Abstract
NASP (nuclear autoantigenic sperm protein) is a member of the N1/N2 family, which is widely conserved among eukaryotes. Human NASP reportedly prefers to bind to histones H3·H4 and the linker histone H1, as compared with H2A·H2B, and is anticipated to function as an H3·H4 chaperone for nucleosome assembly. However, the direct nucleosome assembly activity of human NASP has not been reported so far. In humans, two spliced isoforms, somatic and testicular NASPs (sNASP and tNASP, respectively) were identified. In the present study we purified human sNASP and found that sNASP efficiently promoted the assembly of nucleosomes containing the conventional H3.1, H3.2, H3.3, or centromere-specific CENP-A. On the other hand, sNASP inefficiently promoted nucleosome assembly with H3T, a testis-specific H3 variant. Mutational analyses revealed that the Met-71 residue of H3T is responsible for this inefficient nucleosome formation by sNASP. Tetrasomes, composed of the H3·H4 tetramer and DNA without H2A·H2B, were efficiently formed by the sNASP-mediated nucleosome-assembly reaction. A deletion analysis of sNASP revealed that the central region, amino acid residues 26–325, of sNASP is responsible for nucleosome assembly in vitro. These experiments are the first demonstration that human NASP directly promotes nucleosome assembly and provide compelling evidence that sNASP is a bona fide histone chaperone for H3·H4.
Keywords: Chromatin, Chromatin/Regulation, Chromatin/Remodeling, Chromatin/Epigenetics, Chromosomes/Histones, Chromosomes/Non-histone Proteins, Chromosomes/Nucleosome, Histones
Introduction
The nucleosome is the fundamental unit of eukaryotic chromatin and is composed of a 146-base pair DNA and a histone octamer (1). The histone octamer comprises two H2A·H2B dimers and one H3·H4 tetramer (2). During the nucleosome assembly process, an H3·H4 tetramer is first deposited into chromatin, and H2A·H2B dimers are then incorporated into the H3·H4 tetrasome to form a complete octameric nucleosome. These nucleosome assembly processes are mediated by the histone chaperones, which directly bind to H2A·H2B and/or H3·H4 in cells (3–8).
Histone chaperones are defined by their sequence similarity and histone binding ability. Among the histone chaperone family of proteins, Nap1 (9), Nap2 (10), CAF-1 (11), HIRA (12), ASF1 (13–17), and RbAp46 (18) have been shown to bind to H3·H4. In humans, five nonallelic isoforms of H3 (H3.1, H3.2, H3.3, H3T, and CENP-A) have been identified as H3 variants (19–25), and their specific chaperones were proposed (5, 6, 26–29). Nap1 was the first nucleosome assembly protein identified (9, 30–33). Human Nap1 (hNap1)3 was shown to promote nucleosome assembly with conventional H3.1 (10, 34) but not with a testis-specific histone H3 variant, H3T, in vitro (10). Human Nap2 (35), a paralog of hNap1, promoted the H3T nucleosome assembly in vitro (10) and may function in the assembly of the testis-specific nucleosome containing H3T. CAF-1 is required for the DNA synthesis-dependent nucleosome assembly pathway with H3.1 and H3.2 (11, 26, 36, 37), whereas HIRA may function in the DNA synthesis-independent nucleosome assembly pathway with H3.3 (12, 26). CENP-A is a centromere-specific H3 variant and requires a large number of proteins, including histone chaperones (38–42).
The N1/N2 protein family, which was originally identified in Xenopus laevis (43–45), is widely conserved among eukaryotes and is known to bind to H3·H4, suggesting that it is a chaperone family for H3·H4. In mammals, NASP (nuclear autoantigenic sperm protein) was found to share a high degree of sequence similarity with the N1/N2 family. In mice, disruption of the NASP gene caused early embryonic lethality (46), indicating that NASP is an essential protein for mammals. Two spliced isoforms of NASP are present in humans. One is testicular NASP (tNASP), which was mainly found in the testis and embryonic tissues (47). The other one is somatic NASP (sNASP), which is produced in all dividing cells (47). NASP reportedly possesses chaperone activity for the linker histone H1 (46–50). In addition, tNASP and sNASP were found as subunits in the large histone-chaperone complex containing CAF1, HIRA, ASF1, and H3·H4 (26, 51), suggesting their function in nucleosome assembly. However, direct nucleosome-assembly activity, which may be a common property among the H3·H4 chaperones, has not been reported for the human NASPs so far, although sNASP preferentially binds to H3·H4 over H2A·H2B (52).
In the present study, we purified human sNASP as a recombinant protein and found that sNASP itself promotes nucleosome assembly in vitro. sNASP promotes the assembly of nucleosomes containing human H3 variants, H3.1, H3.2, H3.3, and CENP-A, but not H3T, with different efficiencies. The differences in the sNASP-mediated nucleosome-assembly efficiency among the H3 variants found in the present study may provide important insights into the sNASP function in chromatin organization and dynamics.
EXPERIMENTAL PROCEDURES
Purification of Recombinant Human sNASP
Human sNASP and its deletion mutants were overexpressed in Escherichia coli cells as N-terminal hexahistidine (His6)-tagged proteins. The DNA fragments encoding sNASP were ligated into the NdeI and BamHI sites of the pET15b vector (Novagen), which harbors the His6 tag and the thrombin protease recognition sequence (GE Healthcare) at the N terminus. Freshly transformed E. coli strain BL21(DE3) cells, which also carried an expression vector for the minor tRNAs (Codon(+)RIL; Stratagene), were grown on LB plates containing ampicillin (100 μg/ml) and chloramphenicol (35 μg/ml) at 37 °C. After a 16-h incubation, 5–20 colonies on the LB plates were collected and inoculated into LB medium (5 liters) containing ampicillin (100 μg/ml) and chloramphenicol (35 μg/ml), and the cultures were incubated at 30 °C. When the cell density reached an A600 = 0.4, 1 mm isopropyl-β-d-thiogalactopyranoside was added to induce the expression of sNASP, and the cultures were further incubated at 18 °C for 12 h. The cells producing sNASP were harvested, resuspended in 50 mm Tris-HCl buffer (pH 7.5) containing 2 mm 2-mercaptoethanol, 10% glycerol, and 0.5 m NaCl, and disrupted by sonication. The cell debris was removed by centrifugation (27,216 × g; 20 min), and the lysate was mixed gently with 4 ml (50% slurry) of nickel-nitrilotriacetic acid (Ni-NTA)-agarose resin (Qiagen) at 4 °C for 1 h. The sNASP-bound Ni-NTA beads were then packed into an Econo-column (Bio-Rad) and washed with 100 ml of 50 mm Tris-HCl buffer (pH 7.5) containing 10% glycerol, 500 mm NaCl, and 5 mm imidazole at a flow rate of about 0.8 ml/min. His6-tagged sNASP was eluted by a 100-ml linear gradient of imidazole from 5 to 500 mm in 50 mm Tris-HCl buffer (pH 7.5) containing 10% glycerol and 500 mm NaCl. The His6 tag was removed from the sNASP portion by thrombin protease (3 units/mg of protein). The sample was immediately dialyzed against 20 mm Tris-HCl buffer (pH 7.5) containing 100 mm NaCl, 1 mm EDTA, 10% glycerol, and 2 mm 2-mercaptoethanol. After removing the His6 tag, sNASP was purified using a Mono Q (GE Healthcare) column by elution with a 30-ml linear gradient of 100–600 mm NaCl in 20 mm Tris-HCl buffer (pH 7.5) containing 1 mm EDTA, 10% glycerol, and 2 mm 2-mercaptoethanol. The peak Mono Q fractions were collected, and the protein was further purified by using a Superdex 200 column eluted with 1.2 column volumes of the same buffer containing 100 mm NaCl. After this step, sNASP was again purified and concentrated by Mono Q chromatography by elution with a 30-ml linear gradient of NaCl from 100 to 600 mm in 20 mm Tris-HCl buffer (pH 7.5) containing 1 mm EDTA, 10% glycerol, and 2 mm 2-mercaptoethanol. The sNASP deletion mutants were prepared by the same procedure as the full-length sNASP. The purified sNASP and sNASP deletion mutants were dialyzed against 20 mm Tris-HCl buffer (pH 7.5) containing 150 mm NaCl, 1 mm DTT, 0.5 mm EDTA, 0.1 mm phenylmethylsulfonyl fluoride, and 10% glycerol.
