Abstract
AF9/MLLT3 contributes to the regulation of the gene encoding the epithelial sodium channel α, ENaCα, in renal tubular cells. Specifically, increases in AF9 protein lead to a reduction in ENaCα expression and changes in AF9 activity appear to be an important component of aldosterone signaling in the kidney. Whereas AF9 is found in the nucleus where it interacts with the histone H3 lysine 79 methyltransferase, Dot1, AF9 is also present in the cytoplasm. Data presented in this report indicate that the heat shock protein Hsp90 directly and specifically interacts with AF9 as part of an Hsp90-Hsp70-p60/Hop chaperone complex. Experimental manipulation of Hsp90 function by the inhibitor novobiocin, but not 17-AAG, results in redistribution of AF9 from a primarily nuclear to cytoplasmic location. Knockdown of Hsp90 with siRNA mimics the effect elicited by novobiocin. As expected, a shift in AF9 from the nucleus to the cytoplasm in response to Hsp90 interference leads to increased ENaCα expression. This is accompanied by a decrease in AF9 occupancy at the ENaCα promoter. Our data suggest that the interaction of Hsp90, Hsp70, and p60/Hop with AF9 is necessary for the proper subnuclear localization and activity of AF9. AF9 is among a growing number of nuclear proteins recognized to rely on the Hsp90 complex for nuclear targeting.
Keywords: Chromatin Immunoprecipitation (ChiP), Gene Expression, Heat Shock Protein, Oncogene, Protein Translocation, Sodium Channels, AF9/MLLT3
Introduction
AF9/MLLT3 is a proline-, serine-rich protein of 568 amino acids and contains a KKRKK (amino acids 296–300) nuclear localization sequence, features that are typical of many transcription factors. Nevertheless, the function of AF9 is incompletely characterized. AF9 was first identified as one of the 60+ fusion partners of the mixed lineage leukemia (MLL)2 gene that result from translocations at the 11q23 locus. AF9 is highly homologous to ENL, another MLL fusion partner, and expression of MLL-AF9 or MLL-ENL fusion proteins in hematopoietic precursor cells leads to leukemic transformation (1–3). AF9 shares sequence similarity to TFG3/TAF30/ANC1 in Saccharomyces cerevisiae, a 29-kDa component of SWI/SNF, TFIID, and TFIIF complexes in yeast, which suggests that it may function to promote transcription (4, 5). More recently, AF9 was found in an RNA pol II transcriptional elongation complex comprised of AF4 family member proteins, the histone H3 lysine 79 (H3K79) methyltransferase Dot1, and pTEFb (6–10). In addition to the aforementioned proteins, mass spectroscopy analysis of protein complexes containing the AF9 homolog ENL reveal an even wider array of gene regulatory proteins including BCoR, CBX8/Pc3, and RING1. Rather than forming a large complex comprised of all of the proteins known to associate with ENL, gel filtration experiments indicate that individual ENL-containing complexes range from 230–600 kDa and thus must contain only discrete subsets of these proteins (12). Whether accessory factors facilitate the assembly of specific complexes is not known.
Mice with homozygous deletion of the Af9 gene survive gestation but have homeotic skeletal anomalies and die shortly after birth. The skeletal defects are similar to mice with null mutations of Cdx1, a regulator of Hox gene expression. Thus, it has been suggested that Af9 is also a “master regulator” of Hox gene expression in mice (13). AF9 is highly expressed in human hematopoietic stem cells and experimental manipulation of AF9 in hematopoietic precursors alters erythro- and megakaryopoiesis. Reporter gene assays indicate that AF9 stimulates expression of the GATA1 transcription factor through the HS1 enhancer element (14). Although these findings provide strong support for a role of AF9 in normal hematopoiesis, the hematologic status of Af9-null mice has not been reported.
Whereas AF9 likely serves an important function in regulating the expression of multiple genes, perhaps the best characterized target is the epithelial sodium channel gene α (ENaCα). In this case, AF9 interacts with Dot1 to enhance the H3K79me3 mark at the ENaCα promoter and represses gene expression. Interestingly, this is in contrast to the canonical H3K79 methylation mark, which is typically present within the body of the gene and is associated with transcriptional elongation and active gene expression. Aldosterone treatment of renal collecting duct cells triggers phosphorylation of AF9 impairing the binding of Dot1 and leads to hypomethylation of the ENaCα promoter. Aldosterone thus stimulates ENaCα expression in the renal-collecting duct through an AF9-Dot1-controlled pathway (8).
Data indicate that AF9 alternatively binds to several different proteins and contributes to the activity of distinct gene regulatory complexes. Factors that facilitate the assembly of differing AF9-containing complexes have not been described but here we show that AF9 is a client protein of the molecular chaperone Hsp90. In addition to an important role in nuclear import of steroid receptors, Hsp90 is increasingly recognized as an effector of transcriptional activation through diverse mechanisms (11). In S. cerevisiae, Hsp90 is required for the rapid nucleosome remodeling that occurs with GAL gene induction (15) and pharmacologic inhibition of Hsp90 in Drosophila melanogaster interferes with binding of Trithorax protein at homeotic gene loci (16). In addition to directly affecting transcriptional machinery, Hsp90 is required for the proper function of signaling molecules that, in turn, regulate gene expression (17). We propose that Hsp90 contributes to the normal activity of AF9. Inhibition of Hsp90 leads to mislocalization of AF9 protein and impaired regulation of the AF9 target gene ENaCα.
