
Purified Nk (0.1 mg) was dialyzed into coupling buffer (0.1 m NaHCO3, pH 8.0) and conjugated to 10 ml of a 1:1 mixture of Affi-Gel 10 and Affi-Gel 15 N-hydroxysuccinimide-derivatized agarose beads (Bio-Rad), as described elsewhere (27, 31). To measure binding of venom NGF to Nk-coated beads, a 25% (v/v) suspension of Nk Affi-Gel 10/15 and venom NGF (5 μg/ml, final concentration) purified as described above was suspended in a final volume of 0.2 ml in TS buffer. Parallel assays contained 1 mm ZnCl2, 1 mm CaCl2, 10 mm EDTA, or 1 m NaCl. After 30 min at 22 °C, beads were pelleted by centrifugation (14,000 × g; 5 min; 22 °C), and NGF in the supernatant was analyzed by SDS 5–20% polyacrylamide gel electrophoresis and staining with Coomassie Blue.
Effect of Human NGF on Metalloproteinase-mediated Shedding of Human Platelet GPVI
The shedding of GPVI from washed human platelets (1 × 108/ml) in Tyrode's buffer containing 1 mm ZnCl2, 1 mm CaCl2 was induced by the GPVI-selective agonist, CRP (2.5 μg/ml, final concentration), or by the FcγRIIa agonist, 14A2 (anti-CD151), using previously described methods (14, 20, 28), in the absence or presence of human NGF (0–40 μg/ml, final concentration). After 30 min at 22 °C, samples were made 10 mm in EDTA, platelets were pelleted by centrifugation (14,000 × g; 5 min; 22 °C), and supernatants were analyzed for shed soluble fragment of GPVI (∼55 kDa) by immunoblotting with anti-GPVI antibody (6B12) and visualized using ECL (14, 20).
Surface Plasmon Resonance
The association of human NGF with recombinant human metalloproteinase, ADAM10, was measured by surface plasmon resonance using a Biacore T100 optical biosensor (GE Healthcare, Melbourne, Australia) maintained at 25 °C (20). Aliquots (5–10 μl) of ADAM10 (10 μg/ml in 10 mm HEPES-saline, pH 7.0, containing 0.005% (v/v) surfactant-P20) or buffer alone were immobilized on activated sensor chips (Series S CM5; GE Healthcare) to an equivalent of 200–250 resonance units using a flow rate of 10 μl/min, and then the flow cell was blocked according to manufacturer's instructions. Aliquots (20 μl) of human NGF (10–40 μg/ml) in 10 mm HEPES saline, pH 7.0, containing 1 mm ZnCl2 and 0.005% (v/v) surfactant-P20, were passed over the ADAM10 surface or a control surface at a constant flow rate of 10 μl/min, and resonance changes were recorded. For some analyses, 5 mm EDTA was included in the elution buffer. The response from the control surface was subtracted from that of the ADAM10 surface. Binding analysis was based on published ligand-receptor fragment interactions (32).
