Abstract
Surfactant protein A (SP-A) plays a role in lung innate immunity and surfactant-related functions. Two functional genes, SP-A1 (SFTPA1) and SP-A2 (SFTPA2), are present in humans and primates (rodents have one gene). Single gene SP-A1 or SP-A2 proteins expressed in vitro are functional. To study their role in vivo, we generated humanized transgenic (hTG) C57BL/6 mice, SP-A1(6A4) and SP-A2(1A3). The SP-A cDNA in experimental constructs was driven by the 3.7-kb SP-C promoter. Positive hTG mice were bred with SP-A knock-out mice to generate F8 offspring for study. Epithelial alveolar type II cells were SP-A-positive, and Clara cells were negative by immunohistochemistry in hTG mice. The levels of SP-A in lungs of two hTG lines used were comparable with those in human lungs. Southern blot analysis indicated that two cDNA copies of either SP-A1(6A4) or SP-A2(1A3) were integrated as a concatemer into the genome of each of the two hTG lines. Electron microscopy analysis revealed that hTG mice with a single SP-A1(6A4) or SP-A2(1A3) gene product lacked tubular myelin (TM), but hTG mice carrying both had TM. Furthermore, TM was observed in human bronchoalveolar lavage fluid only if both SP-A1 and SP-A2 gene products were present and not in those containing primarily (>99.7%) either SP-A1 or SP-A2 gene products. In vivo rescue study confirmed that TM can only be restored after administering exogenous SP-A containing both SP-A1 and SP-A2 into the lungs of SP-A knock-out mice. These observations indicate that SP-A1 and SP-A2 diverged functionally at least in terms of TM formation.
Keywords: Gene/Transgene, Genetics/Human, Pulmonary Surfactant Apoproteins, Electron Microscopy (EM), Pathogen-associated Molecular Pattern (PAMP), Surfactant Protein A, Tubular Myelin, Expression of Transgene, Humanized Transgenic Mouse
Introduction
Surfactant protein A (SP-A),2 a member of the C-type lectin protein family, plays an important role in the innate host defense and surfactant-related functions (1, 2). SP-A knock-out (KO) mice, either in the NIH Swiss black strain (3, 4) or in the C57BL/6 background (5, 6), have shown altered surfactant structure and function. These mice exhibited a phenotype deficient in the formation of the extracellular structural form of surfactant, the tubular myelin (TM) (3, 4), as well as increased susceptibility to infection by a variety of pathogens (6–10) and to environmental pollutant factors, such as ozone (11, 12). These findings indicate that SP-A plays a critical role in surfactant structure, and in the first line of host defense in the lung.
The human SP-A (hSP-A) locus consists of two functional genes, SFTPA1 (SP-A1) and SFTPA2 (SP-A2), and a pseudogene. Each of the functional genes has been shown to have several genetic variants (i.e. SP-A1 (6A, 6A2, 6A3, and 6A4) and SP-A2 (1A, 1A0, 1A1, 1A2, 1A3, and 1A5)); these are observed at greater than 1% frequency in the population (13). Human SP-A is expressed in alveolar epithelial type II cells (14) and in tracheal and bronchial submucosal gland cells in lung as well as in other tissues (15–18). SP-A1 and SP-A2 are expressed in the type II cells; SP-A2 is expressed in human tracheal and bronchial submucosal gland cells (15, 18). The presence of SP-A in nonciliated bronchiolar epithelial cells has been inconsistent for human lungs (14, 19, 20), but consistent positive staining of SP-A has been observed in the nonciliated bronchiolar epithelial cells in rodents (21, 22). Although a 2:1 ratio of SP-A1 to SP-A2 was suggested in a model of SP-A oligomerization by Voss et al. (23), experimental data have indicated a range beyond the proposed 2:1 ratio at the mRNA (24) and protein levels (25). The latter showed that the ratio of SP-A1 to total SP-A in bronchoalveolar lavage (BAL) fluid varies as a function of age and health status of the lung (25).
Studies with SP-A1 and SP-A2 variants from in vitro expressed cell systems have demonstrated functional differences between SP-A1 and SP-A2, with SP-A2 variants exhibiting higher activity than SP-A1 variants in numerous assays. These include differences in their ability to stimulate THP-1 cells to produce TNF-α (26–28), inhibit surfactant secretion (29), enhance bacterial phagocytosis by rat and human alveolar macrophages (30–32), and bind carbohydrates (33). The pattern of oligomerization also differs between SP-A1 and SP-A2; oligomers of SP-A1 variants were identified with higher sizes compared with those of SP-A2 under non-reducing conditions (29, 34). One major contributor to the differences observed appears to be amino acid 85 present in the collagen-like domain, where SP-A1 has a cysteine and SP-A2 has an arginine. Site-directed mutagenesis experiments (35) revealed that replacement of the Cys85 of SP-A1 with arginine or the Arg85 of SP-A2 with cysteine was sufficient to reverse the pattern of SP-A oligomerization and the functional activity of both mutants of SP-A1 and SP-A2 (35). Differences have been observed between SP-A1 and SP-A2 in their ability to induce phospholipid aggregation (34) as well as in the characteristics of phospholipid monolayers with surfactant protein B (SP-B) (36). The latter indicates that differences may exist in the interaction of SP-B and human SP-A variants.