EXPERIMENTAL PROCEDURES
Protein Purification
cDNA-encoding residues 475–568 of AF9 was cloned into pFLAG-MAC (Sigma) to generate pFLAG-AF9-ct. This plasmid construct was transformed into BL21 bacterial cells (Invitrogen) for protein expression. Following induction of protein expression with isopropyl β-d-1-thiogalactopyranoside (IPTG), cell pellets were collected and resuspended in bacterial lysis buffer (20 mm Tris, pH 7.4, 1 m NaCl, 1 mm EDTA, and 1 mm dithiothreitol). Cells were lysed using a French Press and debris was removed by centrifugation at 10,000 × g for 20 min. The supernatant was collected and diluted 1:5 with the lysis buffer containing no NaCl (final concentration of NaCl, 200 mm). The FLAG-tagged protein was affinity purified using M2 FLAG agarose resin (Sigma). The bound, purified protein was eluted using 50 mm FLAG peptide and then concentrated using 5-kDa molecular mass cutoff (MWCO) Centricon centrifugal concentrator (Millipore).
Isolation of Mammalian Cell Lysate
THP-1, HeLa (both ATCC), or IMCD3 cells (gift of Samir El-Dahr, Tulane University) were harvested by centrifugation. In some cases, cells were first transfected with Lipofectamine reagent (Invitrogen) to express FLAG-tagged full-length AF9 as previously described (18). Cell pellets were gently resuspended in 250 μl of mammalian lysis buffer (20 mm Tris, pH 7.4, 500 mm NaCl, 1 mm EDTA, 1% Triton X) per 106 cells. Cells were incubated on ice for 1 h and then centrifuged at 10,000 × g for 15 min to remove insoluble debris.
Isolation of AF9 Binding Partners
FLAG-tagged recombinant AF9 proteins were covalently linked to agarose beads using Amino-Link resin (Pierce) according to the manufacturer's instructions. Cell lysate from THP-1 leukemia cells was passed over the column and bound proteins were eluted using 1 m NaCl or 100 mm glycine buffer. Eluent was concentrated using a 3-kDa MWCO spin column (Millipore). Proteins were boiled in Laemmli buffer and then separated on 4–12% Bis-Tris SDS gels (Invitrogen). The gels were stained with Coomassie Brilliant Blue and visible bands were excised and sent for mass spectroscopy analysis at the proteomics core facility at Louisiana State University in New Orleans.
Competitive Enzyme-linked Immunoabsorbent Assay (ELISA)
96-well M2 Plates (Sigma) were blocked with Dulbecco's phosphate-buffered saline (DPBS) containing 5% bovine serum albumin (BSA), and 0.1% Triton-X. 100 μl of a 50 mm solution of recombinant FLAG-AF9 was added to individual wells and incubated overnight at 4 °C. Wells were washed three times in between each step with DPBS containing 0.1% Triton-X. 100 μl of 100 mm purified recombinant Hsp90 (Stressgen) was added to wells with or without 5 mm ATPγS (Sigma) and incubated overnight at 4 °C. Competition was carried out by adding a 0–8 μmol gradient of FLAG-AF9 or FLAG peptide to the wells and incubating overnight at 4 °C. The plate was then washed with blocking buffer before addition of the primary antibody. A 1:2500 dilution of a rabbit-derived anti-Hsp90 antibody (Stressgen) in 1% BSA with 0.1% Triton X was added to the wells and allowed to incubate overnight at 4 °C. A 1:5000 dilution of a secondary horseradish peroxidase antibody (Zymed Laboratories Inc.) was added. Wells were washed three times with phosphate-buffered saline (PBS) with 0.1% Triton X-100. SureBlue (KPL) was added to obtain a colorimetric signal, and the reaction was stopped with 1 n HCl. Absorbance was measured using a plate reader at 650 nm wavelength and normalized to the signal obtained without competitor. Each sample was run in triplicate, and each experiment was repeated three times.
Immunoprecipitation and Western Blotting
Antibodies against Hsp90 α/β, Hsp90 β, Hsp90 α, Hsp70, p60/HOP (all Stressgen), AF9 (Novus), FLAG (Sigma), β-actin (Sigma), and IgG control (Santa Cruz Biotechnology) were employed. A dilution of 1:1000–1:2500 was used for immunoprecipitation. The resulting antibody-protein complex was then recovered using either protein A- or protein G-agarose (Invitrogen). Agarose beads were washed twice in PBS before Laemmli buffer was added. The mixture was boiled and separated on 4–12% Bis-Tris gels. Proteins were transferred to nitrocellulose membranes using iBlot (Invitrogen) and were probed using appropriate primary antibodies as indicated in the text. Following incubation with horseradish peroxidase-conjugated secondary antibodies, signals were detected with enhanced chemiluminescence (GE).
Immunofluorescence Microscopy
IMCD3 cells were seeded into 4-well chamber slides at 5000 cells per well. Cells were treated overnight with either mouse Hsp90 α/β siRNA (Santa Cruz Biotechnology, sc-35610) or control siRNA (Santa Cruz Biotechnology) according to the manufacturer's protocol, or with 2 μm 17-(allylamino)-17-demethoxygeldanamycin (17AAG) (AG Scientific), or with 500 μm novobiocin (Sigma). Cells were fixed using 4% paraformaldehyde (Sigma) and then permeabilized with 50 mm ammonium chloride, 0.1% Triton X. Wells were blocked with DPBS, 5% BSA + 0.1% Triton X before primary antibody was added at a dilution of 1:2500 in PBS, 1% BSA, 0.1% Triton X. Cells were washed three times with DPBS, 0.1% Triton X in between each step. AlexaFluor 488- or 563-conjugated secondary antibodies (Invitrogen) were diluted 1:5000 in DPBS, 1% BSA, 0.1% Triton X. PROLONG Gold antifade with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen) was applied to the slides before being examined using a 3 color Zeiss LSM-510 confocal microscope.
