Abstract
Nucleotide insertions that modify the C terminus of ferritin light chain (FTL) cause neurodegenerative movement disorders named neuroferritinopathies, which are inherited with dominant transmission. The disorders are characterized by abnormal brain iron accumulation. Here we describe the biochemical and crystallographic characterization of pathogenic FTL mutant p.Phe167SerfsX26 showing that it is a functional ferritin with an altered conformation of the C terminus. Moreover we analyze functional and stability properties of ferritin heteropolymers made of 20–23 H-chains and 1–4 L-chains with representative pathogenic mutations or the last 10–28 residues truncated. All the heteropolymers containing the pathogenic or truncated mutants had a strongly reduced capacity to incorporate iron, both when expressed in Escherichia coli, and in vitro when iron was supplied as Fe(III) in the presence of ascorbate. The mutations also reduced the physical stability of the heteropolymers. The data indicate that even a few mutated L-chains are sufficient to alter the permeability of 1–2 of the 6 hydrophobic channels and modify ferritin capacity to incorporate iron. The dominant-negative action of the mutations explains the dominant transmission of the disorder. The data support the hypothesis that hereditary ferritinopathies are due to alterations of ferritin functionality and provide new input on the mechanism of the function of isoferritins.
Keywords: Genetic Diseases, Iron, Neurodegeneration, Protein Assembly, Protein Stability, Ferritin, Iron Homeostasis
Introduction
Ferritins are ubiquitous iron storage molecules with a major role in the control of cellular iron availability. Cytosolic ferritins are heteropolymers composed by tissue-specific proportions of H- and L-chains, whereas mitochondrial ferritins are homopolymers (1). They sequester and incorporate iron in their large cavity by complex reactions that involve Fe(II) oxidation at the ferroxidase site of H-chains, iron hydrolysis, and mineralization, which is facilitated by acidic residues of L-chains (2). The cellular capacity of sequestering iron is mainly related to the level of the H-chain (3), which regulates the size of the cellular labile iron pool, potentially toxic for production of reactive-free radical species. Deletion of H-ferritin in mouse models is embryonically lethal (4), whereas its conditioned deletion after birth strongly increases iron toxicity and oxidative damage (5). Pathogenic mutations of the H-chain have not been reported yet, and all the ferritin genetic disorders so far identified interest L-chain and have dominant transmission. The most common one is associated with mutations of the iron responsive element in the 5′-untranslated region of the transcript, which reduce or abolish its iron-mediated suppression, resulting in a constitutive L-ferritin up-regulation (6, 7). This disorder, named hereditary hyperferritinopathy with cataract syndrome (OMIM code 600886) is characterized by high levels of serum and tissue L-ferritins and by early onset bilateral cataract, caused by the formation of L-ferritin microcrystallines in the lens (8). More severe and rare is the group of diseases named neuroferritinopathies (9) or hereditary ferritinopathies (10) (OMIM code 606159). Clinically these disorders are characterized by abnormal involuntary movements and cognitive decline that often appear in the 3rd to 6th decade of life. The neuropathology of the disorders is characterized by iron accumulation in the basal ganglia of the brain detectable by MR imaging (11) and by intracellular ferritin inclusion bodies in the glia and neurons of central nervous systems and other organs.
Six pathogenic mutations have been identified so far (9, 10, 12–16). They are all private mutations found in single families, except the 460InsA found in several patients of North Anglia. These mutations consist in insertions or duplications that introduce frameshifts with a fully penetrant effect, the exception being the missense Ala96 → Thr. The insertions make the subunit longer than the native subunit: one nucleotide, as 460InsA, introduces an extension at the C terminus of four amino acids, and two nucleotides, as 498InsTC, introduce a 16-amino acid extension. The insertion-duplications modify the L-chain starting from residues 148, 154, and 167 of the protein made of 175 amino acids, and are predicted to alter the protein from the last two turns of the D-helix to the first half of the E-helix (Fig. 1). Most previous mutational studies on ferritin were performed on the H-chain, which is structurally similar to the L-chain. They showed that the extension of a few residues at the C terminus or the fusion to a short peptide, has no major effects on ferritin stability or functionality (17–20). The truncation at the residue equivalent to 158 of the L-chain produced a ferritin that could not retain an iron core (3), whereas the deletion of the last 28 residue (equivalent to 24 residues of the L-chain) inhibited ferritin assembly when expressed in Escherichia coli (21). More recently it was shown that nucleotide insertions in the sites equivalent to those of the L-chain progressively decreased H-ferritin solubility and functionality by increasing the length of the substitution (18). The two pathogenic L-ferritin (FTL)3 mutants corresponding to FTL460InsA and FTL498InsTC (here named L154fs and L167fs) were expressed in E. coli and characterized (22, 23). The mutant L154fs was shown to be poorly soluble, to coassemble with H- and L-ferritins, and to reduce the functionality of the ferritin heteropolymers (23). In contrast, the mutant L167fs was shown to be as soluble as L-wild type (Lwt), to be less thermostable, and to form aggregates when incubated with an excess of Fe(II) in aerobic conditions (22, 24, 25). It was proposed that these iron-rich ferritin aggregates contribute to the formation of ferritin bodies in vivo and alter ferritin functionality (26, 27). Studies on cellular models showed that the expression of L154fs and L167fs caused remarkably similar phenotypes, with the accumulation of endogenous ferritins, an increase of labile iron pool and oxidative damage, joined to a reduction of ferritin half-life and proteasome activity (22). This indicated that the two mutants acted in a similar way to inhibit the activity of isoferritins, and that the formation of ferritin aggregates is not a primary cause of the phenotype. The common property of the two mutants is a major conformational alteration of the C terminus, which was exposed to solvent and accessible to proteolytic enzymes. Moreover, the expression of human L167fs in transgenic mice was shown to cause a phenotype with cytosolic and intranuclear ferritin aggregates analogous to those found in the patients (27), and to cause a deregulation of brain iron with up-regulation of the endogenous ferritin, down-regulation of transferrin receptor, and reduced RNA binding activity of IRP1 (26).
FIGURE 1.
Alignment of the C-terminal sequence of the l-ferritin mutants analyzed. Lwt is the sequence of wild-type L-ferritin. The amino acid numbering above includes the N-terminal methionine. The C-terminal sequence is conserved in LA96T, because the missense mutation Ala96 → Thr is in C-helix. L154fs represents mutant 460InsA, described in Ref. 9, L148fs represents mutant 442InsC described in Ref. 10. L167fs represent mutant 498InsTC described in Ref. 12. L148X, L154X, and L167X are truncated mutants. The mutant produced by the 442 4-nucleotide duplication described in Ref. 13 is the same as L148fs except that the substituted sequence is one amino acid longer. The mutation with a 16-nucleotide duplication in position 469–470 producing a frameshift at residue 162 (15) was not analyzed. The D and E α-helices are indicated by the boxes.
Altogether these data do not establish if the primary cause and dominant transmission of the disorders is linked to the acquisition of a new function of the mutants, such as a decreased solubility and aggregate formation, or if is due to a dominant-negative alteration of ferritin functionality.
In this work we analyzed the structural, crystallographic, and functional properties of recombinant L167fs expressed in E. coli. The properties were similar to those of FTL, except for a disordered conformation of the C terminus around the 4-fold axis. We also produced ferritin heteropolymers made of 20–23 H-chains and 1–4 L-mutant chains. The ones containing the pathogenic mutants caused by nucleotide insertions as well as those truncated at the first residue of the frameshift incorporated less iron than those containing Lwt, under conditions that mimic the physiological ones. In contrast the one containing the A96T point mutations behaved like Lwt. The data demonstrate that modifications of the C terminus act in a dominant-negative manner probably by altering the permeability of the ferritin shell. The results support the hypothesis that hereditary ferritinopathies are caused by alterations of ferritin functionality and provide new data on the mechanism of isoferritin functionality.
