Abstract
Sorting nexin 9 (SNX9) functions in a complex with the GTPase dynamin-2 at clathrin-coated pits, where it provokes fission of vesicles to complete endocytosis. Here the SNX9·dynamin-2 complex binds to clathrin and adapter protein complex 2 (AP-2) that line these pits, and this occurs through interactions of the low complexity domain (LC4) of SNX9 with AP-2. Intriguingly, localization of the SNX9·dynamin-2 complex to clathrin-coated pits is blocked by interactions with the abundant glycolytic enzyme aldolase, which also binds to the LC4 domain of SNX9. The crystal structure of the LC4 motif of human SNX9 in complex with aldolase explains the biochemistry and biology of this interaction, where SNX9 binds near the active site of aldolase via residues 165–171 that are also required for the interactions of SNX9 with AP-2. Accordingly, SNX9 binding to aldolase is structurally precluded by the binding of substrate to the active site. Interactions of SNX9 with aldolase are far more extensive and differ from those of the actin-nucleating factor WASP with aldolase, indicating considerable plasticity in mechanisms that direct the functions of the aldolase as a scaffold protein.
Keywords: Crystallography, Endocytosis, Enzyme Structure, Protein Structure, Protein-Protein Interactions, Sorting Nexin 9, WASP, Aldolase, Low Complexity Domain (LC4)
Introduction
The sorting nexins (SNX)3 are multidomain family proteins that appear to function in the directed delivery (i.e. the “sorting”) of their cargo to selected subcellular compartments, including to transmembrane receptors, endosomes, and clathrin-coated pits that mediate endocytosis. Domains shared among SNX family members include a phosphoinositide-binding phox domain and a Bin-Amphiphysin-Rvs domain, which is required for tubulogenesis and is a hallmark of amphiphysin family proteins that direct the early steps of endocytosis (1). However, two members of the SNX family, SNX9 and SNX18, also harbor Src homology 3 (SH3) domains that may direct specific interactions with a large cast of proteins having proline-rich PXXP motifs (2–4).
A key partner of SNX9 is the large GTPase dynamin-2, which binds to the SH3 domain of SNX9. The SNX9·dynamin-2 complex plays important roles at clathrin-coated pits, where it forms a collar at the base of the pit and its associated GTPase activity is thought to drive fission of these vesicles to complete endocytosis (5–7). The association of the SNX9·dynamin-2 complex with endocytic vesicles is mediated by the N-terminal SH3 domain that also directs its interactions with other partners, including the actin nucleator Wiskott-Aldrich Syndrome protein (WASP) and transmembrane receptors (8). Furthermore, the low complexity (LC4) motif, situated between the SH3 and phosphoinositide-binding phox domains, interacts with adaptor protein complex 2 (AP-2) (9), which binds to and collects transmembrane cargo within the clathrin-coated pit (10), and SNX9 may also directly bind to clathrin (9).
Functional control of the SNX9·dynamin-2 complex is in part mediated by re-localization of the SNX9·dynamin-2 complex from the cytoplasm to the clathrin-coated pits. Intriguingly, a 41-kDa cytosolic protein originally identified as a component of the 14.5 S cytosolic SNX9·dynamin-2 complex was shown to be the intermediary metabolic enzyme fructose-1,6-bisphosphate aldolase (aldolase; EC 4.1.2.13) (11), which catalyzes the reversible cleavage of fructose 1,6-bisphosphate (FBP) to dihydroxyacetone phosphate and glyceraldehyde 3-phosphate in glycolysis and gluconeogenesis.
Aldolase is highly abundant in the cytosol where glycolysis generally occurs, although the pathway may also be active at other locales, for example, at cholesterol and glycosphingolipid-rich caveolae present in the plasma cell membrane (12). Interestingly, noncatalytic roles for aldolase are now evident, where free pools of aldolase can bind to F-actin, α-tubulin, light chain 8 of dynein, WASP, the anion exchanger Band 3, phospholipase D2, transmembrane adhesive proteins, and even the glucose transporter GLUT4 (8, 13–20). Furthermore, all of these interactions are inhibited by the binding of the products or substrates of aldolase (13, 14, 20, 21), indicating that they rely on free, inactive pools of the enzyme. Overall these findings, and those showing that aldolase provides essential links for proteins of pathogens to the actin cytoskeleton (14, 22), suggest that aldolase has important functions as a scaffold protein that coordinates events at the cell membrane with actin and microtubule networks. The findings that aldolase binds to the SNX9·dynamin-2 complex in mutually antagonistic fashion with AP-2 indicate that these functions extend to control of endocytic events, but how the aldolase·AP-2 antagonism is manifest at the molecular level is unclear. To answer this question, as well as how substrates and products of aldolase disable the interaction of the SNX9·dynamin-2 complex with aldolase, we solved the crystal structure of the LC4 domain of SNX9 in complex with aldolase. Our crystal structure and competitive inhibition assays define the aldolase·SNX9 binding mechanism and support a model whereby the regulation of endocytosis is linked to metabolic pathways driven by aldolase.
EXPERIMENTAL PROCEDURES
Purification and Crystallization
Expression and purification of recombinant rabbit muscle aldolase was performed as described (23, 24). The aldolase·SNX9 complex was co-crystallized by the hanging drop vapor-diffusion method at 4 °C using an equal volume of (1:1) of protein:peptide solution (0.09 mm aldolase tetramer and 0.27 mm LC4 peptide 152QAYQGPATGDDDDWDEDWDGPKSSSYFKDSE182 in 20 mm Tris-HCl, pH 7) and precipitant solution (14% PEG-MME 550, 100 mm Tris-HCl, pH 7.1, 10 mm MgCl2) in the drop equilibrated against a reservoir of precipitant solution.
