Abstract
For major histocompatibility complex class I and II molecules, the binding of specific peptide antigens is essential for assembly and trafficking and is at the center of their quality control mechanism. However, the role of lipid antigen binding in stabilization and quality control of CD1 heavy chain (HC)·β2-microglobulin (β2m) complexes is unclear. Furthermore, the distinct trafficking and loading routes of CD1 proteins take them from mildly acidic pH in early endososmal compartments (pH 6.0) to markedly acidic pH in lysosomes (pH 5.0) and back to neutral pH of the cell surface (pH 7.4). Here, we present evidence that the stability of each CD1 HC·β2m complex is determined by the distinct pH optima identical to that of the intracellular compartments in which each CD1 isoform resides. Although stable at acidic endosomal pH, complexes are only stable at cell surface pH 7.4 when bound to specific lipid antigens. The proposed model outlines a quality control program that allows lipid exchange at low endosomal pH without dissociation of the CD1 HC·β2m complex and then stabilizes the antigen-loaded complex at neutral pH at the cell surface.
Keywords: Antigens MHC, Glycolipids, Immunology/Antigen, Immunology/Antigen Processing, Lipid, Protein/Stability, CD1, NKT Cell
Introduction
CD1 molecules represent a distinct lineage of antigen-presenting molecules that are related to major histocompatibility complex (MHC)3 class I proteins sharing limited sequence similarity and domain organization including a heavy chain (HC) composed of α1, α2 and α3 domains associated non-covalently with β2-microglobulin (β2m). Despite these structural similarities, CD1 proteins are functionally distinct because they bind and present lipid antigens to elicit T cell-mediated immunity (1). Lipid antigen binding is determined by the presence of deep hydrophobic grooves in the α1 and α2 antigen binding domains of the CD1 heavy chain. This topology buries the alkyl chains of the antigens below the CD1 surface and exposes the hydrophilic head groups at the CD1 surface, where moieties of the head group and the CD1 α-helices can be recognized by T cell antigen receptors. The assembly of CD1 molecules takes place in the endoplasmic reticulum (ER) with the participation of several chaperones also involved in the formation of the MHC class I peptide loading complex, namely calnexin, calreticulin, and ERp57 (2). In the ER, newly synthesized CD1 molecules are thought to be loaded with self-lipid antigens after which they traffic to the cell surface along the secretory pathway (3). From the plasma membrane CD1 molecules are internalized and recycle through the endocytic system where lipid antigens are bound and exchanged (1). Human CD1a traffics mainly through early endosomes and the early endocytic recycling compartment where it co-localizes with MHC class I (4, 5). In contrast, mouse (m) CD1d and human (h) CD1b are sorted mainly to late endosomes and lysosomes where they co-localize with MHC class II proteins (4, 6–8). Human CD1c and to some extent hCD1d are distributed more extensively in both early and late endocytic compartments (9, 10).
For MHC class I molecules, the binding of specific peptide antigens is essential to complete the assembly of these complexes, regulate their exit from the ER and their integrity on the cell surface. Indeed, in transporter associated with antigen processing-deficient cells, which are unable to deliver peptides to the ER, the stability of MHC class I heavy chain (HC)/β2m complexes is decreased and empty MHC class I proteins are retained in the ER (11, 12). This process functions in part as a quality control mechanism and protects the cell from expression of incompletely assembled or empty antigen-presenting molecules. Similarly, the stability of MHC class II complexes is ensured by the invariant chain (Ii), and the CLIP peptide is replaced by peptide antigens that stabilize MHC class II α/β heterodimers (13). However, it is unclear if CD1 molecules can be stabilized by binding of specific lipid antigens. The ER-localized microsomal triglyceride transfer protein has been implicated in loading self-lipid antigen during assembling of CD1 in the ER (14, 15). In an experimental system, the culture of CD1a expressing cells under serum (lipid)-deficient conditions partially reduces the surface expression of CD1a (16). These studies indirectly point to a protective effect of lipids on CD1 stability, but so far no studies have directly examined the effect of lipid binding on stability of CD1 complexes. Furthermore, the distinct trafficking and loading routes of CD1 proteins take them from markedly acidic pH in lysosomes (pH 5.0) to neutral pH of the cell surface (pH 7.4). Here, we show for the first time that the stability of individual CD1 HC·β2m complexes is determined by a specific pH optimum identical to the intracellular compartment to which the CD1 isoform localizes. Although stable at their optimal intracellular pH, CD1 HC·β2m complexes are only stable at neutral pH when bound by specific lipid antigens. This model outlines a quality control program that allows lipid binding/exchange at low pH where the CD1 HC·β2m heterodimer is stable even without bound lipid and then specific lipid binding stabilizes the complex for expression at the cell surface at neutral pH.
EXPERIMENTAL PROCEDURES
Lipids
α-GalCer, β-GalCer, and C32 glucose monomycolate (GMM) were synthesized by G. Besra as previously described (30, 31). Trehalose monomycolate was isolated in this laboratory from a polar lipid extract obtained from γ-irradiated Mycobacterium tuberculosis (H37Rv, Colorado State University) according to the procedures of Dobson and co-workers (32).
Antibodies
All antibodies used for immunoprecipitation of CD1 and MHC class I and II molecules and their potential cross-reactivity were described previously: anti-CD1a antibodies: 10H3 (mouse, IgG2a) (33), OKT6 (mouse, IgG1) (34), 10D12 (mouse, IgG1) (33); anti-CD1b antibodies: BCD1b3 (mouse, IgG1) (35), BCD1b1 (mouse, IgG1) (36), 4A7.6 (mouse IgG2a) (33); anti-CD1c antibody F10/21.3 (mouse, IgG1) (37); anti-human CD1d antibodies: 42.1.1 (mouse, IgG1), 55.10 (mouse, IgG1), and 75.10 (mouse, IgG1) (17); anti-mouse CD1d antibody 19G11 (rat, IgG2b/κ, kindly provided by A. Bendelac, University of Chicago, Chicago, IL) (38); anti-human β2m Ab BBM.1 (mouse, IgG1) (39); anti-MHC class I antibody W6/32 (mouse, IgG2a) (40); and anti-HLA-DR antibody L243 (mouse, IgG2a) (39). Isotype control antibodies used for experiments were: P3 antibody (mouse IgG1), murine myeloma monoclonal IgG2a (Sigma), and rat isotype IgG2b (BD Biosciences).
CD1-transfected Cell Lines
Previously established cervical carcinoma HeLa cell lines transfected with CD1a (5), CD1b (6), CD1c (41), or CD1d (42) were used. Reagents for cell culture were from Invitrogen. HeLa transfectants were cultured in Dulbecco's modified Eagle's medium containing heat-inactivated 10% fetal calf serum (Gemini Bio-Products), 100 units/ml of penicillin G, 100 μg/ml of streptomycin, 2 mm l-glutamine, and 20 mm HEPES with addition of 1.0 mg/ml of geneticin (G418).
The MHC class I-deficient B lymphoblastoid cell lines, C1R transfected with CD1a, CD1b, CD1c (35, 43), and CD1d (17), were also described previously. C1R cells were cultured in RPMI with heat-inactivated 10% fetal calf serum, 100 units/ml of penicillin G, 100 μg/ml of streptomycin, 2 mm l-glutamine, and 20 mm HEPES. The mouse CD1d-transfected RAW cell line was derived in this laboratory by Dr. Manuela Cernadas.
Protein Labeling and Cell Lysis
HeLa, C1R, or RAW cells (2 × 107) were biotinylated by adding of 10 ml of PBS with 20 mm HEPES (pH 8.0), containing Sulfo-NHS-LC-biotin (Pierce) in a final 0.2 mg/ml concentration. Cells were incubated at 4 °C for 30 or 10 min at room temperature. To quench excess biotin, the cells were incubated for 5 min with 10 ml of 0.4 m glycine in PBS at room temperature and then washed twice with cold PBS.