Preparation of Recombinant Human Histones and Human Nap1
Human H2A, H2B, H4, and all H3 variants (H3.1, H3.2, H3.3, H3T, and CENP-A) were overexpressed in E. coli cells as N-terminal His6-tagged proteins (53). During the protein purification, the His6 tag was removed from the histones, and the H2A·H2B and H3·H4 complexes were prepared by the method described previously (10). Human Nap1 (hNap1) was overexpressed in E. coli cells as an N-terminal His6-tagged protein and was purified by a four-step purification method, including the Ni-NTA column, the His6 tag removal, the heparin-Sepharose column, and the Mono Q column, as described previously (10).
Preparation of a DNA Fragment for Nucleosome Reconstitution
A 195-bp DNA fragment containing the Lytechinus variegates 5 S ribosomal RNA gene was amplified by PCR using following primers: 5 S rDNA forward (5′-CAACGAATAACTTCCAGGGATTTATAAGCCG-3′; 5 S rDNA reverse (5′- AATTCGGTATTCCCAGGCGGTCTCC-3′). After the PCR reaction, the DNA fragment was extracted by phenol/chloroform, precipitated by ethanol, and further purified by Superdex 75 gel filtration chromatography to remove the primers and dNTPs.
Nucleosome Assembly Assay
H2A·H2B (8 ng/μl) and H3.1·H4 (8 ng/μl) were preincubated with sNASP or hNap1 at 23 °C for 10 min. The nucleosome assembly reaction was initiated by the addition of 5 S rDNA (8 ng/μl), and the reaction was continued in 10 μl of 20 mm Tris-HCl buffer (pH 8.0) containing 80 mm NaCl and 1 mm DTT at 23 °C for 60 min. After the reaction, the samples were incubated at 42 °C for 60 min to eliminate nonspecific DNA binding by the histones and then were analyzed by 6% PAGE in 0.2 × TBE buffer (18 mm Tris base, 18 mm boric acid, and 0.4 mm EDTA). The gel was run at 6.25 V/cm for 3 h and stained with ethidium bromide.
Micrococcal Nuclease (MNase) Assay
After the nucleosome assembly reaction with sNASP or hNap1, each sample, containing 40 ng of 5 S rDNA, was treated with 0.8, 0.4, 0.2, 0.1, and 0 unit of MNase (Takara) in 10 μl of 20 mm Tris-HCl buffer (pH 8.0) containing 45 mm NaCl, 5 mm CaCl2, and 0.5 mm DTT. After a 5-min incubation at 23 °C, the reaction was stopped by the addition of 60 μl of a proteinase K solution (20 mm Tris-HCl (pH 8.0), 20 mm EDTA, 0.5% SDS, and 0.5 mg/ml proteinase K (Roche Applied Science)). After a 15-min incubation at 23 °C, the DNA was extracted with phenol/chloroform and precipitated with ethanol. The DNA fragments were then analyzed by 10% PAGE in 0.2 × TBE buffer (21 V/cm for 1 h) and ethidium bromide staining.
Topological Assay for Nucleosome Assembly
H2A·H2B (10 ng/μl) and H3·H4 (10 ng/μl) were preincubated with sNASP or hNap1 at 37 °C for 15 min. The nucleosome assembly reaction was initiated by the addition of relaxed ϕX174 DNA (10 ng/μl), which was previously incubated with 1.7 units of wheat germ topoisomerase I (Promega) per 100 ng of DNA at 37 °C for 150 min. The reaction was continued in 10 μl of 20 mm Tris-HCl buffer (pH 8.0) containing 140 mm NaCl, 2 mm MgCl2, and 5 mm DTT at 37 °C for 60 min. After the reaction, the samples were incubated at 42 °C for 60 min to eliminate nonspecific DNA binding by the histones, and the proteins were then removed by an incubation with 60 μl of a proteinase K solution (20 mm Tris-HCl (pH 8.0), 20 mm EDTA, 0.5% SDS, and 0.5 mg/ml proteinase K) at 37 °C for 15 min followed by phenol-chloroform extraction. The DNA samples were then analyzed by 1% agarose gel electrophoresis in 1 × TAE buffer (40 mm Tris acetate and 1 mm EDTA) (3.3 V/cm for 4 h) with SYBR Gold (Invitrogen) staining.
Gel Electrophoretic Mobility Shift Assay for Binding between sNASP and H3·H4
sNASP (2.4 μg) was mixed with H3·H4 (0.75–3 μg) in 10 μl of 20 mm Tris-HCl buffer (pH 8.0) containing 100 mm NaCl and 1 mm DTT. The samples were then incubated for 1 h at 23 °C. After the incubation, the samples were analyzed by 6% PAGE in 0.5 × TBE buffer (45 mm Tris base, 45 mm boric acid, and 1 mm EDTA). The gel was run at 10.4 V/cm for 110 min, and the bands were visualized by Coomassie Brilliant Blue staining.