EXPERIMENTAL PROCEDURES
Plasmid Construction
Cloning of human L-ferritin cDNA was described in Ref. 28. To generate the L167fs mutant we cloned the cDNA of Lwt with a portion of the 3′-untranslated region and then inserted a thymine and a cytosine in the proper position by oligonucleotide-directed mutagenesis. The construct was subcloned into the pDS20pTrp vector to obtain the pDS-L167fs plasmid for expression in E. coli. The fragment for the H-chain was amplified by PCR from plasmid pUD-Hwt (28) and subcloned into pET12b vector without OmpT (Novagen) digested with NdeI and BamHI (pET-Hwt). The polycystronic vector was constructed by subcloning the L-ferritin cDNA downstream from the cDNA of H-ferritin into the pET-Hwt vector. The cDNA for the Lwt was amplified by PCR from plasmid pDS-Lwt (28), by inserting the restriction site BamHI at the termini and the Shine-Delgarno sequence (AAGGAG) upstream of the ATG. The fragment was subcloned into pET-Hwt digested with BamHI (pET-H/Lwt). For expression of the heteropolymers with the L-ferritin mutants the cDNA of L154fs and L167fs were subcloned in place of Lwt to obtain the pET-H/L154fs and pET-H/L167fs. The vector pET-H/L154fs was then subjected to site-directed mutagenesis to produce the pET-H/L148fs. The vector pET-H/Lwt was subjected to site-directed mutagenesis to introduce the A96T mutation and stop codons in positions 148, 154, and 167.
Recombinant Proteins
Recombinant H- and L-chain ferritins were expressed and purified as before (29). The same procedure was used for the L167fs mutant; the proteins were maintained in buffer containing anti-protease agents (1 mm benzamidine, 1 mm phenylmethylsulfonyl fluoride, 0.1 μm aprotinin, 10 μm leupeptin, 1 μm pepstatin) to avoid degradation. The purity of ferritins was judged by analysis of protein on a 15% acrylamide SDS-PAGE stained with Coomassie Blue. Iron was removed by incubation with 1% thioglycolic acid, pH 5.5, and 2,2-bipyridine followed by dialysis against 0.1 m Hepes buffer, pH 7.0. The protein concentration was determined with a BCA reagent (Pierce) calibrated on bovine serum albumin. Purified ferritin homopolymers were denatured in 0.1 m phosphate, pH 3.0, 6 m guanidine HCl at 4 °C for 16 h. The H- and L-chain heteropolymers were obtained in vitro by mixing the two denatured ferritin chains in different molar ratios and allowed to renature, after at least 10-fold dilution in 0.1 m phosphate buffer, pH 7.4, 1 mm dithiothreitol, for 2 h at room temperature (30). To study iron incorporation of the heteropolymers during expression in E. coli, the cells transformed with the bicistronic vectors were grown in M9 minimal growth medium until they reached 0.6 OD and then induced for 3 h with 0.4 mm isopropyl 1-thio-β-d-galactopyranoside. The cells were harvested, homogenized, and the soluble supernatant heated at 65 °C for 10 min. The ferritins were then analyzed by nondenaturing PAGE. For stability and ferroxidase activity studies, the heteropolymers were purified by gel filtration on Sepharose 6B columns to obtain preparations that were >95% pure, as judged by SDS-PAGE. Dynamic light scattering or photon correlation spectroscopy measurements were performed using DynaPROMS/X on ferritins concentrated up to 10 mg/ml.
Nanospray-ESI-MS Analysis of Intact Proteins
Protein sample solution was acidified to 10% of formic acid for direct nano-ESI-MS analysis on an API QStar PULSAR (PE-Sciex Instruments, Canada) mass spectrometer equipped with a nanoelectrospray ion source (Proxeon Biosystems, Odense, Denmark). Analysis was performed in positive ion mode and the HV potential was set-up around 800 V. Full scan mass spectra ranging from m/z 600 to 2200 Da were collected. Peak deconvolution with the Bayesian Protein Reconstruct was done using BioAnalystTM software 1.1.5 for molecular mass determination.
MALDI-TOF MS Analysis of Digested Proteins
Bands of interest were excised from gels, subjected to reduction by 10 mm dithiothreitol, alkylation by 55 mm iodoacetic acid, and finally digested with trypsin overnight (Roche Applied Science) (31). One-μl aliquots of the supernatant were used for MS analysis on a MALDI-TOF Voyager-DE STR (Applied Biosystems) mass spectrometer using the dried droplet technique and α-cyano-4-hydroxycinnamic acid as matrix. Peak list was obtained by peak de-isotoping. Spectra were accumulated over a mass range of 750–4000 Da with a mean resolution of about 15,000. Spectra were internally calibrated using matrix signals and trypsin autolysis peaks were then processed via Data Explorer software version 4.0.0.0 (Applied Biosystems).
Ferritin Stability
Denaturation of ferritin was evaluated on samples (50 μg/ml) following their incubation for 18 h at 4 °C in 0.1 m phosphate buffer, pH 7.4, 1 mm dithiothreitol, with various guanidine HCl concentrations. The conformational state was estimated from fluorescence spectra (excitation at 295 nm), using as standards, ferritin either in 0.1 m phosphate, pH 7.4 (native state), or in 0.1 m phosphate, pH 3.0, 6 m guanidine HCl (denatured state). Fluorescence spectra were measured with a Kontron SFM25 spectrofluorimeter, collected in the interval of 310–370 nm and used to calculate the fraction of unfolded protein. Chemical stability of ferritin was determined on samples (0.5 mg/ml) incubated in buffer containing 1% SDS for 4 h at room temperature; the presence of denatured ferritin subunits were identified by separation on discontinuous SDS-7.5% polyacrylamide gels in the top layer and 15% in the bottom layer, the samples were not boiled before loading and were stained with Coomassie Blue.
Iron Incorporation
Apoferritins (1 μm, 0.5 mg/ml) were incubated for 2 h at room temperature with 0.5–4.0 mm freshly made ferrous ammonium sulfate in 0.1 m Hepes buffer, pH 7.0. The samples were run on nondenaturing 7% polyacrylamide gels and stained for protein (Coomassie Blue) or iron (Prussian blue). In other experiments, apoferritins (0.2 μm, 0.1 mg/ml) were incubated with 0.1 mm 55Fe (ferric citrate) and 1 mm ascorbic acid in 0.1 m Hepes buffer, pH 7.0, for 2 h at room temperature. The samples were run on nondenaturing 7% polyacrylamide gels, dried, and the presence of 55Fe revealed by autoradiography. To study in vitro iron incorporation in the heteropolymers produced by the bicistronic vectors, equal amounts of the ferritins were separated on nondenaturing PAGE and then treated for iron removal by a 2-h incubation with 1% thioglycolic acid, pH 5.5, and 2,2-bipyridine. The gels were extensively washed with 0.1 m Hepes buffer, pH 6.5, and used to study ferritin iron uptake. In some experiments the gels were incubated in 200 μm ferrous ammonium sulfate for 15 min in Hepes buffer, washed, and stained with Prussian blue. Alternatively the gels were incubated in 20 mm Tris-HCl, pH 7.0, with 1 mm ferric ammonium citrate and 1 mm sodium ascorbate for 15 min, washed, and then stained with Prussian blue. For enhancing after Prussian blue staining, the gels were washed and incubated with 0.05% diaminobenzidine and 0.05% H2O2 for 15 min.