Cells (107) were lysed with 2 ml of lysis buffer (0.5% Triton X-100, 150 mm NaCl, 5 mm EDTA, and 25 mm Tris, pH 7.4, or 50 mm sodium citrate adjusted to pH 7.4, 7.0, 6.5, 6.0, 5.5, 5.0, or 4.5). All buffers contained protease inhibitors (aprotinin, phenylmethylsulfonyl fluoride, antipain dihydrochloride, chymostatin, leupeptin, pepstatin A, sodium orthovanadate, and sodium fluoride; all from Sigma). After 30–45 min of incubation with lysis buffer at 4 °C, lysates were centrifuged (13,000 × g for 10 min at 4 °C) and the supernatants were collected and incubated as described under “Results” and in the figure legends.
Immunoprecipitation
Cell lysates (obtained from 5 × 105–106 cells) were used for each immunoprecipitation (IP). CD1a molecules were immunoprecipitated with monoclonal antibodies (mAbs): 10H3 (1–2 μg/sample), OKT6 (2–4 μg/sample), or 10D12 (2–4 μg/sample). CD1b molecules were precipitated with mAbs: BCD1b3.1 (1–2 μg/sample), BCD1b1.1 (5–10 μg/sample), or 4A7.3 (2–4 μg/sample). CD1c immunoprecipitation was performed with mAb F10/21A3 (1–4 μg/sample). For IP of hCD1d:β2m mAbs 42.1.1 (1–2 μg/sample) or 55.1 (2–4 μg/sample) were used, whereas mAb 75.10 (5–10 μg/sample) was used for IP of free hCD1d heavy chain. mCD1d was precipitated with rat mAb 19G11.2 (0.5–1.0 μg/sample). MHC class I was immunoprecipitated with mAb W6/32 (1–2 μg/sample), β2m with mAb BBM.1 (1–5 μg/sample), and MHC class II with mAb L243 (0.1–0.2 μg/sample). Equivalent amounts of P3 (mouse IgG1), murine myeloma monoclonal IgG2a (Sigma), or rat isotype IgG2b (BD Biosciences) were used as isotype controls. Lysates were incubated with antibodies for 45–60 min on ice and then 20 to 25 μl of GammaBind G-Sepharose (GE Healthcare) were added. After incubating overnight at 4 °C or alternatively 2–3 h with rotation, the beads were washed three times with RIPA buffer (1% Triton X-100, 0.1% SDS, 0.05% deoxycholate, 50 mm Tris, pH 7.4, 150 mm NaCl), then washed once with 50 mm Tris (pH 6.8) and resuspended in SDS sample buffer (62.5 mm Tris, pH 6.8, 10% glycerol, 2% SDS, 0.025% bromphenol blue, and 179 mm β-mercaptoethanol).
Electrophoresis, Immunoblotting, and Protein Detection
Samples were loaded on 15% acrylamide SDS-PAGE and run in a Protean XL apparatus (Bio-Rad) with constant current (50 mAmp per gel). After electrophoresis, proteins were transferred to polyvinylidene fluoride membranes in transfer buffer (25 mm Tris, 192 mm glycine, and 10% methanol) at 25 V overnight in a Trans-Blot Cell (Bio-Rad). After transfer, membranes were blocked with 0.2% Tween 20 in PBS (PBST) and 0.5% bovine serum albumin at room temperature for 1 h, incubated with HRP-conjugated streptavidin (ExtrAvidin, 1:20 000; Sigma;) in PBST for 45 min, washed with PBST, and finally blots were developed with Western Lightning Chemiluminescence Reagent (Perkin Elmer) according to the manufacturer's instructions. The intensity of bands was measured by densitometry using ImageJ software.
RESULTS
CD1 Proteins Are Less Stable at the Cell Surface Than MHC Class I and II Molecules
We first analyzed the stability of CD1 HC·β2m complexes in comparison to MHC class I and II complexes. First, we examined CD1a and CD1b because they differ markedly in the structure of their antigen binding grooves and are localized to different intracellular compartments. Cell surface CD1 molecules were labeled with biotin and cells were lysed with 0.5% Triton X-100 (pH 7.4). We performed IP of CD1a:HeLa cell transfectant cell lysates incubated at 4 or 37 °C. Incubation of lysates at 37 °C for even 2 h dramatically reduced the levels of CD1a HC·β2m complexes immunoprecipitated when compared with incubation at 4 °C. This was observed when we used three different conformation-specific antibodies that recognize the CD1a HC·β2m complex, but not the free CD1a HC without β2m (Fig. 1A, compare lanes 2–4 versus 6–8, and note the loss of both CD1a HC and β2m). These results showing dissociation of the CD1a HC·β2m complex at 37 °C indicate the complex is unstable under these experimental conditions.
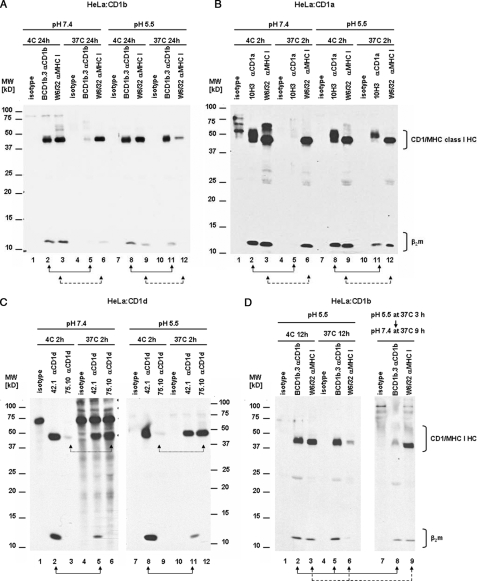
FIGURE 1.
CD1 HC·β2m complexes are unstable at 37 °C. CD1-transfected HeLa cells or C1R cells were surface bioyinylated, lysed with 0.5% Triton X-100 at pH 7.4, and lysates were incubated for 2–24 h at 4 or 37 °C. After immunoprecipitation with specific anti-CD1, anti-MHC class I or II, and anti-β2m antibodies, the immunoprecipitates were analyzed by SDS-PAGE (15%) under reducing conditions and immunoblotted with streptavidin-HRP. A, HeLa:CD1a cell lysates were incubated at 4 (lanes 1–4) or 37 °C (lanes 5–8) for 2 h. CD1a:HC·β2m complexes were immunoprecipitated with anti-CD1a 10H3 (lanes 2 and 6, solid line arrows), OKT6 (lanes 3 and 7, dotted line arrows), and 10D12 (lanes 4 and 8, dashed line arrows) antibodies. B, HeLa:CD1b cell lysates were incubated at 4 (lanes 1–3) or 37 °C (lanes 4–6) for 24 h. CD1b:HC·β2m complexes were immunoprecipitated with anti-CD1b BCD1.3 antibody (lanes 2 and 5) and MHC class I with W6/32 antibody (lanes 3 and 6). As a control, mouse IgG1 was used (isotype). C, C1R:CD1b cell lysates were incubated at 4 (lanes 1–5) or 37 °C (lanes 6–10) for 12 h. CD1b:HC·β2m complexes were immunoprecipitated with anti-CD1b BCD1b.3 (lanes 2 and 7, solid line arrows), anti-CD1b BCD1b.1 (lanes 3 and 8, dotted line arrows) antibodies, anti-β2m antibody BBM.1 (lanes 4 and 9, dashed line arrows), and MHC class II with anti-MHC II antibody L243 (lanes 5 and 10). As a control mouse IgG1 was used (isotype). D, HeLa:CD1d cell lysates were incubated at 4 (lanes 1–5) or 37 °C (lanes 6–10) for 2 h. CD1d HC·β2m complexes were immunoprecipitated with anti-CD1d 42.1 (lanes 2 and 7, solid line arrow) or 55.10 antibodies (lanes 3 and 8, dashed line arrows); free CD1d HC was immunoprecipitated with 75.10 antibody (lanes 4 and 9, dotted line arrows) and MHC class I with antibody W6/32 (lanes 5 and 10). E, C1R:CD1d cell lysates were incubated at 4 (lanes 1–5) or 37 °C (lanes 6–10) for 2 h. CD1d HC·β2m complexes were immunoprecipitated with anti-CD1d 42.1 (lanes 2 and 7, solid line arrows) or anti-β2m BBM.1 antibodies (lanes 4 and 9, dashed line arrows); free CD1d HC with anti-CD1d antibody 75.10 (lanes 4 and 9, dotted line arrows) and MHC class II with anti-MHC II L243 antibody (lanes 5 and 10). In all experiments mouse IgG2a or IgG1 were used as isotype controls. Arrows indicate the lanes to compare.