Nucleosome Reconstitution by the Salt Dialysis Method
Purified H2A·H2B (492 μg) was mixed with H3T·H4 (705 μg) at 4 °C in the presence of a 146-bp DNA fragment (500 μg) in a solution containing 0.5 m KCl (625 μl). The mixture was first dialyzed against 400 ml of dialysis buffer (10 mm Tris-HCl buffer (pH 7.5), 1 mm EDTA, and 1 mm DTT) containing 2 m KCl for 2 h at 4 °C. The concentration of KCl was gradually reduced from 2 to 0.25 m by adding dialysis buffer containing 0.25 m KCl using a peristaltic pump (0.8 ml/min flow rate). The sample was then incubated at 55 °C for 2 h. The H3T nucleosome was purified from the free DNA and histones by non-denaturing polyacrylamide gel electrophoresis using a Prepcell apparatus (Bio-Rad). The nucleosomes were analyzed by 6% PAGE in 0.2 × TBE buffer (18 mm Tris base, 18 mm boric acid, and 0.4 mm EDTA) at 15.6 V/cm for 1 h followed by ethidium bromide staining. The histone contents were also analyzed by 18% SDS-PAGE.
The H3·H4 Deposition Assay
sNASP (2.4 μg) was mixed with H3.1·H4 (1 μg) or H3T·H4 (1.5 μg), incubated for 10 min at 23 °C, and combined with supercoiled plasmid DNA (pGSAT4; 10, 20, 30, 40, 50, 60, 70, 80, 90, or 100 ng). The samples were further incubated for 1 h and analyzed by 5% PAGE in 0.5 × TBE buffer (10.4 V/cm for 1.5 h) followed by Coomassie Brilliant Blue staining.
RESULTS
Purification of Human sNASP
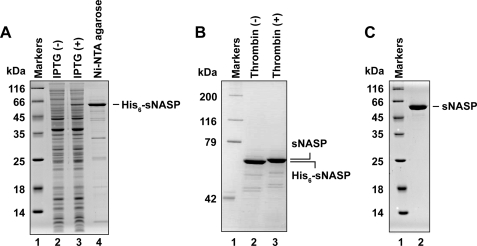
Human sNASP reportedly binds preferentially to histones H3·H4 (52). Our proteome analysis with a HeLa cell extract also showed that human sNASP bound to histones H3·H4 (data not shown). These facts suggest that sNASP may be a histone chaperone for H3·H4. We then purified human sNASP as a recombinant protein (Fig. 1). Human sNASP was overexpressed in E. coli cells as an N-terminal His6-tagged protein (Fig. 1A, lanes 2 and 3). The molecular mass of sNASP is 49 kDa; however, it migrated more slowly on an SDS-polyacrylamide gel, probably because of its extreme acidity (pI = 4.35). The His6-tagged sNASP was purified by Ni-NTA-agarose chromatography (Fig. 1A, lane 4). The His6 tag was removed from sNASP with thrombin protease (Fig. 1B). sNASP without a His6 tag migrated more slowly than His6-tagged sNASP because of the lack of the basic His6 tag (Fig. 1B). After removal of the His6 tag, sNASP was further purified by Mono Q column chromatography and Superdex 200 gel filtration chromatography (Fig. 1C). A mass spectroscopic analysis revealed that the molecular mass of purified recombinant sNASP was 49 kDa, which is identical to the predicted molecular mass of sNASP.
FIGURE 1.
Purification of human sNASP. A, proteins from each purification step were analyzed by 15% SDS-PAGE with Coomassie Brilliant Blue staining. Lane 1 indicates the molecular mass markers. Lanes 2 and 3 are the whole cell lysates before and after induction with isopropyl-β-d-thiogalactopyranoside (IPTG), respectively. Lane 4 indicates the sample from the peak Ni-NTA-agarose fraction. B, an 8% SDS-PAGE image of the Ni-NTA-agarose fraction before (lane 2) and after (lane 3) the removal of the His6 tag by thrombin treatment is shown. C, SDS-PAGE analysis of purified sNASP lacking the His6 tag.
Nucleosome Assembly by sNASP
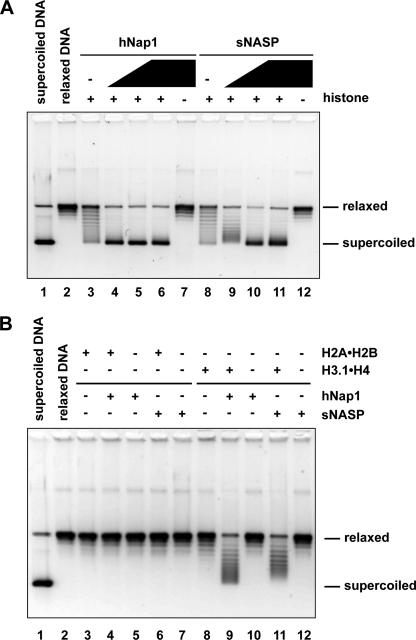
As reported previously, purified sNASP efficiently bound to histones H3.1·H4 (Fig. 2A), suggesting that sNASP may promote nucleosome assembly as the H3·H4 chaperone. However, the nucleosome assembly activity of sNASP has not been reported so far, although sNASP reportedly supported the nucleosome assembly reaction in the presence of a cell extract from yeast (52). We then tested the nucleosome assembly activity of sNASP by the conventional nucleosome formation assay. hNap1, which is known to have robust nucleosome assembly activity, was used for positive controls. In this assay, sNASP or hNap1 was preincubated with H2A·H2B and H3.1·H4, and then the 195-bp 5 S rDNA was added into the reaction mixture. The nucleosome formation was analyzed by a gel electrophoretic mobility shift assay. Intriguingly, we found that substantial amounts of nucleosomes were formed in the presence of sNASP as compared with the control experiments with hNap1 (Fig. 2B). To confirm nucleosome formation by sNASP, we next performed a MNase treatment. In this assay, the 147-bp DNA fragments, which were tightly wrapped around the histone octamer, were detected after the MNase treatment because MNase preferentially digests DNA free from the histone octamer surface. Therefore, if the nucleosomes were properly assembled, then the 147-bp DNA fragment would be detected after MNase treatment, as shown in the positive control experiments with hNap1 (Fig. 2C, lanes 7–11). As shown in Fig. 2C (lanes 12–16), the 147-bp protection from MNase was clearly detected, when the nucleosome assembly reactions were conducted with sNASP. These results strongly supported the conclusion that sNASP actually promotes the formation of nucleosomes.
FIGURE 2.