Immunological Methods
In Western blotting of nondenaturing PAGE the monoclonal antibodies LF03 and rH02 were used for L- and H-ferritin staining, respectively. They were used at a concentration of about 5 μg/ml. For SDS-PAGE we used the polyclonal rabbit anti-ferritin antibody (Sigma) that preferentially recognizes the L-chains. Antibody binding was revealed using horseradish peroxidase-labeled secondary antibody and ECL detection.
Ferroxidase Activity
To study the ferroxidase activity of the ferritins and heteropolymers, 0.1 μm apoferritins in 0.1 m Hepes, pH 6.5, were supplemented with 0.1 mm freshly made ferrous ammonium sulfate, and development of the amber color of Fe(III) was followed at 310 nm. In other experiments, 0.2 μm ferritins in 0.2 m acetate buffer, pH 6.0, containing 4 mg/ml of human apotransferrin were supplemented with 1 mm freshly made ferrous ammonium sulfate and the development of pink Fe(III)-transferrin complex was followed at 460 nm for 5 min.
Crystallization and Structure Determination of L167f
The first crystallization assays of L167fs were performed with a Cartesian Honeybee 961 robot (Genomic Solutions) using Greiner 96-well sitting drop crystallization plates. Different commercial screens were tested (Hampton Research, Qiagen, JenaBioscience, Molecular Dimensions). Then optimization was performed by the hanging drop vapor diffusion method in Linbro plates at room temperature (293 K). Crystals of L167fs grew over a 1-ml reservoir solution (42.5 mm CdSO4, 100 mm Hepes, pH 8.0, 960 mm sodium acetate, 3 mm NaN3) in drops composed of equal volumes (2 μl) of protein solution (9.89 mg/ml in 20 mm Tris-HCl, pH 7.4, 0.1 m sodium azide, 0.1 m phenylmethylsulfonyl fluoride, and 0.1 m EDTA) and reservoir solution. Crystals appeared after 1 or 2 days and grew up to their final size in 1 week. Prior to data collection, single crystals were cryo-protected in the reservoir solution supplemented with increasing concentrations of glycerol up to a final concentration of 35% (v/v). Data sets were collected at 100 K using synchrotron radiation on beam lines ID23-1 at the European Synchrotron Radiation Facility, Grenoble, France. Data were indexed in space group I432, processed with MOSFLM (32), and scaled using SCALA (33) from the CCP4 suite (34). Data collection statistics are given in Table 1.
TABLE 1.
X-ray diffraction data collection and refinement statistics
Values in parentheses correspond to the highest resolution shell.
| Space group | I432 |
| Cell parameter a (Å) | 151.355 |
| Data collection | |
| Resolution range (Å) | 61.78–1.85 (1.95–.85) |
| Number of unique reflections | 25,503 (3,572) |
| Completeness (%) | 98.3 (98.3) |
| Rmerge (%)a | 0.075 (0.407) |
| Multiplicity | 5.1 (5.2) |
| 〈I/σ〉 | 5.8 (2.0) |
| Model and refinement statistics | |
| Resolution range (Å) | 61.78–1.85 (1.90–1.85) |
| Number of used reflections | 23,733 (1,706) |
| Number of non-hydrogen atoms | 1,502 |
| Mean B values (Å2) | 17.97 |
| Root mean square bond lengths (Å) | 0.014 |
| Root mean square bond angles (°) | 1.24 |
| Rwork (%)b | 0.179 (0.226) |
| Rfree (%)c | 0.211 (0.316) |
| Ramachandran plot statistics (%) | |
| Favored regions | 98 |
| Allowed regions | 100 |
aRmerge = ΣhΣiN|Ih,i − 〈Ih〉|/ΣhΣiIh,i, where 〈Ih〉 is the average of symmetry related observations for unique reflections.
bRwork = Σh|Fhobs − Fhcalc|/Σh|Fhobs, where Fhobs and Fhcalc are the observed and calculated structure factor amplitudes, respectively.
c Rfree = as for Rwork, but for 5% of the total reflections chosen at random and omitted from refinement.
Structure Determination
The structure was solved by molecular replacement using the program MOLREP with the atomic coordinates of human L-chain ferritin subunit (35) (Protein Data Bank code 2ffx). Model building and refinements were performed using the programs COOT (36) and REFMAC5 (37) iteratively. Protein stereochemical restraints were taken from Ref. 38. Cadmium ions were positioned with the help of anomalous difference Fourier maps. Water molecules were positioned in well defined positive (mFo − DFc) residual densities with a lower cut-off of 3 σ, if they participated in H-bonds with the protein, cadmium ions, or other water molecules (where m is the figure of merit, and D is the estimate of error in the partial structure derived from coordinate error estimates). The model validity was checked with the program MOLPROBITY (39). The final protein model consists of residues 1–157. Final refinement statistics are given in Table 1. Figures were drawn using the molecular graphics program PyMOL (40).
RESULTS
Biochemical Characterization of the Recombinant L167fs Mutant
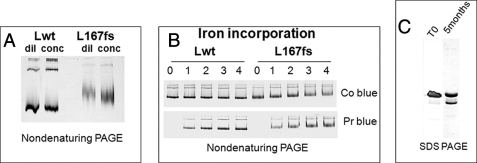
We expressed the full coding sequence of Lwt and L167fs in E. coli. They were recovered at a comparable level in the soluble fraction of the cell extracts (supplemental Fig. S1). The mutants assembled in 24-mer ferritin shells were stable at 70 °C and could be purified with the same procedure used for Lwt. To study the propensity to form aggregates, L167fs and Lwt were concentrated up to 40 mg/ml and then analyzed by nondenaturing PAGE, they did not show signs of precipitation or an evident increase of oligomeric species (Fig. 2A). Also, analyses by dynamic light scattering or photon correlation spectroscopy showed that the average hydrodynamic radius of the mutant (7.39 ± 0.56 nm) was not significantly different from that of Lwt (7.60 ± 0.11 nm). To study the iron effect on protein aggregation, L167fs and Lwt were incubated with ferrous ammonium sulfate at pH 7.0 for 2 h, separated by nondenaturing PAGE, and stained with Coomassie Blue or Prussian blue. With increments from 1,000 to 4,000 Fe(II) atoms per ferritin shell we did not observe evident signs of protein precipitation or aggregation, and the amounts of iron incorporated in L167fs and Lwt were equivalent (Fig. 2B). Denaturation studies showed that L167fs was slightly less stable than Lwt (see below). The purified preparations of L167fs contained a smaller peptide of ∼19 kDa, the proportion of which increased upon storage at 4 °C even in the presence of antiproteases. In a 5-month-old preparation it accounted for about 35% of the L167fs (Fig. 2C). This preparation was analyzed by nanospray ESI-MS and was shown to contain a 21,163-Da species, which corresponds to the full L167fs peptide deleted of the N-terminal methionine (calculated mass = 21,164.8 Da), and a minor component of 18,921 Da corresponding to fragment 2–168 (supplemental Fig. S2A).
FIGURE 2.
Analysis of the homopolymers of L167fs mutant. A, nondenaturing PAGE of the purified Lwt and L167fs ferritin homopolymers before (dil) and after concentration to 40 mg/ml (conc). B, Lwt and L167fs were incubated with Fe(II) increments of 1000, 2000, 3000, and 4000 atoms per molecule (lanes 1–4), pH 7.0, and then run on nondenaturing PAGE and stained for protein with Coomassie Blue (Co blue) or iron with Prussian blue (Pr blue). C, SDS-PAGE of L167fs preparation freshly made (T0) or after 5 months at 4 °C. Protein load was 5 μg, representative of three independent experiments with similar results.