Quantitative Polymerase Chain Reaction (qPCR)
IMCD3 cells were treated overnight with either Hsp90 siRNA, 17AAG or novobiocin. Cells were harvested and RNA was extracted using the RNeasy kit (Qiagen). Total RNA was converted into cDNA using SS III Reverse Transcriptase kit (Invitrogen). 200 ng of total cDNA was used in the qPCR reaction along with primers against GAPDH (F: GCA CCG TCA AGG CTG AGA AC, R: GCC TTC TCC ATG GTG GTG AA) and ENaCα (F: CTA ATG ATG CTG GAC CAC ACC, R: AAA GCG TCT GCT CCG TGA TGC). Reactions were performed with SYBR Green Supermix with ROX according to kit instructions (Bio-Rad).
Chromatin Immunoprecipitation (ChIP)
IMCD3 cells were treated overnight with either Hsp90 siRNA, control siRNA, 17AAG, or novobiocin. Cells were fixed by adding 670 μl of 32% formaldehyde to 10 ml of medium for a final concentration of 1% formaldehyde. Cells were fixed for 10 min and excess formaldehyde was quenched with 1 ml of 10× glycine (4.7 g of glycine in 50 ml of deionized water). Cells were collected, spun, and frozen overnight at −80 °C. Cells were lysed to collect the nuclear fraction which was subjected to sonication of 4 pulses of 15 s each. The resulting lysate was collected and immunoprecipitated using Hsp90 α/β and AF9 antibodies. Primers for PCR amplification of the ENaCα promoter (gift of Wenzheng Zhang, UT-Houston) are described in Ref. 6, and precise sequences were provided by the authors. Following PCR amplification, samples were separated by agarose gel electrophoresis and quantified using Image J software from NIH.
RESULTS
Isolation of AF9 Binding Partners
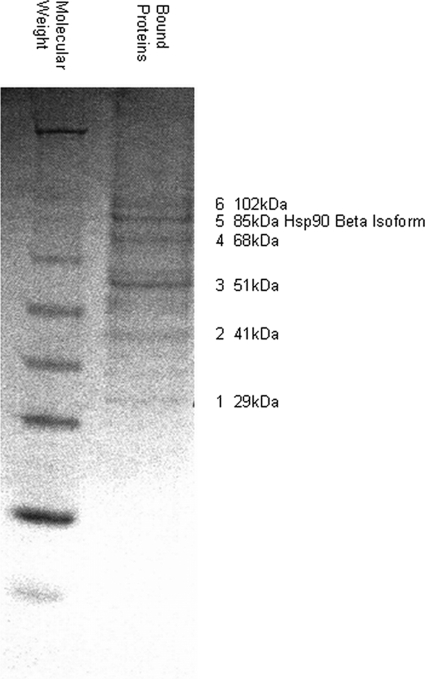
MLL-AF9 chimeric oncoproteins are comprised of the C terminus of AF9 fused to the N-terminal portion of MLL. In experimental systems, the C-terminal ∼90 amino acids of AF9 and the highly homologous protein ENL have been shown to be necessary and sufficient for MLL-mediated leukemic transformation (19, 20). No structural or enzymatic functions have been attributed to the C terminus of either AF9 or ENL although several proteins have been identified that bind this region (7, 21, 22). We used affinity purification to isolate additional factors that associate with immobilized recombinant AF9 C terminus protein (AF9-ct) comprised of amino acids 475–568 of AF9. Total cell lysates were prepared from the leukemia cell line THP-1 that expresses an MLL-AF9 oncogene. Lysates were pre-absorbed on an immobilized FLAG peptide resin and then passed over a FLAG-AF9-ct column. Bound proteins were eluted with either 1 m NaCl or 100 mm glycine buffer and separated by SDS-PAGE.
Fig. 1 shows the resulting Coomassie Blue-stained gel with six distinct bands ranging from ∼40 to 100 kDa. These bands were excised and analyzed by mass spectroscopy. Unexpectedly, four of the bands contained cytoskeleton-associated proteins whose relationship to AF9 is currently being studied. A fifth band, migrating at ∼85 kDa, was identified as the β isoform of Hsp90. Importantly, in an independent yeast two-hybrid screen of AF9-ct-interacting proteins, we previously isolated the closely related α isoform of Hsp90 (data not shown).
FIGURE 1.
Isolation of AF9-binding proteins. THP-1 whole cell extract was pre-adsorbed to a FLAG peptide affinity matrix and was then applied to a FLAG-AF9-ct column (amino acids 475–568 of AF9). Bound proteins were eluted with 1 m NaCl and 100 mm glycine. Eluted proteins were separated by SDS-PAGE and stained with Coomassie Brilliant Blue. Bands 1–6 were excised from the gel and identified by mass spectroscopy. The 85-kDa band is the β isoform of Hsp90. The gel shown is the original and only replicate.