Next, we examined the stability of the CD1b HC·β2m complex under similar conditions. It also dissociates at 37 °C, but with slower kinetics than CD1a, requiring at least 12–24 h after cell lysis (Fig. 1B, lane 2 versus 5, note the nearly complete disappearance of CD1b HC and β2m). As a control, under these same conditions no detectable change in quantity of MHC I HC·β2m complexes was found (Fig. 1B, lane 3 versus 6). We also compared the stability of CD1b with MHC class II in CD1b-transfected C1R cells and found that as in the case of MHC class I, the MHC class II heterodimer was also stable under these conditions (Fig. 1C, lanes 5 versus 10). The instability of CD1b was confirmed using a separate panel of conformation-dependent anti-CD1b HC·β2 mAb (BCD1b.1 and BCD1b3) (Fig. 1C, lanes 2 and 3 versus 7 and 8, respectively) and mAb 4A7.6 (data not shown). To complement the analysis using CD1 HC·β2m conformation-dependent mAb, next we used anti-β2m mAb BBM.1, which recognizes β2m whether free or bound to CD1/MHC I heavy chains. In C1R cells, MHC I is essentially absent so that on this cell line the mAb recognizes only free or CD1 bound β2m. Strikingly, mAb BBM.1 confirmed the reduced stability of the CD1b HC·β2m complex at 37 °C compared with 4 °C as the quantity of co-immunoprecipitated CD1b HC was markedly reduced at 37 °C, whereas the total amount of β2m was comparable (Fig. 1C, lane 4 versus 9). The difference noted between lanes where CD1b HC·β2m complexes were immunoprecipitated with anti-CD1b antibodies versus lanes precipitated with β2m-specific antibody indicates that the antibodies recognize slightly different pools of molecules. However, in every case the mAb revealed a similar pattern of HC·β2m instability at 37 °C (Fig. 1, A–C).
Next, we analyzed the stability of CD1d using three different anti-hCD1d mAb (42.1, 55.1, and 75.10) that allowed us to distinguish the free CD1d HC from the CD1d HC·β2m complex. mAb 42.1 is conformation-dependent and recognizes only CD1d HC·β2m complexes, whereas mAb 75.10 selectively binds the free CD1d HC (17). mAb 55.1 recognizes both pools of molecules under different conditions because it recognizes the CD1d HC·β2m complex (by IP), but it can also recognize the free CD1d HC (by Western blotting, data not shown). In accordance with the results obtained for CD1a and CD1b, we observed a reduction in immunoprecipitated CD1d HC·β2m complexes and an increase in free CD1d HC in lysates incubated for 2 h at 37 °C (Fig. 1, D and E). When anti-CD1d 42.1 mAb was used we saw a significant reduction of signal for the CD1d HC both in HeLa:CD1d and C1R:CD1d transfectant cells (Fig. 1D, lane 2 versus 7). When the IP was performed with anti-CD1d 55.1, which recognizes CD1d HC in a conformation-independent manner, we observed a decrease in the β2m signal at 37 °C but no change in the signal for CD1d HC (Fig. 1D, lane 3 versus 8). This suggests that the CD1d HC·β2m complex disassembles without degradation of the HC. If true, then the proportion of free CD1d HC should increase. This prediction was confirmed by noting the significant increase in free CD1d HC signal by IP when we used anti-CD1d free HC-specific mAb 75.10 in samples incubated 37 °C (Fig. 1D, lanes 4 versus 9). In fact, the results obtained with anti-β2m BBM1 mAb were also consistent with this interpretation, because the CD1d HC signal decreased, although no change in the β2m signal was noted after incubating cell lysates for 2 h at 37 °C (Fig. 1E, lanes 4 versus 9). Importantly, signals for MHC class I and II were relatively unchanged under these same experimental conditions (Fig. 1, E, lanes 5 versus 10 and D, lanes 5 versus 10) although MHC I-associated β2m labeled only weakly, therefore limiting its analysis in this experiment. Together, these data outline conditions under which CD1 molecules labeled at the cell surface dissociate after lysis in 0.5% Triton X-100 at 37 °C compared with MHC I and II molecules under the same conditions.
The different times required for disassembly of CD1a HC·β2m compared with CD1b HC·β2m complexes at 37 °C suggests that each CD1 isoform may vary in its stability. To address this possibility, we used HeLa cells transfected with different CD1 proteins. As shown in Fig. 2A, CD1a displayed the lowest stability after 2 h at 37 °C with over 95% dissociation, followed by CD1d (75%) and CD1c (50%) (Fig. 2, D and C, respectively). CD1b was the most stabile CD1 isoform, with only 10% dissociation after 2 h at 37 °C (Fig. 2B). We also confirmed that MHC class I is very stable, showing no significant reduction in signal after 2 h at 37 °C (Fig. 2, A–D).
FIGURE 2.
Rank order of CD1 isoform stability: CD1b > CD1c > CD1d > CD1a. HeLa:CD1a, CD1b, CD1c, or CD1d transfectants were surface biotinylated, lysed with 0.5% Triton X-100 (pH 7.4), and then lysates were incubated for 2 h at 4 or 37 °C. CD1 heavy chain/β2m complexes were immunoprecipitated with 10H3 (anti-CD1a, A), BCD1b.3 (anti-CD1b, B), F10/23.A.1 (anti-CD1c, C), or 42.1 (anti-CD1d, D) antibodies, whereas MHC class I:HC·β2m complexes were precipitated with antibody W6/32 (A–D). Mouse IgG1 and IgG2a were used as isotype controls. Immunoprecipitates were analyzed by SDS-PAGE (15%) in reducing conditions and immunoblotted with streptavidin-HRP. Arrows indicate lanes to compare. Quantification of band intensity was measured by densitometry using ImageJ software and results were calculated as proportion of HC or β2m signals at 37 °C compared with HC or β2m signals at 4 °C (A–D).
Stabilization of CD1a and CD1b HC·β2M Complexes by Low pH
Following biosynthesis in the ER and delivery to the plasma membrane, CD1 proteins are then internalized to survey endocytic compartments for lipid antigens. CD1a and CD1b are exposed to different environments because CD1a localizes to the endocytic recycling pathway (pH ∼ 6), whereas CD1b localizes to lysosomes (pH below 5.5). To investigate if pH influences CD1 stability, CD1-transfected HeLa cells were lysed in 0.5% Triton X-100 at pH 7.4 or 5.5 and then incubated at 37 °C for various times. As expected, incubation for 24 h at 37 °C at pH 7.4 resulted in significant CD1b HC·β2m complex disassembly (Fig. 3A, lane 2 versus 5). Surprisingly, when lysis was performed at pH 5.5 the complex was markedly more stable than at pH 7.4 with CD1b HC 30 versus 90% and β2m 50 versus 100% of signal detected at pH 7.4 versus 5.5 (Fig. 3, A lane 8 versus 11). Similar results were also obtained using two other anti-CD1b mAb, 4A7.6 and BCD1b.1 (not shown). In striking contrast to CD1b stability at acidic pH, the MHC class I HC·β2m complex was much less stable at pH 5.5 (Fig. 3A, lane 9 versus 12). Thus, CD1b HC·β2m complex stability is greater in low pH, whereas the opposite is true for the MHC class I HC·β2m.