Nucleosome assembly activity of sNASP. A, electrophoretic mobility shift assay for histone H3·H4 binding by sNASP is shown. hNap1 (2.3 μg) or sNASP (2.4 μg) was incubated with different amounts of H3.1·H4, and the hNap1·H3.1·H4 or sNASP·H3.1·H4 complex was detected by non-denaturing 5% PAGE with Coomassie Brilliant Blue staining. The amounts of H3.1·H4 were 0 μg (lanes 1 and 7), 0.375 μg (lanes 2 and 8), 0.75 μg (lanes 3 and 9), 1.5 μg (lanes 4 and 10), 2.25 μg (lanes 5 and 11), and 3 μg (lanes 6 and 12). B, nucleosome assembly assay is shown. A 195-bp 5 S DNA fragment was incubated with histones, and the nucleosome assembly reactions were performed with hNap1 (lane 2) or sNASP (lane 3). Lane 1 indicates a negative control experiment without histone chaperone. The samples were analyzed by non-denaturing 6% PAGE. C, shown is an MNase assay. The nucleosome assembly reactions were performed with hNap1 (lanes 7–11) or sNASP (lanes 12–16). For negative control experiments, the reactions were also performed without histone chaperone (lanes 2–6). The samples were then treated with MNase, and the resulting DNA fragments were analyzed by non-denaturing 10% PAGE. The amounts of MNase were 0.8 unit (lanes 2, 7, and 12), 0.4 unit (lanes 3, 8, and 13), 0.2 unit (lanes 4, 9, and 14), 0.1 unit (lanes 5, 10, and 15), and 0 unit (lanes 6, 11, and 16). NCP indicates DNA fragments tightly wrapped around histone octamers.
We next tested sNASP-mediated nucleosome assembly by a topological assay. When nucleosomes are formed on relaxed circular DNA, negative supercoils are introduced, and the superhelicity can be analyzed on an agarose gel. As shown in Fig. 3A, a progressive increase in the number of negative superhelical turns introduced into DNA was observed with increasing amounts of sNASP (lanes 9–11). The nucleosome assembly efficiency of sNASP was comparable with that of hNap1, which is known to have robust nucleosome assembly activity (Fig. 3A). Like hNap1, sNASP also promoted the formation of tetrasomes, in which the DNA is wrapped around the H3·H4 tetramer without H2A·H2B (Fig. 3B). Therefore, we concluded that sNASP itself possesses the nucleosome assembly activity, as an H3·H4 chaperone.
FIGURE 3.
Topological assay for nucleosome assembly by sNASP. A, relaxed ϕX174 DNA (10 ng/μl), which was previously treated with wheat germ topoisomerase I (lane 2), was incubated with hNap1 (lanes 4–6) or sNASP (lanes 9–11) in the presence of core histones. The reaction products were then analyzed by 1% agarose gel electrophoresis in 1 × TAE buffer. Lanes 3 and 8 indicate negative control experiments without histone chaperone in the presence of core histones. Lanes 7 and 12 indicate the other negative control experiments without core histones in the presence of hNap1 (1 μm) or sNASP (1 μm). The concentrations of hNap1 and sNASP were 0.25 μm (lanes 4 and 9), 0.5 μm (lanes 5 and 10), and 1 μm (lanes 6 and 11). B, tetrasome assembly is shown. Reactions were conducted as described for the experiments shown in panel A, except H2A·H2B or H3.1·H4 were used instead of the four core histones, H2A·H2B/H3.1·H4. Lanes 3 and 8 indicate negative control experiments without histone chaperone in the presence of H2A·H2B or H3.1·H4, respectively. Lanes 5, 7, 10, and 12 indicate the other negative control experiments without core histones in the presence of either hNap1 (1 μm) or sNASP (1 μm). Lanes 3, 4, and 6 represent experiments with H2A·H2B, and lanes 8, 9, and 11 represent experiments with H3.1·H4.
Specificity of sNASP Binding to the H3 Variants
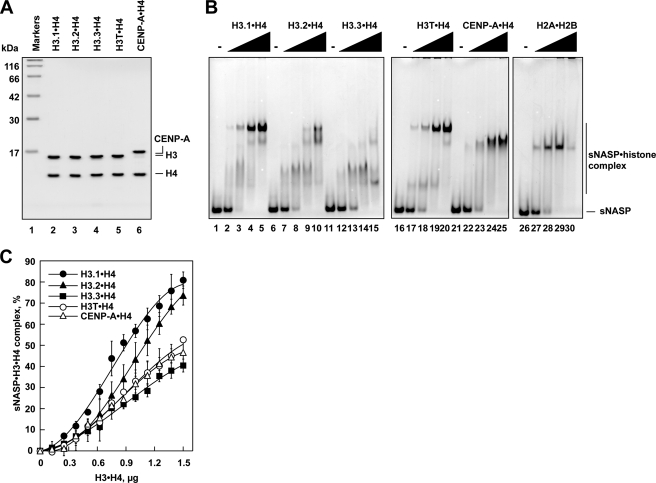
In humans, five non-allelic H3 variants, H3.1, H3.2, H3.3, H3T, and CENP-A, have been identified. All five H3 variants were prepared as recombinant proteins in the complex form with H4 (Fig. 4A). The electrophoretic mobility shift assay revealed that sNASP bound to all of the H3·H4 complexes, although each H3·H4·sNASP complex migrated differently on the non-denaturing polyacrylamide gel (Fig. 4B). Careful titration experiments revealed that sNASP binds to H3.1·H4 with the highest apparent binding affinity and also efficiently binds to H3.2·H4 with slightly reduced affinity, as compared with H3.1·H4 (Fig. 4C). H3.3·H4 exhibited the lowest apparent binding affinity to sNASP among the human H3 variants, and H3T·H4 and CENP-A·H4 exhibited moderate sNASP binding, which was slightly higher than that of H3.3·H4 (Fig. 4C).
FIGURE 4.
Interactions between sNASP and human H3 variants. A, shown is a 16% SDS-PAGE analysis of purified human H3 variants complexed with H4. B, shown is an electrophoretic mobility shift assay. sNASP (2.4 μg) was incubated with different amounts of H3.1·H4 (lanes 2–5), H3.2·H4 (lanes 7–10), H3.3·H4 (lanes 12–15), H3T·H4 (lanes 17–20), CENP-A·H4 (lanes 22–25), and H2A·H2B (lanes 27–30), and the complexes were detected by non-denaturing 5% PAGE with Coomassie Brilliant Blue staining. The amounts of histones were 0 μg (lanes 1, 6, 11, 16, 21, and 26), 0.75 μg (lanes 2, 7, 12, 17, 22, and 27), 1.5 μg (lanes 3, 8, 13, 18, 23, and 28), 2.25 μg (lanes 4, 9, 14, 19, 24, and 29), and 3 μg (lanes 5, 10, 15, 20, 25, and 30). C, shown is a graphic representation of the sNASP binding to H3.1·H4, H3.2·H4, H3.3·H4, H3T·H4, and CENP-A·H4. The relative amounts of sNASP in the complex with H3.1·H4 (closed circles), H3.2·H4 (closed triangles), H3.3·H4 (closed squares), H3T·H4 (open circles), and CENP-A·H4 (open triangles) are plotted as the averages of three independent experiments with the S.D. values.