RESULTS
Isolation of Snake Venom NGF
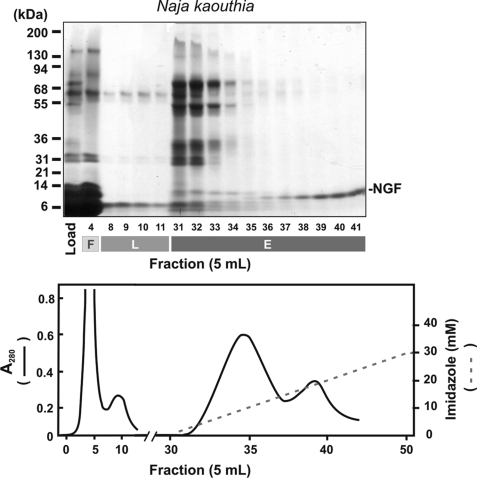
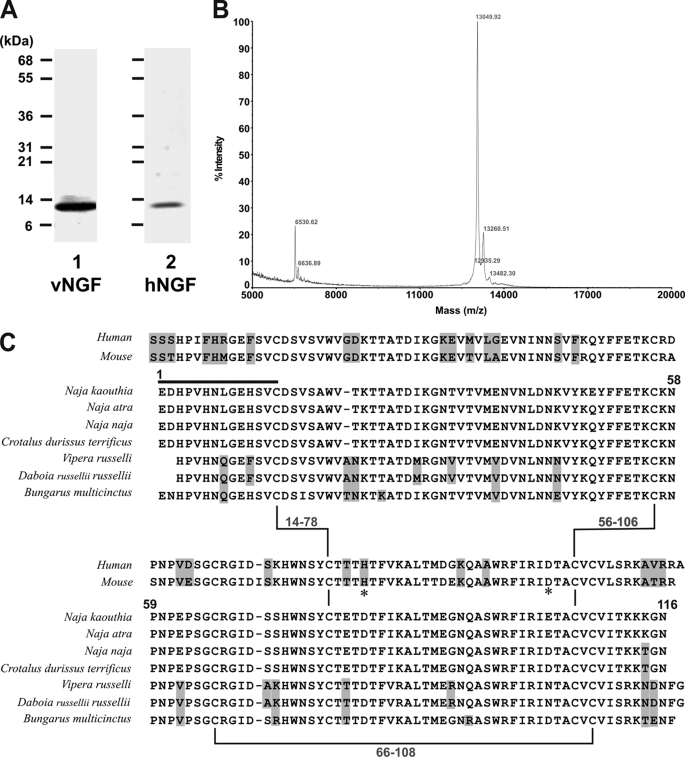
Ni2+-agarose chromatography of N. kaouthia venom revealed a protein of ∼13 kDa under reducing conditions (Fig. 1A) that eluted after the main metalloproteinase peak at ∼15 mm imidazole. This protein was not immunoreactive to anti-mocarhagin (a snake venom metalloproteinase-disintegrin) antibody (data not shown), suggesting that it was not a metalloproteinase-derived fragment. The identity of the purified protein (Fig. 2A), which co-migrated on SDS-polyacrylamide gels with human NGF, was confirmed as venom NGF by mass spectrometry and N-terminal sequence analysis. A single species with mass of ∼13,000 was detected (Fig. 2B), and the N-terminal 14 amino acid residues were identical to NGF from N. kaouthia and other venoms (Fig. 2C).
FIGURE 1.
Ni2+-Agarose chromatography of N. kaouthia venom. Ni2+-Agarose chromatography of venom (0.1 mg/10 ml TS buffer) analyzed by SDS 5–20% polyacrylamide gel electrophoresis (reducing conditions) and Coomassie Blue staining is shown, demonstrating load, flow-through (F), lag (L), and eluted (E) fractions. The A280 elution profile (solid line) and the linear 0–30 mm imidazole gradient (dashed line) are shown in the lower panel.
FIGURE 2.
Characterization of snake venom NGF. A, SDS 5–20% polyacrylamide gel (reducing conditions) stained with Coomassie Blue of venom NGF (vNGF, lane 1) from N. kaouthia venom or recombinant human NGF (hNGF, lane 2). B, analysis by matrix-assisted laser desorption/ionization-time of flight mass spectrometry reveals an abundant species with mass of 13,049. Peaks at 13,260 and 13,482 presumably represent single (“+206”) or double (“+412”) sinapinic acid matrix adducts within acceptable limits of accuracy. A minor peak at 6,530 (with no corresponding adduct at +206) may indicate the presence of a minor low abundance protein, and the peak at 6,636 may indicate a doubly charged species, although further analysis was not performed in detail. No higher mass peaks were detected up to 40,000 (not shown). C, comparison of human NGF, mouse NGF, and venom NGF from N. kaouthia or other species. Residue numbers refer to the N. kaouthia NGF, the black line is the N-terminal sequence determined in this study, and highlighted residues are non-identical or non-conserved in N. kaouthia NGF. Zn2+-binding sites in murine NGF involving residues His84 and Asp105 (33) are marked by an asterisk. Accession numbers/gi numbers are: human NGF, 1SG1B/49258981; mouse NGF, P01139; N. kaouthia, P61899/48429019; Naja atra, A58566/7438538; Naja naja, P01140/128164; Crotalus durissus terrificus, AAG30924/11120560; Vipera russelli, AAA03282/263157; Daboia russellii russellii, P30894/400499; Bungarus multicinctus, AAB25729/266299.
