Abstract
Telomeres are repetitive nucleoprotein structures that cap the ends of chromosomes. Without telomerase, telomeres shorten with replication and eventually signal cell cycle arrest (cell senescence). Homologous recombination (HR)-based mechanisms slow senescence, and distinct HR mechanisms support the growth of the rare survivors of senescence. Here, we report novel roles for the post-translational modification of small ubiquitin-like modifier (SUMO) in regulating the rate of senescence in Saccharomyces cerevisiae telomerase mutants. We identify Mms21 as the relevant SUMO E3 ligase and demonstrate that cells lacking Mms21-dependent sumoylation accumulate HR intermediates selectively at telomeres during senescence. One target of Mms21-dependent sumoylation is the cohesin- and condensin-related Smc5-Smc6 complex (Smc5/6). We show that hypomorphic smc5 or smc6 alleles exhibit phenotypes similar to mms21 sumoylation-deficient mutants with regard to senescence and the accumulation of unresolved HR intermediates. Further, we provide evidence that Mms21 and Smc5/6 prevent aberrant recombination between sister telomeres and also globally facilitate resolution of sister chromatid HR intermediates.
Keywords: DNA Recombination, DNA Replication, DNA Structure, Sumoylation, Telomere, Smc5/6, Senescence, Telomerase
Introduction
Telomeres comprise the repeated DNA sequences and associated proteins that protect the ends of linear chromosomes. Telomeres can be elongated via the ribonucleoprotein telomerase, which promotes de novo telomere addition and, by doing so, allows cells to combat the loss of terminal sequences that occur during telomere replication and processing. If telomerase is down-regulated or genetically inactivated, telomere length decreases with each cell division until a critical length is reached that signals cell cycle arrest, termed replicative senescence (1, 2). The yeast Saccharomyces cerevisiae expresses telomerase constitutively, but the genetic inactivation of telomerase (e.g. tlc1Δ mutants, which lack the telomerase RNA template) causes telomere loss leading to senescence (3, 4). Homologous recombination (HR)2-dependent mechanisms slow senescence and also enable the emergence of rare telomerase-independent “survivors” of senescence (4–7). A mechanism of telomere maintenance similar to that used by yeast survivors is seen in a small but significant subset of human cancers termed ALT (for alternative lengthening of telomeres) (8–10). Examination of ALT cells has revealed that the mammalian homologues of several of the proteins first identified as important in S. cerevisiae telomerase-independent survivors appear to function in the mammalian ALT mechanism, making S. cerevisiae an attractive model for studying HR-based telomere maintenance (5, 11–15). In yeast, although the HR mechanisms that slow senescence and those that support survivor growth use shared factors (e.g. Rad52), the mechanisms are nonetheless distinct. For example, survivors use a break-induced replication mechanism that depends on Pol32 (a nonessential subunit of DNA polymerase δ) and that involves inter-telomere recombination, but pol32 deletion does not affect the rate of senescence in telomerase mutants (16). The rate of senescence instead appears to be modulated by HR mechanisms that involve replication-associated template switch events (5, 17, 18). Therefore, HR factors may affect telomere maintenance differently in senescing cells and survivors.
Several years ago we identified a role for the Slx5 and Slx8 complex of proteins in slowing the senescence of yeast tlc1Δ mutants in a fashion that appeared likely to depend on HR (19). Subsequently, it has become clear that Slx5/8 regulate the level of proteins that are modified by the SUMO (small ubiquitin-like modifier) protein (20–22). In addition, several of the proteins that are important in HR-based telomere maintenance are posttranslationally modified by SUMO (23–25). These observations raised the possibility that SUMO regulates the pace of senescence in tlc1Δ mutants.
The addition of SUMO to target proteins (sumoylation) is essential for viability in S. cerevisiae and higher eukaryotes (26–28). In S. cerevisiae, there is a single SUMO protein, Smt3, which is conjugated to protein targets in a manner similar to that of ubiquitination. Activated SUMO is first conjugated to the E1 heterodimer (Aos1/Uba2), which then transfers the SUMO moiety to the E2 ligase, Ubc9, which in turn mediates the specific conjugation of SUMO to target proteins, with or without the assistance of an E3 ligase (Siz1, Siz2, or Mms21) (29–31). Sumoylation has been shown to have important roles in protein localization, protein-protein interactions, enzyme kinetics, and protein turnover (29). Although a large portion of the yeast proteome (∼5%) is modified by sumoylation, for most of these proteins the functional consequences of sumoylation remain unknown (32–35).
Recently, Potts and Yu (36) reported a role for the SMC5/6 complex in the maintenance of telomeres in human ALT cells. They uncovered a role for the complex in sumoylating components of the telomere shelterin complex, including TRF1, TRF2, RAP1, and TIN1, and apparently thus facilitating telomere recruitment or retention to ALT-associated promyelocytic leukemia bodies (APBs), which are thought to be the sites of HR-based telomere maintenance in ALT cells (36–38). Although this report establishes a role for the SMC5/6 complex in recruiting telomeres to promyelocytic leukemia bodies, whether that is the only function the complex plays in recombination based telomere maintenance is unknown.
The SMC5/6 complex is one of three structural maintenance of chromosome (SMC) complexes within eukaryotic cells (39, 40). Although the SMC5/6 complex is conserved and essential in eukaryotes, it is arguably the least well understood of the SMC complexes and has been the recent subject of intensive investigation. SMC5/6 promotes the HR-dependent repair of DNA damage, including DNA replication lesions that arise spontaneously or secondary to DNA damage caused by exogenous compounds such as the alkylating agent methyl methanesulfonate (MMS) (41, 42). Cells with hypomorphic smc5 or smc6 alleles fail to properly separate chromatids at mitosis, leading to their fragmentation and missegregation, and this defect is partially suppressed by inactivation of HR (e.g. via rad51 or rad52 deletion in yeast) (41, 43–46). These observations are consistent with a role for the SMC5/6 complex in the resolution of interchromatid HR intermediates, but this model has not been tested directly. Increased levels of such apparent intermediates have indeed been observed in hypomorphic smc6 mutants, although the precise character of the intermediates, i.e. whether they are convergent replication forks, Holliday junctions, or hemicatenane-related species, has not been examined (43, 45, 46). The SMC5/6 complex comprises eight subunits, including a SUMO E3 ligase (MMS21) whose activity is likely assisted by the integrity of the complex (31; for review, see Ref. 40). In S. cerevisiae, Mms21 has been shown to sumoylate Smc5 along with several other targets, although the functional significance of this sumoylation remains unclear (31, 47).
Here we utilize S. cerevisiae to determine the roles of sumoylation in tlc1Δ mutants during senescence and demonstrate that sumoylation-deficient telomerase-null cells senesce at an elevated rate. We further show that Mms21 is the key E3 ligase mediating the effect of sumoylation on senescence, and that during senescence cells deficient in Mms21-dependent sumoylation exhibit elevated levels of recombination intermediates selectively at their telomere ends. Finally we provide evidence that the Smc5/6 complex is a target of Mms21-dependent sumoylation during DNA repair throughout the genome, including telomeres during senescence, and serves to ensure the faithful completion of template switch recombination.