The 18-kDa peptide was further analyzed for C-terminal sequencing by MALDI-TOF, obtaining the LGGPEAGLGEYLSS sequence, which corresponds to residues 155–168 of the mutant. This peptide results from cleavage of L167fs after Ser168, two residues downstream the site of the mutation (supplemental Fig. S2B). Lwt is not cleaved under the same conditions, indicating it has a more compact structure, in agreement with observations that the C terminus of L167fs is accessible to proteolytic enzymes, whereas that of Lwt is not (24) and with evidence that a FLAG tag epitope attached to the C terminus of the L154fs mutants is exposed (22). Thus the C termini of the two mutants have abnormal conformations.
Crystallographic Structure of 167fs
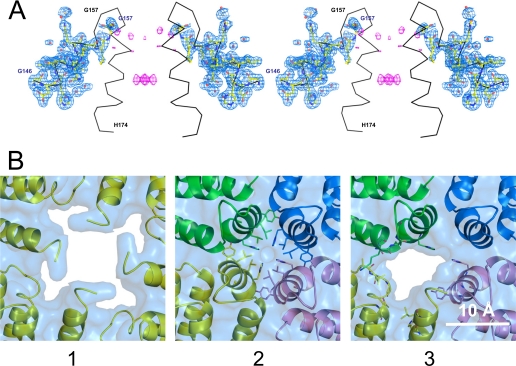
For further characterization, the purified protein was crystallized and analyzed by x-ray diffraction and its structure solved at 1.85-Å resolution. It fully overlapped the Lwt structure from the N terminus up to residue Gly157, two residues after the C-terminal end of helix D. Cadmium cations, used in crystallization conditions, were found to bind the protein at the usual metal binding sites, i.e. the residues of the 3-fold axes funnel (His115, Cys127, Asp128, and Glu131) and the nucleation center (His50, Glu54, Glu57, Glu58, and Glu61). As a whole, the L167fs structure is very close to that of Lwt as well as of the mutant structure determined by Baraibar et al. (25). When superimposing these structures onto L167fs, one obtains a root mean square deviation on Cα positions of 0.36 and 0.29 Å, respectively. However, a common feature with the previously published mutant structure (25) is observed: residues Gly156 and Gly157 display a similar conformation, which differs from that adopted by the D-E loop in the Lwt structure. This is illustrated in Fig. 3A where the 4-fold axis of the L167fs structure is shown together with the (2mFo − DFc) and (mFo − DFc) electron density maps and the Cα trace of the Lwt structure. The (mFo − DFc) map shows that the residual density at the 4-fold axis is very weak and does not allow one to add up any more atoms to the model. Moreover, six well ordered water molecules have been positioned in direct contact with B and D chains, in place of some side chains of the missing helix E. These observations indicate that there is no disordered chain within the 4-fold channel. To check the integrity of the crystallized protein at the C-terminal part, we performed an SDS-PAGE analysis of the washed and dissolved crystals and the protein solution used for crystallization (supplemental Fig. S4). Both samples migrated to a molecular mass of ∼21 kDa with a very small quantity of truncated protein detected around 19 kDa. These data altogether suggest that, in the vicinity of the 4-fold molecular symmetry axes, the C-terminal part of L167fs is turned outside toward the solvent, as indicated before (25), leading to the disruption of the tightly packed hydrophobic pore, and to the formation of a large permeable opening. This disruption occurs also in heteropolymers in which a single mutant subunit is present in the 4-fold axes as in the models shown in Fig. 3B. Fig. 3B, panels 1 and 2, display the 4-fold axis of the L167fs and Hwt structures, respectively. Panel 3 exhibits a model corresponding to three H-chains plus one L167fs chain. The absence of a single helix E in the heteropolymer H/L167fs renders the pore hydrophilic, exposing to solvent some residues of both L167fs and H-chain E helices.
FIGURE 3.
Crystal structure of the L167fs mutant. A, stereo view of two opposite chains of the 4-fold axis of the L167fs mutant structure (shown as balls and sticks) together with the (2mFo − DFc) electron density map (blue) contoured at 1.0 σ and the (mFo − DFc) electron density map (pink) contoured at 3.0 σ. The Cα trace of the Lwt structure is superimposed and shown in black. B, 4-fold axes representation: 1, L167fs crystal structure (yellow); 2, human H-chain crystal structure; and 3, a heteropolymer model composed of one L167fs chain (yellow) and three H-chains. The surfaces represent the solvent accessible areas. In panel 2, the residues represented as sticks are those which line the 4-fold axis. In panel 3, the residues represented as sticks are those that polar atoms have their solvent accessible surface increased due to the absence of the C-terminal part of the L167fs subunit within the pore, i.e. Ala44, Leu45, Gly47, Thr96, His148, and Leu152 for the L167fs subunit, and Asn154, Lys157, Met158, His173, Tyr168, Lys172, and His173 for the H-chain subunits that are in close contact with the L167fs subunit.
Hybrid Ferritins
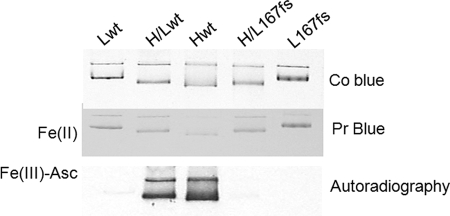
The data showed that L167fs homopolymers do not exhibit major differences from Lwt, except for lower stability due to the disordered structure of the C terminus. However, this might not be physiologically relevant, because L-ferritin homopolymers are not found inside the cells and the role of the L-chain is to assist the functionality of the H-chain in heteropolymers (1, 2). Thus, we produced in vitro hybrid ferritin molecules by renaturing L167fs or Lwt in the presence of H-chain WT. We used a ratio of 1 L-chain with 3–4 H-chains. The efficiency of L167fs assembly was comparable with that of Lwt (not shown), and the electrophoretic mobility of the hybrids was related to the subunit composition (H > H/Lwt ∼ H/L167fs > Lwt > L167fs) (Fig. 4). The renatured ferritins were incubated with 1,000 Fe(II) atom increments at pH 7.0, separated on nondenaturing PAGE, and stained with Prussian blue. Iron incorporation was comparable in all the samples (Fig. 4). However, these conditions are far from the physiological ones, because inside the cell iron is expected to be less available and loosely bound to small molecules such as citrate or phosphate. Therefore we used other conditions that were previously found to distinguish the functionality of H-ferritin mutants with altered permeability at the 3-fold channels (41). The ferritin solutions (0.2 μm) were incubated at pH 7.0 with 0.1 mm 55Fe(III)-citrate complex in the presence of 1 mm ascorbate as reducing agent, and then exposed to autoradiography. Under these conditions the Lwt and L167fs homopolymers did not incorporate radioactive iron, whereas H-homopolymer was active (Fig. 4). Interestingly, the H/Lwt heteropolymers incorporated radioactive iron, whereas the H/L167fs heteropolymers did not. It remained to verify that these conditions that distinguish the functionality of H/L167fs heteropolymers are physiologically relevant, and if the same applies to other pathogenic mutants associated with neurodegeneration.
FIGURE 4.
In vitro assembled ferritin heteropolymers. The purified ferritins were denatured and renatured alone to produce Lwt, Hwt, or L167fs homopolymers, or renatured together to produce H/Lwt and H/L167fs heteropolymers with a H:L ratio of 3:1. The protein were incubated with 1000 Fe(II) atoms at pH 7.0, 5 μg was run on nondenaturing PAGE and stained for protein (Co Blue) or Prussian blue (Pr Blue). Alternatively the proteins were incubated with radioactive 55Fe(III) in the presence of 1 mm ascorbate at pH 7.0, 1.5 μg of protein was loaded on nondenaturing PAGE and exposed to autoradiography for iron incorporation detection (Autoradiography). Representative of three independent experiments with similar results.