X-ray Data Collection and Processing
The aldolase·SNX9 complex crystals were transferred into 14% PEG-MME 550, 100 mm Tris-HCl, pH 7.1, 10 mm MgCl2, and 25% (v/v) glycerol prior to flash freezing in liquid nitrogen. Diffraction data were collected at the SER-CAT beamline 22-BM at the Advanced Photon Source (Argonne National Laboratory, Argonne, IL). Data sets were processed with HKL2000 (25) and the results are summarized in Table 1.
TABLE 1.
Data reduction statistics of the aldolase·SNX9 complex
| Space group | P212121 |
| Unit cell dimensions | |
| a | 87.03 Å |
| b | 118.18 Å |
| c | 175.9 Å |
| α = β = γ | 90° |
| Resolution | 50 Å-2.2 Å |
| Last shell | 2.28 Å-2.2 Å |
| Total measurements | 594,924 |
| Number of unique reflections | 91,624 |
| Last shell | 8,482 |
| Wavelength | 1.0 Å |
| Rsyma | 0.142 |
| Last shell | 0.491 |
| I/σ(I) | 4.9 |
| Last shell | 2.0 |
| Completeness | 0.99 |
| Last shell | 0.928 |
| Redundancy | 6.5 |
| Last shell | 3.2 |
aRsym = ΣhklΣi|Ii(hkl) − |/ΣhklΣiIi(hkl).
Structure Determination and Crystallographic Refinement
The aldolase·SNX9 complex crystals belong to space group P212121 and were not isomorphous with native aldolase crystals (space group P21). Therefore, we solved the crystal structure by molecular replacement using the program MOLREP (26) and the aldolase tetramer (PDB code 2OT0) as the search model (27). After initial rigid body refinement, the model was subjected to iterative cycles of restrained refinement using REFMAC5 (28) and model building using COOT (29). We used COOT (29) and BUSTER-TNT (30) to add water molecules and performed crystallographic refinement to 2.2 Å with the REFMAC5 program (28). Non-crystallographic symmetry restraints were applied using local structure similarity restraints implemented in autoBUSTER (31, 32). The stereochemical geometry of the model was determined by PROCHECK (33). The final refinement statistics are provided in Table 2.
TABLE 2.
Crystallographic refinement statistics of the aldolase·SNX9 complex
| Resolution | 32.21 Å-2.2 Å |
| Last shell | 2.26 Å-2.2 Å |
| No. of reflections (working set) | 86,911 |
| No. of reflections (test set) | 4,580 |
| R-factora | 0.1531 |
| Last shell | 0.2268 |
| Rfreeb | 0.1894 |
| Last shell | 0.2778 |
| No. of protein residues | 1,402 |
| No. of ligand residues | 52 |
| No. of solvent molecules | 1,715 |
| Average B-factor | |
| Protein | 22.2 Å2 |
| Solvent | 37.4 Å2 |
| Ligand | 41.8 Å2 |
| Root mean square deviation from ideal geometry | |
| Bond lengths | 0.01 Å |
| Bond angles | 0.97° |
aRcryst = Σhkl‖Fobs(hkl)| − |Fcalc(hkl)‖Σhkl|Fobs(hkl)|.
b The free R-factor is a cross-validation residual calculated by using 5% reflections, which were randomly chosen and excluded from the refinement.
Peptide Inhibition Assay
Aldolase activity was measured using a coupled assay system of triose-phosphate isomerase and glycerol-3-phosphate dehydrogenase by following NADH oxidation at 340 nm at 22 °C (34). Reactions were initiated by the addition of FBP substrate to complete a solution containing aldolase made up in Tris buffer (100 mm Tris-HCl, pH 7.1, 10 mm Mg-acetate, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride) and 0.3 mm NADH and containing coupling enzymes (10 μg/μl of glycerol-3-phosphate dehydrogenase and 2 μg/μl of triose-phosphate isomerase) to a final volume of 250 μl. The rate of substrate cleavage was determined by measuring the decrease in absorbance per minute at 340 nm. Aldolase was dialyzed at 4 °C against the Tris buffer before use, and the protein concentration was determined by spectrophotometry using an extinction coefficient of ϵ280 = 0.91 ml mg−1 cm−1.
Inhibition of aldolase activity by 7-mer (wild type, 165WDEDWDG171; W165A mutant, 165ADEDWDG171) and 20-mer (wild type, 163DDWDEDWDGPKSSSYFKDSE182; W165A mutant, 163DDADEDWDGPKSSSYFKDSE182) LC4 peptides was determined from initial rates measured in the presence of 12.5–500 μm FBP and 10–300 μm LC4 peptide, 400–1300 μm WASP peptide (496EDDEWDD502), or a 350–1500 μm random sequence peptide (DNYEFDG) at a constant enzyme concentration (0.175 μg/ml). LC4 and WASP peptides were solubilized in the inhibition assay buffer to the desired concentration. The random sequence peptide was solubilized in 5% dimethyl sulfoxide and then diluted to the desired concentration in inhibition assay buffer. The addition of 5% dimethyl sulfoxide did not modify Michaelis parameters characterizing aldolase kinetics. Peptide concentrations were determined from molar extinction coefficients at 280 nm calculated on the basis of their respective amino acid sequence (35). The inhibitor dissociation constant (Ki) corresponding to 7- and 20-mer peptides was then calculated from the initial velocities using GraFit 6 (36).