EXPERIMENTAL PROCEDURES
Yeast Strains
The BY4741/2 background was used for all experiments. The smc5-6 and smc6-9 alleles were backcrossed four times into the BY4741/2 background before being used in experiments. Details of individual strains are provided in supplemental Table 3.
Senescence Assays
All senescence assays except the tlc1Δ UBA2 versus tlc1Δ uba2-ts10 set (details provided in supplemental Materials and Methods) were performed by sporulating diploid cells heterozygous for mutations of interest and then selecting haploid strains with the desired genotype. This ensures that all haploids inherit a similar starting telomere length and are derived from the same epigenetic environment. For all senescence assays shown, at least three and in most cases four or more haploid spore products of each genotype were senesced in liquid culture. Senescence assays were performed as described previously (14).
X-structures
Preparation of DNA, probing for ARS304 and ARS305, branch migration, and mung bean nuclease assays were performed as described (48). Analysis of rDNA and telomere recombination structures were performed as reported previously (17). Variability in X-structure levels exists between two-dimensional agarose gel electrophoresis experiments (likely due to differences in sample preparation). To control for this variability, quantitative comparisons between genotypes were made between samples grown and prepared in parallel. For the mms21-sp GAL1-MMS21 experiment, mms21-sp cells containing pRS413-based GAL1-MMS21 were arrested with nocodazole and released into YPA + 2% raffinose medium containing 0.033% MMS for 2.5 h before glucose or galactose were added to a final concentration of 2%. Cells were then allowed to cycle for an additional 3 h prior to harvest.
Statistical Analysis
Two-tailed unpaired t-tests were performed for all comparisons.
RESULTS
Sumoylation Slows the Rate of Senescence
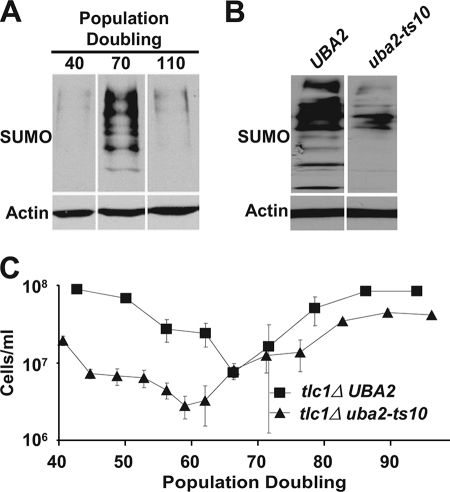
To begin probing the role sumoylation might play in yeast lacking telomerase, we examined bulk levels of sumoylated proteins after deletion of TLC1. We observed a striking increase in the global levels of sumoylated proteins within cells near peak senescence (70 population doublings (PD) after loss of telomerase), which returned to base-line levels in survivors (Fig. 1A).
FIGURE 1.
Sumoylation deficiency increases senescence rate. A, global levels of protein sumoylation in tlc1Δ cells corresponding to early senescence (PD40), peak senescence (PD70), and early survivor formation (PD110), left to right. Proteins ≥60 kDa are shown. All lanes are from the same immunoblot and were exposed for the same length of time. B, global levels of protein sumoylation shown for UBA2 and uba2-ts10 cells grown at 30 °C. Proteins ≥26 kDa are shown. C, senescence assay performed at 30 °C comparing the rate of senescence between tlc1Δ UBA2 and tlc1Δ uba2-ts10 cells. Each point shows the mean ± S.E. (error bars) of four independent spore products.
To determine the functional significance of the increased sumoylation, we compared rates of senescence in tlc1Δ cells with normal or reduced levels of sumoylation. Because sumoylation is essential for viability, a temperature-sensitive mutant of UBA2, a component of the E1 SUMO ligase, was utilized (uba2-ts10). Consistent with previous findings, this allele confers decreases in bulk sumoylation at the semipermissive temperature of 30 °C (Fig. 1B) (49). Diploid TLC1/tlc1Δ UBA2/uba2Δ cells containing plasmid-based UBA2 or a temperature-sensitive uba2-ts10 allele were sporulated, and the haploid tlc1Δ uba2Δ cells containing either plasmid-based UBA2 or uba2-ts10 (tlc1Δ UBA2 and tlc1Δ uba2-ts10, respectively) were senesced via serial passaging in liquid culture at a semipermissive temperature (30 °C). tlc1Δ uba2-ts10 mutants senesced faster than the tlc1Δ UBA2 control, with peak senescence at 59.0 versus 66.2 PD, respectively (Fig. 1C and supplemental Fig. 1). We concluded that sumoylation slows senescence, although the relevant sumoylated targets remained to be identified.
We wish to emphasize three aspects of our experimental approach. First, because dysfunction within the sumoylation pathway in S. cerevisiae has been shown to lead to clonal lethality through the up-regulation of 2-μm plasmid levels, all experiments involving alterations in the SUMO-conjugation pathway were performed with strains cured of the plasmid (50, 51). Second, senescence data were plotted as the extent of growth versus PDs from spore germination, as we have done previously (14, 17) and as is standard in senescence assays of human cultured cells (52). PD rather than time is used as a metric because telomere shortening mediated by the end replication problem and other replication associated events is related to cell division and not time, and furthermore using PD prevents mutations that merely alter the rate of cell division (such as sumoylation deficiency) from being mistakenly viewed as modulating the senescence rate. Finally, we note that the absolute lifespan of a strain of any given genotype varies slightly between experiments. However, this variability does not affect our analyses because all senescence experiments (except Fig. 1C; see “Experimental Procedures”) involved cells of different genotypes that were inherited from the same diploid parent and compared within the same experiment. For this reason, we stress that reliable comparisons of senescence rates can only be made within a given experiment and not between experiments.