In vitro studies indicated that SP-A is an essential component for the formation of a surfactant-specific lattice-like structure, named tubular myelin (TM) (37, 38). Through mixing SP-A and lipids in the presence of SP-B, a lattice-like TM structure could be produced, and the structure and dimensions of TM were similar to those of natural TM found in pulmonary alveoli (37). The role of SP-A in TM formation could not be replaced by other surfactant protein, such as SP-D or SP-C (37–39). Furthermore, two distinct SP-A populations (APP I and APP II) from BAL of a patient with alveolar proteinosis were separated by gel filtration chromatography (40). The APP I and APP II consisted primarily of multimeric and octadecameric forms, respectively, and exhibited distinct effects in the in vitro formation of TM with lipids (APP II fraction formed TM but not APP I) (41), indicating that a certain oligomeric form of SP-A is required for a lattice-like TM structure. SP-A was found to be localized at the corners of the TM lattice (42). Although the physiological importance of TM is not clear under either health or disease, an increase of TM-associated SP-A was recently observed to correlate with improved lung preservation in ischemia/reperfusion lung injury (43). However, until the present study, it was unknown whether either SP-A1 or SP-A2 or both are necessary for TM formation.
Humanized mouse techniques have provided a powerful tool in the study of many human biological processes and in the preclinical testing (44, 45). Humanized mice include two types of mice: 1) immunodeficient mice engrafted with human hematopoietic cells and tissues and 2) mice that transgenically express human gene(s) (i.e. humanized transgenic (hTG) mice) (44). hTG mouse models have helped overcome a severe limitation of the study of human biology in vivo. By means of the hTG mouse method, physiological and pathological functions of human genetic gene/variants (alleles) could be investigated in complex biological processes, so that in vivo difference (even subtle) on phenotypes caused by the transgenic gene/allele can be elucidated (46–48). Thus, hTG mice may be an ideal in vivo system to study biologically functional differences of human SP-A1 and SP-A2.
SP-A KO mice have been generated by two independent groups, either in the NIH Swiss black strain (3) or in the C57BL/6 background (5, 6). These SP-A KO mice have been used for investigation of overexpressing the rat SP-A gene (49) and truncated rat SP-A (4). Information from these studies indicated that the collagen-like domain of rat SP-A is required for the correction of structural and functional defects of surfactant in the SP-A KO mouse (4). Collectively, these studies have provided both the technical and biological basis for generating hTG SP-A1 and SP-A2 mice described in the present study.
In the present study, we investigated the hypothesis that human SP-A1 and SP-A2 exhibit differences in their biological effects in vivo with emphasis on surfactant structure. Toward this goal, we first generated humanized TG in the wild-type (WT) background of the C57BL/6 and then backcrossed these with the SP-A KO C57BL/6 mice to obtain mice that were negative for mouse SP-A, and each was positive for the human SP-A1(6A4) or SP-A2(1A3) cDNA or positive for both human SP-A1(6A4) and SP-A2(1A3). These hTG mice were characterized with regard to the copy number of the transgene, expression of the transgene SP-A1 or SP-A2, oligomerization of SP-A1 and SP-A2 in vivo, and functional effects of the transgene with regard to their role in the tubular myelin (TM) formation. Furthermore, by studying surfactant large aggregates from human BAL fluid, we confirmed a novel observation regarding TM formation via the study of this humanized transgenic mouse model.
MATERIALS AND METHODS
Mice and Animal Husbandry
Wild-type C57BL/6 mice used in this study were purchased from the Jackson Laboratory (Bar Harbor, ME). SP-A KO mice were kindly provided by Dr. Samuel Hawgood (University of California, San Francisco) and propagated in the animal core facility of the Pennsylvania State University College of Medicine. These SP-A KO mice contained the C57BL/6 background after a more than 10-generation backcross with C57BL/6, although the original SP-A KO founder was created with F1 BL6/129Sv embryonic stem cells (5, 6). hTG SP-A1 and SP-A2 mice generated in the present study were on the SP-A KO C57BL/6 background (see below). Mice used in this study were handled using protocols approved by the institutional animal care and use committee at the Pennsylvania State University College of Medicine. All mice were maintained in bubble facilities under pathogen-free conditions or barrier containment facilities.