RESULTS
The Structure of Aldolase·SNX9 Complex
The SNX9·dynamin-2 complex associates with AP-2 and clathrin and clathrin-coated pits of the cell membrane to direct endocytosis (5, 9). This interaction is held in check through cytosolic interactions of the SNX9·dynamin-2 complex with the glycolytic enzyme aldolase (aldolase; EC 4.1.2.13), preventing its association with cell membranes (11). The domain of SNX9 that directs its interaction with aldolase is the low complexity region called LC4 (residues 152–183) (9, 11), which also directs its interactions with AP-2 and clathrin at sites of endocytosis, and mutation of the two tryptophan residues found in the LC4 motif completely abolish the binding of SNX9 to aldolase (11). To define this interaction, we crystallized the LC4 segment of SNX9 with rabbit muscle aldolase for structure determination.
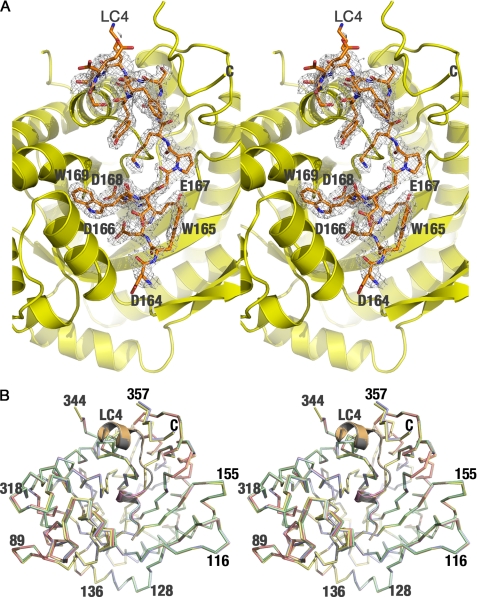
Although all previous aldolase data sets show a large mosaic spread (∼0.6 to over 1 degree), aldolase·SNX9 crystals grown at room temperature using the known aldolase conditions showed too large a mosaic spread that prevented data collection. However, by generating crystals at 4 °C and reducing the concentrations of the protein and precipitant, the mosaic spread was ∼0.1 degree, the lowest ever observed for any rabbit muscle aldolase data set. This allowed data collection and structure determination with final Rcryst and Rfree values of 0.153 and 0.189, respectively (Table 2). Difference maps (Fobs − Fcalc) calculated after refinement with REFMAC5 defined the LC4 peptide in the complex, which was found near the active site of aldolase (Fig. 1A), consistent with the fact that the substrate (FBP) of the enzyme and its products (dihydroxyacetone phosphate and glyceraldehyde 3-phosphate) impair SNX9 binding (11). Each asymmetric unit contains one aldolase homotetramer, with each subunit binding to one LC4 molecule. The bound LC4 fragment in aldolase subunits A and D had visible density only for residues 165–171 having the sequence WDEDWG. In contrast, the LC4 fragment in subunits B and C are well defined and extend up to 19 residues from the C terminus of the 31-residue SNX9 LC4 peptide. In the final aldolase model, residues 345–358 (subunits A and D) and 345–355 (subunits B and C) could not be modeled due to poor electron density.
FIGURE 1.
Structure of the aldolase·SNX9 complex. A, stereo view of aldolase in complex with the LC4 domain of SNX9 is shown, where the aldolase subunit (yellow) is represented as a schematic, and the LC4 peptide is represented as sticks (pale orange). The final model of LC4 is superposed onto the Fobs − Fcalc difference map contoured at 2.5 σ. Representative residues of the LC4 peptide adjacent to Trp-169 are labeled. B, superposition of four aldolase·SNX9 complex subunits in the asymmetric unit is shown to depict the identical nature of the subunits. The aldolase subunit and LC4 peptide are labeled for clarity and the extended LC4 peptide is only observed in two subunits. Some residues are labeled.
All four subunits of the aldolase tetramer were identical, with root mean square deviations of 0.16 Å (subunits A and D) and 0.15 Å (subunits B and C) for 344 Cα atoms. All other combinations yielded an average root mean square deviation of 0.306 Å for the same number of Cα atoms (Fig. 1B). Although the average temperature factors for the aldolase subunits were similar (∼20 Å2), those for the corresponding bound LC4 peptide showed significant differences. Although the observed values were higher for LC4 peptides bound to subunits A (∼69 Å2) and subunit D (∼45 Å2), the values for the peptide bound to subunits B and C (∼23 Å2) are comparable with that of the aldolase subunits (∼20 Å2), suggesting a stable binding as previously described (11).