Inhibition of Snake Venom Metalloproteinases by Venom NGF
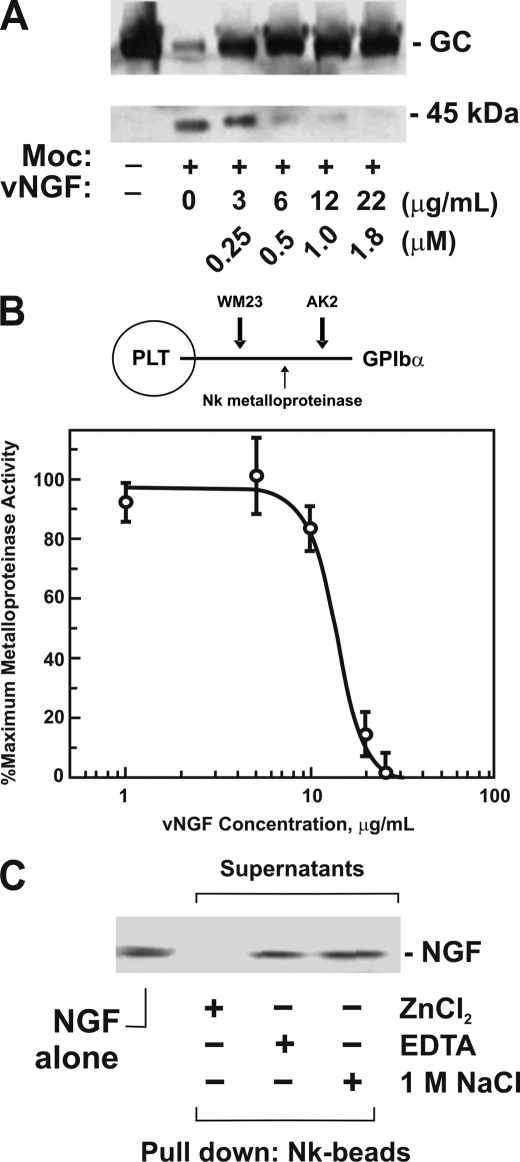
The finding that both venom NGF (this study) and metalloproteinase-enriched fractions of snake venom (27) associated with Ni2+-agarose led us to test whether venom NGF may affect metalloproteinase activity. Initially, we tested this possibility using a snake venom metalloproteinase, mocarhagin, which was previously shown to inactivate platelet GPIbα by selective proteolysis of the von Willebrand factor receptor (GPIbα) at Glu282/Asp283, thereby eliminating the von Willebrand factor-binding domain (17, 18). Mocarhagin-dependent proteolysis of glycocalicin, a purified soluble ectodomain fragment of GPIbα containing the Glu282/Asp283 cleavage site (17, 18), was inhibited by venom NGF in a dose-dependent manner (Fig. 3A). Second, we tested whether NGF inhibited a metalloproteinase isolated from venom of the same species, N. kaouthia (27). A microtiter assay was used to measure the cleavage of platelet GPIbα by cobra Nk metalloproteinase. Like mocarhagin, Nk cleaves GPIbα ectodomain to generate the same digestion pattern (27). In this assay, two previously characterized anti-GPIbα monoclonal antibodies, AK2 and WM23, with epitopes either N-terminal or C-terminal, respectively, of the proteolytically sensitive sequence encompassing Glu282/Asp283, were immobilized on plastic, and adhesion of washed platelets was measured. GPIbα-dependent platelet adhesion was discriminated from nonspecific binding using wells coated with control antibody (anti-FLAG) or BSA. We previously showed that binding to AK2-coated wells, but not wells coated with WM23, anti-FLAG, or BSA, is inhibited either by soluble AK2 or by pretreating platelets with Nk metalloproteinase (27). Using this assay, it was shown that Nk-dependent cleavage of platelet GPIbα was inhibited by venom NGF (Fig. 3B).
FIGURE 3.