Expression and Activity of Hybrid Ferritins in E. coli
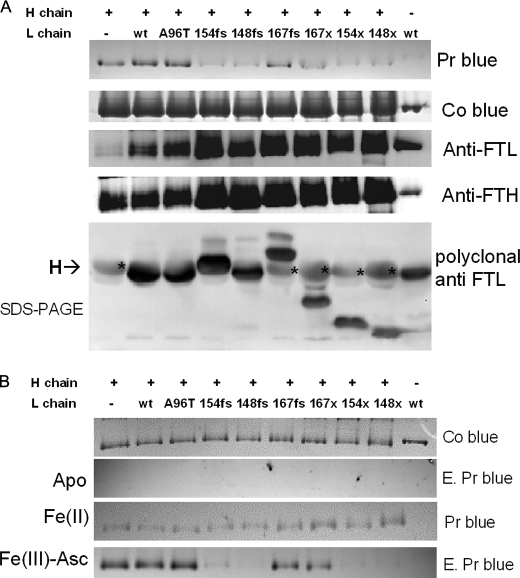
The study of the functionality of hybrid ferritin molecules in mammalian cells is complex, for example, it is still unclear if the L-rich ferritins expressed in the liver are more functional than the H-rich molecules expressed in the heart or brain. Moreover, analysis of mutant ferritins expressed in transfected cells is confused by coassembly with endogenous ferritin subunits. E. coli can be useful for cellular models, because their endogenous ferritins incorporate iron with a mechanism similar to the one used by mammalian cells and their subunits do not coassemble with the mammalian ones. We made a bicistronic vector (pET-H/Lwt) in which the human H- and L-ferritin cDNAs were consecutively subcloned into a pET vector, downstream of the same, unique promoter (42). The E. coli were transformed with pET-H and pET-H/Lwt, and ferritin expression was induced. The ferritins were purified and analyzed. The one expressed by the pET-H/Lwt vector contained 90–95% H and 5–10% Lwt, which corresponds to about 1–2 L-chains per 24-mer shell (supplemental Fig. S5). This ratio is probably close to the one found in the brains, and particularly in the neurons, which contain H-rich ferritins (43). Next we introduced the pathogenic mutations in the L-ferritin, to express L148fs, L154fs, L167fs, and LA96T (Fig. 1). The transformed cells were grown in an iron-poor medium, induced, and the cell homogenates were heated at 65 °C to enrich ferritins. Nondenaturing PAGE showed that the different strains expressed a similar level of the hybrid ferritin shells, and that they had similar electrophoretic mobility. Enhanced Prussian blue staining showed that H-homopolymer and heteropolymers of H/Lwt and H/LA96T incorporated similar amounts of iron, whereas L-homopolymer bound only trace amounts of iron. More interestingly, the heteropolymers containing pathogenic mutants L148fs and L154fs incorporated little iron and behaved similarly to the Lwt homopolymer. L167fs with the shortest modification of E-helix incorporated slightly more iron but still less than H/Lwt. To study the role of the sequences mutated by the insertions, we produced mutants with a stop codon at the site of the frameshift. The heteropolymers were expressed at the same levels as the other ones and their iron incorporation was as impaired as that of the frameshift mutants, and similar to that of L-homopolymer (Fig. 5A). Staining protein with Coomassie Blue or anti-H and anti-L ferritin monoclonal antibodies confirmed that the loads were properly calibrated and the samples contained similar proportions of H- and L-chains. As a further verification, the same amounts of preparations were separated on SDS-PAGE and analyzed by Western blotting with a polyclonal antibody with higher affinity for the L-chain than the H-chain. The dark band of the L-chain had similar intensity in the different heteropolymers, and the deletion mutants had the expected molecular weights (Fig. 5B, lower panel). The faster mobility of mutant L148fs may be due to a proline-rich region, which is absent in the other proteins (Fig. 1). Also the paler band of the H-ferritin had a similar intensity. Thus, the hybrids contained a similar H:L ratio. In other experiments the different ferritins were separated on nondenaturing PAGE and then initially stained with Coomassie Blue to compare the loads (Fig. 5B, upper panel). They were treated for iron removal, and enhanced Prussian blue staining confirmed the absence of iron. The gels containing the proteins were then incubated aerobically either with 200 μm Fe(II) or 1 mm Fe(III) in the presence of 1 mm ascorbate for 15 min. After the reactions the gels were washed and stained with Prussian blue. The results show that iron incorporation was analogous for the various ferritins when iron was supplied as Fe(II), on the contrary, when Fe(III)-ascorbate was used H-homopolymer, H/Lwt and H/LA96T incorporated a large amount of iron, L-homopolymer was inactive, the heteropolymers with mutations at 154 and 148 (fs or stop) did not incorporate iron, and those at 167 had an intermediate activity (Fig. 5B), with an activity pattern that overlapped that shown in Fig. 5A.
FIGURE 5.
Analysis of the heteropolymers with the pathogenic and truncated l-subunits expressed by E. coli. A, the E. coli transformed with the bicistronic vector to express the heteropolymers were grown in iron poor medium and the homogenates heated at 65 °C for ferritin enrichment. The protein were run on nondenaturing PAGE and stained with enhanced Prussian blue (Pr Blue), Coomassie Blue (Co Blue), or Western blotted and stained with monoclonal antibodies for L-ferritin (Anti-FTL) or H-ferritin (Anti-FTH). The bottom panel shows an SDS-PAGE blotted with a polyclonal antibody that has higher affinity for the L-chain (darker bands), but that recognizes also H-chains (paler bands indicated with an asterisk). The + above the lanes indicates the molecules containing H-chain. B, the ferritins were separated on nondenaturing PAGE and stained with Coomassie Blue (Co Blue). They were then treated for iron removal and colored with enhanced Prussian blue to verify the absence of iron (E. Pr Blue). The gels containing the apoproteins were incubated for 15 min with 200 μm ferrous ammonium sulfate at pH 6.5. Fe(II) was then washed and stained with Prussian blue (Pr Blue) or incubated for 15 min with 1 mm ascorbate and 1 mm ferric ammonium citrate in 20 mm Tris HCl, pH 7.0 (Fe(III)-Asc), extensively washed, and stained with enhanced Prussian blue (E. Pr Blue). This is representative of three independent experiments.
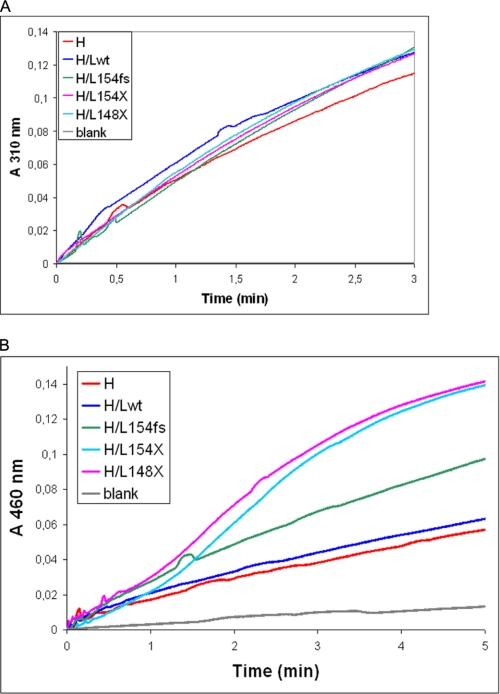
Ferritin Iron Oxidation
To study the reason of the lower iron incorporation activity of the heteropolymers, the ferritins were purified and analyzed for rate of iron oxidation at pH 6.5, i.e. under conditions in which ferritins without ferroxidase centers are inactive. The progression plots of the formation of the amber colored Fe(III) at 310 nm for the hybrids with mutated or truncated C terminus were analogous to those of H/Lwt heteropolymers and to those of H-homopolymers (Fig. 6A), indicating that the ferroxidase activity was not modified in the heteropolymers containing the mutants. Then we studied iron oxidation in the presence of apotransferrin, which binds the oxidized iron not incorporated into the ferritin shell and forms a pink complex. Under these conditions the activity of H-homopolymer and H/Lwt were comparable, whereas that of the H/L154fs was faster, and that of the heteropolymers with the truncated L-chains (H/L148X and H/L154X) was even faster with a sigmoid progression plot (Fig. 6B). Thus the heteropolymers with mutated or truncated C termini oxidize iron at a similar rate as H- or H/Lwt, but a higher proportion of the oxidized iron is made available to transferrin, indicating that it is not efficiently retained inside the cavity. In other experiments the ferritins (1 μm) were incubated for 2 h with 0.1 mm ferrous ammonium sulfate to load 100 iron atoms per molecule, and then the rate of iron release was analyzed after the addition of 5 mm NADPH and 2.5 mm bipyridyl. The ones containing the mutated L-chains showed rates of iron release about 2-fold faster than H/Lwt and H-homopolymers (not shown). This further supports the hypothesis that the mutants make ferritin shells with higher permeability.