Binding Mode of LC4 to Aldolase
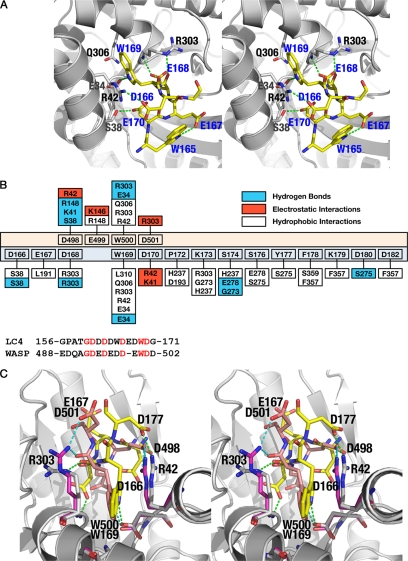
The binding of the SNX9 LC4 peptide in the aldolase·SNX9 complex is well ordered in subunits B and C and consists of a well defined α-helix and a small helical turn near the N terminus. The C-terminal helix of LC4 is sandwiched between the C terminus and one of the peripheral α-helices of the aldolase triose-phosphate isomerase-barrel fold. The indole ring of Trp-169 of SNX9, which is part of the helical turn, is accommodated within a hydrophobic pocket generated by the side chains of aldolase residues Arg-42 and Arg-303 (Fig. 2A). The hydrophobic interactions are complemented by hydrophilic bonding exerted by the NE atom of the indole ring of Trp-169 of SNX9 with the side chain oxygen of Glu-34 of aldolase. Furthermore, the aliphatic portion of the Gln-306 side chain of aldolase contributes to the formation of the base of the pocket. Although aldolase residues Ala-307 and Leu-310 that are present in the binding pocket do not participate in van der Waals interactions with the indole ring of SNX9 Trp-169, they indirectly contribute to interaction by maintaining the hydrophobic environment of the binding pocket.
FIGURE 2.
Interaction of aldolase with the LC4 peptide of SNX9. A, the major interactions observed between the LC4 peptide and aldolase, common to all subunits in the asymmetric unit, is depicted. The aldolase subunit is shown as a schematic (gray) and the LC4 peptide represented as sticks (yellow). The residues of aldolase subunits involved in the LC4 interaction are also represented as sticks (gray). The residues corresponding to the LC4 peptide are labeled in blue, whereas those of aldolase are in black. B, top panel, shows the schematic representation of the interactions found between aldolase·SNX9 and aldolase·WASP complexes. The residues of the WASP peptide are depicted with peach background, whereas those of LC4 are pale cyan. The aldolase residues exhibiting hydrophobic (plain), electrostatic (red), and hydrogen bond (blue) interactions with either WASP or LC4 are also shown. Lower panel is the sequence alignment of the binding regions of LC4 and WASP peptides, showing the similarities and differences found around the tryptophan residue. Identical residues are in red. C, stereo image of the superposition of aldolase·SNX9 and aldolase·WASP complexes. The interactions observed between aldolase (gray) and the peptide LC4 (yellow) or WASP (peach) residues are shown by dotted lines.
Apart from Trp-169, acidic residues Asp-166, Asp-168, and Asp-170 of SNX9 also contribute additional interactions. Here the side chain oxygen of Asp-168 of SNX9 is involved in hydrogen bonding interaction with the main chain nitrogen of Arg-303 of aldolase, Asp-170 of SNX9 engages in electrostatic interactions with the NH2 side chain of aldolase Arg-42, and Asp-166 of SNX9 forms hydrogen bonding interactions with the side chain oxygen of Ser-38 of aldolase. In addition, a hydrogen bond is observed between the backbone oxygen of Asp-168 of SNX9 and the ϵ-NH of Arg-303 of aldolase. This interaction differs significantly from that observed of aldolase with WASP, a regulatory factor that promotes actin nucleation (27), wherein the backbone oxygen of Trp-500 of WASP interacts with NH1 of Arg-303. Indeed, salient features of the aldolase·SNX9 structure are the extensive numbers of interactions that are observed between the SNX9 and aldolase versus the rather limited interactions of WASP and aldolase (Fig. 2B). At a minimum, this indicates a higher affinity interaction between SNX9 and aldolase than between WASP and aldolase, in accord with measurements of their relative affinities (27, 37).
In addition to Trp-169 and its surrounding residues, the C terminus of the LC4 peptide directs several stable interactions with the aldolase. Here the nitrogen of Ser-174 of SNX9 engages in electrostatic interactions with the oxygen of aldolase Gly-273, and the Ser-174 Oγ and Asp-180 Oδ2 of SNX9 form hydrogen bonding interactions with Glu-278 Oϵ1 and Ser-275 Oγ of aldolase, respectively. Additionally, the C terminus of the LC4 motif of SNX9 exhibits hydrophobic interactions with the C terminus of aldolase, where the aliphatic side chain of SNX9 Lys-179 makes a hydrophobic interaction with the aromatic ring of aldolase Phe-357, and where the aromatic side chain of SNX9 Phe-179 has a stacking interaction with the side chain of aldolase Ser-359.
Although the tryptophan-mediated mode of binding of the LC4 region of SNX9 to aldolase is similar to the interaction of aldolase with the C-terminal peptide of WASP (27), where there are shared hydrophobic interactions of these tryptophan residues with Gln-306, Arg-303, and Arg-42 of aldolase, the structure of the aldolase·SNX9 complex indicates a distinct mechanism of interaction between this regulator of endocytosis and aldolase. Specifically residues that direct electrostatic interactions are not shared, and the hydrogen bonding and hydrophobic interactions of SNX9 with aldolase are much more extensive (Fig. 2B), and together these features define the unique structure of the low complexity region of this endocytic regulator in complex with the C-terminal region of the aldolase (Fig. 2C). Interestingly, in both SNX9 and WASP peptides, the N-terminal region is disordered in the crystal structures.