FIGURE 3.
Stabilization of CD1a and -b HC·β2m complexes by low pH. CD1-transfected HeLa cells were surface biotinylated and lysed with 0.5% Triton X-100 (pH 7.4 or 5.5). Then lysates were incubated for 2–24 h at 4 or 37 °C. After immunoprecipitation with specific anti-CD1 and anti-MHC class I antibodies, immunoprecipitates were analyzed by SDS-PAGE (15%) in reducing conditions and immunoblotted with streptavidin-HRP. A, HeLa:CD1b cell lysates at pH 7.4 (lanes 1–6) or 5.5 (lanes 7–12) were incubated at 4 (lanes 1–3 and 7–9) or 37 °C (lanes 4–6 and 10–12) for 24 h. CD1b:HC·β2m complexes were immunoprecipitated with BCD1b.3 (lanes 2, 5, 8, and 11, solid line arrows) and MHC class I:HC·β2m complexes with W6/32 antibodies (lanes 3, 6, 9, and 12, dashed line arrows). B, HeLa:CD1a cell lysates at pH 7.4 (lanes 1–6) or 5.5 (lanes 7–12) were incubated at 4 (lanes 1–3 and 7–9) or 37 °C (lanes 4–6 and 10–12) for 2 h. CD1a:HC·β2m complexes were immunoprecipitated with 10H3 (lanes 2, 5, 8, and 11, solid lines arrows) and MHC class I:HC·β2m complexes with W6/32 antibodies (lanes 3, 6, 9, and 12, dashed line arrows). C, HeLa:CD1d cell lysates at pH 7.4 (lanes 1–6) or 5.5 (lanes 7–12) were incubated at 4 °C (lanes 1–3 and 7–9) or 37 °C (lanes 4–6 and 10–12) for 2 h. CD1d:HC·β2m complexes were immunoprecipitated with mAb 42.10 (lanes 2, 5, 8, and 11, solid line arrows) and the free CD1d HC was detected with mAb 75.10 (lanes 3, 6, 9, and 12, dotted line arrows). D, HeLa:CD1b cell lysates at pH 5.5 were incubated at 4 °C for 12 h (lanes 1–3) or 37 °C for 12 (lanes 4–6) or 3 h followed by pH neutralization to pH 7.4 with 1.5 m Tris-HCl buffer (pH 8.8) and a further 9-h incubation at 37 °C (lanes 7–9). CD1b HC·β2m complexes were immunoprecipitated with mAb BCD1b.3 (lanes 2, 5, and 8, solid line arrows) and MHC class I with mAb W6/32 (lanes 3, 6, and 9, dashed line arrows). In all experiments isotype-matched mouse IgG2a or IgG1 were used as controls. Arrows indicate lanes to compare.
In a similar way, we found that low pH also protects CD1a HC·β2m complexes from disassembling during a period of 2 h at 37 °C (Fig. 3B, lane 2 versus 5 and 8 versus 11). Incubation of lysates at pH 7.4 markedly decreased the signal of immunoprecipitated CD1a HC·β2m complexes (reduction over 95%), whereas incubation at pH 5.5 partially protected CD1a from disassembly (reduction of 50–60%). The same result was also obtained using two other anti-CD1a mAb, 10D12 and OKT6 (not shown).
In contrast to CD1a and CD1b, very little if any protection from disassembly was found at pH 5.5 for human CD1d HC·β2m complexes (Fig. 3C, lane 2 versus 5 and 8 versus 11). Consistent with disassembly of the complex, we simultaneously detected an increase in free CD1d HC immunoprecipitated with mAb 75.10 (Fig. 3C, lane 3 versus 6 and 9 versus 12). pH does not affect antibody binding efficiency because the amount of immunoprecipitated CD1 and MHC class I was similar at 4 °C (Fig. 3, A–D). Also, an additional background band at ∼75 kDa was noted in the isotype control at 37 °C (pH 7.4) in this experiment (Fig. 3C, lane 4).
Given that internalization followed by recycling from endocytic compartments would expose CD1 molecules to higher and lower pH in sequence, we next determined if the protective effect of low pH on CD1 stability is permanent or is reversed by pH neutralization. Thus, we lysed CD1b-transfected HeLa cells in pH 5.5 buffer, incubated the lysates for 3 h at 37 °C, and then rapidly neutralized the cells to pH 7.4. Then, after further incubation at 37 °C (pH 7.4) for 9 h, we immunoprecipitated CD1b and MHC class I with specific mAb. As a control, we analyzed samples incubated at 4 or 37 °C for 12 h, without the neutralization step. The incubation of lysates in low pH for 3 h followed by 9 h at neutral pH revealed instability of CD1b HC·β2m complexes (about 85% reduction of CD1b HC) when compared with lysates incubated at low pH throughout the experiment (Fig. 3D, lane 2 versus 8). These results suggest that the effect of pH on stability of the CD1 HC·β2m complex is reversible and does not cause a permanent change in protein structure. In summary, the evidence suggests that pH 5.5 markedly stabilizes of CD1a and CD1b HC·β2m complexes but does not have a significant influence on CD1d. This pH-dependent protective effect is reversible.
pH-dependent Protection of CD1 Molecules Corresponds with Their Locations in Intracellular Compartments
Given the protection of CD1a and CD1b HC·β2m complexes noted at pH 5.5, we tested a range of pH to determine the optimum for stability of each CD1 isoform. We lysed cells at pH 7.4, 7.0, 6.5, 6.0, 5.5, 5.0, or 4.5 and then incubated lysates at 4 or 37 °C for different times. This pH range was selected to cover the typical pH encountered at the plasma membrane (pH 7.4) or in the early endosomes (pH 6.0) and lysosomes (pH 5.5) where CD1 isoforms traffic.
Remarkably, each CD1 isoform displayed a different optimum pH for stability (Fig. 4, A–C). A maximum protective effect for CD1a was achieved at pH 6.0, with some protection occurring also at pH 5.5. In the case of the CD1b HC·β2m complex, a broader range of protection from disassembling by pH was observed with maximal protection seen at a pH between 5.0 and 5.5. CD1c achieves the maximum stability at pH 6.5 with a second peak of protection at pH 5.0 and 5.5 (Fig. 4, A and B). These results confirm that low pH stabilizes CD1a, -b, and -c HC·β2m complexes from disassembly at 37 °C. Strikingly, the maximum protection for each CD1 isoform is equivalent to the pH of the intracellular compartment where that particular CD1 molecule localizes physiologically. CD1a, which localizes to early recycling endosomes that are typically between of pH 6.0 and 6.5, achieves its maximum protection in pH 6.0. CD1b, which localizes to lysosomes, shows maximum protection at pH 5 to 5.5. Remarkably, CD1c, which is broadly distributed from early to late endosomes and lysosomes, shows significant protection across a broader pH range than the other CD1 isoforms.