Modulation of Senescence by Sumoylation Depends on the E3 Ligase Mms21
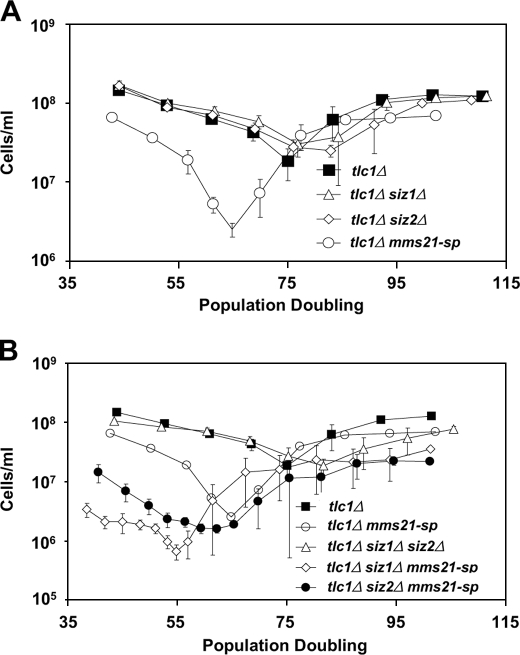
To confirm and further define a functional role for sumoylation in slowing senescence, we investigated which, if any, of the SUMO E3 ligases that are active in vegetative cells, Siz1 Siz2, or Mms21, prevents rapid senescence. Unlike SIZ1 and SIZ2, MMS21 is required for viability, but a sumoylation defective allele of MMS21, mms21-sp, supports viability (this allele encodes a protein containing two point mutations within the RING domain, C200S and H202A; it is also referred to as mms21-CH, and is distinct from the mms21-SP allele) (30, 53, 54). Upon loss of telomerase, only a single E3 ligase mutant, mms21-sp, recapitulated the rapid senescence phenotype of uba2-ts10 cells. Despite performing the bulk of sumoylation within cells, Siz1 and Siz2 had no apparent impact on senescence rate, either alone or in combination (Fig. 2 and supplemental Figs. 2 and 3). Even when combined with the mms21-sp allele, siz1Δ or siz2Δ deletion had only moderate effects on senescence rates (Fig. 2B). These data suggest that Mms21 is the E3 ligase through which sumoylation functions to modulate the rate of senescence.
FIGURE 2.
Mms21 is the SUMO E3 ligase responsible for preventing rapid senescence. A, comparisons of senescence rates among single SUMO E3 ligase mutants. B, comparisons of senescence rates among double SUMO E3 ligase mutants (the triple mutant is inviable). Data in A and B are from one senescence experiment, with all genotypes derived from the same parental diploid. The data are displayed in the two panels for clarity, but data for tlc1Δ mms21-sp mutants are presented in both panels to aid comparisons. Each point represents the mean ± S.E. (error bars) of at least three and in most cases four independent spore products of the indicated genotype.
Mms21 Prevents the Accumulation of Telomere Recombination Intermediates
Having determined that MMS21 was the key E3 ligase responsible for preventing rapid senescence, we sought to uncover its mechanism of action. Previous data have shown that mms21-sp mutants, similar to sgs1Δ mutants, accumulate unresolved recombination intermediates when replicating damaged templates (23). Recent work from our laboratory suggests that Sgs1 prevents rapid senescence by resolving similar recombination intermediates that accumulate at telomeres during senescence (17). Thus, we hypothesized that Mms21, similar to Sgs1, would play a role in the resolution of telomere recombination intermediates during senescence. We do not believe that Mms21 functions via sumoylation of Sgs1 during senescence because Sgs1 sumoylation was reported to be independent of Mms21, and we found that one key indicator of Sgs1 function, the inhibition of growth in top3Δ mutants, was not affected by diminished sumoylation (supplemental Fig. 4) (23, 55). Given these data, Mms21 probably functions in parallel with Sgs1 during senescence.
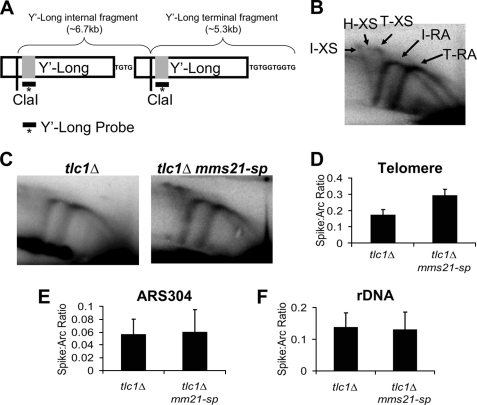
To examine a possible role for Mms21 in the resolution of telomere recombination intermediates, we used two-dimensional agarose gel electrophoresis followed by Southern blotting with probes directed against telomere Y′ elements to visualize replication and recombination intermediates at telomeres. As controls, we also examined genomic regions near a non-telomeric origin of replication (ARS304) or within the rDNA. A brief explanation of telomere two-dimensional Southern patterns is shown in Fig. 3, with supplemental Fig. 5 providing a more in-depth explanation; but succinctly stated, the rightmost replication arc corresponds to the terminal Y′ element, whereas the leftmost arc corresponds to internal Y′ elements. Near senescent tlc1Δ mms21-sp mutants showed a 70% increase in X-shaped molecules at telomere ends versus PD-matched tlc1Δ controls (Fig. 3, C and D). This increase in X-shaped molecules was specific for the telomeres because no increase in the ARS304 region or the rDNA was observed (Fig. 3, E and F). The role for Mms21 in preventing accumulation of X-shaped molecules in senescing cells was not shared with Siz1 or Siz2 because tlc1Δ siz1Δ siz2Δ mutants were found to have levels of X-shaped molecules equal to that of tlc1Δ controls (supplemental Fig. 6). Additional experiments also revealed that the increase in X-shaped molecules seen in tlc1Δ mms21-sp cells is dependent upon loss of telomerase because telomerase-positive mms21-sp mutants displayed no change in the levels of X-structures at telomere ends (supplemental Fig. 7). These data argue that the accumulation of X-shaped intermediates caused by defects in sumoylation may promote senescence, although this remains to be formally tested. We have previously observed a similar specific increase in X-shaped molecules at telomeres during senescence, where it correlated with an elevated utilization of HR-based repair at shortened telomeres (17).
FIGURE 3.
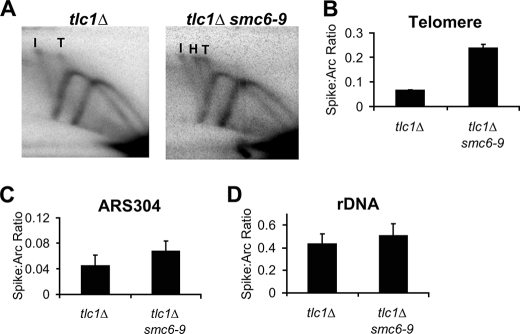
Mms21 regulates the level of telomere recombination intermediates during senescence. A, example of the structure of a S. cerevisiae telomere showing Y′-Long elements and demonstrating the difference in fragment length between internal and terminal Y′-Long elements. Because of the existence of two distinct Y′-Long elements (internal and terminal), analysis of replication intermediates using the Y′-Long probe leads two distinct replication arcs (RA) and X-spikes (XS) being visualized during two-dimensional gel analysis. B, migration pattern of the RA and X-shaped molecules corresponding to the internal (I) and terminal (T) Y′-Long elements within two-dimensional gels are shown. The location of a hybrid (H) X-spike present in smc5/6 mutants is also indicated. C, representative two-dimensional gel analysis of replication intermediates accumulated in tlc1Δ and tlc1Δ mms21-sp strains at the telomere visualized with a probe specific for Y′-Long elements. D–F, comparisons of the ratio of X-shaped molecules to the replication arc within the telomere, near ARS304, and the rDNA locus for tlc1Δ and tlc1Δ mms21-sp mutants at 55 PD after spore germination. The respective p values are p < 0.05, p = 0.97, and p = 0.61 for differences between genotypes. Data represent the mean ± S.E. (error bars) of three to four independent samples/genotype.