SNX9 Competes with Substrate Binding to Aldolase and the Intramolecular Interaction of the C Terminus of Aldolase with Its Active Site
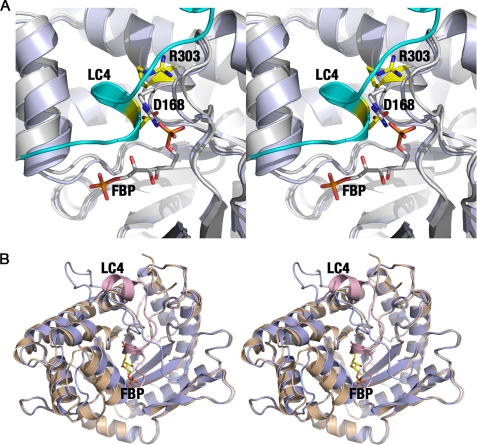
The crystal structure of the aldolase·SNX9 complex predicts that binding of SNX9 in the region juxtaposed with the aldolase active site would compete with binding by its substrate FBP. Indeed, binding of the LC4 motif of SNX9 to aldolase is impaired by FBP (11), suggesting that the natural substrate has a higher affinity for aldolase in binding to the active site than that of SNX9 in binding to the adjacent hydrophobic pocket. Indeed, the side chain of Asp-168 of SNX9 clashes with the P1 phosphate of FBP (Fig. 3A) and with the side chain of Arg-303 of aldolase that comprises the P1 phosphate binding pocket.
FIGURE 3.
Overlapping binding sites and C-terminal rearrangement of aldolase. A, stereo view of the overlapping binding sites for aldolase substrate (FBP; pale blue) and the LC4 domain of SNX9 (cyan and gray) indicate a competition between the LC4 peptide and FBP. B, stereo representation of the C-terminal region of aldolase depending on the type of ligand bound in the same orientation as shown in Fig. 1B. The substrate FBP (pale blue) accommodates the C terminus in the active site, whereas the LC4 peptide (wheat and pink) precludes FBP binding and the intramolecular active site-C terminus interaction.
The superposition of the aldolase·FBP (PDB code 1ZAI) and aldolase·SNX9 complex structures also establishes overlaps in binding of the C terminus of the SNX9 LC4 peptide to aldolase and the intramolecular interactions manifest in the substrate- or product-bound conformation of aldolase, where the aldolase C terminus interacts with its active site (Fig. 3B). Clashes between the two C-terminal regions underscore the fact that only one C-terminal region can be accommodated within a single subunit.
LC4 Competitively Inhibits Aldolase Activity
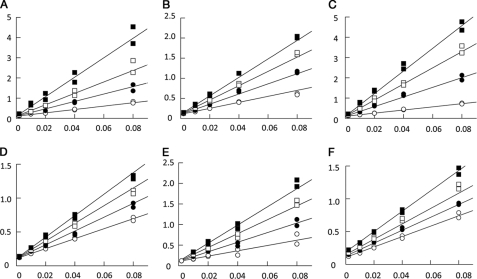
To investigate LC4 binding affinity at the aldolase active site, inhibition kinetics were carried out with peptides corresponding to the residues of LC4 observed in the crystal structure. Enzyme activity was determined in crystallization buffer and inhibition profiles were analyzed using 7- and 20-mer peptide sequences corresponding to LC4 residues 165–171 and 163–182. In addition, to assess the role of Trp-165 whose indole ring stabilizes a helical turn in LC4, the 7- and 20-mer peptides having Trp-165 substituted with alanine were also assessed for their ability to inhibit aldolase activity. The double-reciprocal plots clearly indicate competitive inhibition of aldolase activity consistent with active binding by all four peptide sequences and in agreement with the observed active site binding for LC4 residues 165–171 (subunits A and D) and 164–182 (subunits B and C) in the crystal structure (Fig. 4, A–D). The inhibition dissociation constants for these peptides ranged between 15 and 150 μm (Table 3), and their binding affinity was independent of peptide length, as Ki values were very similar for both 7- and 20-mer peptides. Furthermore, binding affinity by these peptides was reduced 7–8-fold upon loss of Trp-165, supporting the notion that only the 7-mer core sequence of LC4 residues 165–171 is important for aldolase interaction. Residues 163–164 and 172–182 of the 20-mer peptide do not enhance binding affinity. In the B and C subunits, where residues 172–182 are observed in the crystal structure, interactions of these peptide residues with the aldolase C-terminal region are due to crystal packing. Thus, the C-terminal region of aldolase snares the LC4 peptide and induces ordering of residues 172–183.
FIGURE 4.
Competitive inhibition of aldolase by LC4 and WASP peptides. Aldolase shows reciprocal reaction velocity (1/velocity in mg/unit) when plotted against the reciprocal substrate concentration (1/[FPB] in μm−1) for the cleavage of FBP in the presence of 7- and 20-mer LC4 peptides of SNX9. Duplicates are shown. A, 0 (open circle), 5 (closed circle), 10 (open square), and 20 μm (closed square) 7-mer peptide (LC4 residues 165–171); B, 0 (open circle), 100 (closed circle), 200 (open square), and 300 μm (closed square) 7-mer peptide except W165A; C, 0 (closed circle), 35 (closed circle), 70 (open square), and 105 μm (closed square) 20-mer peptide (LC4 residues 163–183); D, 0 (open circle), 50 (closed circle), 100 (open square), and 150 μm (closed square) 20-mer W165A mutated peptide; E, 0 (open circle), 431 (closed circle), 862 (open square), and 1293 μm (closed square) 7-mer peptide (WASP residues 496–502); and F, 0 (open circle), 363 (closed circle), 726 (open square), and 1452 μm (closed square) 7-mer random sequence peptide (DNYEFDG) as a negative control. Inhibition kinetics was performed using crystallization buffer conditions.