Hsp90 Directly Binds AF9 in Vitro
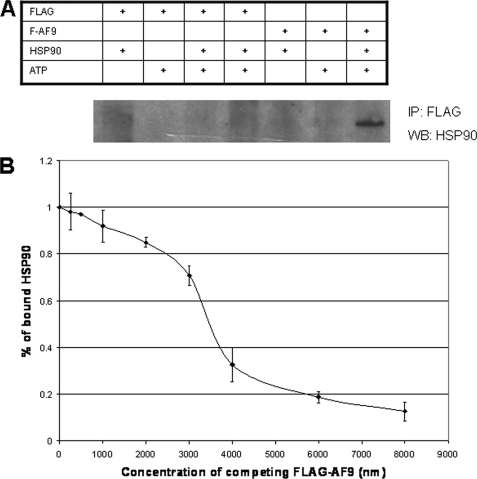
To test if the interaction between AF9 and Hsp90 is direct and not mediated through members of a multiprotein complex, purified AF9-ct and Hsp90 were used in a direct binding assay. Bacterially expressed FLAG-AF9-ct was added to purified recombinant human Hsp90β in the presence and absence of ATPγS. Protein complexes were recovered by anti-Hsp90 immunoprecipitation. FLAG-AF9-ct was found only in immunoprecipitates of reactions containing Hsp90 and ATPγS (Fig. 2A). These data provide evidence of a direct interaction between Hsp90 and AF9 that does not require additional protein factors. Consistent with other Hsp90 client proteins, binding does require ATP.
FIGURE 2.
In vitro binding of AF9 and Hsp90. A, purified recombinant FLAG-AF9-ct and Hsp90 were used to determine if the proteins interacted directly and in the absence of other factors. FLAG peptide was included as a control. Proteins were incubated in the presence or absence of the non-hydrolyzable ATP analog ATPγS. Proteins were then recovered by anti-FLAG immunoprecipitation and analyzed by Western blot for Hsp90. The Table indicates the composition of the incubation mixture. Hsp90 co-precipitates with FLAG-AF9-ct but only when ATP is present. The figure is representative of three experiments. B, competitive ELISA was used to determine the Kd of the Hsp90 and AF9 interaction. The assay is described under “Experimental Procedures” and uses purified recombinant proteins as in Fig. 2A. Data are derived from three independent experiments. The Kd is ∼3.5 μm. Error bars are the standard deviation from the mean of three replicates.
Using the purified recombinant proteins described above, we measured the dissociation constant of Hsp90 and AF9. FLAG-AF9-ct was immobilized in 96-well plates and an ELISA was performed with anti-Hsp90 antibodies to detect Hsp90 binding. Hsp90 was displaced from the immobilized FLAG-AF9-ct by free FLAG-AF9-ct allowing a determination of the Kd. With this competitive ELISA system, the Kd was calculated to be ∼3.5 μm a value comparable to other Hsp90-client protein complexes (Fig. 2B) (25–28). The Kd calculation takes into account Hsp90 homodimerization as Hsp90 has been shown to bind to client proteins as homodimer (29).
Hsp90 and AF9 Comprise a Protein Complex in Vivo
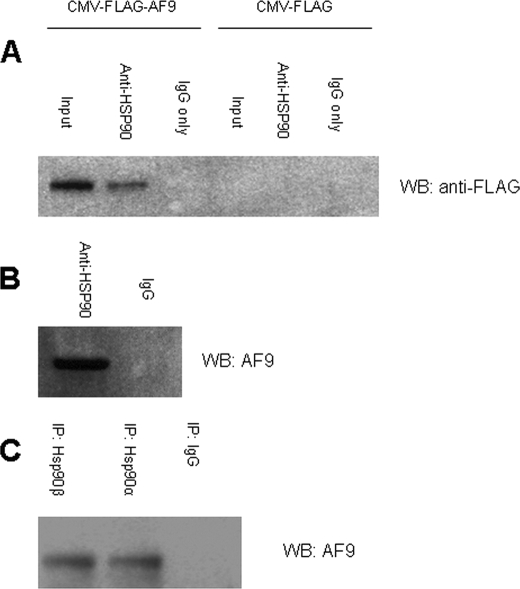
To verify the interaction between Hsp90 and a full-length AF9 molecule in vivo, we performed immunoprecipitation experiments using HeLa cells expressing either FLAG-tagged AF9 or the FLAG tag alone. When endogenous Hsp90 is precipitated with anti-Hsp90 antibodies recognizing both α and β isoforms, FLAG-tagged AF9 is recovered in the immune complexes (Fig. 3A). This indicates that full-length AF9 interacts with Hsp90 in living cells. However, as the FLAG-AF9 in this experiment was produced from a CMV promoter-driven expression vector, the observed association with Hsp90 was further confirmed with physiologic levels of AF9 protein. Endogenous AF9 and Hsp90 were co-precipitated with a polyclonal Hsp90 antibody. In both HeLa and IMCD3 cell extracts, endogenous AF9 was detected in immune complexes with endogenous Hsp90 (Fig. 3B). These findings reveal that Hsp90 and AF9 interact under physiological conditions.
FIGURE 3.
Co-precipitation of AF9 and Hsp90 in vivo. A, HeLa cells were transfected with CMV-FLAG-AF9 vector expressing full-length AF9 or with the empty vector (CMV-FLAG) as a control. Cell lysates were immunoprecipitated with an antibody that recognizes both Hsp90 isoforms (or IgG control). The precipitated complexes were analyzed by Western blot for FLAG to detect FLAG-tagged AF9. The figure is representative of three experiments. B, non-transfected whole cell lysates were prepared from IMCD3 cells and immunoprecipitated as in Fig. 3A. Endogenous AF9 was detected by Western blot with an AF9 antibody. The figure is representative of three experiments. C, antibodies specific for the α and β isoforms of Hsp90 were used to immunoprecipitate proteins from IMCD3 cells. IgG served as a negative control. Endogenous AF9 was detected by Western blot. The figure is representative of three experiments.
From our initial experiments, we found both isoforms of Hsp90 are able to interact with AF9; the α isoform was found in a yeast two hybrid screen of AF9-interacting proteins and the β isoform was recovered using AF9 column purification. Immunoprecipitation using isoform-specific antibodies was performed to confirm these preliminary results and to verify that AF9 binds both isoforms in living cells. These experiments showed that both isoforms stably associate with AF9 under the conditions used for immunoprecipitation (Fig. 3C).