Effect of NGF on snake venom metalloproteinase activity. A, venom NGF inhibits mocarhagin-dependent digestion of glycocalicin. Glycocalicin (GC; ∼30 μg/ml, final concentration) was treated with mocarhagin (Moc; 5 μg/ml or ∼0.1 μm, final concentration) in TS buffer containing 1 mm ZnCl2, 1 mm CaCl2 for 60 min at 22 °C in the absence or presence of venom NGF (vNGF, 0–22 μg/ml or 0–1.8 μm for NGF monomer, final concentration). Samples were analyzed on SDS-polyacrylamide gels (reducing conditions) and blotted with rabbit anti-glycocalicin IgG visualized using horseradish peroxidase-conjugated secondary antibody and ECL. B, venom NGF blocks Nk-dependent inhibition of GPIbα-dependent platelet (PLT) adhesion to AK2. Washed platelets (1 × 108/ml) were treated with Nk metalloproteinase (5 μg/ml, final concentration) in the absence or presence of venom NGF (0–40 μg/ml) for 60 min at 22 °C. Platelet adhesion to microtiter wells coated with anti-GPIbα monoclonal antibody, AK2 (with epitope lost following Nk treatment, upper diagram), was measured using anti-glycocalicin IgG as described under “Experimental Procedures.” Nk-dependent cleavage was confirmed by measuring platelet adhesion to WM23-coated wells, where the epitope is unaffected by Nk (27). The error bars represent 1× S.D. C, binding of venom NGF to Nk-coated beads. Purified venom NGF (5 μg/ml, final concentration) incubated with Nk-agarose beads (25% (v/v) suspension) in TS buffer for 30 min at 22 °C in the presence of EDTA, 1 mm ZnCl2, 1 mm CaCl2, or 1 m NaCl was analyzed after pelleting the resin by centrifugation and SDS-polyacrylamide gel electrophoresis and Coomassie Blue staining.
Binding of Venom NGF to Nk-coated Beads
Purified venom NGF bound directly to Nk metalloproteinase immobilized on agarose beads. Binding in the presence of Zn2+/Ca2+ (represented by loss of NGF in the supernatant) was inhibited either by EDTA or by the inclusion of 1 m NaCl in the binding assay (Fig. 3C). EDTA and NaCl were also shown to dissociate mammalian NGF from the oligomeric complex with esterase (7, 8).
Effect of Human NGF on Metalloproteinase-dependent Shedding of Platelet GPVI
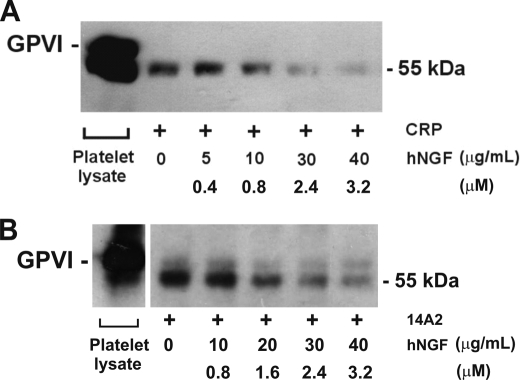
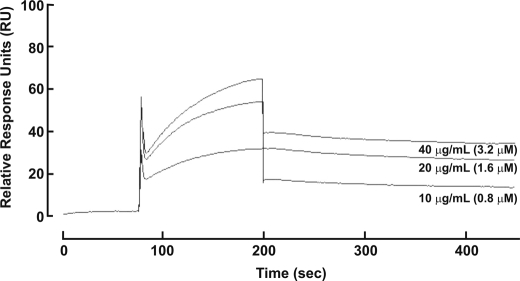
Because venom NGF was able to inhibit activity of venom metalloproteinases, we addressed whether human NGF could inhibit human metalloproteinase activity. To do this, we utilized an assay for ectodomain shedding of platelet GPVI, previously shown to involve activation of endogenous platelet metalloproteinases (20, 24). Treating washed human platelets with the GPVI-selective ligand, CRP, resulted in release into the supernatant of a soluble ∼55-kDa ectodomain fragment detectable using an anti-GPVI antibody, 6B12 (Fig. 4). CRP-induced shedding involves platelet metalloproteinases because it is inhibited by EDTA or a broad specificity metalloproteinase inhibitor, GM6001 (20). Recombinant human NGF (Fig. 2A) inhibited ectodomain shedding of platelet GPVI induced by CRP in a dose-dependent manner (Fig. 4), with >90% inhibition when compared with shedding in the absence of NGF under the same conditions. Human NGF (40 μg/ml, final concentration) had no effect on CRP-induced GPVI-dependent platelet aggregation (data not shown), suggesting that NGF was not competing for CRP binding to GPVI nor interfering by an unknown mechanism with GPVI signaling, required for aggregation and ligand-induced shedding (20). In support of this, NGF also inhibited GPVI shedding induced by activation of the Fc receptor, FcγRIIa, using the anti-platelet antibody, 14A2 (28), with a similar dose response as observed for CRP (Fig. 4). Human NGF also associated with immobilized recombinant ADAM10 in a dose-dependent manner by surface plasmon resonance (Fig. 5). No association of NGF was observed using a control surface, and binding to ADAM10 was divalent cation-dependent and inhibitable by inclusion of 5 mm EDTA to the analysis buffer (not shown). These data indicate an association and rapid dissociation of the isolated components in a cell-free system.