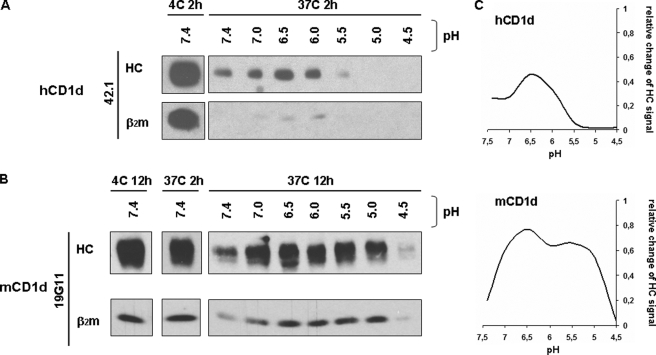
FIGURE 4.
pH-dependent protection of CD1 molecules corresponds to their locations in intracellular compartments. HeLa:CD1a, -CD1b, -CD1c, and -CD1d transfectants were surface biotinylated and lysed with 0.5% Triton X-100 at pH 7.4, 7.0, 6.5, 6.0, 5.5, 5.0, or 4.5. Lysates were incubated for 2 (CD1a and CD1d), 12 (CD1c), or 24 h (CD1b) at 37 °C. CD1 heavy chain·β2m complexes were immunoprecipitated with 10H3 (CD1a), BCD1b.3 (CD1b), F10/23.A.1 (CD1c), 42.1 or 75.10 (CD1d) antibodies. Immunoprecipitates were analyzed by SDS-PAGE (15%) in reducing conditions and immunoblotted with streptavidin-HRP. A, immunoprecipitated CD1 HC and β2m from lysates incubated at 37 °C at the indicated pH. B, quantification of band intensities for particular CD1 HC at different pH values by densitometry using ImageJ software. Curves indicate the ratio of signal for CD1 HC in lysates incubated at 37 °C at different pH in comparison to the incubation at 4 °C. C, the schematic shows the physiological pH of different intracellular compartments (ovals; ERC, endocytic recycling compartment; EE, early endosomes; LE, late endosomes), the intracellular location of CD1a, CD1b, and CD1c (black bars), and maximal protective effect of pH on the stability of CD1:HC·β2m complexes (gray triangles).
To further test the hypothesis that each CD1 isoform is most stable at the pH of its steady state endosomal localization site, we compared the pH dependence of human (h) and mouse (m) CD1d HC·β2m complex stability. As noted above, the effect of pH on hCD1d was distinct compared with that observed for the group I CD1 molecules. Human CD1d only partially localizes to LAMP-1 compartments (18), whereas mouse CD1d strongly associates with AP3 (7, 8) and predominantly localizes to LAMP-1 compartments (19). Thus mCD1d is more similar in steady state localization to hCD1b than hCD1d. To examine the effect of pH on hCD1d and mCD1d HC·β2m complex stability, we used hCD1d-transfected HeLa cells and mCD1d-transfected RAW cells. First, we found that mCD1d is more stable after cell lysis than hCD1d, requiring at least 12 h for a significant reduction in the signal of the HC·β2m complex (Fig. 5, A versus B). This higher stability of mCD1d is also more similar to CD1b than hCD1d. When we incubated lysates from CD1d-transfected cells at different pH at 37 °C (Fig. 5 and data not shown), we found pH-dependent protection from disassembly for both human and mouse CD1d HC·β2m complexes. However, the pH that protects mCD1d was significantly lower than in the case of hCD1d and closer to CD1b (Fig. 5). The effect of pH on the stability of hCD1d was less dramatic than observed in the case of the human CD1a, -b, and -c isoforms (Figs. 5A versus 4), but nevertheless, the hCD1 HC·β2m complex was still more stable at endosomal pH (between 6.5 and 6.0) than at neutral pH. Thus, the pH at which each CD1 isoform HC·β2m complex is protected from disassembly remarkably corresponds to the pH of the compartment where each isoform is predominantly localized in the cell at steady state.
FIGURE 5.
Effect of low pH on stability of human and mouse CD1d. HeLa:hCD1d and RAW:mCD1d transfectants were surface biotinylated and lysed with 0.5% Triton X-100 at pH 7.4, 7.0, 6.5, 6.0, 5.5, 5.0, or 4.5. Lysates were incubated for 2 h (hCD1d) or 2, 6, and 12 h (mCD1d) at 4 or 37 °C. Human CD1d:HC·β2m complexes were immunoprecipitated with anti-CD1d mAb 42.1 and 75.10, whereas mouse CD1d:HC·β2m complexes were immunoprecipitated with antibody 19G11. Immunoprecipitates were analyzed by SDS-PAGE (15%) in reducing conditions and immunoblotted with streptavidin-HRP. A and B, immunoprecipitated hCD1d and mCD1d HC and β2m from lysates incubated at 37 °C at the indicated pH. C, quantification of CD1d HC band intensities at different pH values by densitometry using ImageJ software. Curves indicate the ratio of signal for CD1d HC in lysates incubated at 37 °C at different pH values in comparison to the incubation at 4 °C.
CD1 HC·β2M Complexes Are Stabilized by Specific Lipid Antigens
A major factor in the stability of MHC molecules is peptide binding. In fact, in the absence of high affinity binding peptides, MHC class I and II heterodimers either fail to complete folding and/or are prone to dissociate. To study the possible role of specific lipid antigens in the stabilization of CD1 HC·β2m complexes we determined if α-galactosylceramide (α-GalCer) could protect hCD1d from disassembling at 37 °C. Eighty percent of the hCD1d HC·β2m complexes disassemble when the lysates are incubated at 37 °C (pH 7.4) for 2 h (see Figs. 1 and 2) and in this experiment over 95% dissociation occurred (Fig. 6A, compare β2m in lanes 2 versus 6). To determine whether α-GalCer can protect CD1d HC·β2m complexes from disassembly, we lysed CD1d-transfected HeLa cells and incubated the lysates at 4 or 37 °C for 2 h at pH 7.4 in the presence or absence of α-GalCer. The addition of α-GalCer to the lysates during incubation at 37 °C markedly stabilized the CD1d HC·β2m complexes immunoprecipitated by conformation-dependent mAb 42.1 (Fig. 6A, lane 6 versus 22). Importantly, this effect was dose-dependent and α-GalCer had a greater effect at higher concentrations (Fig. 6A, lanes 10, 14, 18, and 22). Consistent with this conclusion, the incubation with α-GalCer reduced the quantity of free CD1d HC as detected by IP with mAb 75.10 (Fig. 6A, compare lane 7 with lanes 11, 15, 19, and 23). In contrast, the addition of α-GalCer had no effect on MHC class I HC·β2m complex stability (Fig. 6A, compare lane 8 versus lanes 12, 16, 20, and 24). The quantification of CD1d HC·β2m complex (42.1 mAb) and free CD1d HC (75.10 mAb) underscores the protective effect of α-GalCer on CD1d HC·β2m disassembly as shown in Fig. 6B. In samples incubated without or with low concentrations of α-GalCer, the ratio of CD1d HC·β2m complex to free CD1d HC is lower than 0.7, whereas this ratio increases progressively with rising concentrations of lipid antigen until it reaches a value of 1.47 at 50 μg/ml of α-GalCer (Fig. 6B). Similar results were obtained with C1R:CD1d transfectants (supplemental Fig. S1). It should be noted that protection from dissociation of the CD1d HC·β2m complex was not complete, even at the highest concentrations of α-GalCer tested (100 μg/ml) (Fig. 6B and data not shown). This could be due to the incomplete ability of α-GalCer to prevent HC·β2m dissociation or perhaps more likely insufficient loading under our experimental conditions. Nevertheless, the data strongly indicates that dose-dependent binding of a specific lipid antigen protects the CD1d HC·β2m complex from disassembling.