Several transcription factors including the glucocorticoid receptor, STAT3, and p53 (35–37) are found within heteromeric complexes containing Hsp90 as well as Hsp70 and p60/Hop. These multiprotein complexes collaborate in the cellular trafficking and nuclear import of the client transcription factors. In S. cerevisiae, Hsp90, Hsp70, and the yeast homolog of p60/Hop, Sti1, promote removal of nucleosomes to activate GAL1 gene expression (15). Accordingly, the Hsp90-Hsp70-p60 complex facilitates a variety of dynamic nuclear processes.
Below, we show that the subcellular distribution of AF9 is affected by manipulating Hsp90 thus raising the possibility that AF9 exists within Hsp90-Hsp70-p60 heteromeric complexes that help direct the protein to specific subcellular compartments. To test whether AF9 is a component of Hsp70-p60-containing complexes, we performed immunoprecipitation experiments with Hsp70 and p60/Hop antibodies using IMCD3 cell lysates (Fig. 4). When the resulting Hsp70 and p60/Hop immune precipitates were analyzed by Western blot, we detected AF9. Although this finding does not exclude the possibility that AF9 individually and mutually exclusively binds Hsp90, Hsp70, and p60/Hop, we believe that the most likely explanation is that AF9 is part of an Hsp90-Hsp70-p60 chaperone complex.
FIGURE 4.
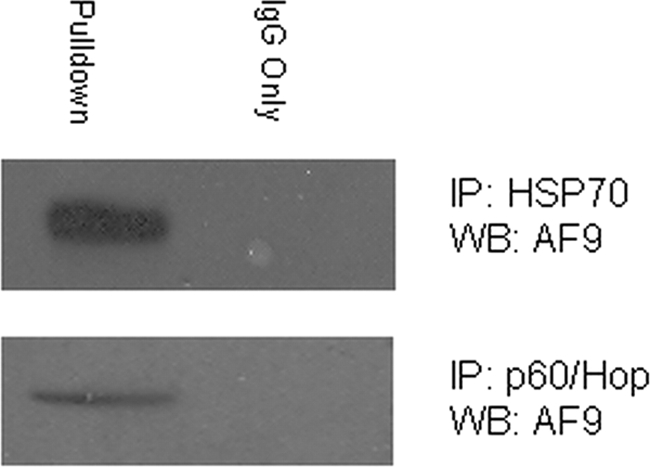
Co-precipitation of AF9 and Hsp70 and the co-chaperone p60/Hop. IMCD3 cell lysates were immunoprecipitated with an Hsp70 antibody (upper panel) or p60/Hop antibody (lower panel). AF9 was detected in the immune complexes by Western blot. The figure is representative of three experiments.
Hsp90 Affects the Subcellular Localization of AF9
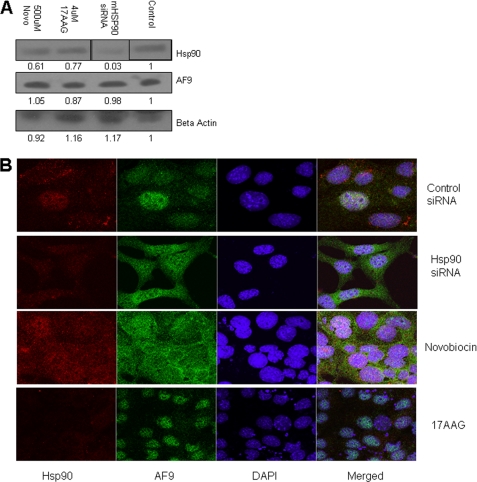
To test the significance of the Hsp90-AF9 interaction, we used Hsp90 siRNA directed against both isoforms of mouse Hsp90 to deplete the protein, or the Hsp90 inhibitors 17AAG and Novobiocin to perturb Hsp90 function (15, 30, 31). 17AAG is a derivative of geldanamycin, a well-studied N-terminal ATPase inhibitor that has been shown to inhibit Hsp90's chaperonin function (32). Novobiocin inhibits the C-terminal ATPase of Hsp90 and prevents co-chaperones from binding the molecule (33). We tested whether depletion of Hsp90 with siRNA or chemical inhibition of Hsp90 function leads to AF9 destabilization and/or degradation. IMCD3 cells were treated with increasing concentrations of 17AAG or novobiocin and whole cell lysates were then analyzed for Hsp90 and AF9 protein by Western blot. Similarly, cells were treated with siRNA directed against both Hsp90 isoforms. siRNA treatment did diminish total Hsp90 while the chemical inhibitors had no effect on Hsp90 protein abundance. Remarkably, under all experimental conditions tested, levels of AF9 protein were not changed (Fig. 5A). In contrast to some Hsp90 client proteins, AF9 is not readily degraded despite interfering with normal Hsp90 activity with siRNA, 17AAG, or novobiocin.
FIGURE 5.
Effects of pharmacologic inhibition and siRNA knock-down of Hsp90 on AF9. A, IMCD3 cells were treated with Hsp90 inhibitors novobiocin (Novo) and 17AAG as well as siRNA directed against both isoforms of mouse Hsp90. Scrambled siRNA served as a control. Whole cell lysates were prepared, and the relative abundance of proteins was assesses by Western blot. siRNA reduces Hsp90 protein abundance. AF9 protein abundance is not affected by any of the interventions. The vertical lines in the Hsp90 Western blot indicate that the position of the lanes was digitally manipulated. Experiment was repeated in triplicate with similar results. B, IMCD3 cells were treated as in Fig. 5A. Cells were then examined by immunofluorescence microscopy with the antibodies and stains indicated in the figure. AF9 shifts from a primarily nuclear localization to a nuclear and cytoplasmic distribution in cells treated with Hsp90 siRNA or novobiocin. The experiment was repeated in triplicate, and data from one set of experiments are shown. Additional images can be furnished upon request.