Mms21 May Function through Smc5 to Modulate Senescence Rate
Our two-dimensional gel data suggested that Mms21 influences the rate of senescence by allowing for the efficient resolution of telomere recombination intermediates which might impede progression through the cell cycle or lead to nondysjunction events. To identify the protein target of Mms21-mediated sumoylation that prevents the accumulation of telomere X-structures during senescence, we performed a screen of proteins with known roles in telomere biology or HR to identify sumoylated candidates. Of the 17 proteins screened (supplemental Table 1), four (Rad59, Rfa1, Rfa2, and Rfa3) were found to be sumoylated (24). Of note, sumoylation of all four proteins was dramatically elevated upon the addition of the DNA-alkylating agent MMS, but sumoylation of these proteins proved to be primarily dependent upon Siz2 and independent of Mms21 (supplemental Table 2). Having no novel target of Mms21-dependent sumoylation, we focused on to two previously identified targets, Yku70 and Smc5. Yku70 functions in nonhomologous end joining and also at telomeres where it facilitates capping and extension by telomerase (56). Epistasis analysis, detailed in supplemental Fig. 8, did not reveal a role for Mms21 in Yku70-dependent telomere biology.
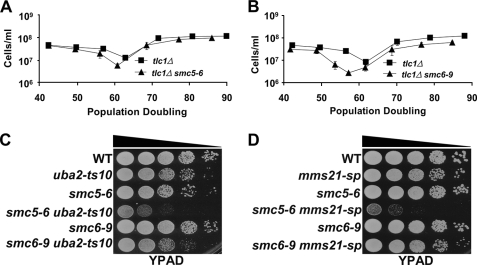
We next turned our attention to Smc5, a member of the essential Smc5/6 octameric protein complex, of which Mms21 is itself a member. Smc5/6 functions in the HR-based repair of double-strand breaks and confers resistance to a variety of genotoxic agents (41, 44, 57–59). Roles for the complex in replication fork stability and/or restart have also been suggested recently, although its precise role in these pathways remains unclear (43, 44, 59, 60). In addition, previous work has found that mms21-sp mutants, similar to hypomorphic smc5-6 and smc6-9 alleles, confer sensitivity to the genotoxic agents MMS and hydroxyurea (supplemental Fig. 9) (39, 40, 46, 54). These data prompted us to investigate whether other members of the Smc5/6 complex function similarly to Mms21 to prevent rapid senescence. Similar to tlc1Δ mms21-sp cells, tlc1Δ smc5-6 and tlc1Δ smc6-9 cells senesced more rapidly than corresponding tlc1Δ controls when grown at a semipermissive temperature, 30 °C (Fig. 4, A and B, and supplemental Fig. 10). Therefore, Smc5/6 functions to slow senescence.
FIGURE 4.
Mms21 may function through the sumoylation of Smc5 to prevent rapid senescence. A and B, senescence assays performed at 30 °C comparing the senescence rate of tlc1Δ smc5-6 and tlc1Δ smc6-9 mutants versus tlc1Δ controls. Each data point represents the mean ± S.E. (error bars) of six independent spore colonies. C and D, spot assay performed at 30 °C comparing the growth rates of uba2-ts10 and mms21-sp mutants alone or in combination with smc5-6 and smc6-9 alleles. For spot assays, 100,000 cells, and 10-fold serial dilutions, were grown on YPAD plates. WT, wild type.
The similar effects of the smc5-6, smc6-9, and mms21-sp alleles suggested that sumoylation of the Smc5/6 complex might explain the rapid senescence of mms21-sp mutants. Alternatively, because the full activity of Mms21 likely depends on the integrity of the Smc5/6 complex, the smc5-6 and smc6-9 alleles might affect Mms21 function (31). Given that Smc5, but not Smc6, is sumoylated in an Mms21-dependent fashion and Mms21 associates with Smc5/6 via a direct interaction with Smc5 (44, 47, 61), these data suggest that Smc5 might be a key target of Mms21. Proof of this would require a smc5 allele encoding a mutant form of Smc5 that cannot be sumoylated by Mms21. No such allele yet exists, despite significant effort to generate one.3 Although they do not constitute definite proof, the following observations nonetheless indicate that Mms21 is likely to exert important effects via sumoylation of Smc5. First, double mutants between uba2-ts10 and smc5-6 or smc6-9 were generated. Because Smc5 and Smc6 are essential proteins, if their optimal function depends on sumoylation, then double mutants between uba2-ts10 and these hypomorphic alleles of SMC5 and SMC6 might show a synergistic enhancement of temperature sensitivity. Interestingly, although smc6-9 mutants are more severely compromised for growth at 37 °C and in the presence of DNA damaging agents than smc5-6 mutants (46), smc5-6 uba2-ts10 cells showed a much stronger growth defect than smc6-9 uba2-ts10 cells at 30 °C (Fig. 4C). Second, double mutants between each of the SUMO E3 ligases and smc5-6 or smc6-9 were generated. Consistent with our senescence data and the smc5-6 uba2-ts10 result, only smc5-6 mms21-sp cells showed a synergistic growth defect (Fig. 4D and supplemental Fig. 11). These epistasis analyses argue that with regard to cell growth, sumoylation must function partly through the Smc5/6 complex. If it instead functioned largely in parallel to Smc5/6, then both smc5-6 and smc6-9 mutants ought to have shown synthetic sickness when combined with sumoylation mutants, contrary to what we observe. Because the Mms21-sp protein has no SUMO ligase activity, the findings also argue that the effect of the smc5-6 allele in this assay is not due to defects in Mms21-dependent sumoylation of targets outside of the Smc5/6 complex. Third, although smc5-6 and smc6-9 alleles contain mutations targeted within the C-terminal Walker-B motif present on each protein, it is possible that the observed synergistic growth defect between mms21-sp and smc5-6 but not smc6-9 is due to the hypomorphic smc6-9 allele already being defective for the biochemical function provided by Mms21, whereas smc5-6 is not (46). If this were in fact true, then one might expect to observe additional differential sickness between smc5-6 and smc6-9 alleles when combined with mutations in genes involved in various DNA damage repair pathways. To address this possibility, epistasis analyses between smc5-6 or smc6-9 and >30 genes involved in DNA damage repair, including a hypomorphic allele of the Smc5/6 complex member NSE5 were performed.4 In all cases the mutations affected smc5-6 and smc6-9 mutants equally, with none showing the preferential sickness observed for the mms21-sp allele. We also point out that in all other cases tested, the smc5-6 and smc6-9 alleles exhibit similar phenotypes, with the smc6-9 allele consistently showing a stronger phenotype (46, 62). Together, these new and previous observations suggest that sumoylation modulates Smc5/6 at least in part by enhancing Smc5 activity or stability. Furthermore when sumoylation of Smc5 was investigated, it was found to increase during senescence, in an Mms21-dependent fashion (supplemental Fig. 12). As a whole, these data are consistent with Smc5 being a key SUMO target of Mms21 in senescing cells. However, even though the selective synthetic growth defect of smc5-6 mms21-sp mutants argues that the smc5-6 mutation affects functions contributing to growth rate that are distinct from Mms21-dependent sumoylation, it is still possible that the senescence-specific function of Smc5 is to support Mms21-dependent sumoylation. Thus, until a nonsumoylatable allele of SMC5 can be tested, the model that Smc5 is the central target of Mms21 during senescence remains speculative.