FIGURE 6.
Ferroxidase activity of the ferritin heteropolymers. A, the purified apoferritins (0.1 μm) were incubated with 0.1 mm ferrous ammonium sulfate at pH 6.5 and the iron oxidation reaction was followed at 310 nm. B, the apoferritins (0.2 μm) were incubated with 4 mg/ml of apotransferrin, in 0.2 m sodium acetate, pH 6.0, supplemented with 1 mm ferrous ammonium sulfate, and the formation of pink holotransferrin followed at 460 nm. This is representative of three independent experiments.
Ferritin Stability
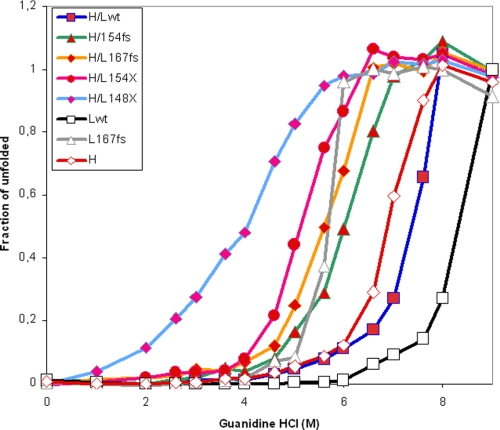
To study protein physical stability, the purified ferritins were treated with 1% SDS at room temperature, and analyzed on discontinuous SDS-PAGE. The Lwt homopolymer released a minor proportion of disassembled subunits (about 5%), whereas the L167fs more than 20%. Heating at 100 °C in 1% SDS fully dissociated both ferritins (supplemental Fig. S6A). Under the same conditions the heteropolymers behaved similarly to the Lwt homopolymers, except the ones containing the mutant with the longest deletion (L148X), which showed a 20% dissociation (supplemental Fig. S6B). Next we performed unfolding by increasing guanidine hydrochloride concentrations, and monitored the change of intrinsic fluorescence of the single tryptophan (44). The Lwt homopolymer started unfolding at concentrations of 6 m with a midpoint above 8 m, the H-homopolymer had a midpoint at 6.75 m, whereas the L167fs homopolymer showed a sharp transition between 5 and 6 m, similar to that of H-ferritin (Fig. 7). The H/Lwt heteropolymer plot was intermediate between those of H- and Lwt homopolymers, and those with L154fs and L167fs were shifted to the left. The polymers containing the truncated subunits were the least stable, particularly that with L148X.
FIGURE 7.
Unfolding plots in guanidine chloride of the purified heteropolymers. The ferritins (50 μg/ml) were incubated for 18 h at 4 °C in 0.1 m phosphate buffer, pH 7.4, 1 mm dithiothreitol, with the different guanidine HCl concentrations. Unfolding was determined by fluorescence spectroscopy (excitation at 295 nm), using as standards, ferritin either in 0.1 m phosphate, pH 7.4 (native state), or in 0.1 m phosphate, pH 3.0, and 6 m guanidine HCl (denatured state). Data were plotted as fraction of unfolded shells. H, Lwt, and L167fs are the corresponding homopolymers, H/Lwt, H/L154fs, H/L167Fs, H/L148X, and H/L154X are the heteropolymers. This is representative of three independent experiments.
DISCUSSION
Hereditary ferritinopathies have similar phenotypic expressions, with brain iron accumulation, movement disorders, and the formation of ferritin bodies in the brain and other tissues (15, 16, 45). However, the ferritin mutations associated with the disorders are rather different: they modify different lengths of the C terminus, and one- or two-nucleotide frameshifts produce different sequences with extensions of 4 or 16 residues. The characterization of two of these mutants showed remarkably different biochemical properties. Although 154fs was hardly soluble, difficult to purify and analyze (23), L167fs was highly soluble, could be easily purified and even crystallized (this work and Refs. 24 and 25). The crystals we obtained had a different space group (I432) from that reported in Ref. 25 (P1) and diffracted at higher resolution, but the solved structures are very similar. The main common feature of the two structures is the observation of a strong disorganization at the 4-fold molecular axis. In the P1 structure, the authors could model 13 chains up to residue Phe165, of the 144 chains that build the asymmetrical unit. The five last residues, Gly161–Phe165, point toward the cavity, through the 4-fold axis pore. It seems therefore, that such a conformation must be a very minority. Moreover, such a chain conformation needs an important rearrangement, to construct a heteropolymer model as the one shown on panel 3 of Fig. 3B, because fragment Gly161–Phe165 in its actual conformation would overlap with the E helix of a neighboring H-chain (not shown). In our single chain model, the protein chain stops at residue Gly157, and the absence of the residual electron density within the 4-fold axis pore leads us to consider that the C terminus end is turned outside toward the solvent, certainly a common feature of the two mutants 154fs and 167fs.
This characteristic is expected to have two major effects: a reduced physical stability due to the loss of stabilizing interactions around the 4-fold axis, and the formation of a large opening in place of the tight hydrophobic channel, which might affect ferritin functionality. A reduced physical stability of L167fs was demonstrated in the present and previous studies (24). The same probably occurs for the pathogenic mutations with frameshifts at residues 148 or 154, where the modified sequences are even longer and interest up to the last two turns of the D-helix, which contain important stabilizing interactions. This could not be explored because of difficulties obtaining a sufficient amount of these proteins, due to their folding problems and low solubility. In fact, the H-chain truncated from the last 28 residues (equivalent to 24 residues of the L-chain) could not assemble when expressed in E. coli (21). However, patients with L148fs or L154fs showed onset of tremors, gait disturbances, and cognitive decline at an age earlier than with L167fs (13), suggesting that upstream insertions have more severe effects than downstream insertions. This may be paradoxical if we consider only the stability/folding properties, because the less stable subunits should have problems assembling in ferritin shells, and should form homopolymeric aggregates or be degraded. However, the ferritin bodies found in patients with the L154fs and L167fs mutations were iron-rich and contained H- and L-mutant chains, implying that they are made of assembled shells. Aggregates made only of mutant chains have not been observed in the patients. This apparent contradiction can be explained by the strong tendency of ferritin to form heteropolymers even with subunits with major folding problems. For example, L-ferritin mutants with a constructed ferroxidase center were totally insoluble when expressed in E. coli, and could not be renatured in vitro. However, when renatured together with WT H- or L-chains they formed stable heteropolymers that could be analyzed (46). Our data show that all the pathogenic mutants, including the ones with upstream insertions, expressed in E. coli together with H-chain, formed heteropolymers with similar L proportions, and produced soluble and stable ferritins that could be easily purified and analyzed. This occurred also with the truncated mutants, even the ones deleted of the last 28 residues, which were predicted not to assemble when alone. This indicates that in vivo the pathogenic mutants can also assemble with the endogenous ferritin subunits, and that this process is independent from the length of the mutation for these pathogenic mutants. Thus, the pathogenicity of the mutants is more likely due to abnormal properties of heteropolymers containing even a low proportion of the mutant, rather than to the mutant homopolymers. Moreover, the finding that the heteropolymers are highly soluble and present a physical stability only marginally lower than that of the H/Lwt heteropolymers suggests that mutant stability and aggregations are not the primary causes of the disease. It should be noted that the mutants are particularly toxic in the brains and neurons, which are known to have H-rich ferritins with an H/L ratio probably similar to that of the heteropolymers we expressed in E. coli.