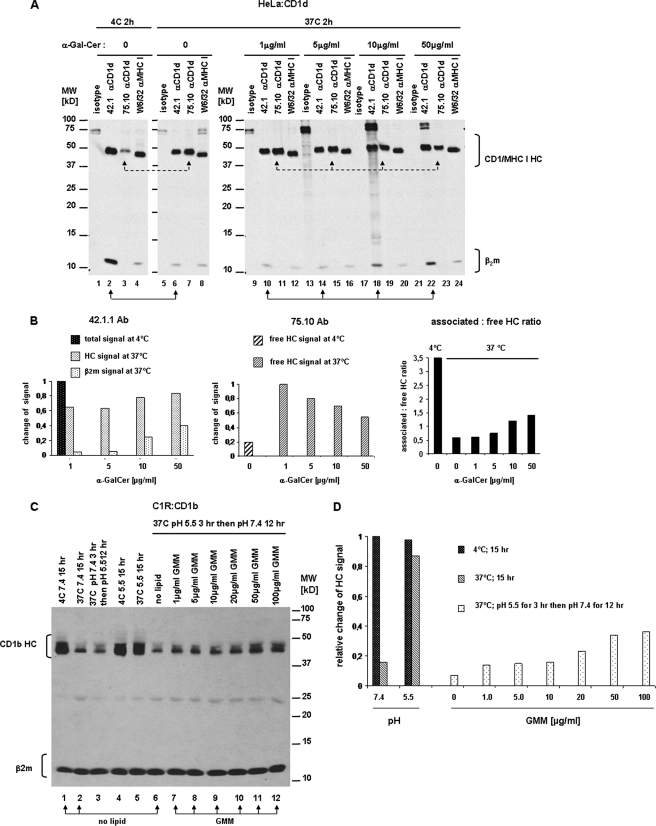
FIGURE 6.
Specific lipid antigens protect CD1d and CD1b HC·β2m complexes from disassembly. A, CD1d-transfected HeLa cells were surface biotinylated and lysed with 0.5% Triton X-100 (pH 7.4). Lysates were incubated for 2 h at 4 (lanes 1–4) or 37 °C (lanes 5–24) with different concentrations of α-GalCer (lanes 9–24) or without lipid (lanes 1–8). CD1d HC·β2m complexes were immunoprecipitated with anti-CD1d mAb 42.1 (lanes 2, 6, 10, 14, 18, and 22; solid line arrows), free CD1d HC were precipitated with mAb 75.10 (lanes 3, 7, 11, 15, 19, and 23; dashed line arrows), and MHC class I with antibody W6/32 (lanes 4, 8, 12, 16, 20, and 24). Mouse IgG1 and IgG2a were used as isotype controls. Immunoprecipitates were analyzed by SDS-PAGE (15%) under reducing conditions and immunoblotted with streptavidin-HRP. B, quantification of intensities of CD1d HC bands immunoprecipitated with 42.1 and 75.10 antibodies. Bars indicate the relative change of CD1d HC signal in samples incubated for 2 h at 37 °C in the presence of α-GalCer compared with incubation at 4 °C without lipid. Right graph shows the ratio between the associated and free form of CD1d HC in lysates incubated with increasing concentrations of α-GalCer at 37 or 4 °C without lipid. All measures were done by densitometry with ImageJ software. C, CD1b-transfected C1R cells were surface biotinylated and lysed with 0.5% Triton X-100 (pH 7.4) (lanes 1–3) or pH 5.5 (lanes 4–12). C32 GMM was added in increasing concentrations to samples run in lanes 7–12. Lysates were incubated for 15 h at 4 °C (lane 1 and 4), 15 h at 37 °C (lane 2 and 5), or 3 h at 37 °C followed by change in pH to 5.5 (lane 3) or 7.4 (lanes 6–12) and an additional 12-h incubation at the same temperature. CD1d HC·β2m complexes were immunoprecipitated with anti-β2m mAb BBM.1. Immunoprecipitates were analyzed by SDS-PAGE (15%) under reducing conditions and immunoblotted with streptavidin-HRP. D, quantification of intensities of bands corresponding to CD1b HC immunoprecipitated with BBM.1 antibody. Bars indicate the relative change of CD1d HC signal in samples incubated at different conditions (different temperatures and pH, in presence or absence of C32 GMM) compared with incubation at 4 °C without lipid. Intensities of bands were measured by densitometry with ImageJ software and relative change of signal was calculated as proportion of CD1b HC signal in samples incubated at 37 °C with or without GMM to intensity of CD1b HC signal at 4 °C.
Next, we sought to determine whether specificity of the lipid was important in CD1d complex stability. It was possible that stabilization of CD1d seen with α-GalCer results from a nonspecific effect of lipid that is not related to specific binding of α-GalCer to CD1d. To examine this, we tested the effect of C80 trehalose monomycolate because it is predicted to be too long to pack into the hCD1d binding groves (20). First, we confirmed CD1d HC·β2m complex stabilization at 37 °C in the presence of α-GalCer in a dose-dependent manner (supplemental Fig. S2). In contrast, trehalose monomycolate had no effect across a broad concentration range on CD1d HC·β2m complex stability (supplemental Fig. S2D). This result supports the specific stabilization of the CD1d HC·β2m complex by α-GalCer.
Next, we further confirmed the role of lipid antigen binding on stability of a second CD1 isoform, CD1b, and evaluated how the dual factors, pH and lipid antigen binding, might cooperate in stabilizing this HC·β2m complex. We tested if C32 (GMM), a CD1b-presented antigen isolated from M. tuberculosis, could stabilize CD1b molecules. We surface biotinylated CD1b-transfected C1R cells, lysed them in 0.5% Triton X-100 adjusted to pH 5.5, and incubated the cells at 37 °C for 15 h in the presence or absence of C32 GMM, in increasing concentrations. This first incubation was performed at pH 5.5 as this pH favors lipid exchange (21) and we also showed this pH stabilizes the CD1b HC·β2m complex. After a 3-h incubation at 37 °C, the pH of samples was then neutralized and the lysates were incubated for another 12 h at 37 °C, and stability of the CD1b HC·β2m complexes was assessed by quantitating the CD1b HC that was co-immunoprecipitated with anti-β2m-specific mAb BBM.1. In addition, similar aliquots of lysate were incubated at 4 °C as baseline controls for the 37 °C sample (Fig. 6, C and D). Confirming the previous result, we found that the CD1b HC signal was reduced in samples incubated at 37 °C compared with 4 °C at neutral pH (Fig. 6C, compare lane 1 versus 2). Furthermore, samples maintained at acidic pH 5.5 were also stable even at 37 °C (Fig. 6C, lane 5). Importantly, we found that addition of GMM to the cell lysates substantially protected the CD1b HC·β2m complexes from disassembly when the pH was raised to 7.4 (Fig. 6C, lane 6 versus 7–12) and this protective effect was dose-dependent (Fig. 6D). In contrast, incubation of lysates with equivalent concentrations of α-GalCer did not alter the stability of the CD1b HC·β2m complex (supplemental Fig. S3).
Together these results show that the loading of specific lipid antigens, such as C32 GMM, stabilizes the CD1b complex in a manner similar to the protection of the CD1d complex by α-GalCer. It suggests a model for the sequence of events during CD1 intracellular trafficking (Fig. 7). We propose that when CD1 molecules localize to the endocytic compartment in which they survey lipids, the CD1 HC·β2m complex is stabilized by endosomal or lysosomal pH. This low pH-mediated stabilization of the CD1 HC·β2m complex allows HC·β2m complexes to remain intact during lipid exchange when the groove might be temporarily empty. Then, when properly loaded with an appropriate lipid antigen, the CD1 HC·β2m complex traffics to the cell surface. At the cell surface, CD1 complexes encounter neutral pH but no longer require low pH stabilization because the presence of lipid in the binding groove provides stability at neutral pH. This model also supports a potential quality control mechanism in which CD1 HC·β2m complexes without stabilizing lipid antigens that reach the cell surface are prone to dissociate and have a shortened half-life (Fig. 7).
FIGURE 7.