Smc5/6 Prevents the Accumulation of Telomere Recombination Intermediates during Senescence
If Mms21 slows senescence by stimulating the activity of the Smc5/6 complex and thereby preventing the accumulation of telomere HR intermediates (X-structures), then mutants in Smc5 and Smc6 should accumulate telomere X-structures during senescence. To test this hypothesis, tlc1Δ and tlc1Δ smc6-9 mutants were senesced at 30 °C and harvested at equivalent population doublings, and recombination intermediates at telomeres and other loci were examined via two-dimensional Southern analysis. Given that the smc6-9 allele had a stronger phenotype than smc5-6 in the senescence and DNA damage sensitivity assays, we used it to observe more readily subtle alterations in the level of telomeric X-structures caused by dysfunction within the Smc5/6 complex. As predicted, tlc1Δ smc6-9 double mutants show an increase (250%) in X-shaped molecules at telomere ends compared with controls (Fig. 5, A and B). We were unable to detect significant elevations in telomere X-structures in tlc1Δ smc5-6 double mutants (data not shown), likely because of the weak nature of the smc5-6 allele, but tlc1Δ mutants carrying the stronger smc5-31 allele (63) senesced rapidly and accumulated high levels of telomere X-structures (supplemental Fig. 13). Similar to our tlc1Δ mms21-sp results, no significant increase in X-structures at other genomic loci, such as the region near ARS304 or the rDNA, was observed in tlc1Δ smc6-9 cells compared with controls (Fig. 5, C and D), and no increase in telomere X-spikes was observed in telomerase-positive smc6-9 mutants (supplemental Fig. 7). Of note, smc6-9 mutants have been previously shown to accumulate X-structures in the rDNA locus when grown at 37 °C in the presence of hydroxyurea or nocodazole (46, 62). Our experiments were conducted at 30 °C where these mutants cycle at a near normal rate and do not accumulate significant levels of rDNA X-structures.
FIGURE 5.
Smc6 prevents the accumulation of X-structures during senescence. A, representative two-dimensional gel analysis of replication intermediates accumulated in tlc1Δ and tlc1Δ smc6-9 strains at the telomere visualized with a Y′-Long-specific probe. I and T refer to X-spikes coming from internal and terminal replicating telomere fragments, respectively; H refers to an apparent hybrid X-spike. B–D, comparisons of the ratio of X-shaped molecules to the replication arc within the telomere, near ARS304 and the rDNA locus for tlc1Δ and tlc1Δ smc6-9 cells at 45 PD after spore germination. The means ± S.E. (error bars) from three independent samples/genotype are shown. The respective p values are p < 0.0001, p = 0.15, and p = 0.38.
In addition, tlc1Δ smc6-9 mutants also showed the appearance of a third X-spike. We hypothesize that this new X-spike reflects recombination between regions of homology in the same chromatid, between misaligned sister chromatid regions or between telomere regions present on other chromosomes (further explained under “Discussion” and illustrated in supplemental Figs. 14 and 15). Careful examination revealed that this third spike is also the most prominent seen in the tlc1Δ mms21-sp and tlc1 smc5-31 samples (Fig. 3C, supplemental Fig. 13A, and data not shown).
The Smc5/6 Complex Functions in the Resolution of Recombination Intermediates throughout the Genome
Given our data demonstrating that Mms21 and the Smc5/6 complex prevent the accumulation of telomere recombination intermediates during senescence, we sought to determine whether this role for Smc5/6 in recombination resolution was specific to telomeres or was indicative of a more general role for the complex throughout the genome. Smc5/6 mutants were synchronized with nocodazole and released into fresh medium containing MMS at the nonpermissive temperature, 37 °C. After 2.5 h of growth, cells were harvested and subjected to two-dimensional Southern analysis using probes specific to the nontelomeric regions ARS304 and ARS305. In agreement with previous analysis of mms21 alleles and data concerning smc6-9 published recently but after we had conducted our experiments (23, 45), mms21-sp, smc5-6, and smc6-9 mutants showed a robust increase in X-shaped molecules during replication in the presence of MMS near ARS304 (Fig. 6, A and B); similar results were obtained with a probe for ARS305 (data not shown). The accumulation of these X-shaped molecules depends on HR because rad52 deletion prevented their accumulation (supplemental Fig. 16). Further characterization of the X-structures revealed that mms21-sp, smc5-6, and smc6-9 mutants accumulate recombination intermediates with properties similar to those observed previously in sgs1Δ and ubc9-1 cells (supplemental Fig. 17 summarizes the biochemical properties of different X-shaped molecules) (23, 64). These include the ability to branch migrate in the absence or presence of Mg+2 and sensitivity to mung bean nuclease (Fig. 6C, and supplemental Figs. 18 and 19, and data not shown). Therefore, the X-structures are neither hemicatenanes, convergent replication forks, Holliday junctions nor regressed forks. Rather, they appear to be so-called Rec-X-structures, which are thought to arise from hemicatenane-facilitated template switching between sister chromatids at stalled replication forks, thus enabling bypass of the stall-inducing lesion (18). After replicating past the lesion point, the replicating strand is thought to return to its original parental template, and normal replication resumes (model illustrated in supplemental Fig. 20). If not resolved, the sister chromatids that remain linked are visualized as X-structures by two-dimensional Southern analysis. Because the X-structures that accumulate in smc5/6 and sgs1Δ mutants are similar and because Sgs1 facilitates the resolution of recombination intermediates at telomeric and nontelomeric loci, it is likely that X-structures that accumulate in smc5/6 and mms21-sp mutants are caused by their lack of resolution, rather than by their increased formation. In support of this interpretation, we found that X-structures disappeared rapidly in MMS-treated mms21-sp cells after inducible expression of Mms21 (Fig. 6D). Of note, a role for SMC proteins in template switch recombination appears to be specific for the Smc5/6 complex because mutants in the Smc1/3 cohesin or the Smc2/4 condensin complex did not accumulate similar levels of X-structures when replicating through damaged DNA templates (data not shown).
FIGURE 6.