sNASP Specificity for the H3 Variants in Nucleosome Assembly
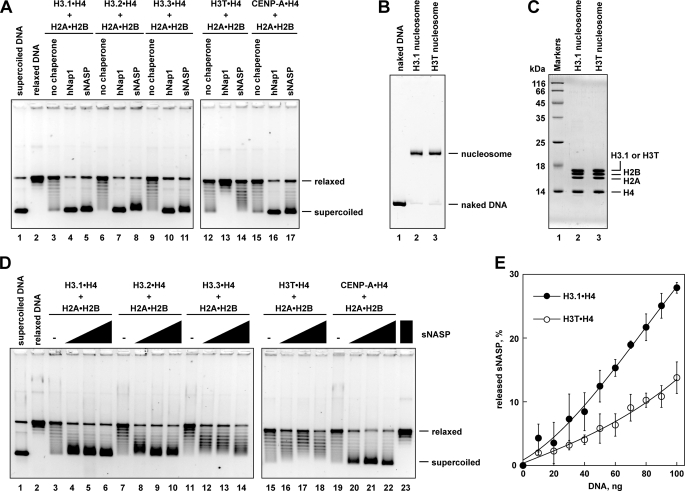
We then tested sNASP-mediated nucleosome assembly with these H3·H4 complexes containing the human H3 variants by a topological assay. As shown in Fig. 5A (lanes 3–11), sNASP exhibited comparable nucleosome assembly activity to that of hNap1 with H3.1, H3.2, and H3.3. sNASP also efficiently formed the nucleosome containing the centromere-specific H3 variant, CENP-A (Fig. 5A, lane 17). In contrast, sNASP was significantly defective in the H3T nucleosome assembly (Fig. 5A, lane 14), although it bound to H3T·H4 (Fig. 4, B, lanes 16–20, and C). The amount of the H3T nucleosome formed by sNASP was below the background level (Fig. 5A, lanes 12 and 14). Consistent with our previous results, hNap1 did not promote the assembly of the H3T nucleosome (Fig. 5A, lane 13). It should be noted that hNap1 inhibited the spontaneous H3T nucleosome assembly, which was detected in the absence of histone chaperone (Fig. 5A, lane 12). On the other hand, unlike hNap1, sNASP may not inhibit the spontaneous H3T nucleosome assembly (Fig. 5A, lanes 12–14). The H3T·H4 complex used in this study was perfectly competent for nucleosome formation, as assessed by salt dialysis (Fig. 5, B and C), indicating that the defective H3T nucleosome formation by sNASP and hNap1 is not due to the inability of H3T to participate in nucleosome formation.
FIGURE 5.
Nucleosome assembly with human H3 variants by sNASP. A, nucleosome assembly with H3 variants by sNASP is shown. The topological assay was employed. Relaxed ϕX174 DNA (10 ng/μl), which was previously treated with wheat germ topoisomerase I (lane 2), was incubated with hNap1 (lanes 4, 7, 10, 13, and 16) or sNASP (lanes 5, 8, 11, 14, and 17) in the presence of core histones. Lanes 3–5, 6–8, 9–11, 12–14, and 15–17 indicate experiments with H3.1·H4, H3.2·H4, H3.3·H4, H3T·H4, and CENP-A·H4, respectively. The reaction products were then analyzed by 1% agarose gel electrophoresis in 1 × TAE buffer. Lanes 3, 6, 9, 12, and 15 indicate negative control experiments without histone chaperone in the presence of core histones. The concentration of hNap1 or sNASP was 1 μm. B, the H3.1 and H3T nucleosomes reconstituted by salt dialysis were fractionated using a Prepcell apparatus and were analyzed by non-denaturing 6% PAGE. C, histone compositions of the purified H3.1 and H3T nucleosomes were analyzed by 18% SDS-PAGE. D, shown are protein titration experiments. The reactions were conducted as described in panel A. Lanes 3–6, 7–10, 11–14, 15–18, and 19–22 indicate experiments with H3.1·H4, H3.2·H4, H3.3·H4, H3T·H4, and CENP-A·H4, respectively. The concentrations of sNASP were 0 μm (lanes 3, 7, 11, 15, and 19), 0.1 μm (lanes 4, 8, 12, 16, and 20), 0.2 μm (lanes 5, 9, 13, 17, and 21), and 0.4 μm (lanes 6, 10, 14, 18, and 22). Lane 23 represents a negative control experiment without core histones. E, shown is deposition of H3.1·H4 or H3T·H4 from the sNASP·H3.1·H4 or sNASP·H3T·H4 complex onto DNA, analyzed by non-denaturing 5% PAGE with Coomassie Brilliant Blue staining. sNASP (2.4 μg) was incubated without or with H3.1·H4 (1 μg) or H3T·H4 (1.5 μg) to form the complex; about half of the sNASP remained free under these conditions. After the incubation with supercoiled DNA, the samples were analyzed. The amounts of competitor DNA were 0, 10, 20, 30, 40, 50, 60, 70, 80, 90, and 100 ng. The amounts of sNASP released from sNASP·H3.1·H4 (closed circles) or sNASP·H3T·H4 complex (open circles) are plotted as the averages of three independent experiments with the S.D. values.
Because negative supercoils were introduced as a consequence of nucleosome formation, the longer migration distance on the gel corresponded to the higher number of nucleosomes assembled by sNASP. Careful sNASP titration experiments revealed that sNASP efficiently promoted nucleosome assembly with H3.1 and H3.2 (Fig. 5D, lanes 3–10). H3.3 was incorporated into nucleosomes by sNASP with reduced efficiency, as compared with H3.1 and H3.2 (Fig. 5D, lanes 11–14). Interestingly, sNASP robustly promoted the assembly of nucleosomes containing the centromere-specific H3, CENP-A (Fig. 5D, lanes 19–22). These specificities for the H3 variants in the sNASP-mediated nucleosome formation are almost consistent with the apparent affinity of sNASP to the H3 variants complexed with H4, except for H3T (Fig. 4C). This discrepancy with H3T may be due to the defective H3T·H4 deposition onto DNA by sNASP.
To test this possibility, we performed an H3·H4 deposition assay (10). When the sNASP·H3T·H4 or sNASP·H3.1·H4 complex was incubated in the presence of DNA, sNASP may be released from the complex with the histones, if the histones are deposited onto DNA. As shown in Fig. 5E, sNASP was efficiently released from the complex with H3.1·H4 in the presence of DNA. In contrast, sNASP was not efficiently released from the sNASP·H3T·H4 complex in the presence of DNA (Fig. 5E). These results support the hypothesis that sNASP binds to H3T·H4, but it is defective in the H3T·H4 deposition onto DNA.