Constructs for Generation of hTG SP-A1 and SP-A2 Mice
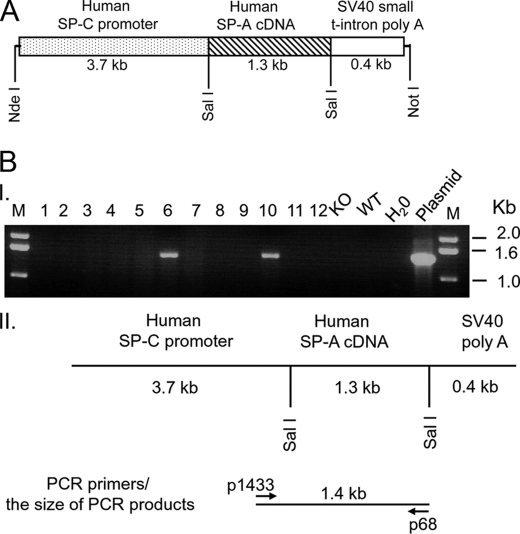
A 5.4-kb DNA fragment shown in Fig. 1A was used for DNA microinjection. This DNA fragment was excised from a recombinant plasmid with two restriction enzymes, NotI and NdeI, and consisted of a 3.7-kb human SP-C promoter, 1.3-kb human SP-A cDNA (i.e. either SP-A1(6A4) or SP-A2(1A3)), and a 0.4-kb SV40 small t-intron poly(A) sequence. In order to prepare this recombinant construct, we used a basic 3.7-hSP-C/SV40 vector, which was kindly provided by Drs. Stephan W. Glasser and Jeffrey A. Whitsett (Cincinnati Children's Research Foundation, Cincinnati, OH) (50). It has been demonstrated that the vector could drive alveolar type II cell-specific expression of the transgene in mice (4, 50–53). Because two restriction enzyme sites (NotI and NdeI) were chosen to excise the 5.4-kb DNA fragment from the recombinant construct, the restriction enzyme site of either NotI and NdeI in human SP-A cDNA sequence had to be removed before the human SP-A cDNA fragment was cloned into the vector. Thus, first, the NotI site was eliminated in the coding region of hSP-A cDNA using a site-directed mutagenesis (Stratagene, La Jolla, CA) with primers 1244/1245, and then with primers 1246/1247, the NdeI site of the hSP-A cDNA sequence was eliminated according to a previous method (35). The primers used in this study are shown in Table 1. These mutations only changed one nucleotide at each site of either NotI or NdeI and did not change the amino acid of hSP-A. Following site-directed mutagenesis, an hSP-A cDNA fragment (1.3 kb), containing about 0.06 kb of 5′-UTR, 0.74 kb of SP-A coding region, and 0.5 kb of 3′-UTR, was amplified through PCR with primers 1242/1243, and the PCR products were then digested with SalI. The 1.3-kb hSP-A cDNA fragment was cloned into 3.7SP-C/SV40 vector. Recombinant constructs were sequenced, and the orientation of human SP-A cDNA was confirmed by DNA sequencing. The processes of recombinant DNA were carried out according to standard molecular cloning methods.
FIGURE 1.
Construct of the recombinant DNA and identification of positive transgenic mice. A, diagrammatic representation of the recombinant DNA fragment used for the generation of hTG mice. The original vector (3.7-hSP-C/SV40) contained a fragment that included the 3.7 kb of the human SP-C promoter region and a 0.4-kb fragment of the SV40 poly(A) sequence. This vector has been used previously for foreign gene expression in alveolar epithelial type II cells in TG mice. The cDNA of human SP-A variants was first cloned into a pGEM vector, and prior to subcloning of the human SP-A cDNA into the SalI site of the 3.7-hSP-C/SV40 vector, two restriction sites (NotI and NdeI) within the coding region of the hSP-A cDNA were eliminated using site-directed mutagenesis (Stratagene, CA). A single nucleotide of the restriction enzyme recognition motif of either NotI or NdeI was changed, but the amino acid of SP-A was not changed. Then the 1.3-kb cDNA of hSP-A2(1A3) or SP-A1(6A4), containing a 0.1-kb 5′-UTR, the entire coding region of human SP-A (0.74-kb), and 0.5-kb of the 3′-UTR, was inserted into the SalI site of the 3.7hSP-C/SV40 vector. The 5.4-kb DNA fragment (3.7-kb SP-C promoter region, 1.3-kb cDNA of hSP-A, and the 0.4-kb SV40 poly-A terminator) was isolated and purified from the construct with both restriction enzymes NdeI and NotI. The pure DNA fragment was used for microinjection into fertilized C57BL/6 oocytes. B, identification of positive hTG mice by PCR. Positive hTG mice were identified using PCR with genomic DNA extracted from a tail fragment of potential hTG mice. The top section (I) of B shows the results from one representative experiment in which DNAs from 12 potential hTG mice along with the respective negative and positive control DNAs were analyzed. Positive hTG mice (lanes 6 and 10) exhibit a 1.4 kb band of PCR products with primer pair 1433 and 68, whereas the SP-A KO and WT mice lack this PCR band. M, DNA molecular marker; lanes 1–12 show 12 DNA samples from potential hTG mice; KO, SP-A KO mouse DNA; WT, wild-type mouse DNA; H2O, negative control for PCR (H2O was used instead of DNA template); plasmid, positive control for PCR (the plasmid used for positive control is the recombinant plasmid used for the generation of the hTG mouse). The bottom section (II) of B depicts a diagram of the transgene and location of primers used for PCR as well as the size of the PCR products. Primers 1433 (sense) and 68 (antisense) are located on the human SP-C promoter region and the human SP-A coding region, respectively.