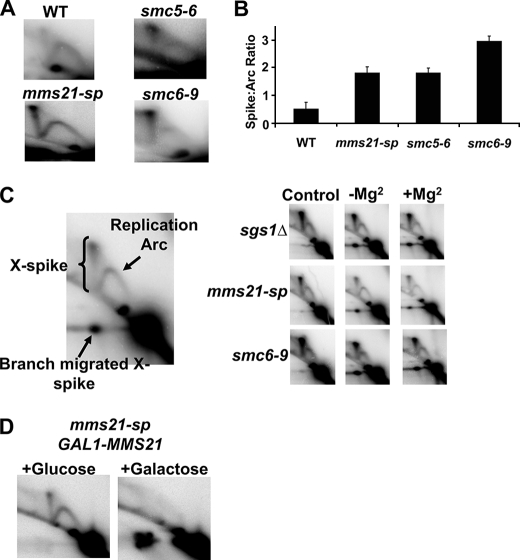
Defects in Smc5/6 lead to elevated levels of unresolved recombination intermediates near the genomic ARS304 region after MMS treatment. A, representative two-dimensional gel analysis of replication intermediates at ARS304 (similar results were obtained at ARS305). WT, wild type. B, quantification of the ratio of X-shaped molecules to structures running within the replication arc. Data represent the mean ± S.E. (error bars) of three or more independent samples per genotype. Comparisons between wild type and mms21-sp, smc5-6, and smc6-9 values were all significant, p < 0.002. C, replication intermediates from two-dimensional gel analysis are shown, and the various species of interest are labeled. Branch migration assay shows control (nonbranch migrated), −Mg2+, and +Mg2+ (samples migrated in the absence or presence of magnesium) reactions. D, expression of Mms21 resolves X-spikes that have accumulated in mms21-sp mutants exposed to MMS (for details, see “Experimental Procedures”).
Given our results showing that the X-shaped molecules formed at genomic loci in smc5/6 mutants represent Rec-X species we sought to determine whether the telomeric X-structures in senescing tlc1Δ smc6-9 cells were similar. The X-structures were able to branch migrate in the absence and presence of Mg+2, indicating that they are indeed Rec-X species (supplemental Fig. 21). Thus, the template switch recombination pathway appears to be active throughout the genome and dependent upon the Smc5/6 complex for its completion.
DISCUSSION
In this work we address the role of sumoylation and the Smc5/6 complex in preventing the accumulation of HR-dependent X-shaped molecules at telomeres and throughout the genome. We propose that the unresolved telomeric X-structures, which accumulate in telomerase-null cells with hypomorphic alleles of the Smc5/6 complex members MMS21, SMC5, and SMC6, explain the rapid senescence of these strains. A previous report identified a role for the human SMC5/6 complex (including MMS21) in the maintenance of telomere length in a different setting, that of ALT cancer cells (36), and our findings extend roles for Smc5/6 in telomere maintenance to the setting of senescence. Potts and Yu (36) proposed that the primary role of SMC5/6 SUMO ligase activity is to mediate the efficient recruitment/retention of telomeres to APBs to properly engage recombination-based pathways of telomere maintenance. Our study suggests that the SMC5/6 complex does not simply function to recruit telomeres to APBs because yeast have no known APB-like structures and because even in the human context APBs are only present in immortalized ALT cells but not in primary cells undergoing senescence. Rather, our findings suggest that Smc5/6 assists in the proper formation and resolution of recombination intermediates between telomeres during senescence. Although the telomere shortening seen following depletion of the SMC5/6 complex in ALT cells was argued to be due to a lack of recruitment of telomeres to APBs, the possibility that the lack of SMC5/6 also directly compromised telomere HR was left open, and our findings indicate that this is indeed the case. The importance of properly functioning HR mechanisms in telomere biology is clear when considering the unidirectional replication of telomeres (39). In contrast to internal genomic sites, where incomplete replication caused by a damaged fork can be rescued by convergent replication from a neighboring fork, no such rescue can occur at the telomere. Telomerase could elongate a shortened telomere arising from the collapse and breakage of a fork within a telomere, but in its absence cells become dependent upon HR mechanisms of fork repair to prevent the loss of telomeric sequence due to fork collapse.
Although mms21-sp mutants have previously been shown to accumulate HR-based intermediates at genomic loci (23), the relevant target of Mms21-dependent sumoylation was not identified, and whether the accumulation of intermediates was due to increased formation or decreased resolution was not fully explored. Here, we attempted to identify the target of Mms21-dependent sumoylation that impacts the accumulation of recombination intermediates, and we provide direct evidence that Mms21 facilitates their resolution.
Recently, Sollier et al. (45) reported that HR-dependent X-structures accumulate at nontelomeric loci in S. cerevisiae smc6-9 mutants and not in other Smc complex mutants after damage with MMS, consistent with our findings. Importantly, the biochemical nature of the X-shaped structures that accumulated in smc5-6 and smc6-9 mutants has not been previously performed. This information is essential for an understanding of the Smc5/6 complex because the pathways involved in the generation of different X-shaped molecules are distinct. For example, Holliday junctions occur during the repair of double-strand breaks, whereas Rec-Xs are believed to arise due to template switching during replication to bypass a stall-inducing lesion. Furthermore, a portion of the X-shaped molecules that accumulate in the rDNA in smc6 mutants under more extreme conditions than those explored in our studies (i.e. 37 °C) were interpreted to be a mixture of recombination intermediates and convergent forks nearing the termination of replication, which are at least in part different from the RecXs that we observed (46, 62). Therefore, Smc5/6 may perform related functions in different settings, and a full characterization of these activities is required to understand the roles of this enigmatic complex.
Recently, Smc5/6 was shown not only to function in resolution of HR intermediates and the completion of DNA replication, but also in the recruitment of Rad52 to stably stalled forks (60, 62). Here, we provide evidence for an additional function of Smc5/6 during replication, which relates to the existence of a third X-spike at telomere ends in tlc1Δ Smc5/6 complex mutants that is not seen in tlc1Δ cells. This X-spike has a size that is intermediate to the X-spikes arising from intersister telomere recombination within terminal or subtelomeric fragments. Smc5/6 mutants are deficient in both equal and unequal sister recombination events, but not in intrachromatid HR (57, 65). Therefore, the simplest explanation for the origin of the third X-spike is from an intrachromatid recombination event, in which a stalled terminal nascent strand invades into an internal homologous region to bypass the stall-inducing lesion. This would create a Rec-X linking a terminal and an internal Y′ fragment, and we therefore refer to it as a hybrid H X-spike (Fig. 7 and supplemental Fig. 14). Alternatively, similar aberrant recombination events between sister chromatids (supplemental Fig. 15) or between different chromosomes could form the hybrid species. We hypothesize that Smc5/6 prevents such abnormal events, either directly or indirectly, by enhancing cohesion between sister telomeres. Such a mechanism would allow the Smc5/6 complex to assist the HR machinery in favoring the selection of nearby homologous sequences on the sister chromatid and thus inhibiting inappropriate recombination reactions.
FIGURE 7.