FIGURE 4.
Human NGF (hNGF) inhibits metalloproteinase-dependent shedding of human platelet GPVI. Metalloproteinase-dependent GPVI shedding from washed human platelets was induced by CRP (2.5 μg/ml, final concentration) (A) or the monoclonal antibody, 14A2 (5 μg/ml, final concentration (B); it activates platelets via FcγRIIa) in the absence or presence of human NGF after 30 min at 22 °C. Supernatant fractions were analyzed by SDS-polyacrylamide gel electrophoresis (reducing conditions), immunoblotting with anti-GPVI antibody (6B12 or 1A5), and visualizing with horseradish peroxidase-conjugated secondary antibody and ECL. Platelet lysate containing intact GPVI is run as a blotting control.
FIGURE 5.
Human NGF interacts with recombinant human ADAM10. Aliquots of purified hNGF (0.8–3.2 μm) were passed over CM5 sensor chip flow cells containing immobilized ADAM10 or a control surface as described under “Experimental Procedures.” The response from the control surface was subtracted from that of the ADAM10 surface.
DISCUSSION
In this study, we show that (i) snake venom NGF can be isolated from N. kaouthia venom by a single-step purification on Ni2+-agarose, elutable by imidazole; (ii) venom NGF inhibits venom metalloproteinase-dependent proteolysis of platelet glycoprotein (GP)Ibα; and (iii) human NGF inhibits human metalloproteinase-mediated ectodomain shedding of GPVI from platelets. The combined results suggest that NGF can inhibit metalloproteinases.
We initially investigated the use of Ni2+-agarose chromatography for isolating snake venom metalloproteinases based on the presence of a conserved metal ion coordination sequence in venom metalloproteinases and the preliminary finding that a purified metalloproteinase, Nk, bound Ni2+-agarose and was eluted by ∼10 mm imidazole (27). In the course of these studies, it was observed that an ∼13-kDa protein was purified from N. kaouthia venom run on Ni2+-agarose, eluting at imidazole concentrations ≥15 mm. Mass spectrometry and N-terminal sequence analysis (100% identity of 14 residues) confirmed that this protein was venom NGF. Binding of venom NGF to Ni2+-agarose is consistent with the presence of Zn2+-binding sites in murine NGF (involving residues His84 and Asp105) (33) (Fig. 2C), and Zn2+ stabilizes the oligomeric complex involving mammalian NGF (NGF β subunit) and esterase (γ subunit) (7, 8). The N-terminal sequence of venom NGF also contains a conserved His-rich sequence, 3HXXHXXGXH11 (Fig. 2C), similar but not identical to the metal ion coordination motif, HEXXHXXGXXH, in metalloproteinase-disintegrins.