TABLE 1.
Primers used in this study
Lowercase type in primers 1242 and 1243 indicates an additional sequence present in the given primers. This sequence is the recognition site of restriction enzyme SalI. Underlined letters in primers 1244, 1245, 1246, and 1247 indicate the changed nucleotides compared with hSP-A cDNA sequences.
| Primer | Sequence (5′ to 3′) | Comments |
|---|---|---|
| 1458 | GGAGGGCCAGGAACAAACAGG | Human SP-C promoter, sense |
| 1459 | CCAGGAGTGCCGGGGATACCAG | Human SP-A exon I, antisense |
| 1433 | CACATATAAGACCCTGGTCACACCT | Human SP-C promoter, sense |
| 292 | CCATTATTCCCAGGAGGACTAGGTG | Human SP-A2 antisense at exon II |
| 293 | CCATCATTTCCAGGAGGACTAGGCA | Human SP-A1 antisense at exon II |
| 68 | CAGGTCGACTGCCACAGAGACCTCAGAGT | Human SP-A antisense at 3′-UTR |
| 1549 | CCTGTCTGGTAGCAAGTAGAGCT | Mouse SP-A sense |
| 1542 | CCAGTGACATCGGGAGGAAATCC | Mouse SP-A antisense |
| 1280 | GTGGGGTGGGATTAGATAAATGC | Mouse SP-A for SP-A (−/−) KO mouse |
| 1242 | GGCCCGGGgtcgacTGGAGGCTCTGTGTGTGGG | Human SP-A, sense with SalI site |
| 1243 | GGCCCGGGgtcgacTGCCACAGAGACCTCAGAGT | Human SP-A, antisense with SalI site |
| 1244 | ATGTGCCAGAGCAGGAGGCCGCATTGCTGTC | Human SP-A, sense with a mutated nucleotide |
| 1245 | GACAGCAATGCGGCCTCCTGCTCTGGCACAT | Human SP-A, antisense with a mutated nucleotide |
| 1246 | GTGAAGAAGTACAACACCTATGCCTATGTAGGC | Human SP-A, sense with a mutated nucleotide |
| 1247 | GCCTACATAGGCATAGGTGTTGTACTTCTTCAC | Human SP-A, antisense with a mutated nucleotide |
| 1634 | CCTTGCCCCGTGCCTATGAG | Human SP-C 5′-flanking region, antisense |
| 1636 | TGAGCAACTCTGGCCAAGCATAA | Human SP-A 3′-UTR, sense |
Generation of hTG Mice by Microinjection into Fertilized Oocytes
The generation of hTG mice was carried out via contractual agreement in the transgenic mouse facility of the University of California, Irvine. DNA was prepared for microinjection into fertilized oocytes as follows. A 5.4-kb DNA fragment (see Fig. 1A) was excised from the construct with both restriction enzymes NotI and NdeI. The DNA fragment was subjected to electrophoresis of lower melting agarose gel, and the appropriate DNA band at the gel was purified according to the protocol for DNA to be used for the generation of hTG mice. The quality and quantity of DNA was examined by an agarose gel and QC 2100 bioanalyzer method, and DNA was diluted to 20 ng/μl. Fertilized oocytes were prepared from wild-type C57BL/6 mice. The DNA was directly microinjected into fertilized C57BL/6 oocytes, and the injected oocytes were immediately delivered into the uterus of WT C57BL/6 mice. The transgenic status of the pups was determined by genotyping analysis of DNA from mouse tails.
Genotyping Analysis of the Human Transgenes, hSP-A1(6A4) and SP-A2(1A3), as Well as the Mouse SP-A Gene
For identification of positive hSP-A TG mice, genomic DNA was extracted from mouse tail (about 0.6 cm) according to the protocol of the Qiagen DNeasy tissue kit (Qiagen, Valencia, CA). The quality of the genomic DNA (5 μl) was assessed by the pattern seen on 0.8% agarose gel electrophoresis. λDNA/HindIII digest was used as a molecular marker. Only high quality genomic DNA was used further for genotyping. Several PCR amplifications were performed with total genomic DNA and appropriate primers (see Table 1), and the PCR products were examined by 1 or 2% agarose gel electrophoresis to determine whether the potential TG mice were positive or negative for the transgene. For example, primer pair 1458/1459 was used to screen transgenic mice for both SP-A1 and SP-A2, and primer pairs 1433/292 and 1433/293 were used to identify SP-A2(1A3)- and SP-A1(6A4)-specific TG mice, respectively. The primers used are shown in Table 1. A positive TG mouse had one DNA band of PCR products of about 340 bp in a 2% agarose gel. For the above three primer pairs, the PCR conditions were as follows: 1) 94 °C for 2 min; 2) 35 cycles at 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 1 min; and 3) the final extension step at 72 °C for 7 min.