Mutational Analysis of the Amino Acid Residue(s) Responsible for the Defective H3T Nucleosome Formation by sNASP
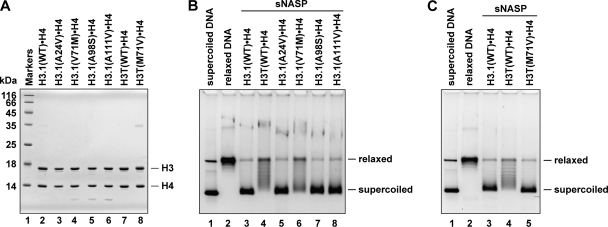
To identify the amino acid residue(s) responsible for the defective formation of the H3T nucleosome by sNASP, we prepared the H3·H4 complexes containing the H3.1 mutants, H3.1(A24V), H3.1(V71M), H3.1(A98S), and H3.1(A111V), in which each H3.1 residue, Ala-24, Val-71, Ala-98, or Ala-111, was replaced by the corresponding amino acid residue of H3T (Fig. 6A). We then tested these H3.1 mutants for the sNASP-mediated nucleosome formation. When the H3.1(V71M) mutant was used for the experiment, the sNASP-mediated nucleosome formation was significantly decreased and resembled that of H3T (Fig. 6B). In contrast, sNASP efficiently promoted nucleosome formation with the H3.1(A24V), H3.1(A98S), and H3.1(A111V) mutants (Fig. 6B). A reciprocal experiment with the H3T(M71V) mutant (Fig. 6A) confirmed that the M71V mutation of H3T significantly improved the efficiency of the sNASP-mediated formation of the H3T nucleosome (Fig. 6C). Therefore, we conclude that the Met-71 residue of H3T is responsible for the defective incorporation of H3T into nucleosomes by sNASP.
FIGURE 6.
Mutational analyses of the amino acid residue(s) required for H3T incorporation into nucleosomes. A, shown is SDS-PAGE analysis of the H3.1 and H3T mutants complexed with H4. Lane 1, molecular mass markers. Lanes 2–6, H3.1 mutants complexed with H4; lane 2, H3.1(WT); lane 3, H3.1(A24V); lane 4; H3.1(V71M); lane 5; H3.1(A98S); lane 6; H3.1(A111V). Lanes 7 and 8, H3T mutants complexed with H4; lane 7, H3T(WT), lane 8, H3T(M71V). B, nucleosome reconstitution with the H3.1 mutants by sNASP is shown. The topological assay was employed. Relaxed ϕX174 DNA (10 ng/μl), which was previously treated with wheat germ topoisomerase I (lane 2), was incubated with sNASP in the presence of core histones. Lanes 3–8 indicate experiments with H3.1(WT)·H4, H3T(WT)·H4, H3.1(A24V)·H4, H3.1(V71M)·H4, H3.1(A98S)·H4, and H3.1(A111V)·H4, respectively. The reaction products were then analyzed by 1% agarose gel electrophoresis in 1 × TAE buffer. The sNASP concentration was 1 μm. C, nucleosome reconstitution with the H3T(M71V) mutant by sNASP is shown. The topological assay was employed. Relaxed ϕX174 DNA (10 ng/μl), which was previously treated with wheat germ topoisomerase I (lane 2), was incubated with sNASP in the presence of core histones. Lanes 3–5 indicate experiments with H3.1(WT)·H4, H3T(WT)·H4, and H3T(M71V)·H4, respectively.
Mapping of the Functional Domain of sNASP for the Nucleosome Assembly Activity
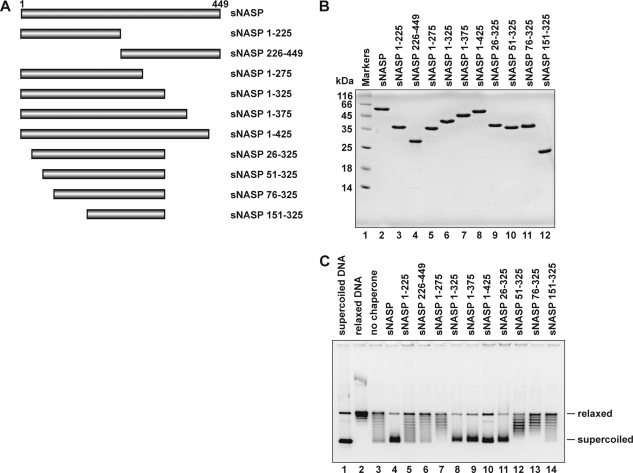
To identify the functional domain of sNASP responsible for the nucleosome assembly activity, we designed 10 sNASP fragments (Fig. 7A). These sNASP fragments contained amino acid residues 1–225, 226–449, 1–275, 1–325, 1–375, 1–425, 26–325, 51–325, 76–325, and 151–325. We purified these sNASP fragments by the same method, including the His6-tag removal step, as the full-length sNASP purification (Fig. 7B). We then tested the nucleosome assembly activity by a topological assay. As shown in Fig. 7C, significant nucleosome assembly activity was detected with the 1–325, 1–375, 1–425, and 26–325 fragments, but the 1–225, 226–449, 1–275, 51–325, 76–325, and 151–325 fragments were defective in the nucleosome assembly. Therefore, the region of amino acids 26–325 contains the nucleosome assembly domain of sNASP.
FIGURE 7.
Deletion analysis of sNASP. A, shown is a schematic representation of the sNASP fragments. B, shown is a 15% SDS-PAGE analysis of the purified sNASP fragments (lanes 3–12). These sNASP fragments lack the His6 tag. Lane 1 indicates the molecular mass markers. Lane 2 represents full-length sNASP. C, nucleosome assembly by the sNASP fragments is shown. Relaxed ϕX174 DNA (10 ng/μl), which was previously treated with wheat germ topoisomerase I (lane 2), was incubated with sNASP or the sNASP fragments in the presence of core histones. Lane 3 indicates a negative control experiment without sNASP or the sNASP fragments in the presence of core histones. The reaction products were then analyzed by 1% agarose gel electrophoresis in 1 × TAE buffer. The concentrations of sNASP and the sNASP fragments were 1 μm.
DISCUSSION
In mammalian cells, two spliced forms of NASP, tNASP and sNASP, were identified (47). tNASP is highly expressed in testis, suggesting its function during spermatogenesis. On the other hand, sNASP is ubiquitously found in somatic cells. Several studies suggested that the NASP proteins may be chaperones specific for the linker histone H1 (46–50). However, significant sequence similarity has been found between sNASP and the N1/N2 family of proteins, whose members are histone H3·H4-specific chaperones. Parthun and co-workers (52) elegantly showed that sNASP preferentially binds to both histones H1 and H3·H4 in vitro and in vivo. This finding suggests that sNASP may function as a chaperone for both the linker histone H1 and core histones H3·H4. However, the H3·H4 chaperone activity of sNASP alone was not reported so far, although sNASP reportedly supported nucleosome assembly in the presence of a yeast cytosolic extract (52). The requirement of the yeast cytosolic extract in this sNASP-dependent nucleosome formation implied that sNASP may not be able to generate stable nucleosomes by itself (52). Therefore, whether sNASP indirectly deposits histones onto DNA through other factors that directly assemble nucleosomes or whether sNASP is more directly involved in histone deposition remains to be determined.