Venom NGF inhibited mocarhagin-dependent cleavage of a purified soluble GPIbα fragment (glycocalicin) containing the mocarhagin cleavage site (17, 18), inhibited cleavage of GPIbα on platelets by Nk metalloproteinase isolated from the same venom as NGF (N. kaouthia) (27), and bound directly to Nk metalloproteinase-coated beads. One of the functions of NGF in venom may be to act as a metalloproteinase inhibitor to prevent autodigestion (19), a function previously reported for other venom constituents (34). The NGF concentration in snake venom is high relative to the amount of metalloproteinase in the same venom (1, 27) and consistent with a role for NGF as an inhibitor in venom. NGF from other snake venoms is highly conserved (Fig. 2C), suggesting that this may be a common role. The prevalence of NGF in venom glands is consistent with their derivation from salivary glands, a potent source of NGF in other animals (2). In prey, mast cell proteases, such as carboxypeptidase A and others, play an important role in defense of envenomation (35), and venom (metallo)proteinase inhibitors may also potentially contribute to venom toxicity.
These results obtained with venom NGF raise the possibility that mammalian NGF may also function as a metalloproteinase inhibitor. Human NGF is ∼80% identical to venom NGF based on primary sequence, and 6 Cys residues forming three disulfide bonds are conserved (Fig. 2C). In this study, the effect of human NGF on the activity of mammalian metalloproteinases was assessed using a previously described assay involving metalloproteinase-mediated shedding of human platelet GPVI (20, 24, 28). Ligand-induced shedding of GPVI was inhibited by human NGF in a dose-dependent manner, consistent with the inhibition of platelet metalloproteinases by human NGF. GPVI-dependent platelet reactivity is proportional to GPVI surface levels in a graded fashion (reviewed in Refs. 25 and 26), suggesting that if shedding was only partially blocked by low concentrations of NGF, this could have a significant effect on platelet reactivity. The specific platelet metalloproteinase implicated in GPVI shedding is ADAM10 of the ADAM family of sheddases (24–26), and human NGF was also shown to associate with ADAM10 by surface plasmon resonance. Although we did not determine in our study whether NGF can inhibit other members of the metalloproteinase family, it is interesting to note that NGF can also inhibit the proteolytic activity of esterases (7, 8), and determining the broader inhibition profile of NGF could be worthwhile in future studies.
Surface expression of two major NGF receptors expressed on nerve cells, p75NTR and TrkA, is also regulated by ADAM17-dependent ectodomain shedding, thereby controlling cellular responses to NGF-mediated cross-linking of TrkA-TrkA or TrkA-p75NTR (5, 36, 37). Activation of ADAM family metalloproteinases (such as ADAM9, ADAM10, and/or ADAM17) also controls proteolytic processing of amyloid precursor protein (39), a process that also occurs on human platelets (40, 41). The relative activity of different metalloproteinases may therefore control normal (neuroprotective) versus pathological (neurotoxic) processing of this protein. NGF, acting as a metalloproteinase inhibitor, may at least in part regulate this processing under certain conditions and/or regulate other pathophysiology involving NGF. Furthermore, our results provide a potential functional link between NGF and GPVI-dependent platelet function in that NGF-dependent inhibition of GPVI shedding could increase platelet reactivity. Clinical data from Alzheimer disease patients (n = 30) indicate decreased levels of soluble GPVI in plasma (38). The potential role of NGF in regulating metalloproteinase-mediated events, parameters such as physiological, pathological, and therapeutic concentrations of NGF, relative localization of binding partners, and the possible regulation of platelet reactivity through inhibition of GPVI/sheddase activity warrants further investigation.
Acknowledgments
We thank C. Llerena, C. Berndt, A. Aprico, and J. Jing for technical assistance, Dr. A. I. Smith and S. Reeve for N-terminal sequence analysis, and Dr. S. Harris for mass spectrometry.
This work was supported by grants from the National Health and Medical Research Council of Australia and Monash University.
- NGF
- nerve growth factor
- ADAM
- a disintegrin and metalloproteinase
- GP
- glycoprotein
- TS
- Tris-saline
- CRP
- collagen-related peptide
- BSA
- bovine serum albumin.