To examine whether intact 1.3-kb hSP-A cDNA was integrated in the genome of potential transgenic mice, the primer pair 1433/68 for PCR was utilized. This PCR product (about 1.35 kb) should contain the entire hSP-A cDNA as well as a partial region of the human SP-C promoter. The conditions for PCR amplification were as follows: 94 °C for 2 min and 35 cycles at 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 1.5 min and then the final extension step at 72 °C for 7 min. Positive PCR results indicated that the genome of the TG mouse contained an intact hSP-A cDNA, and the 1.35-kb DNA PCR products were confirmed as hSP-A sequence by sequencing. The conditions for the above PCR amplification were as follows. The 50-μl reaction buffer contained 35.25 μl of distilled H2O, 2 μl of mouse DNA, 1 μl of primer 1, and 1 μl of primer 2, 2.5 μl of PCR 10× buffer 1, 2.5 μl of PCR 10× buffer 2, 5 μl of dNTP, and 0.75 μl of Taq enzyme. Four controls were also included in every PCR series: 1) DNA from the SP-A knock-out mouse, 2) DNA from wild-type mouse, 3) recombinant plasmid DNA used for the generation of the TG mice, 4) water as negative control (no DNA).
Because TG founders were generated in the WT C57BL/6 background (i.e. mouse SP-A (+/+)), these TG founders and their hSP-A positive offspring were backcrossed with SP-A KO mice to generate a mouse background of SP-A KO. Therefore, offspring from these backcrossing breeding have two types of mouse SP-A background (i.e. mouse SP-A KO or heterozygous mouse SP-A (+/−)). Two PCRs were used to identify these two mouse backgrounds using different primer pairs at the following conditions: 94 °C for 2 min; 33 cycles at 94 °C for 30 s, 61 °C for 30 s, and 72 °C for 33 s; and then the final extension step at 72 °C for 5 min. Primer pairs a (1549/1542) and b (1280/1542) were used for identification of SP-A (+/+) WT mice and SP-A KO background, respectively. The SP-A (+/+) wild-type mouse with primers 1549/1542 showed a band of 290 bp, the SP-A KO mouse had a 520 bp band, and the heterozygous SP-A (+/−) mouse exhibited both bands of 290- and 520-bp PCR products.
Western Blot Analysis
To examine the level of SP-A1 and SP-A2 expression in lung and other tissues of TG mice, lung and other tissues from PCR-positive TG mice were homogenized and analyzed by Western blot analysis. To determine secreted hSP-A in the lung of the TG, the lungs were lavaged, and hSP-A in the BAL fluid was detected by Western blot analysis, using a specific SP-A antibody as described previously (29). In brief, total protein (100 μg/lane) from lung tissues of TG mice and SP-A KO mice as well as BAL protein (15 μg per lane) from TG mice was subjected to gel electrophoresis (10% SDS-PAGE or 4–20% gradient SDS-PAGE) under reducing conditions. The protein in the gel was transferred onto polyvinylidene difluoride membrane. SP-A was detected using a rabbit antibody (IgG) to human SP-A at a 1:2000 dilution and then goat anti-rabbit IgG (horseradish peroxidase-conjugated) antibody. The blot was exposed to XAR film following enhanced chemiluminescent detection. Human SP-A protein from the BAL of a patient with alveolar proteinosis was used as a positive control in the immunoblotting assay. Endogenous mouse SP-B, SP-C, and SP-D proteins were detected in the BAL fluid of hTG mice (n = 3 for each type of hTG mice) and KO mice (n = 3) using commercial SP-B, SP-C, and SP-D antibodies, respectively (Santa Cruz Biotechnology, Inc., Santa Cruz, CA).
Lung Morphology and Immunohistochemistry with hSP-A Antibody and SP-A1-specific Antibody
Lungs from 12-week-old TG mice, SP-A1(6A4), SP-A2(1A3), and SP-A2/SP-A1(1A3/6A4), were fixed at 25 cm of water pressure with Bouin's solution (Sigma) for 24 h. For human lung fixation, Bouin's solution was first delivered into a lobe of human lung, and after 30 min, the whole lobe of lung was transferred into a container with the same fix solution for 24 h. Lung tissues were then transferred into 70% ethanol solution and processed into paraffin blocks. About 5-μm-thick sections were prepared from these blocks with the fixed lung tissues and were stained with hematoxylin and eosin. Immunohistochemical analysis using an ABC kit (Vector Laboratories, Inc., Burlingame CA) was performed with a rabbit polyclonal antibody (purified IgG) against human SP-A protein or a chicken polyclonal antibody (purified IgY) to SP-A1-specific peptides (25). In addition, SP-C antibody (Santa Cruz Biotechnology, Inc.) was used to detect type II cells of lung tissues.