TABLE 3.
LC4 peptide inhibition dissociation constants
The values shown for the random peptide (DNYEFDG) correspond to dissociation constants Ki and K′i, respectively, used to characterize the mixed inhibition exhibited by this peptide and is different from competitive inhibition displayed by LC4 and WASP peptides, which is characterized by a single dissociation constant Ki.
| Peptide | Ki | Ki′ |
|---|---|---|
| LC4 (165–171) | 17 ± 2 μm | |
| LC4 (165–171) W165A | 144 ± 7 μm | |
| LC4 (163–183) | 18 ± 2 μm | |
| LC4 (163–183) W165A | 130 ± 5 μm | |
| WASP (496–502) | 532 ± 33 μm | |
| DNYEFDG | 1,807 ± 258 μm | 1,821 ± 130 μm |
To gain further insight into the interactions of aldolase with SNX9 versus WASP, and the role of Trp-165 of SNX9 in the interaction, the binding affinity was assayed using a 7-mer peptide corresponding to WASP residues 496–502. The 7-mer amino acid sequence of LC4 (165WDEDWDG171) is homologous with the WASP 7-mer peptide (EDDEWDD). The double-reciprocal plots indicate competitive inhibition of aldolase activity by the WASP 7-mer peptide (Fig. 4E), consistent with active binding as observed in the crystal structure of the aldolase·WASP complex (27). However, the dissociation constants established that the WASP peptide interaction is considerably weaker (31-fold) than that of LC4 peptides (Table 3). Thus, the additional glutamic acid residue in the WASP peptide appears to impair binding and the tryptophan residue is important for binding of both SNX9 as well as WASP to aldolase. A random sequence peptide (DNYEFDG) used as a negative control displayed mixed inhibition kinetics (Fig. 4F), indicating inhibition of aldolase activity through very weak nonspecific binding (38), consistent with its millimolar dissociation constants (Table 3). Thus, the specific and competitive inhibition by the LC4 and WASP peptides demonstrate that they indeed specifically and functionally target the active site of aldolase.
DISCUSSION
The functions of aldolase in glycolysis and gluconeogenesis have been known for over 70 years, yet its non-catalytic roles as a scaffold protein that binds to many proteins to coordinate cell membrane processes with the actin and microtubule networks are only recently appreciated. The finding that pools of free, inactive aldolase interact with this cast of proteins suggests active roles for this enzyme in these processes, but the precise roles of these interactions are generally not clear. An exception is aldolase-mediated control of the SNX9·dynamin-2 complex that controls the final step of endocytosis, the fission of clathrin-coated pits (5). Here there is a clear mutual antagonism between aldolase and AP-2 in binding to and controlling the locale, and thus the activity, of the SNX9·dynamin-2 complex (11).
Our crystal structure of the LC4 motif of human SNX9 in complex with aldolase explains both the biochemistry and the biology of the aldolase·SNX9 interaction. First, the interactions of the LC4 motif of SNX9 with aldolase occur in overlapping regions immediately adjacent to the active site of the enzyme, explaining the competitive nature of the binding of SNX9 and the substrate and its products of aldolase. Here SNX9 binds to a hydrophobic pocket adjacent to where FBP binds in the active site and steric clashes prevent mutual binding; thus, the SNX9-bound aldolase complex must be catalytically inactive. Furthermore, the LC4 affinity interaction for aldolase (Table 3) is comparable with FBP (39). Thus, the SNX9·aldolase complex is likely stable until the cell receives strong metabolic cues that ATP via glycolysis is needed.
Second, the binding of SNX9 to aldolase, like several other described interactions with the enzyme (8, 13–19), involves interactions mediated in part by tryptophan residues on the aldolase binding partner. For SNX9 these interactions are directed by Trp-169. Importantly, Trp-169 of SNX9 is also required for its binding to AP-2 at clathrin-coated pits (9), thus explaining the mutual antagonism of aldolase and AP-2 in binding to SNX9, as well as the ability of aldolase to also abolish membrane and clathrin binding of SNX9 (11). Mutational analyses have suggested that the double mutant of the two tryptophan residues found in the LC4 motif of SNX9, Trp-165 and Trp-169, are required for binding to both AP-2 and aldolase (9, 11). Accordingly, binding of AP-2 and aldolase to SNX9 is mutually exclusive. However, in both studies the effects of mutation of the individual tryptophan residues were not addressed. The roles of these two tryptophans are clearly delineated by our aldolase·SNX9 structure, which establishes that Trp-169 contributes to aldolase binding, whereas SNX9 Trp-165 stabilizes the helical turn at SNX9 Trp-169 through its intrapeptide interaction with SNX9 Glu-167. Our structure refines this notion, in that for aldolase this residue does not directly participate in the interaction. Rather, stabilization of the helical turn by LC4 Trp-165 facilitates entry of SXN9 Trp-169 into the hydrophobic binding pocket of aldolase. The loss of the indole side chain in the alanine-modified 7- and 20-mer peptides confer greater disorder in solution, augmenting entropy loss and decreasing binding affinity (Table 3). This scenario is also supported by the even weaker affinity of the WASP peptide, where the indole side chain is replaced by an acidic side chain and where the peptide lacks a helical turn in the crystal structure. These findings suggest that Trp-165 is important to enhance the binding affinity of LC4 but is not essential for enforcing binding to aldolase. We cannot, however, rule out that mutation of this residue perturbs proper folding and/or expression of SNX9.