The in vitro conditions we initially used to study L-homopolymers were not particularly informative: they showed a low solubility of mutant L148fs and a high solubility of L167fs, which did not show signs of aggregation in the absence of iron. In our hands L167fs exhibited only a minor, if any, decrease in solubility when exposed to high iron increments (Fig. 2B), which contrasts with the data of Refs. 24 and 25 showing the massive precipitation of the mutant when incubated with 3000–4000 irons/molecule. Baraibar et al. (25) used a protein with an altered N terminus, which differs from the native one of our proteins, and this might affect protein solubility. More important, L-homopolymers lack the ferroxidase activity, which is necessary for in vivo iron incorporation (47), thus the in vitro conditions used to study their functionality are far from the physiological ones. We did not observe the difference in rate of iron oxidation between Lwt and L167fs described in Ref. 25, and under those conditions the kinetics at 310 nm of the two proteins were much slower and overlapping (not shown). The L-chains assist the H-ones in the mechanism of ferritin iron incorporation, thus their functionality must be analyzed when both are together in the same molecule. In fact the heteropolymer study was more informative. It was shown earlier that the reconstituted H/L154fs heteropolymers incorporated in vitro less 55Fe than H/Lwt, using conditions in which Fe(III) was reduced to Fe(II) by ascorbate (23). The same occurred with the reconstituted H/L167fs heteropolymers, which were incompetent in taking up Fe(III) in the presence of ascorbate, while H/Lwt were active (Fig. 4). A more direct approach to study all the mutants that have been associated with hereditary ferritinopathies was to make the heteropolymers in E. coli and analyze their iron incorporation during expression in the bacteria. This showed that H, H/Lwt, and H/LA96T were much more active than the other heteropolymers and Lwt alone. The level of iron incorporation was inversely related to the length of the E-helix modification, with L154fs being less active than L167fs. The same order of iron incorporation was followed by the apoferritins exposed to Fe(III) in the presence of ascorbate. Moreover, the truncated mutants behaved similarly to the frameshift ones, indicating that the presence of an altered sequence at the C terminus leads to a loss of interactions in the 4-fold axis, and thus to the opening of a large pore that modifies the ferritin functionality.
These data demonstrate that mutations on the C-terminal sequence have a dominant-negative effect on the ferritin capacity to incorporate iron, and that even 1 or 2 mutant subunits per 24-mer shells are sufficient for this. One subunit is enough to alter the conformation of one of the six 4-fold channels in the molecule by flipping outside its C terminus and therefore abolishing local interactions, as shown in the model of Fig. 3B. The same interactions are also lost by the truncation of the C terminus, and the stability of the proteins shells is altered by both types of mutations. This changes the permeability of the shell, with possible effects on the microenvironment of the cavity that contributes to iron deposition. This is demonstrated by the finding that although the rate of iron oxidation of the heteropolymers is similar to that of the H-homopolymer, the rate of iron supplied to transferrin is faster, because some of the oxidized iron cannot be retained in the ferritin cavity. We concluded that the full integrity of the shell contributes to efficient iron incorporation. These data also contribute to the understanding why the pathogenic mutations are insertions confined to the C-terminal part of the molecule: if they were further upstream, they might inhibit assembly in the heteropolymers, and missense mutations would not alter the 4-fold channels. The mutant LA96T behaved like Lwt, thus it is unlikely that it has pathogenic effects. Moreover, the results indicate that altered ferritin functionality is due to local unfolding of the C terminus rather than to the properties of the altered sequence, and this is consistent with the observation that insertion of one or two nucleotides has similar effects.
Our data confirm previous hypothesis that hereditary ferritinopathies are caused by reduced activity of iron incorporation in the ferritin (22). This opens a vicious circle in the cell: the iron not retained by ferritin induces the synthesis of more ferritin chains, both WT and mutant, which form inefficient heteropolymers. Indeed an increase of endogenous ferritin and iron was found in the brain of the mouse model for ferritinopathy (26). The iron-induced ferritin precipitation occurs at protein and iron concentrations that are far from the physiological one (25) and has been described for homopolymers but not heteropolymers, this relevance for the in vivo formation of ferritin aggregates is unclear. We observed that cells expressing two pathogenic mutants have reduced proteasome activity (22), and this, together with the high level of ferritin expression, may explain the formation of the ferritin bodies found in the brain and other tissues of the affected patients. It is noteworthy that aggregates made of Lwt ferritin were observed in various cells types (48).
In conclusion, our data show that the major common property of the pathogenic L-mutants involved in hereditary ferritinopathies is the reduction of iron incorporation activity when assembled in heteropolymers. This reduction is linked to frameshifts or deletions in the C-terminal sequence. The finding that all the mutants act in a dominant-negative manner explains the dominant transmission of the disorders. The role of the A96T mutation in neuroferritinopathy should be revised because it does not have these properties and it does not affect ferritin iron incorporation.
Supplementary Material
Acknowledgments
We thank Dr. Massimo Degano for dynamic light scattering analyses. We also thank the European Synchrotron Radiation Facility for provision of synchrotron radiation facilities and the staff of the ID23-1 beam line for assistance during data collections.
This work was supported in part by Telethon-Italia Grant GGP05141 (to S. L. and P. A.), Fondazione Grant CARIPLO-2007 (to S. L. and A. C.), and European Community Grant Euroiron1 (to P. A.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
The atomic coordinates and structure factors (code 2KXU) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- FTL
- ferritin light chain
- WT and wt
- wild-type
- Lwt
- wild-type L-ferritin
- Hwt
- wild-type H-ferritin
- LA96T
- L-ferritin mutant Ala96 → Thr
- L148fs
- L154fs, and L167fs, L-ferritin mutant with frameshift mutations in residues 148, 154, and 167, respectively
- L148X
- L154X, and L167X, L-ferritin mutants with stop codon mutation in residues 148, 154 and 167, respectively
- MALDI-TOF
- matrix-assisted laser desorption ionization time-of-flight
- ESI-MS
- electrospray ionization-mass spectrometry.