Regulation of CD1 HC·β2m complex stability and quality control. A, mechanisms of the CD1 stability. At neutral pH in the absence of lipid antigen the CD1 HC·β2m complex is unstable (upper panel). At endosomal (acidic) pH, the CD1 HC·β2m complex is relatively more stable (middle panel). At endosomal pH, lipid antigen binding or exchange is facilitated, resulting in formation of the CD1 HC·β2m/lipid complex that is very stable even in neutral pH (lower panel). B, quality control model based on regulation of CD1 HC·β2m complex stability. In endosomal compartments, empty CD1 molecules are stabilized by low pH providing a pool of molecules available for binding lipid antigen. CD1 molecules loaded with proper lipid antigens in endosomal compartments form stable HC·β2m/lipid complexes that can traffic to the cell surface and remain stable at neutral pH. On the other hand, CD1 molecules that are empty or loaded with poor quality lipid are prone to dissociate at neutral pH, which would result in a shortened half-life at the cell surface. Relatively unstable complexes are depicted in blue and stable complexes are black.
DISCUSSION
For both MHC class I and II, the binding of high affinity peptides completes assembly and favors expression of the most stable MHC complexes on the cell surface (22) and increases the chances of recognition by T cells (23). On the other hand, unloaded MHC molecules disassemble after a few hours at 4 °C (24). Thus, their quality control mechanism focuses on peptide binding dependent stability.
However, the role of lipid binding by CD1 molecules and the conditions that result in stable or unstable CD1 HC·β2m complexes are not known. Previously, an important role for pH was identified as a factor influencing antigen loading or exchange (25, 26), and two types of accessory molecules, microsomal triglyceride transfer protein (14) and saposins were shown to facilitate the loading of lipids onto CD1 molecules (27–29). It is generally assumed that self-lipid antigens are first bound to CD1 in the ER and then foreign lipid antigens may be exchanged for self-lipid antigens in endosomal compartments. Several studies have indirectly implicated lipid loading of CD1 molecules as important for CD1 HC·β2m complex stability. For example, decreased surface expression of CD1 in dendritic cells was observed if microsomal triglyceride transfer protein function was impaired (15) and decreased surface recognition of CD1a by mAb was noted in cells cultured under serum/lipid-deficient conditions and could be reversed by adding of CD1a-specific sulfatide antigen (16).
Here under various conditions tested, CD1 molecules were found to be less stable than MHC class I and II molecules after lysis at neutral pH. To examine CD1 HC·β2m complex stability, we developed experimental systems controlling pH, temperature, and detergent to model critical factors that influence complex stability. CD1 HC·β2m complexes were noted to disassemble after a period of several hours at 37 °C, under conditions where MHC I and II complexes were very stable. Although CD1 molecules (with the exception of hCD1d) are stable at 4 °C or room temperature for up to 3 days (not shown), the complexes readily dissociate after cell lysis at neutral pH when warmed to 37 °C. Because the CD1 isoforms localize to different endocytic compartments before they recycle to the plasma membrane, we realized that they encounter markedly different pH in each location. This led us to investigate the possibility that pH influences the stability of CD1 HC·β2m complexes at 37 °C. We found that each CD1 isoform is optimally stable in vitro at a different pH. CD1a and hCD1d are maximally stable at pH 6.0, whereas the effect on CD1b is maximal at a pH between 5.0 and 5.5. For CD1c stabilization was noted from pH 6.5 to 5.5. Interestingly, the pH at which each isoform is maximally protected corresponds remarkably well to the pH of the intracellular compartment where the particular CD1 isoform localizes at steady state. The pH of the endocytic recycling compartment, where CD1a is found, is estimated between 6.0 and 6.5. Lysosomes, where CD1b localizes, have a pH lower than 5.5. CD1c and hCD1d show a broader intracellular distribution with some accumulation in early endosomes and endocytic recycling compartments, as well as in late endosomes and lysosomes. Moreover, mouse CD1d, which contrary to its human ortholog, accumulates almost exclusively in LAMP-1 positive compartments, is protected across a range of lower pH, down to pH 5.0, like hCD1b, which has a similar lysosomal intracellular localization. Therefore, our results suggest that the physiologic pH where CD1 molecules are normally localized stabilizes their subunit structure. This is likely to be important because it would be desirable for CD1 HC·β2m complexes to remain intact at endosomal pH to provide a pool of molecules ready for lipid antigen binding. We suggest that the pH-dependent protection mechanism shown here could represent a physiological stabilization mechanism for molecules awaiting lipid binding (if empty) or lipid exchange (Fig. 7). Furthermore, we demonstrated that specific lipid antigens can stabilize CD1 HC·β2m complexes in detergent lysates at pH 7.4 and 37 °C, conditions under which they would otherwise disassemble. For example, the hCD1d HC·β2m complex is protected from disassembly by α-GalCer in a dose-dependent manner and the integrity of the CD1b HC·β2m complex is protected by GMM (Fig. 6). Thus, successful lipid loading generates stable CD1 HC·β2m·lipid complexes that are no longer dependent on endosomal pH stabilization. It is also likely that lipids with varying affinities for binding to CD1 may correspondingly provide varying levels of complex stability.
Together, these results suggest the existence of two complementary and cooperative mechanisms for stabilizing CD1 protein complexes. Namely, CD1 HC·β2m localized in endocytic compartments at steady state are stabilized by the lower pH encountered in these compartments. When lipid loading/exchange occurs in endosomal compartments the CD1 HC·β2m·lipid complexes are further stabilized so that they are competent to withstand the “higher” pH found at the cell surface where T cell recognition occurs. Quality control is inherently suggested by this model because CD1 HC·β2m complexes that are not stabilized by lipid antigen binding would be predicted to disassemble and have a shorter half-life at the cell surface (Fig. 7).
Supplementary Material
Acknowledgments
We thank Dr. M. Cernadas for providing mCD1d-transfected RAW cells and Dr. A. Bendelac for anti-mCD1d mAb 19G11. We are also grateful to Drs. B. Moody, J. Higgins, and S. Behar for critical comments and suggestions.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 AI 028973 and AI 063428 (to M. B. B.). This work was also supported by a personal research chair from Mr. James Badrick, Royal Society Wolfson Research Merit Award, as a former Lister Institute-Jenner Research Fellow, the Medical Council, and the Wellcome Trust (084923/B/08/7) (to G. S. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- MHC
- major histocompatibility complex
- HC
- heavy chain
- β2m
- β2-microglobulin
- ER
- endoplasmic reticulum
- GMM
- glucose monomycolate
- PBS
- phosphate-buffered saline
- IP
- immunoprecipitation
- mAb
- monoclonal antibody
- HRP
- horseradish peroxidase
- GalCer
- galactosylceramide.