Southern Blot and Hybridization
To study the copy number of the transgene, total DNA was extracted and purified from the tails of experimental hTG and control mice and human lung tissue with the Wizard genomic DNA purification kit (Promega, Madison, WI). Eight μg of pure DNA of each sample were digested overnight at 37 °C by restriction enzyme EcoRI, and the digested DNA samples were subjected to an 0.8% agarose gel electrophoresis. Then the DNA fragments in the gel were transferred onto a nylon membrane for Southern blot analysis. Two DNA probes (0.6 and 1.1 kb) amplified from human SP-A cDNA of SP-A2(1A3) were used for DNA hybridization in order to detect TG DNA. Probes were labeled with [32P]dATP by a DNA probe labeling kit (Stratagene, La Jolla, CA), and the labeled probe was then added into Hybrisol I solution (Chemicon, Temecula, CA). Southern blot hybridization analysis was carried out in a rolling tube with the Hybrisol I solution at 48 °C overnight. The blot membrane was exposed to XAR film after washing three times (15 min each) at 55 °C.
Analysis of Human Genomic DNA Sequences from the Human Genome Data Base
Human genomic DNA sequences (about 15 kb) containing SP-A1 and SP-A2 were downloaded from the National Institutes of Health human genome data base. The sequences of each SP-A1 and SP-A2 were analyzed by the MapDraw and MegAlign programs of DNAstar to determine recognition sites of EcoRI and the resulting fragments by Southern genomic blot analysis to predict both the size and the number of bands on the Southern blot of human genomic DNA used as control.
Analysis of Total Phospholipids of Mouse BAL Fluid
hTG, SP-A1(6A4), SP-A2(1A3), SP-A1/SP-A2(6A4/1A3), KO, and WT mice were killed, and the lung was lavaged three times, each time with 0.5 ml of saline buffer. The BAL was centrifuged at 150 × g to remove cells, and the supernatant was used for analysis. The content of total phospholipids in the BAL fluid from mice (n = 5 per type of mouse) was determined quantitatively by an in vitro phospholipids C assay kit according to the manufacturer's protocol (Wako Diagnostics, Richmond, VA).
Preparation of Surfactant Large Aggregates from Mouse BAL Fluid
BAL fluid was obtained from hTG mice (n = 5) carrying a single gene, SP-A1(6A4) or SP-A2(1A3), and both SP-A genes SP-A2/SP-A1(1A3/6A4) as well as from the same genetic background SP-A KO and WT mice. To generate a sufficient amount of the large aggregate fraction for the electron microscopy assay, BAL fluids from five mice were pooled for each sample studied. Large aggregates were prepared from BAL using sucrose gradient centrifugation, and the large aggregate fraction of surfactant was collected from the interface following centrifugation (53).
Preparation of Surfactant Large Aggregates from Human BAL Fluid
Human BAL fluids were prepared from non-diseased human lungs (n = 5) obtained from the Gift of Life Donor Program (Philadelphia, PA) and stored at −80 °C from a previous study (25). The protocol was approved by the Penn State College of Medicine Institutional Review Board. Each donor lung (routinely the left lung) was lavaged with 1 liter of 0.9% saline, and the lavage fluid was collected. BALs were centrifuged at 150 × g for 10 min to obtain cell-free BAL. The ratios of SP-A1 to total SP-A of these BAL samples had been assessed with SP-A1-specific and SP-A antibodies by an enzyme-linked immunosorbent assay in our previous work (25). BAL samples containing either a high (i.e. 0.999) (n = 1) or a low (i.e. 0.003) (n = 2) SP-A1/total SP-A ratio or a ratio of about 0.5 (n = 2) were used for preparation of surfactant large aggregates in this study (53). The ratio was used as a tool to select BAL fluids with potentially very high SP-A1 content and a very low SP-A2 and vice versa as well as select samples where both SP-A1 and SP-A2 products were readily present. BAL fluid was separated at 8% sucrose gradient centrifugation with 40,000 × g at 4 °C for 30 min. The fraction of large aggregates of BAL was removed from the interface to a new tube and resuspended in saline buffer. Pellets of large aggregates were obtained from centrifugation with 40,000 × g at 4 °C for 10 min (53) and fixed using fixing buffer with 2.5% paraformaldehyde, 4% glutaraldehyde.
In Vivo Study of TM “Rescue” in SP-A KO Mice
SP-A KO mice (10–12 weeks old) were used for in vivo TM rescue experiments because SP-A KO mice lack TM structures in the lung. To test whether or not exogenous SP-A is able to restore TM formation in vivo, pure SP-A (2.5 or 5 μg) in 50 μl of saline containing 2 mm Ca2+ was administered into the lung of SP-A KO mice intratracheally. One negative control used 50 μl of saline with 2 mm Ca2+ but without SP-A. Three SP-A KO mice were used for studies of each condition, or a total of 36 mice for this in vivo study. For SP-A1, due to protein sample limitation, we tested only the 6 h time point and used 5 μg of SP-A1 protein/mouse. The mice were killed at 6 and 12 h after treatment, and the lung tissues were fixed immediately with Karnovsky's fixative solution for 24 h. The fixed lung tissues were used for analysis by electron microscopy (see “Electron Microscopy Analysis”). To determine TM structures in the lung tissues of these mice, at least 1000 fields from several sections of each mouse were examined.