REFERENCES
- 1.Arosio P., Ingrassia R., Cavadini P. (2009) Biochim. Biophys. Acta 1790, 589–599 [DOI] [PubMed] [Google Scholar]
- 2.Harrison P. M., Arosio P. (1996) Biochim. Biophys. Acta 1275, 161–203 [DOI] [PubMed] [Google Scholar]
- 3.Levi S., Luzzago A., Cesareni G., Cozzi A., Franceschinelli F., Albertini A., Arosio P. (1988) J. Biol. Chem. 263, 18086–18092 [PubMed] [Google Scholar]
- 4.Ferreira C., Bucchini D., Martin M. E., Levi S., Arosio P., Grandchamp B., Beaumont C. (2000) J. Biol. Chem. 275, 3021–3024 [DOI] [PubMed] [Google Scholar]
- 5.Darshan D., Vanoaica L., Richman L., Beermann F., Kühn L. C. (2009) Hepatology 50, 852–860 [DOI] [PubMed] [Google Scholar]
- 6.Beaumont C., Leneuve P., Devaux I., Scoazec J. Y., Berthier M., Loiseau M. N., Grandchamp B., Bonneau D. (1995) Nat. Genet. 11, 444–446 [DOI] [PubMed] [Google Scholar]
- 7.Levi S., Girelli D., Perrone F., Pasti M., Beaumont C., Corrocher R., Albertini A., Arosio P. (1998) Blood 91, 4180–4187 [PubMed] [Google Scholar]
- 8.Brooks D. G., Manova-Todorova K., Farmer J., Lobmayr L., Wilson R. B., Eagle R. C., Jr., St. Pierre T. G., Stambolian D. (2002) Invest. Ophthalmol. Vis. Sci. 43, 1121–1126 [PubMed] [Google Scholar]
- 9.Curtis A. R., Fey C., Morris C. M., Bindoff L. A., Ince P. G., Chinnery P. F., Coulthard A., Jackson M. J., Jackson A. P., McHale D. P., Hay D., Barker W. A., Markham A. F., Bates D., Curtis A., Burn J. (2001) Nat. Genet. 28, 350–354 [DOI] [PubMed] [Google Scholar]
- 10.Mancuso M., Davidzon G., Kurlan R. M., Tawil R., Bonilla E., Di Mauro S., Powers J. M. (2005) J. Neuropathol. Exp. Neurol. 64, 280–294 [DOI] [PubMed] [Google Scholar]
- 11.Gregory A., Polster B. J., Hayflick S. J. (2009) J. Med. Genet. 46, 73–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vidal R., Ghetti B., Takao M., Brefel-Courbon C., Uro-Coste E., Glazier B. S., Siani V., Benson M. D., Calvas P., Miravalle L., Rascol O., Delisle M. B. (2004) J. Neuropathol. Exp. Neurol. 63, 363–380 [DOI] [PubMed] [Google Scholar]
- 13.Kubota A., Hida A., Ichikawa Y., Momose Y., Goto J., Igeta Y., Hashida H., Yoshida K., Ikeda S., Kanazawa I., Tsuji S. (2009) Mov. Disord. 24, 441–445 [DOI] [PubMed] [Google Scholar]
- 14.Maciel P., Cruz V. T., Constante M., Iniesta I., Costa M. C., Gallati S., Sousa N., Sequeiros J., Coutinho P., Santos M. M. (2005) Neurology 65, 603–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohta E., Nagasaka T., Shindo K., Toma S., Nagasaka K., Ohta K., Shiozawa Z. (2008) Neurology 70, 1493–1494 [DOI] [PubMed] [Google Scholar]
- 16.Vidal R., Delisle M. B., Rascol O., Ghetti B. (2003) J. Neurol. Sci. 207, 110–111 [DOI] [PubMed] [Google Scholar]
- 17.Gerardi G., Biasiotto G., Santambrogio P., Zanella I., Ingrassia R., Corrado M., Cavadini P., Derosas M., Levi S., Arosio P. (2005) Blood Cells Mol. Dis. 35, 177–181 [DOI] [PubMed] [Google Scholar]
- 18.Ingrassia R., Gerardi G., Biasiotto G., Arosio P. (2006) J. Biochem. 139, 881–885 [DOI] [PubMed] [Google Scholar]
- 19.Jappelli R., Luzzago A., Tataseo P., Pernice I., Cesareni G. (1992) J. Mol. Biol. 227, 532–543 [DOI] [PubMed] [Google Scholar]
- 20.Luzzago A., Cesareni G. (1989) EMBO J. 8, 569–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levi S., Luzzago A., Franceschinelli F., Santambrogio P., Cesareni G., Arosio P. (1989) Biochem. J. 264, 381–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cozzi A., Rovelli E., Frizzale G., Campanella A., Amendola M., Arosio P., Levi S. (2010) Neurobiol. Dis. 37, 77–85 [DOI] [PubMed] [Google Scholar]
- 23.Cozzi A., Santambrogio P., Corsi B., Campanella A., Arosio P., Levi S. (2006) Neurobiol. Dis. 23, 644–652 [DOI] [PubMed] [Google Scholar]
- 24.Baraibar M. A., Barbeito A. G., Muhoberac B. B., Vidal R. (2008) J. Biol. Chem. 283, 31679–31689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baraibar M. A., Muhoberac B. B., Garringer H. J., Hurley T. D., Vidal R. (2010) J. Biol. Chem. 285, 1950–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barbeito A. G., Garringer H. J., Baraibar M. A., Gao X., Arredondo M., Núñez M. T., Smith M. A., Ghetti B., Vidal R. (2009) J. Neurochem. 109, 1067–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vidal R., Miravalle L., Gao X., Barbeito A. G., Baraibar M. A., Hekmatyar S. K., Widel M., Bansal N., Delisle M. B., Ghetti B. (2008) J. Neurosci. 28, 60–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cozzi A., Corsi B., Levi S., Santambrogio P., Albertini A., Arosio P. (2000) J. Biol. Chem. 275, 25122–25129 [DOI] [PubMed] [Google Scholar]
- 29.Santambrogio P., Cozzi A., Levi S., Rovida E., Magni F., Albertini A., Arosio P. (2000) Protein Expr. Purif. 19, 212–218 [DOI] [PubMed] [Google Scholar]
- 30.Santambrogio P., Levi S., Cozzi A., Rovida E., Albertini A., Arosio P. (1993) J. Biol. Chem. 268, 12744–12748 [PubMed] [Google Scholar]
- 31.Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 32.Leslie A. G. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 48–57 [DOI] [PubMed] [Google Scholar]
- 33.Evans P. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 34.(1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 35.Wang Z., Li C., Ellenburg M., Soistman E., Ruble J., Wright B., Ho J. X., Carter D. C. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 800–806 [DOI] [PubMed] [Google Scholar]
- 36.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 37.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 38.Engh R. A., Huber R. (1991) Acta Crystallogr. Sect. A 47, 392–400 [Google Scholar]
- 39.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 41.Santambrogio P., Pinto P., Levi S., Cozzi A., Rovida E., Albertini A., Artymiuk P., Harrison P. M., Arosio P. (1997) Biochem. J. 322, 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rucker P., Torti F. M., Torti S. V. (1997) Protein Eng. 10, 967–973 [DOI] [PubMed] [Google Scholar]
- 43.Connor J. R., Boeshore K. L., Benkovic S. A., Menzies S. L. (1994) J. Neurosci. Res. 37, 461–465 [DOI] [PubMed] [Google Scholar]
- 44.Santambrogio P., Levi S., Arosio P., Palagi L., Vecchio G., Lawson D. M., Yewdall S. J., Artymiuk P. J., Harrison P. M., Jappelli R. (1992) J. Biol. Chem. 267, 14077–14083 [PubMed] [Google Scholar]
- 45.Chinnery P. F., Crompton D. E., Birchall D., Jackson M. J., Coulthard A., Lombès A., Quinn N., Wills A., Fletcher N., Mottershead J. P., Cooper P., Kellett M., Bates D., Burn J. (2007) Brain 130, 110–119 [DOI] [PubMed] [Google Scholar]
- 46.Levi S., Corsi B., Rovida E., Cozzi A., Santambrogio P., Albertini A., Arosio P. (1994) J. Biol. Chem. 269, 30334–30339 [PubMed] [Google Scholar]
- 47.Cozzi A., Corsi B., Levi S., Santambrogio P., Biasiotto G., Arosio P. (2004) Blood 103, 2377–2383 [DOI] [PubMed] [Google Scholar]
- 48.Infante A. A., Infante D., Chan M. C., How P. C., Kutschera W., Linhartová I., Müllner E. W., Wiche G., Propst F. (2007) Exp. Cell Res. 313, 1602–1614 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.