REFERENCES
- 1.Cohen N. R., Garg S., Brenner M. B. (2009) Adv. Immunol. 102, 1–94 [DOI] [PubMed] [Google Scholar]
- 2.Kang S. J., Cresswell P. (2002) J. Biol. Chem. 277, 44838–44844 [DOI] [PubMed] [Google Scholar]
- 3.De Silva A. D., Park J. J., Matsuki N., Stanic A. K., Brutkiewicz R. R., Medof M. E., Joyce S. (2002) J. Immunol. 168, 723–733 [DOI] [PubMed] [Google Scholar]
- 4.Sugita M., Grant E. P., van Donselaar E., Hsu V. W., Rogers R. A., Peters P. J., Brenner M. B. (1999) Immunity 11, 743–752 [DOI] [PubMed] [Google Scholar]
- 5.Barral D. C., Cavallari M., McCormick P. J., Garg S., Magee A. I., Bonifacino J. S., De Libero G., Brenner M. B. (2008) Traffic 9, 1446–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sugita M., Jackman R. M., van Donselaar E., Behar S. M., Rogers R. A., Peters P. J., Brenner M. B., Porcelli S. A. (1996) Science 273, 349–352 [DOI] [PubMed] [Google Scholar]
- 7.Lawton A. P., Prigozy T. I., Brossay L., Pei B., Khurana A., Martin D., Zhu T., Späte K., Ozga M., Höning S., Bakke O., Kronenberg M. (2005) J. Immunol. 174, 3179–3186 [DOI] [PubMed] [Google Scholar]
- 8.Cernadas M., Sugita M., van der Wel N., Cao X., Gumperz J. E., Maltsev S., Besra G. S., Behar S. M., Peters P. J., Brenner M. B. (2003) J. Immunol. 171, 4149–4155 [DOI] [PubMed] [Google Scholar]
- 9.Briken V., Jackman R. M., Watts G. F., Rogers R. A., Porcelli S. A. (2000) J. Exp. Med. 192, 281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sugita M., van Der Wel N., Rogers R. A., Peters P. J., Brenner M. B. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8445–8450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Townsend A., Elliott T., Cerundolo V., Foster L., Barber B., Tse A. (1990) Cell 62, 285–295 [DOI] [PubMed] [Google Scholar]
- 12.Ljunggren H. G., Stam N. J., Ohlén C., Neefjes J. J., Höglund P., Heemels M. T., Bastin J., Schumacher T. N., Townsend A., Kärre K. (1990) Nature 346, 476–480 [DOI] [PubMed] [Google Scholar]
- 13.Wubbolts R., Neefjes J. (1999) Immunol. Rev. 172, 189–208 [DOI] [PubMed] [Google Scholar]
- 14.Dougan S. K., Salas A., Rava P., Agyemang A., Kaser A., Morrison J., Khurana A., Kronenberg M., Johnson C., Exley M., Hussain M. M., Blumberg R. S. (2005) J. Exp. Med. 202, 529–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaser A., Hava D. L., Dougan S. K., Chen Z., Zeissig S., Brenner M. B., Blumberg R. S. (2008) Eur. J. Immunol. 38, 2351–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manolova V., Kistowska M., Paoletti S., Baltariu G. M., Bausinger H., Hanau D., Mori L., De Libero G. (2006) Eur. J. Immunol. 36, 1083–1092 [DOI] [PubMed] [Google Scholar]
- 17.Spada F. M., Borriello F., Sugita M., Watts G. F., Koezuka Y., Porcelli S. A. (2000) Eur. J. Immunol. 30, 3468–3477 [DOI] [PubMed] [Google Scholar]
- 18.Sugita M., Cao X., Watts G. F., Rogers R. A., Bonifacino J. S., Brenner M. B. (2002) Immunity 16, 697–706 [DOI] [PubMed] [Google Scholar]
- 19.Jayawardena-Wolf J., Benlagha K., Chiu Y. H., Mehr R., Bendelac A. (2001) Immunity 15, 897–908 [DOI] [PubMed] [Google Scholar]
- 20.Koch M., Stronge V. S., Shepherd D., Gadola S. D., Mathew B., Ritter G., Fersht A. R., Besra G. S., Schmidt R. R., Jones E. Y., Cerundolo V. (2005) Nat. Immunol. 6, 819–826 [DOI] [PubMed] [Google Scholar]
- 21.Relloso M., Cheng T. Y., Im J. S., Parisini E., Roura-Mir C., DeBono C., Zajonc D. M., Murga L. F., Ondrechen M. J., Wilson I. A., Porcelli S. A., Moody D. B. (2008) Immunity 28, 774–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerundolo V., Elliott T., Elvin J., Bastin J., Rammensee H. G., Townsend A. (1991) Eur. J. Immunol. 21, 2069–2075 [DOI] [PubMed] [Google Scholar]
- 23.Paulsson K. M., Wang P. (2004) FASEB J. 18, 31–38 [DOI] [PubMed] [Google Scholar]
- 24.Schumacher T. N., Heemels M. T., Neefjes J. J., Kast W. M., Melief C. J., Ploegh H. L. (1990) Cell 62, 563–567 [DOI] [PubMed] [Google Scholar]
- 25.Ernst W. A., Maher J., Cho S., Niazi K. R., Chatterjee D., Moody D. B., Besra G. S., Watanabe Y., Jensen P. E., Porcelli S. A., Kronenberg M., Modlin R. L. (1998) Immunity 8, 331–340 [DOI] [PubMed] [Google Scholar]
- 26.Gumperz J. E., Roy C., Makowska A., Lum D., Sugita M., Podrebarac T., Koezuka Y., Porcelli S. A., Cardell S., Brenner M. B., Behar S. M. (2000) Immunity 12, 211–221 [DOI] [PubMed] [Google Scholar]
- 27.Kang S. J., Cresswell P. (2004) Nat. Immunol. 5, 175–181 [DOI] [PubMed] [Google Scholar]
- 28.Yuan W., Qi X., Tsang P., Kang S. J., Illarionov P. A., Besra G. S., Gumperz J., Cresswell P. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 5551–5556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winau F., Schwierzeck V., Hurwitz R., Remmel N., Sieling P. A., Modlin R. L., Porcelli S. A., Brinkmann V., Sugita M., Sandhoff K., Kaufmann S. H., Schaible U. E. (2004) Nat. Immunol. 5, 169–174 [DOI] [PubMed] [Google Scholar]
- 30.Moody D. B., Briken V., Cheng T. Y., Roura-Mir C., Guy M. R., Geho D. H., Tykocinski M. L., Besra G. S., Porcelli S. A. (2002) Nat. Immunol. 3, 435–442 [DOI] [PubMed] [Google Scholar]
- 31.Leadbetter E. A., Brigl M., Illarionov P., Cohen N., Luteran M. C., Pillai S., Besra G. S., Brenner M. B. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 8339–8344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minnikin D. E., Dobson G., Draper P. (1985) J. Gen. Microbiol. 131, 2007–2011 [DOI] [PubMed] [Google Scholar]
- 33.Olive D., Dubreuil P., Mawas C. (1984) Immunogenetics 20, 253–264 [DOI] [PubMed] [Google Scholar]
- 34.McMichael A. J., Pilch J. R., Galfré G., Mason D. Y., Fabre J. W., Milstein C. (1979) Eur. J. Immunol. 9, 205–210 [DOI] [PubMed] [Google Scholar]
- 35.Behar S. M., Porcelli S. A., Beckman E. M., Brenner M. B. (1995) J. Exp. Med. 182, 2007–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melián A., Watts G. F., Shamshiev A., De Libero G., Clatworthy A., Vincent M., Brenner M. B., Behar S., Niazi K., Modlin R. L., Almo S., Ostrov D., Nathenson S. G., Porcelli S. A. (2000) J. Immunol. 165, 4494–4504 [DOI] [PubMed] [Google Scholar]
- 37.Grant E. P., Degano M., Rosat J. P., Stenger S., Modlin R. L., Wilson I. A., Porcelli S. A., Brenner M. B. (1999) J. Exp. Med. 189, 195–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roark J. H., Park S. H., Jayawardena J., Kavita U., Shannon M., Bendelac A. (1998) J. Immunol. 160, 3121–3127 [PubMed] [Google Scholar]
- 39.Smith L. M., Petty H. R., Parham P., McConnell H. M. (1982) Proc. Natl. Acad. Sci. U.S.A. 79, 608–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barnstable C. J., Bodmer W. F., Brown G., Galfre G., Milstein C., Williams A. F., Ziegler A. (1978) Cell 14, 9–20 [DOI] [PubMed] [Google Scholar]
- 41.Jackman R. M., Stenger S., Lee A., Moody D. B., Rogers R. A., Niazi K. R., Sugita M., Modlin R. L., Peters P. J., Porcelli S. A. (1998) Immunity 8, 341–351 [DOI] [PubMed] [Google Scholar]
- 42.Spada F. M., Koezuka Y., Porcelli S. A. (1998) J. Exp. Med. 188, 1529–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beckman E. M., Melián A., Behar S. M., Sieling P. A., Chatterjee D., Furlong S. T., Matsumoto R., Rosat J. P., Modlin R. L., Porcelli S. A. (1996) J. Immunol. 157, 2795–2803 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.