Third, the significantly stronger interaction of aldolase with SNX9 versus WASP suggests that the interactions of aldolase with the SNX9·dynamin-2 complex that control endocytosis are preferred over that of its interactions with WASP to regulate actin dynamics. The sequence alignment of WASP with LC4 (Fig. 2B) shows that SNX9 Trp-165 is an aspartate residue in WASP. This charged carboxylate moiety cannot participate in hydrogen bond formation with another aspartate or glutamate residue in WASP to stabilize a helical turn at Trp-500. Indeed, in the crystal structure of WASP in complex with aldolase, this equivalent charged residue is disordered (27). Thus, the presence of the additional Trp-165 residue locally orders LC4 configuration inducing helical turn formation at Trp-169 that promotes the extensive active site binding and reduces the entropic penalty incurred upon binding. Finally, the aspartate residues of SNX9 that interact with the Arg-42 and Arg-303 of aldolase are swapped when compared with the binding modes of WASP (Fig. 2B) and TRAP (19). This fact, and Trp-165 function as a hydrogen bond donor, provides the molecular mechanisms that distinguish SNX9 and WASP binding.
Finally, the crystal structures of aldolase in complex with SNX9 versus WASP establish that different modes of interactions can occur between aldolase and its binding partners, suggesting considerable plasticity of the enzyme in its role as a scaffolding protein. Indeed, the aldolase·SNX9 interaction involves a totally different cast of electrostatic interactions, and a host of new hydrophobic interactions and hydrogen bonding that are not evident in the WASP·aldolase interaction (Fig. 2B).
The signal(s) that disrupts the interaction of aldolase with the SNX9·dynamin-2 complex is not clear. At one level this may be directed by effective displacement of aldolase by AP-2 in situ at the cell membrane. Indeed, aldolase has been observed at localized domains in the cell membrane (40, 41), suggesting exchange of these two partners of the SNX9·dynamin-2 complex. However, at the cell membrane active aldolase interacts with caveolin-1 and is found at cholesterol- and glycosphingolipid-rich caveolae (12). If such a scenario applies, it would seem more likely that caveolin-1 disrupts the interactions of aldolase with the SNX9·dynamin-2 complex to facilitate its localization to caveolae.
Alternatively, phosphorylation of SNX9 in its LC4 domain may trigger displacement of the aldolase·SNX9·dynamin-2 complex. Indeed, phosphorylation of LC4 has been hypothesized to disable SNX9·aldolase interactions and direct recruitment of the SNX9·dynamin-2 complex to clathrin-coated pits (11). The effects of the tyrosine phosphatase inhibitor orthovanadate support this notion (11), yet there are no data indicating that SNX9 is differentially tyrosine phosphorylated in resting versus active complexes, nor is it clear which of the three serines or one tyrosine residue present in the LC4 motif are phosphorylated in vivo.
The fact that aldolase interacts with both SNX9 and the actin nucleator WASP suggests a potential role for the enzyme in coordinating endocytosis and actin dynamics. Indeed, these are linked processes where regulation of actin nucleation by the Arp2/3 complex by proteins such as WASP contributes to the formation of endocytic vesicles, as well as to their movement in the cytoplasm following fission by the SNX9·dynamin-2 complex. The scaffolds provided by aldolase are predicted to facilitate these interactions, where the homotetrameric enzyme may simultaneously bind to both of these partners, thus providing micro-compartments poised for executing endocytosis.
Acknowledgments
We are indebted to John Cleveland (Scripps, FL) for discussions and critical review of the manuscript and Andrew Sharff and Gerard Bricogne (Global Phasing Ltd.) for help with BUSTER refinements. We thank Dr. Miguel St-Jean (University of Montréal) for generously providing purified rabbit muscle aldolase for the structural studies. We are grateful to the staff at the Advanced Photon Source, SER-CAT, for synchrotron support.
This work was supported, in whole or in part, by a National Institutes of Health grant (to T. I.). This is Publication Number 20157 from The Scripps Research Institute.
- SNX9
- sorting nexin 9
- AP-2
- adaptor protein complex-2
- FBP
- fructose 1,6-bisphosphate
- LC4
- low complexity domain 4
- SH3
- Src homology 3 domain
- WASP
- Wiskott-Aldrich syndrome protein.