REFERENCES
- 1.Cohen S. (1959) J. Biol. Chem. 234, 1129–1137 [PubMed] [Google Scholar]
- 2.Cohen S. (2004) Ann. N.Y. Acad. Sci. 1038, 98–102 [DOI] [PubMed] [Google Scholar]
- 3.Salehi A., Delcroix J. D., Swaab D. F. (2004) J. Neural. Transm. 111, 323–345 [DOI] [PubMed] [Google Scholar]
- 4.Dechant G., Barde Y. A. (2002) Nat. Neurosci. 5, 1131–1136 [DOI] [PubMed] [Google Scholar]
- 5.He X. L., Garcia K. C. (2004) Science 304, 870–875 [DOI] [PubMed] [Google Scholar]
- 6.Tuszynski M. H., Thal L., Pay M., Salmon D. P., U. H. S., Bakay R., Patel P., Blesch A., Vahlsing H. L., Ho G., Tong G., Potkin S. G., Fallon J., Hansen L., Mufson E. J., Kordower J. H., Gall C., Conner J. (2005) Nat. Med. 11, 551–555 [DOI] [PubMed] [Google Scholar]
- 7.Palmer T. E., Neet K. E. (1980) J. Biol. Chem. 255, 5170–5176 [PubMed] [Google Scholar]
- 8.Rao A. G., Neet K. E. (1984) J. Biol. Chem. 259, 73–79 [PubMed] [Google Scholar]
- 9.Harrington A. W., Leiner B., Blechschmitt C., Arevalo J. C., Lee R., Mörl K., Meyer M., Hempstead B. L., Yoon S. O., Giehl K. M. (2004) Proc. Natl Acad. Sci. 101, 6226–6230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villoslada P., Genain C. P. (2004) Prog. Brain Res. 146, 403–414 [DOI] [PubMed] [Google Scholar]
- 11.Kawamoto K., Matsuda H. (2004) Prog. Brain Res. 146, 369–384 [DOI] [PubMed] [Google Scholar]
- 12.Freund-Michel V., Frossard N. (2008) Pharmacol. Ther. 117, 52–76 [DOI] [PubMed] [Google Scholar]
- 13.Wijeyewickrema L. C., Berndt M. C., Andrews R. K. (2005) Toxicon 45, 1051–1061 [DOI] [PubMed] [Google Scholar]
- 14.Arthur J. F., Gardiner E. E., Matzaris M., Taylor S. G., Wijeyewickrema L., Ozaki Y., Kahn M. L., Andrews R. K., Berndt M. C. (2005) Thromb. Haemost. 93, 716–723 [DOI] [PubMed] [Google Scholar]
- 15.Shen Y., Romo G. M., Dong J. F., Schade A., McIntire L. V., Kenny D., Whisstock J. C., Berndt M. C., López J. A., Andrews R. K. (2000) Blood 95, 903–910 [PubMed] [Google Scholar]
- 16.Kamiguti A. S. (2005) Toxicon 45, 1041–1049 [DOI] [PubMed] [Google Scholar]
- 17.Andrews R. K., Berndt M. C. (2004) in Handbook of Proteolytic Enzymes (Barrett A. J., Rawlings N. D., Woessner J. F. eds) 2nd Ed., pp. 696–699, Elsevier, London [Google Scholar]
- 18.Ward C. M., Andrews R. K., Smith A. I., Berndt M. C. (1996) Biochemistry 35, 4929–4938 [DOI] [PubMed] [Google Scholar]
- 19.Andrews R. K., Gardiner E. E., Asazuma N., Berlanga O., Tulasne D., Nieswandt B., Smith A. I., Berndt M. C., Watson S. P. (2001) J. Biol. Chem. 276, 28092–28097 [DOI] [PubMed] [Google Scholar]
- 20.Gardiner E. E., Arthur J. F., Kahn M. L., Berndt M. C., Andrews R. K. (2004) Blood 104, 3611–3617 [DOI] [PubMed] [Google Scholar]
- 21.Aktas B., Pozgajova M., Bergmeier W., Sunnarborg S., Offermanns S., Lee D., Wagner D. D., Nieswandt B. (2005) J. Biol. Chem. 280, 39716–39722 [DOI] [PubMed] [Google Scholar]
- 22.Bergmeier W., Burger P. C., Piffath C. L., Hoffmeister K. M., Hartwig J. H., Nieswandt B., Wagner D. D. (2003) Blood 102, 4229–4235 [DOI] [PubMed] [Google Scholar]