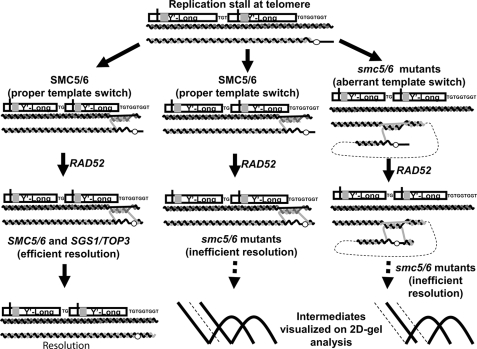
Model depicting roles of the Smc5/6 complex at telomeres. During DNA replication, a damaged base or aberrant DNA structure leads to replication fork stalling and the initiation of template switch recombination to bypass the lesion. Smc5/6 helps ensure the proper selection of template and thus prevents abnormal linkages during the template switch. After replicating past the lesion using a complementary region of the sister chromatid, Rad52-dependent HR returns replication to the original parental template past the lesion. Smc5/6 along with Sgs1 and Top3 then assist in the resolution of the formed Rec-X recombination intermediate (left pathway). In the absence of a properly functioning Smc5/6 complex, telomeres remain linked preventing proper chromatid segregation (center pathway), and in addition are also more likely to use improper templates for template switch events (right pathway). An intrachromatid template switch is depicted, but aberrant events between sister chromatids or different chromosomes are also possible.
Our new findings highlight important roles for Smc5/6 during the replication of damaged DNA or of telomeres that have become dysfunctional due to telomerase inactivation. These mechanistic insights into the role of the Smc5/6 complex in DNA repair and telomere maintenance thus may provide novel avenues by which age-related diseases and cancer might be targeted for therapy.
Supplementary Material
Acknowledgments
We thank Susana Garcia, Sri Bandhakavi, James Morley, and Avinash Bhandoola for outstanding mentorship (to A. C.). We thank Amy Tsou, Brian Keith, and members of the Brown, Greenberg, and Johnson laboratories for helpful discussions; Matt Porteus for comments on the manuscript; and Xiaolan Zhao for communicating unpublished information about smc5 alleles. We thank Erica Johnson, Marc Gartenberg, Charles Boone, Aaron Gitler, Gregory Cost, Jörg Heierhorst, and Luis Aragón for gifts of yeast strains and/or plasmids.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-AG021521 and P01-AG031862 (to F. B. J.) and T32-AG000255 (to A. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Materials and Methods, additional references, Figs. 1–21, and Tables 1–3.
X. Zhao, unpublished data.
A. Chavez and F. B. Johnson, unpublished data.
- HR
- homologous recombination
- ALT
- alternative lengthening of telomeres
- APB
- ALT-associated promyelocytic leukemia bodies
- SMC
- structural maintenance of chromosome
- MMS
- methyl methanesulfonate
- PD
- population doubling.
REFERENCES
- 1.Bertuch A. A., Lundblad V. (2006) Curr. Opin. Cell Biol. 18, 247–253 [DOI] [PubMed] [Google Scholar]
- 2.Blackburn E. H. (2001) Cell 106, 661–673 [DOI] [PubMed] [Google Scholar]
- 3.Lundblad V., Szostak J. W. (1989) Cell 57, 633–643 [DOI] [PubMed] [Google Scholar]
- 4.Singer M. S., Gottschling D. E. (1994) Science 266, 404–409 [DOI] [PubMed] [Google Scholar]
- 5.Le S., Moore J. K., Haber J. E., Greider C. W. (1999) Genetics 152, 143–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lundblad V., Blackburn E. H. (1993) Cell 73, 347–360 [DOI] [PubMed] [Google Scholar]
- 7.Teng S. C., Zakian V. A. (1999) Mol. Cell. Biol. 19, 8083–8093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muntoni A., Reddel R. R. (2005) Hum. Mol. Genet. 14, R191–R196 [DOI] [PubMed] [Google Scholar]
- 9.Bryan T. M., Englezou A., Dalla-Pozza L., Dunham M. A., Reddel R. R. (1997) Nat. Med. 3, 1271–1274 [DOI] [PubMed] [Google Scholar]
- 10.Bryan T. M., Englezou A., Gupta J., Bacchetti S., Reddel R. R. (1995) EMBO J. 14, 4240–4248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhong Z. H., Jiang W. Q., Cesare A. J., Neumann A. A., Wadhwa R., Reddel R. R. (2007) J. Biol. Chem. 282, 29314–29322 [DOI] [PubMed] [Google Scholar]
- 12.Tsai H. J., Huang W. H., Li T. K., Tsai Y. L., Wu K. J., Tseng S. F., Teng S. C. (2006) J. Biol. Chem. 281, 13717–13723 [DOI] [PubMed] [Google Scholar]
- 13.Temime-Smaali N., Guittat L., Wenner T., Bayart E., Douarre C., Gomez D., Giraud-Panis M. J., Londono-Vallejo A., Gilson E., Amor-Guéret M., Riou J. F. (2008) EMBO J. 27, 1513–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson F. B., Marciniak R. A., McVey M., Stewart S. A., Hahn W. C., Guarente L. (2001) EMBO J. 20, 905–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stavropoulos D. J., Bradshaw P. S., Li X., Pasic I., Truong K., Ikura M., Ungrin M., Meyn M. S. (2002) Hum. Mol. Genet. 11, 3135–3144 [DOI] [PubMed] [Google Scholar]
- 16.Lydeard J. R., Jain S., Yamaguchi M., Haber J. E. (2007) Nature 448, 820–823 [DOI] [PubMed] [Google Scholar]
- 17.Lee J. Y., Kozak M., Martin J. D., Pennock E., Johnson F. B. (2007) PLOS Biol. 5, e160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liberi G., Maffioletti G., Lucca C., Chiolo I., Baryshnikova A., Cotta-Ramusino C., Lopes M., Pellicioli A., Haber J. E., Foiani M. (2005) Genes Dev. 19, 339–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Azam M., Lee J. Y., Abraham V., Chanoux R., Schoenly K. A., Johnson F. B. (2006) Nucleic Acids Res. 34, 506–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prudden J., Pebernard S., Raffa G., Slavin D. A., Perry J. J., Tainer J. A., McGowan C. H., Boddy M. N. (2007) EMBO J. 26, 4089–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uzunova K., Göttsche K., Miteva M., Weisshaar S. R., Glanemann C., Schnellhardt M., Niessen M., Scheel H., Hofmann K., Johnson E. S., Praefcke G. J., Dohmen R. J. (2007) J. Biol. Chem. 282, 34167–34175 [DOI] [PubMed] [Google Scholar]
- 22.Xie Y., Kerscher O., Kroetz M. B., McConchie H. F., Sung P., Hochstrasser M. (2007) J. Biol. Chem. 282, 34176–34184 [DOI] [PubMed] [Google Scholar]
- 23.Branzei D., Sollier J., Liberi G., Zhao X., Maeda D., Seki M., Enomoto T., Ohta K., Foiani M. (2006) Cell 127, 509–522 [DOI] [PubMed] [Google Scholar]