Electron Microscopy Analysis
Pellets from the large aggregate fraction of hTG mice were prepared from mice with a single gene, SP-A1(6A4) and SP-A2(1A3), and both SP-A1/SP-A2(6A4/1A3) genes as well as from the same genetic background SP-A KO and WT mice. Pellets from the large aggregate fraction were also prepared from BAL fluid of human lungs. Samples were fixed with 2.5% paraformaldehyde, 4% glutaraldehyde. After 24 h, the fixed pellets were used for electron microscopy. Samples were stained with 1% osmium tetroxide, 1.5% potassium ferrocyanide. The fixed pellets were embedded in Embed812 resin (Electron microscopy science). Ultrathin sections (90 nm) were prepared and stained with 2% aqueous uranyl acetate and lead citrate for electron microscopy analysis. Each sample was examined at various magnifications from ×10,000 to ×35,000. For each sample, at least 200 fields from several sections were examined. The photographs were taken from 10 randomly selected grids (of 50 grids) of each section. The random number generator program was used to select the 10 grids.
Statistical Analysis
The data were analyzed by one-way analysis of variance or Student's t test using the standard program software Sigmastat (Version 3.5, SPSS). Differences were considered statistically significant at p < 0.05.
RESULTS
Generation of hTG SP-A1 and SP-A2 Mice and Transmission of SP-A1 and SP-A2 in hTG Mice
Before beginning the DNA microinjection into mouse fertilized oocytes, we examined whether the constructs could express hSP-A protein in H441 cells by transient transfection as described previously (54). Western blotting results from transfected cells indicated that the two constructs, SP-A1(6A4) and SP-A2(1A3), could express hSP-A protein in H441 cells that express SP-A but not in CHO cells that do not express SP-A, indicating the ability of constructs to express SP-A in a cell-specific manner (data not shown). Then the 5.4-kb DNA fragment (Fig. 1A) was used for microinjection into fertilized oocytes, which were then transferred into the uterus of C57BL/6 mice. Initially, we used fertilized oocytes from SP-A KO C57BL/6 mice, but these failed to generate positive TG mice, as assessed by genotyping of more than 150 potential TG mice. Therefore, fertilized oocytes from WT C57BL/6 mice were used. In this study, four positive TG founder mice were obtained for each of the SP-A1(6A4) and SP-A2(1A3) constructs from a total of 56 potential TG mice from three independent microinjection experiments. This represents an about 14% success rate in obtaining positive TG mice. An example of the genotyping analysis with 12 potential TG mice is shown in Fig. 1B. Of the 12 mice, two mice (numbers 6 and 10) carrying the 1.4 kb PCR band were positive for the transgene; the other 10 mice (without the 1.4-kb PCR products) were all negative, as also were the negative controls (WT and KO mice) (Fig. 1B).
To remove the endogenous mouse SP-A background and to study transmission of transgene, four TG founder mice of each SP-A1(6A4) and SP-A2(1A3) construct were backcrossed with SP-A KO C57BL/6 mice to generate F1 generation. Of the four SP-A1(6A4) founder mice, three (6A4-T1, -T3, and -T4) were positive TG F1 mice, and one 6A4-T2 (male) was infertile. All four SP-A2(1A3) founder mice were fertile when bred with SP-A KO mice, but only two founder mice (1A3-T2 and T4) were positive TG F1 mice. No positive TG F1 mice were obtained with the other two founder mice (1A3-T1 and T3) even after several breeding attempts. Positive transgenic F1 mice carrying either SP-A1(6A4) or SP-A2(1A3) were subsequently backcrossed with SP-A KO C57BL/6 mice to generate F2 mice. For F2 mice, in addition to genotyping human SP-A1(6A4) and SP-A2(1A3), the mouse SP-A background (i.e. heterozygous mouse SP-A (+/−) or homozygous mouse SP-A KO) was also analyzed as described under “Materials and Methods.” The processes of breeding and genotype analysis were performed through F7-F8 generations. Then hTG mice carrying either SP-A1(6A4) or SP-A2(1A3) with mouse SP-A KO background were characterized and used in the present study.