- 23.Bergmeier W., Piffath C. L., Cheng G., Dole V. S., Zhang Y., von Andrian U. H., Wagner D. D. (2004) Circ. Res. 95, 677–683 [DOI] [PubMed] [Google Scholar]
- 24.Gardiner E. E., Karunakaran D., Shen Y., Arthur J. F., Andrews R. K., Berndt M. C. (2007) J. Thromb. Haemost. 5, 1530–1537 [DOI] [PubMed] [Google Scholar]
- 25.Berndt M. C., Karunakaran D., Gardiner E. E., Andrews R. K. (2007) J. Thromb. Haemost. 5, Suppl. 1, 212–219 [DOI] [PubMed] [Google Scholar]
- 26.Andrews R. K., Karunakaran D., Gardiner E. E., Berndt M. C. (2007) Arterioscler. Thromb. Vasc. Biol. 27, 1511–1520 [DOI] [PubMed] [Google Scholar]
- 27.Wijeyewickrema L. C., Gardiner E. E., Shen Y., Berndt M. C., Andrews R. K. (2007) Toxicon 50, 1064–1072 [DOI] [PubMed] [Google Scholar]
- 28.Gardiner E. E., Karunakaran D., Arthur J. F., Mu F. T., Powell M. S., Baker R. I., Hogarth P. M., Kahn M. L., Andrews R. K., Berndt M. C. (2008) Blood 111, 165–174 [DOI] [PubMed] [Google Scholar]
- 29.Andrews R. K., Booth W. J., Gorman J. J., Castaldi P. A., Berndt M. C. (1989) Biochemistry 28, 8317–8326 [DOI] [PubMed] [Google Scholar]
- 30.De Luca M., Dunlop L. C., Andrews R. K., Flannery J. V., Jr., Ettling R., Cumming D. A., Veldman G. M., Berndt M. C. (1995) J. Biol. Chem. 270, 26734–26737 [DOI] [PubMed] [Google Scholar]
- 31.Andrews R. K., Harris S. J., McNally T., Berndt M. C. (1998) Biochemistry 37, 638–647 [DOI] [PubMed] [Google Scholar]
- 32.Christou C. M., Pearce A. C., Watson A. A., Mistry A. R., Pollitt A. Y., Fenton-May A. E., Johnson L. A., Jackson D. G., Watson S. P., O'Callaghan C. A. (2008) Biochem. J. 411, 133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holland D. R., Cousens L. S., Meng W., Matthews B. W. (1994) J. Mol. Biol. 239, 385–400 [DOI] [PubMed] [Google Scholar]
- 34.Huang K. F., Hung C. C., Wu S. H., Chiou S. H. (1998) Biochem. Biophys. Res. Commun. 248, 562–568 [DOI] [PubMed] [Google Scholar]
- 35.Metz M., Piliponsky A. M., Chen C. C., Lammel V., Abrink M., Pejler G., Tsai M., Galli S. J. (2006) Science 313, 526–530 [DOI] [PubMed] [Google Scholar]
- 36.Weskamp G., Schlöndorff J., Lum L., Becherer J. D., Kim T. W., Saftig P., Hartmann D., Murphy G., Blobel C. P. (2004) J. Biol. Chem. 279, 4241–4249 [DOI] [PubMed] [Google Scholar]
- 37.Díaz-Rodríguez E., Esparís-Ogando A., Montero J. C., Yuste L., Pandiella A. (2000) Biochem. J. 346, 359–367 [PMC free article] [PubMed] [Google Scholar]
- 38.Laske C., Leyhe T., Stransky E., Eschweiler G. W., Bueltmann A., Langer H., Stellos K., Gawaz M. (2008) J. Psychiatr. Res. 42, 746–751 [DOI] [PubMed] [Google Scholar]
- 39.Allinson T. M., Parkin E. T., Turner A. J., Hooper N. M. (2003) J. Neurosci. Res. 74, 342–352 [DOI] [PubMed] [Google Scholar]
- 40.Evin G., Zhu A., Holsinger R. M., Masters C. L., Li Q. X. (2003) J. Neurosci. Res. 74, 386–392 [DOI] [PubMed] [Google Scholar]
- 41.Colciaghi F., Marcello E., Borroni B., Zimmermann M., Caltagirone C., Cattabeni F., Padovani A., Di Luca M. (2004) Neurology 62, 498–501 [DOI] [PubMed] [Google Scholar]