In the present study we found that (i) sNASP itself promotes nucleosome assembly with four core histones, (ii) sNASP promotes tetrasome assembly with H3·H4 in the absence of H2A·H2B, and (iii) sNASP mediates nucleosome assembly with the human histone H3 variants, H3.1, H3.2, H3.3, and CENP-A, but not with H3T, with different efficiencies. (iv) The Val-71 residue, which is not conserved in H3T, is essential for the sNASP-mediated nucleosome assembly. In addition, we found that (v) the central region, amino acid residues 26–325, of sNASP is responsible for the nucleosome assembly in vitro. These new results provide a compelling answer for the previously unsolved question of whether sNASP is able to directly promote nucleosome assembly. Therefore, we conclude that sNASP may function as a bona fide histone chaperone for H3·H4, in addition to the linker histone H1. It should be noted that the purified sNASP used in this study lacked the His6 tag, whereas the sNASP used in the previous biochemical analysis (52) contained the His6 tag. This difference between the sNASP preparations may have caused the difference in the nucleosome assembly activity, although other experimental differences, such as the composition of the reaction mixtures, may also have contributed. sNASP is an extremely acidic protein (pI = 4.35). The addition of the basic His6 tag to the sNASP sequence may affect the nucleosome assembly activity of sNASP because the binding of sNASP to the basic histones could largely depend on its extreme acidity. A yeast factor(s), which complements the negative effect of the His6 tag, may be required to detect the nucleosome assembly activity of the His6-tagged sNASP protein.
In the present study we found that sNASP is inefficient in the nucleosome assembly with a testis-specific histone variant, H3T. Previously, we reported that a conventional histone chaperone, hNap1, is defective in nucleosome assembly with H3T and that the Val-111 residue of H3T is responsible for the defective H3T nucleosome assembly by hNap1 (10). These results suggest that both sNASP and hNap1 discriminate H3T from other histone H3 variants. However, interestingly, our mutational analyses revealed that sNASP and hNap1 require different amino acid residues, Met-71 and Val-111, respectively, for the H3T discrimination. This difference may reflect variations in the nucleosome assembly mechanisms between sNASP and hNap1. Structural analyses of sNASP and hNap1 complexed with H3·H4 are awaited to address this issue.
Intriguingly, an Schizosaccharomyces pombe homologue of sNASP, Sim3, was found to be important for S. pombe CENP-A localization at the centromere region of chromosomes (54). This suggested that sNASP may also function in the formation of the centromere-specific nucleosome containing CENP-A. Sim3 reportedly interacted with non-chromosomal CENP-A (54); however, human NASPs were found only in the cytosolic H3 complex (26, 51). In the present study we found that sNASP binds to the human CENP-A·H4 complex and promotes the assembly of the CENP-A nucleosome in vitro. Interestingly, CENP-A was one of the efficient substrates for nucleosome assembly by sNASP. These findings support the idea that sNASP may function as a chaperone for the centromere-specific nucleosome formation. A histone chaperone, HJURP, for the CENP-A nucleosome, has been identified in humans (41, 42). Given that sNASP, like HJURP, functions as a CENP-A chaperone, it would be intriguing to determine how these two CENP-A chaperones participate in the formation of the centromere-specific chromatin.
This work was supported in part by grants-in-aid from the Japanese Society for the Promotion of Science and the Ministry of Education, Culture, Sports, Science, and Technology, Japan.
- hNap1
- human Nap1
- NASP
- nuclear autoantigenic sperm protein
- sNASP
- somatic NASP
- tNASP
- testicular NASP
- MNase
- micrococcal nuclease
- Ni-NTA
- nickel-nitrilotriacetic acid
- DTT
- dithiothreitol
- TBE
- Tris-buffered EDTA.
REFERENCES
- 1.Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 2.Arents G., Burlingame R. W., Wang B. C., Love W. E., Moudrianakis E. N. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 10148–10152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akey C. W., Luger K. (2003) Curr. Opin. Struct. Biol. 13, 6–14 [DOI] [PubMed] [Google Scholar]
- 4.Loyola A., Almouzni G. (2004) Biochim. Biophys. Acta 1677, 3–11 [DOI] [PubMed] [Google Scholar]
- 5.Henikoff S., Ahmad K. (2005) Annu. Rev. Cell Dev. Biol. 21, 133–153 [DOI] [PubMed] [Google Scholar]
- 6.Polo S. E., Almouzni G. (2006) Curr. Opin. Genet. Dev. 16, 104–111 [DOI] [PubMed] [Google Scholar]
- 7.Park Y. J., Luger K. (2008) Curr. Opin. Struct. Biol. 18, 282–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henikoff S. (2008) Nat. Rev. Genet. 9, 15–26 [DOI] [PubMed] [Google Scholar]
- 9.McBryant S. J., Park Y. J., Abernathy S. M., Laybourn P. J., Nyborg J. K., Luger K. (2003) J. Biol. Chem. 278, 44574–44583 [DOI] [PubMed] [Google Scholar]
- 10.Tachiwana H., Osakabe A., Kimura H., Kurumizaka H. (2008) Nucleic Acids Res. 36, 2208–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaufman P. D., Kobayashi R., Kessler N., Stillman B. (1995) Cell 81, 1105–1114 [DOI] [PubMed] [Google Scholar]
- 12.Ray-Gallet D., Quivy J. P., Scamps C., Martini E. M., Lipinski M., Almouzni G. (2002) Mol. Cell 9, 1091–1100 [DOI] [PubMed] [Google Scholar]