Hsp90 is also required for nuclear-cytoplasmic shuttling of specific protein complexes (34). Previous studies have revealed that AF9 localizes to the nucleus, therefore we tested whether Hsp90 is required for efficient transport of AF9 into the nucleus. In cells exposed to either novobiocin or Hsp90 siRNA, confocal imaging showed differences in AF9 distribution within the cell (Fig. 5). As has been previously reported (22), in control-treated cells AF9 is localized primarily to the nucleus within discrete subnuclear speckles. Upon treatment of IMCD3 cells with novobiocin or exposure of these cells to Hsp90 siRNA, AF9 proteins are redistributed. Nuclear AF9 is shifted into DAPI-staining pockets or, more dramatically, is delocalized from the nucleus into the cytoplasm. In contrast, treatment with 17AAG causes no observable change. These observations are consistent with a novobiocin-sensitive role for Hsp90 in directing and/or maintaining AF9 in precise sites within the nucleus. Furthermore, the observations suggest that disruption of Hsp90-AF9 interactions may affect AF9-modulated gene expression by depleting AF9 from the nucleus.
Hsp90 Is Required for Normal Expression of the AF9-regulated Gene ENaCα
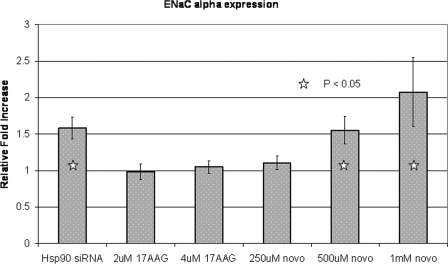
To determine if expression of AF9-responsive genes would be affected by altering Hsp90 activity, we used quantitative RT-PCR to measure ENaCα mRNA. We predicted that by diminishing AF9 abundance within the nucleus, Hsp90 inhibition would lead to increased ENaCα gene expression. Indeed, following treatment with novobiocin or Hsp90 siRNA, ENaCα mRNA levels increased. Moreover, in the case of novobiocin, the response was dose-dependent (Fig. 6). Conversely, treatment with 17AAG had no effect on gene expression consistent with the finding that 17AAG treatment shows no delocalization of AF9 from the nucleus. Enhanced ENaCα gene expression is consistent with diminished AF9 activity. Thus, pharmacologic or genetic manipulation of Hsp90 affects ENaCα transcript abundance possibly by preventing the proper subnuclear localization and gene targeting of AF9.
FIGURE 6.
qPCR for ENaCα gene expression after Hsp90 perturbation. IMCD3 cells were treated with Hsp90 inhibitors or siRNA as described in the text. Total RNA was isolated, reverse transcribed, and quantitative PCR was performed for the mouse ENaCα cDNA. The experiment was repeated in triplicate. Error bars are the standard deviation of the mean from three replicates.
Perturbation of Hsp90 Affects AF9 Binding to the ENaCα Gene Promoter
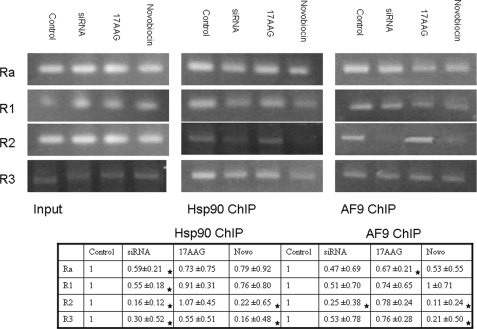
Previous studies of the ENaCα promoter region have shown that AF9 is required to recruit the H3K79 methyltransferase Dot1. Dot1-mediated H3K79 methylation, in turn, helps repress ENaCα expression (6, 8, 9). Changes in AF9 localization as well as the qPCR data of ENaCα expression presented in the previous sections suggest that impaired Hsp90 function interferes with AF9 binding in the promoter region. To assess this possibility, chromatin immunoprecipitation was performed with Hsp90 and AF9 antibodies and regions of the ENaCα promoter were analyzed by semi-quantitative PCR. The gel images were quantified using Image J software from NIH and normalized to input for relative intensity quantification. The most dramatic change detected was diminished association of both Hsp90 and AF9 with the R2 region of the ENaCα promoter after cells were treated with either novobiocin or Hsp90 siRNA (Fig. 7). As expected from localization and gene expression experiments, Hsp90 and AF9 binding to the R2 region is unchanged with 17AAG treatment. In sum, alterations in ENaCα gene expression with experimental manipulation of Hsp90 are associated with changes in Hsp90 and AF9 binding at the R2 region of the ENaCα promoter.
FIGURE 7.
Analysis of the ENaCα promoter region by ChIP. ChIP was performed with Hsp90 and AF9 antibodies and precipitated DNA was amplified by primers spanning the region −1372 to +494 relative to the start site. The primers are described in the text and are derived from the work of Zhang et al. (6) and personal communication with Dr. Zhang. The experiment was repeated in triplicate and fold difference relative to control was calculated using densitometry from the NIH program Image J. The combined data are presented as 95% confidence intervals (CI) of a t distribution of unknown variance. CIs that fall outside the control value of 1.0 are indicated with a star.