REFERENCES
- 1.Worby C. A., Dixon J. E. (2002) Nat. Rev. Mol. Cell Biol. 3, 919–931 [DOI] [PubMed] [Google Scholar]
- 2.Alto N. M., Weflen A. W., Rardin M. J., Yarar D., Lazar C. S., Tonikian R., Koller A., Taylor S. S., Boone C., Sidhu S. S., Schmid S. L., Hecht G. A., Dixon J. E. (2007) J. Cell Biol. 178, 1265–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Håberg K., Lundmark R., Carlsson S. R. (2008) J. Cell Sci. 121, 1495–1505 [DOI] [PubMed] [Google Scholar]
- 4.Mayer B. J. (2001) J. Cell Sci. 114, 1253–1263 [DOI] [PubMed] [Google Scholar]
- 5.Soulet F., Yarar D., Leonard M., Schmid S. L. (2005) Mol. Biol. Cell 16, 2058–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hinshaw J. E. (2000) Annu. Rev. Cell Dev. Biol. 16, 483–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmid S. L., McNiven M. A., De Camilli P. (1998) Curr. Opin. Cell Biol. 10, 504–512 [DOI] [PubMed] [Google Scholar]
- 8.Worby C. A., Simonson-Leff N., Clemens J. C., Kruger R. P., Muda M., Dixon J. E. (2001) J. Biol. Chem. 276, 41782–41789 [DOI] [PubMed] [Google Scholar]
- 9.Lundmark R., Carlsson S. R. (2003) J. Biol. Chem. 278, 46772–46781 [DOI] [PubMed] [Google Scholar]
- 10.Collins B. M., McCoy A. J., Kent H. M., Evans P. R., Owen D. J. (2002) Cell 109, 523–535 [DOI] [PubMed] [Google Scholar]
- 11.Lundmark R., Carlsson S. R. (2004) J. Biol. Chem. 279, 42694–42702 [DOI] [PubMed] [Google Scholar]
- 12.Raikar L. S., Vallejo J., Lloyd P. G., Hardin C. D. (2006) J. Cell. Biochem. 98, 861–871 [DOI] [PubMed] [Google Scholar]
- 13.Wang J., Morris A. J., Tolan D. R., Pagliaro L. (1996) J. Biol. Chem. 271, 6861–6865 [PubMed] [Google Scholar]
- 14.Kusakabe T., Motoki K., Hori K. (1997) Arch. Biochem. Biophys. 344, 184–193 [DOI] [PubMed] [Google Scholar]
- 15.Volker K. W., Knull H. (1997) Arch. Biochem. Biophys. 338, 237–243 [DOI] [PubMed] [Google Scholar]
- 16.Navarro-Lérida I., Martínez Moreno M., Roncal F., Gavilanes F., Albar J. P., Rodríguez-Crespo I. (2004) Proteomics 4, 339–346 [DOI] [PubMed] [Google Scholar]
- 17.Steck T. L. (1978) J. Supramol. Struct. 8, 311–324 [DOI] [PubMed] [Google Scholar]
- 18.Kim J. H., Lee S., Kim J. H., Lee T. G., Hirata M., Suh P. G., Ryu S. H. (2002) Biochemistry 41, 3414–3421 [DOI] [PubMed] [Google Scholar]
- 19.Bosch J., Buscaglia C. A., Krumm B., Ingason B. P., Lucas R., Roach C., Cardozo T., Nussenzweig V., Hol W. G. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7015–7020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kao A. W., Noda Y., Johnson J. H., Pessin J. E., Saltiel A. R. (1999) J. Biol. Chem. 274, 17742–17747 [DOI] [PubMed] [Google Scholar]
- 21.Buscaglia C. A., Coppens I., Hol W. G., Nussenzweig V. (2003) Mol. Biol. Cell 14, 4947–4957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schindler R., Weichselsdorfer E., Wagner O., Bereiter-Hahn J. (2001) Biochem. Cell Biol. 79, 719–728 [PubMed] [Google Scholar]
- 23.Morris A. J., Tolan D. R. (1993) J. Biol. Chem. 268, 1095–1100 [PubMed] [Google Scholar]
- 24.Penhoet E., Kochman M., Valentine R., Rutter W. J. (1967) Biochemistry 6, 2940–2949 [DOI] [PubMed] [Google Scholar]
- 25.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 26.Vagin A., Teplyakov A. (1997) J. Appl. Cryst. 30, 1022–1025 [Google Scholar]
- 27.St-Jean M., Izard T., Sygusch J. (2007) J. Biol. Chem. 282, 14309–14315 [DOI] [PubMed] [Google Scholar]
- 28.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 29.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 30.Blanc E., Roversi P., Vonrhein C., Flensburg C., Lea S. M., Bricogne G. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2210–2221 [DOI] [PubMed] [Google Scholar]
- 31.Bricogne G. (1997) Methods Enzymol. 276, 361–423 [DOI] [PubMed] [Google Scholar]
- 32.Smart O. S., Brandl M., Flensburg C., Keller P., Paciorek W., Vonrhein C., Womack T. O., Bricogne G. (2008) Annual Meeting Americal Crystallography Association, Knoxville, TN, May 31 – June 5, 2008, Abstr. TP139, ACA Publications, Buffalo, NY [Google Scholar]
- 33.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Cryst. 26, 283–291 [Google Scholar]
- 34.Racker E. (1947) J. Biol. Chem. 167, 843–854 [PubMed] [Google Scholar]
- 35.Gill S. C., von Hippel P. H. (1989) Anal. Biochem. 182, 319–326 [DOI] [PubMed] [Google Scholar]
- 36.Leatherbarrow R. J. (2007) GraFit, version 6, 6.0 Ed., Erithacus Software Limited, Horley, UK [Google Scholar]
- 37.Buscaglia C. A., Penesetti D., Tao M., Nussenzweig V. (2006) J. Biol. Chem. 281, 1324–1331 [DOI] [PubMed] [Google Scholar]
- 38.Mathews C. K., van Holde K. E., Ahern K. G. (2000) Biochemistry, 3rd Ed., pp. 383–388, Addison-Wesley Publishing Co., San Francisco, CA [Google Scholar]
- 39.Midelfort C. F., Gupta R. K., Rose I. A. (1976) Biochemistry 15, 2178–2185 [DOI] [PubMed] [Google Scholar]
- 40.Campanella M. E., Chu H., Low P. S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 2402–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chu H., Low P. S. (2006) Biochem. J. 400, 143–151 [DOI] [PMC free article] [PubMed] [Google Scholar]