- 24.Burgess R. C., Rahman S., Lisby M., Rothstein R., Zhao X. (2007) Mol. Cell. Biol. 27, 6153–6162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sacher M., Pfander B., Hoege C., Jentsch S. (2006) Nat. Cell Biol. 8, 1284–1290 [DOI] [PubMed] [Google Scholar]
- 26.Seufert W., Futcher B., Jentsch S. (1995) Nature 373, 78–81 [DOI] [PubMed] [Google Scholar]
- 27.Nacerddine K., Lehembre F., Bhaumik M., Artus J., Cohen-Tannoudji M., Babinet C., Pandolfi P. P., Dejean A. (2005) Dev. Cell 9, 769–779 [DOI] [PubMed] [Google Scholar]
- 28.Fraser A. G., Kamath R. S., Zipperlen P., Martinez-Campos M., Sohrmann M., Ahringer J. (2000) Nature 408, 325–330 [DOI] [PubMed] [Google Scholar]
- 29.Geiss-Friedlander R., Melchior F. (2007) Nat. Rev. Mol. Cell Biol. 8, 947–956 [DOI] [PubMed] [Google Scholar]
- 30.Johnson E. S., Gupta A. A. (2001) Cell 106, 735–744 [DOI] [PubMed] [Google Scholar]
- 31.Zhao X., Blobel G. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4777–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Panse V. G., Hardeland U., Werner T., Kuster B., Hurt E. (2004) J. Biol. Chem. 279, 41346–41351 [DOI] [PubMed] [Google Scholar]
- 33.Denison C., Rudner A. D., Gerber S. A., Bakalarski C. E., Moazed D., Gygi S. P. (2005) Mol. Cell. Proteomics 4, 246–254 [DOI] [PubMed] [Google Scholar]
- 34.Wohlschlegel J. A., Johnson E. S., Reed S. I., Yates J. R., 3rd. (2004) J. Biol. Chem. 279, 45662–45668 [DOI] [PubMed] [Google Scholar]
- 35.Zhou W., Ryan J. J., Zhou H. (2004) J. Biol. Chem. 279, 32262–32268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Potts P. R., Yu H. (2007) Nat. Struct. Mol. Biol. 14, 581–590 [DOI] [PubMed] [Google Scholar]
- 37.Cesare A. J., Reddel R. R. (2008) Mech. Ageing Dev. 129, 99–108 [DOI] [PubMed] [Google Scholar]
- 38.Henson J. D., Neumann A. A., Yeager T. R., Reddel R. R. (2002) Oncogene 21, 598–610 [DOI] [PubMed] [Google Scholar]
- 39.Murray J. M., Carr A. M. (2008) Nat. Rev. Mol. Cell Biol. 9, 177–182 [DOI] [PubMed] [Google Scholar]
- 40.Potts P. R. (2009) DNA Repair 8, 499–506 [DOI] [PubMed] [Google Scholar]
- 41.Lehmann A. R., Walicka M., Griffiths D. J., Murray J. M., Watts F. Z., McCready S., Carr A. M. (1995) Mol. Cell. Biol. 15, 7067–7080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taylor E. M., Moghraby J. S., Lees J. H., Smit B., Moens P. B., Lehmann A. R. (2001) Mol. Biol. Cell 12, 1583–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ampatzidou E., Irmisch A., O'Connell M. J., Murray J. M. (2006) Mol. Cell. Biol. 26, 9387–9401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pebernard S., Wohlschlegel J., McDonald W. H., Yates J. R., III, Boddy M. N. (2006) Mol. Cell. Biol. 26, 1617–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sollier J., Driscoll R., Castellucci F., Foiani M., Jackson S. P., Branzei D. (2009) Mol. Biol. Cell 20, 1671–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Torres-Rosell J., Machín F., Farmer S., Jarmuz A., Eydmann T., Dalgaard J. Z., Aragón L. (2005) Nat. Cell Biol. 7, 412–419 [DOI] [PubMed] [Google Scholar]
- 47.Takahashi Y., Dulev S., Liu X., Hiller N. J., Zhao X., Strunnikov A. (2008) PLoS Genet. 4, e1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liberi G., Cotta-Ramusino C., Lopes M., Sogo J., Conti C., Bensimon A., Foiani M. (2006) Methods Enzymol. 409, 442–462 [DOI] [PubMed] [Google Scholar]
- 49.Johnson E. S., Schwienhorst I., Dohmen R. J., Blobel G. (1997) EMBO J. 16, 5509–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen X. L., Reindle A., Johnson E. S. (2005) Mol. Cell. Biol. 25, 4311–4320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsalik E. L., Gartenberg M. R. (1998) Yeast 14, 847–852 [DOI] [PubMed] [Google Scholar]
- 52.Martens U. M., Chavez E. A., Poon S. S., Schmoor C., Lansdorp P. M. (2000) Exp. Cell Res. 256, 291–299 [DOI] [PubMed] [Google Scholar]
- 53.Reindle A., Belichenko I., Bylebyl G. R., Chen X. L., Gandhi N., Johnson E. S. (2006) J. Cell Sci. 119, 4749–4757 [DOI] [PubMed] [Google Scholar]
- 54.Andrews E. A., Palecek J., Sergeant J., Taylor E., Lehmann A. R., Watts F. Z. (2005) Mol. Cell. Biol. 25, 185–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gangloff S., McDonald J. P., Bendixen C., Arthur L., Rothstein R. (1994) Mol. Cell. Biol. 14, 8391–8398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Downs J. A., Jackson S. P. (2004) Nat. Rev. Mol. Cell Biol. 5, 367–378 [DOI] [PubMed] [Google Scholar]
- 57.Potts P. R., Porteus M. H., Yu H. (2006) EMBO J. 25, 3377–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Verkade H. M., Bugg S. J., Lindsay H. D., Carr A. M., O'Connell M. J. (1999) Mol. Biol. Cell 10, 2905–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lindroos H. B., Ström L., Itoh T., Katou Y., Shirahige K., Sjögren C. (2006) Mol. Cell 22, 755–767 [DOI] [PubMed] [Google Scholar]
- 60.Irmisch A., Ampatzidou E., Mizuno K., O'Connell M. J., Murray J. M. (2009) EMBO J. 28, 144–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sergeant J., Taylor E., Palecek J., Fousteri M., Andrews E. A., Sweeney S., Shinagawa H., Watts F. Z., Lehmann A. R. (2005) Mol. Cell. Biol. 25, 172–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Torres-Rosell J., De Piccoli G., Cordon-Preciado V., Farmer S., Jarmuz A., Machin F., Pasero P., Lisby M., Haber J. E., Aragón L. (2007) Science 315, 1411–1415 [DOI] [PubMed] [Google Scholar]
- 63.Cost G. J., Cozzarelli N. R. (2006) Genetics 172, 2185–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Branzei D., Vanoli F., Foiani M. (2008) Nature 456, 915–920 [DOI] [PubMed] [Google Scholar]
- 65.De Piccoli G., Cortes-Ledesma F., Ira G., Torres-Rosell J., Uhle S., Farmer S., Hwang J. Y., Machin F., Ceschia A., McAleenan A., Cordon-Preciado V., Clemente-Blanco A., Vilella-Mitjana F., Ullal P., Jarmuz A., Leitao B., Bressan D., Dotiwala F., Papusha A., Zhao X., Myung K., Haber J. E., Aguilera A., Aragón L. (2006) Nat. Cell Biol. 8, 1032–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.