DISCUSSION
These studies reveal that the interaction between Hsp90 and AF9 is specific and is required for normal AF9 function. Our data indicate that Hsp90 regulates AF9's distribution in the cell thus affecting the function of AF9 on a downstream target gene, ENaCα. We examined this interaction using purified protein components and verified that Hsp90 and AF9 are able to interact directly and that protein binding requires ATP. Under a variety of assay conditions using different Hsp90 and AF9 antisera, Hsp90 and AF9 are found together within immune complexes isolated from living cells. Consequently, we propose that AF9 is a bona fide client protein of Hsp90. Strengthening this proposal, we have also shown that AF9 associates with both Hsp90 and the cooperating chaperones Hsp70 and p60/Hop. Of note, affinity purification of the AF9 homolog, ENL, and its associated proteins (“EAPs”) using immobilized ENL antibodies recovered Hsp70, but neither Hsp90 nor Hop were described in this report by Slany and co-workers (12).
However, we are surprised by the lack of effect of the chemical inhibitor 17AAG. Treatment with novobiocin and Hsp90 knockdown gave similar results indicating a mechanism that is specific for Hsp90. Novobiocin acts through inhibition of the C terminus of Hsp90 while 17AAG inhibits the N-terminal ATPase. Nevertheless, the explanation for differences in the disposition of AF9 in cells treated with novobiocin versus 17AAG is still not clear.
In silico sequence analysis of the AF9 C terminus, the region that associates with Hsp90, suggests that it is highly unstructured. In our experiments, perturbations of Hsp90 by various methods did not result in changes in the steady-state level of AF9 as is observed for proteins that require Hsp90 for proper folding. Under these circumstances, disrupting the Hsp90-client protein interaction would normally cause ubiquitination of the client protein, signaling its eventual proteosomal degradation (38). Thus, our data suggest that Hsp90 is not facilitating the proper folding or stabilization of AF9. Rather, Hsp90 transports AF9 to specific domains within the nucleus where it gains access to target genes and the macromolecular assemblies that regulate these genes. This is similar to several other nuclear proteins and is consistent with a model proposed by DeZwaan and Freeman (39). In this model, Hsp70 and Hsp90 serve to promote the active and dynamic assembly of higher order structures including those involved in telomere maintenance, DNA repair, and gene transcription.
In addition to the well studied role of Hsp90 in cytoplasmic-nuclear transport of glucocorticoid receptors (reviewed in Ref. 40), Hsp90 appears to be required for the trafficking of a heterogeneous collection of proteins. Hsp90 travels to the nucleus in complex with influenza virus RNA polymerase and stimulates viral RNA synthesis (41). As another example, incorporation of the Argonaute RNA-binding proteins into cytoplasmic P-bodies is mediated by Hsp90 and inhibition of Hsp90 interferes with post-transcriptional gene silencing (23). We propose that AF9 is yet another protein whose proper subcellular localization, hence function, requires Hsp90. These results further emphasize the importance of Hsp90 in a wide range of cellular processes including the modulation of factors participating in regulated gene expression. Similarly, given the many binding partners that have been identified, it is likely that AF9 variably associates with differing proteins in distinct complexes. We can speculate that Hsp90 may help direct AF9 to one or more of these multiprotein complexes to facilitate its assembly.
Acknowledgments
We thank Drs. Richard Schultz, Nancy Zeleznik-Le, and Charles Miller III for input and advice.
This work was supported by National Institutes of Health Grant CA098459. This work was also supported by funds from the Cardinal Bernardin Cancer Center.
- MLL
- mixed lineage leukemia
- ATPγS
- adenosine 5′-[γ-thio]triphosphate
- ELISA
- enzyme-linked immunosorbent assay
- BSA
- bovine serum albumin
- PBS
- phosphate-buffered saline
- qPCR
- quantitative polymerase chain reaction
- ChIP
- chromatin immunoprecipitation assay.
REFERENCES
- 1.Krivtsov A. V., Twomey D., Feng Z., Stubbs M. C., Wang Y., Faber J., Levine J. E., Wang J., Hahn W. C., Gilliland D. G., Golub T. R., Armstrong S. A. (2006) Nature 442, 818–822 [DOI] [PubMed] [Google Scholar]
- 2.Chen W., Kumar A. R., Hudson W. A., Li Q., Wu B., Staggs R. A., Lund E. A., Sam T. N., Kersey J. H. (2008) Cancer Cell 13, 432–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DiMartino J. F., Miller T., Ayton P. M., Landewe T., Hess J. L., Cleary M. L., Shilatifard A. (2000) Blood 96, 3887–3893 [PubMed] [Google Scholar]
- 4.Cairns B. R., Henry N. L., Kornberg R. D. (1996) Mol. Cell Biol. 16, 3308–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nie Z., Yan Z., Chen E. H., Sechi S., Ling C., Zhou S., Xue Y., Yang D., Murray D., Kanakubo E., Cleary M. L., Wang W. (2003) Mol. Cell Biol. 23, 2942–2952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang W., Xia X., Reisenauer M. R., Hemenway C. S., Kone B. C. (2006) J. Biol. Chem. 281, 18059–18068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bitoun E., Oliver P. L., Davies K. E. (2007) Hum. Mol. Genet. 16, 92–106 [DOI] [PubMed] [Google Scholar]