- 13.English C. M., Maluf N. K., Tripet B., Churchill M. E., Tyler J. K. (2005) Biochemistry 44, 13673–13682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mousson F., Lautrette A., Thuret J. Y., Agez M., Courbeyrette R., Amigues B., Becker E., Neumann J. M., Guerois R., Mann C., Ochsenbein F. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 5975–5980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antczak A. J., Tsubota T., Kaufman P. D., Berger J. M. (2006) BMC Struct. Biol. 6, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.English C. M., Adkins M. W., Carson J. J., Churchill M. E., Tyler J. K. (2006) Cell 127, 495–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Natsume R., Eitoku M., Akai Y., Sano N., Horikoshi M., Senda T. (2007) Nature 446, 338–341 [DOI] [PubMed] [Google Scholar]
- 18.Murzina N. V., Pei X. Y., Zhang W., Sparkes M., Vicente-Garcia J., Pratap J. V., McLaughlin S. H., Ben-Shahar T. R., Verreault A., Luisi B. F., Laue E. D. (2008) Structure 16, 1077–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahmad K., Henikoff S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 16477–16484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malik H. S., Henikoff S. (2003) Nat. Struct. Biol. 10, 882–891 [DOI] [PubMed] [Google Scholar]
- 21.Henikoff S., Furuyama T., Ahmad K. (2004) Trends Genet. 20, 320–326 [DOI] [PubMed] [Google Scholar]
- 22.Kamakaka R. T., Biggins S. (2005) Genes Dev. 19, 295–310 [DOI] [PubMed] [Google Scholar]
- 23.Hake S. B., Allis C. D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 6428–6435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loyola A., Almouzni G. (2007) Trends Biochem. Sci. 32, 425–433 [DOI] [PubMed] [Google Scholar]
- 25.Black B. E., Bassett E. A. (2008) Curr. Opin. Cell Biol. 20, 91–100 [DOI] [PubMed] [Google Scholar]
- 26.Tagami H., Ray-Gallet D., Almouzni G., Nakatani Y. (2004) Cell 116, 51–61 [DOI] [PubMed] [Google Scholar]
- 27.Jin J., Cai Y., Li B., Conaway R. C., Workman J. L., Conaway J. W., Kusch T. (2005) Trends Biochem. Sci. 30, 680–687 [DOI] [PubMed] [Google Scholar]
- 28.Sarma K., Reinberg D. (2005) Nat. Rev. Mol. Cell Biol. 6, 139–149 [DOI] [PubMed] [Google Scholar]
- 29.Sanchez-Pulido L., Pidoux A. L., Ponting C. P., Allshire R. C. (2009) Cell 137, 1173–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishimi Y., Yasuda H., Hirosumi J., Hanaoka F., Yamada M. (1983) J. Biochem. 94, 735–744 [DOI] [PubMed] [Google Scholar]
- 31.Ishimi Y., Hirosumi J., Sato W., Sugasawa K., Yokota S., Hanaoka F., Yamada M. (1984) Eur. J. Biochem. 142, 431–439 [DOI] [PubMed] [Google Scholar]
- 32.Mazurkiewicz J., Kepert J. F., Rippe K. (2006) J. Biol. Chem. 281, 16462–16472 [DOI] [PubMed] [Google Scholar]
- 33.Andrews A. J., Downing G., Brown K., Park Y. J., Luger K. (2008) J. Biol. Chem. 283, 32412–32418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okuwaki M., Kato K., Shimahara H., Tate S., Nagata K. (2005) Mol. Cell. Biol. 25, 10639–10651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu R. J., Lee M. P., Johnson L. A., Feinberg A. P. (1996) Hum. Mol. Genet. 5, 1743–1748 [DOI] [PubMed] [Google Scholar]
- 36.Gaillard P. H., Martini E. M., Kaufman P. D., Stillman B., Moustacchi E., Almouzni G. (1996) Cell 86, 887–896 [DOI] [PubMed] [Google Scholar]
- 37.Kamakaka R. T., Bulger M., Kaufman P. D., Stillman B., Kadonaga J. T. (1996) Mol. Cell. Biol. 16, 810–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Izuta H., Ikeno M., Suzuki N., Tomonaga T., Nozaki N., Obuse C., Kisu Y., Goshima N., Nomura F., Nomura N., Yoda K. (2006) Genes Cells 11, 673–684 [DOI] [PubMed] [Google Scholar]
- 39.Foltz D. R., Jansen L. E., Black B. E., Bailey A. O., Yates J. R., 3rd, Cleveland D. W. (2006) Nat. Cell Biol. 8, 458–469 [DOI] [PubMed] [Google Scholar]
- 40.Okada M., Cheeseman I. M., Hori T., Okawa K., McLeod I. X., Yates J. R., 3rd, Desai A., Fukagawa T. (2006) Nat. Cell Biol. 8, 446–457 [DOI] [PubMed] [Google Scholar]
- 41.Dunleavy E. M., Roche D., Tagami H., Lacoste N., Ray-Gallet D., Nakamura Y., Daigo Y., Nakatani Y., Almouzni-Pettinotti G. (2009) Cell 137, 485–497 [DOI] [PubMed] [Google Scholar]
- 42.Foltz D. R., Jansen L. E., Bailey A. O., Yates J. R., 3rd, Bassett E. A., Wood S., Black B. E., Cleveland D. W. (2009) Cell 137, 472–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kleinschmidt J. A., Franke W. W. (1982) Cell 29, 799–809 [DOI] [PubMed] [Google Scholar]
- 44.Kleinschmidt J. A., Fortkamp E., Krohne G., Zentgraf H., Franke W. W. (1985) J. Biol. Chem. 260, 1166–1176 [PubMed] [Google Scholar]
- 45.Kleinschmidt J. A., Dingwall C., Maier G., Franke W. W. (1986) EMBO J. 5, 3547–3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richardson R. T., Alekseev O. M., Grossman G., Widgren E. E., Thresher R., Wagner E. J., Sullivan K. D., Marzluff W. F., O'Rand M. G. (2006) J. Biol. Chem. 281, 21526–21534 [DOI] [PubMed] [Google Scholar]
- 47.Richardson R. T., Batova I. N., Widgren E. E., Zheng L. X., Whitfield M., Marzluff W. F., O'Rand M. G. (2000) J. Biol. Chem. 275, 30378–30386 [DOI] [PubMed] [Google Scholar]
- 48.Alekseev O. M., Bencic D. C., Richardson R. T., Widgren E. E., O'Rand M. G. (2003) J. Biol. Chem. 278, 8846–8852 [DOI] [PubMed] [Google Scholar]
- 49.Alekseev O. M., Widgren E. E., Richardson R. T., O'Rand M. G. (2005) J. Biol. Chem. 280, 2904–2911 [DOI] [PubMed] [Google Scholar]
- 50.Finn R. M., Browne K., Hodgson K. C., Ausió J. (2008) Biophys. J. 95, 1314–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loyola A., Tagami H., Bonaldi T., Roche D., Quivy J. P., Imhof A., Nakatani Y., Dent S. Y., Almouzni G. (2009) EMBO Rep. 10, 769–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang H., Walsh S. T., Parthun M. R. (2008) Nucleic Acids Res. 36, 5763–5772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanaka Y., Tawaramoto-Sasanuma M., Kawaguchi S., Ohta T., Yoda K., Kurumizaka H., Yokoyama S. (2004) Methods 33, 3–11 [DOI] [PubMed] [Google Scholar]
- 54.Dunleavy E. M., Pidoux A. L., Monet M., Bonilla C., Richardson W., Hamilton G. L., Ekwall K., McLaughlin P. J., Allshire R. C. (2007) Mol. Cell 28, 1029–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]