- 8.Zhang W., Xia X., Reisenauer M. R., Rieg T., Lang F., Kuhl D., Vallon V., Kone B. C. (2007) J. Clin. Invest. 3, 773–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang D., Yu Z. Y., Cruz P., Kong Q., Li S., Kone B. C. (2009) Kidney Int. 75, 260–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steger D. J., Lefterova M. I., Ying L., Stonestrom A. J., Schupp M., Zhuo D., Vakoc A. L., Kim J. E., Chen J., Lazar M. A., Blobel G. A., Vakoc C. R. (2008) Mol. Cell Biol. 28, 2825–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Issacs J. S., Xu W., Neckers L. (2003) Cancer Cell 3, 23–27 [DOI] [PubMed] [Google Scholar]
- 12.Mueller D., Bach C., Zeisig D., Garcia-Cuellar M. P., Monroe S., Sreekumar A., Zhou R., Nesvizhskii A., Chinnaiyan A., Hess J. L., Slany R. K. (2007) Blood 110, 4445–4454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins E. C., Appert A., Ariza-McNaughton L., Pannell R., Yamada Y., Rabbitts T. H. (2002) Mol. Cell Biol. 22, 7313–7324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pina C., May G., Soneji S., Hong D., Enver T. (2008) Cell Stem Cell 6, 264–273 [DOI] [PubMed] [Google Scholar]
- 15.Floer M., Bryant G. O., Ptashne M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2975–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tariq M., Nussbaumer U., Chen Y., Beisel C., Paro R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 1157–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Truman A. W., Millson S. H., Nuttall J. M., King V., Mollapour M., Prodromou C., Pearl L. H., Piper P. W. (2006) Eukaryot. Cell 5, 1914–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hemeway C. S., de Erkenez A. C., Gould G. C. (2001) Oncogene 20, 3798–3805 [DOI] [PubMed] [Google Scholar]
- 19.Corral J., Lavenir I., Impey H., Warren A. J., Forster A., Larson T. A., Bell S., McKenzie A. N., King G., Rabbitts T. H. (1996) Cell 85, 853–861 [DOI] [PubMed] [Google Scholar]
- 20.Marcu M. G., Chadli A., Bohouche I., Catelli B., Neekers L M. (2001) J. Biol. Chem. 276, 37181–37186 [DOI] [PubMed] [Google Scholar]
- 21.Bushnell D. A., Bamdad C., Kornberg R. D. (1996) J. Biol. Chem. 271, 20170–20174 [DOI] [PubMed] [Google Scholar]
- 22.Srinivasan R. S., de Erkenez A. C., Hemenway C. S. (2003) Oncogene 22, 3395–3406 [DOI] [PubMed] [Google Scholar]
- 23.Pare J. M., Tahbaz N., Lopez-Orozco J., LaPointe P., Lasko P., Hobman T. C. (2009) Mol. Cell Biol. 20, 3273–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deleted in proof
- 25.Blatch G. L. (2007) Networking of Chaperones by Co-chaperones, Vol. 55, Landes Biosciences [Google Scholar]
- 26.Gooljarsingh L. T., Fernandes C., Yan K., Zhang H., Grooms M., Johanson K., Sinnamon R. H., Kirkpatrick R. B., Kerrigan J., Lewis T., Arnone M., King A. J., Lai Z., Copeland R. A., Tummino P. J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 7625–7630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma Y., Bogatcheva N. V., Gusev N. B. (2000) Biochim. Biophys. Acta 1476, 300–310 [DOI] [PubMed] [Google Scholar]
- 28.Ali M. M., Roe M. S., Vaughan C. K., Meyer P., Panaretou B., Piper P. W., Prodromou C., Pearl L. H. (2006) Nature 440, 1013–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wayne N., Bolon D. N. (2007) J. Biol. Chem. 282, 35386–35395 [DOI] [PubMed] [Google Scholar]
- 30.Kamphausen T., Fanghänel J., Neumann D., Schulz B., Rahfeld J. U. (2002) Plant J. 32, 263–276 [DOI] [PubMed] [Google Scholar]
- 31.Freeman B. C., Yamamoto K. R. (2002) Science 296, 2232–2235 [DOI] [PubMed] [Google Scholar]
- 32.Zeng Y., Feng H., Graner M. W., Katsanis E. (2003) Blood 101, 4485–4491 [DOI] [PubMed] [Google Scholar]
- 33.Schulte T. W., Neckers L. M. (1998) Cancer Chemother. Pharmacol. 42, 273–279 [DOI] [PubMed] [Google Scholar]
- 34.Marcu M. G., Chadli A., Bouhouche I., Catelli M., Neckers L. M. (2000) J. Biol. Chem. 275, 37181–37186 [DOI] [PubMed] [Google Scholar]
- 35.Selkirk J. K., Merrick B. A., Stackhouse B. L., He C. (1994) Appl. Theor. Electrophor. 4, 11–18 [PubMed] [Google Scholar]
- 36.Dittmar K. D., Hutchison K. A., Owens-Grillo J. K., Pratt W. B. (1996) J. Biol. Chem. 271, 12833–12839 [DOI] [PubMed] [Google Scholar]
- 37.Longshaw V. M., Baxter M., Prewitz M., Blatch G. L. (2009) Eur. J. Cell Biol. 88, 153–166 [DOI] [PubMed] [Google Scholar]
- 38.Echeverría P. C., Mazaira G., Erlejman A., Gomez-Sanchez C., Piwien Pilipuk G., Galigniana M. D. (2009) Mol. Cell Biol. 29, 4788–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dezwaan D. C., Freeman B. C. (2008) Cell Cycle 7, 1006–1012 [DOI] [PubMed] [Google Scholar]
- 40.Grad I., Picard D. (2007) Mol. Cell Endocrinol. 275, 2–12 [DOI] [PubMed] [Google Scholar]
- 41.Naito T., Momose F., Kawaguchi A., Nagata K. (2007) J. Virol. 81, 1339–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]