Expression of Human SP-A1 and SP-A2 and Mouse SP-B, SP-C, and SP-D in hTG Mice Examined by Western Blot Analysis
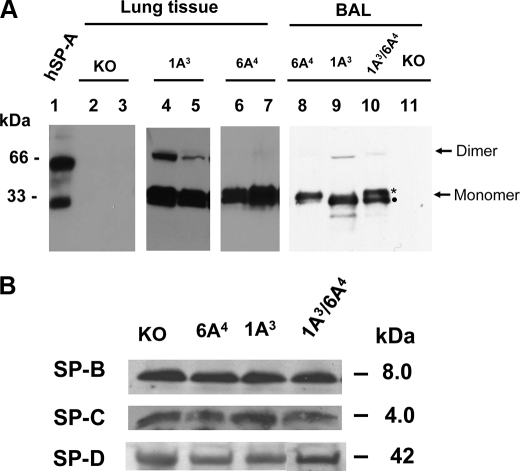
To examine transgene SP-A1(6A4) and SP-A2(1A3) expression in hTG mice, lung tissue and BAL fluid were prepared from hTG mice, SP-A1(6A4), SP-A2(1A3), and SP-A1/SP-A2(6A4/1A3), as well as negative control from the same litter of mice, and the samples were analyzed by Western blot with human SP-A antibody (27). The results showed that lung tissue and BAL fluid in the hTG mice have positive SP-A bands (major band of about 33 kDa (monomer) and a low intensity band of about 66 kDa (SP-A dimer) (Fig. 2A). The presence of SP-A protein in BAL indicated that human SP-A1 and SP-A2 protein could be secreted into alveolar space of the lung. As expected, no SP-A was detected in other tissues, such as heart, large intestine, and kidney (data not shown). Based on the signal on the Western blot, SP-A protein levels in the lung of two founder lines (T2 (lane 4) and T4 (lane 5)) of SP-A2(1A3) and one founder line (T1; two sublines of this line are shown in lanes 6 and 7) of SP-A1(6A4) were comparable with that of human lung. The levels of SP-A1(6A4) and SP-A2(1A3) protein expression in mice carrying both SP-A1 and SP-A2 genes are nearly equal; SP-A1(6A4) has a slightly larger size than SP-A2(1A3) when proteins are separated by 4–20% gradient SDS-PAGE (Fig. 2A, lane 10). The difference in the size between SP-A1 and SP-A2 precursors from in vitro translation has been observed previously (55). Furthermore, the mouse endogenous SP-B, SP-C, and SP-D expression in the hTG mice was similar to that in SP-A KO mice (Fig. 2B). Another two founder lines (T3 and T4) of SP-A1(6A4) had lower SP-A protein expression (data not shown).
FIGURE 2.
Western blot analysis of surfactant protein expression in the lung of hTG mice. A, hSP-A expression in the lung of hTG mice. Total protein (100 μg/lane) from lung tissues of hTG mice and SP-A KO mice or BAL protein (15 μg per lane) from hTG mice were subjected to gel electrophoresis (10% SDS-PAGE for lung tissues and 4–20% gradient SDS-PAGE for BAL fluid) under reducing conditions. The protein in the gel was transferred onto a polyvinylidene difluoride membrane, and SP-A was detected using a rabbit antibody (IgG) to human SP-A at a 1:2000 dilution and then a goat anti-rabbit IgG (horseradish peroxidase-conjugated) antibody. The blot was exposed to XAR film following enhanced chemiluminescent detection. Lung tissues were prepared from the following mice: two SP-A KO (lanes 2 and 3), two SP-A2(1A3) hTG mice (lanes 4 and 5), and two SP-A1(6A4) hTG mice (lanes 6 and 7). BAL fluids were isolated from the hTG mice carrying a single human SP-A transgene, i.e. SP-A1(6A4) (lane 8), SP-A2(1A3) (lane 9), or both human SP-A1 and SP-A2 transgenes, SP-A2/SP-A1(1A3/6A4) (lane 10) and from the control KO mouse (lane 11). Two bands with nearly equal intensity were observed at the position of monomeric SP-A in the BAL from the hTG mice carrying human SP-A1 and SP-A2 transgenes (SP-A2/SP-A1(1A3/6A4) (lane 10)). The band with the slightly larger size is SP-A1 (marked by an asterisk), and the other is SP-A2 (marked by a filled circle). All hTG mice were generated in the SP-A KO background. Purified human SP-A protein (lane 1) used as positive control was from the BAL of an alveolar proteinosis patient. Monomer and dimer of SP-A are pointed out by arrows on the right. B, SP-B, SP-C, and SP-D expression in the lung of hTG mice. Total BAL proteins (15 μg/lane) from hTG mice and SP-A KO mice were subjected to gel electrophoresis (4–15% gradient SDS-PAGE) under reducing conditions. The protein in the gel was transferred onto a polyvinylidene difluoride membrane; SP-B, SP-C, and SP-D were detected by SP-B, SP-C, and SP-D antibody, respectively. The blot was exposed to XAR film following enhanced chemiluminescent detection. No significant differences were observed in the levels of each SP-B, SP-C, and SP-D among hTG mice and KO mice.
Oligomerization Pattern of in Vivo Expressed SP-A1 and SP-A2
Oligomerization differences between SP-A1 and SP-A2 expressed in vitro in mammalian CHO and insect cells have been observed (26, 29, 34, 35). To examine whether the in vivo oligomeric pattern of SP-A1 differs from that of SP-A2, BAL fluid from hTG mice, SP-A1(6A4) and SP-A2(1A3) were analyzed by Western blot analysis (Fig. 3). Under non-reducing conditions, the SP-A2(1A3) oligomer pattern includes four major bands: dimer (2×), trimer (3×), tetramer (4×), and hexamer (6×), as well as several minor bands (>6×). The SP-A1(6A4) oligomer pattern differs from that of SP-A2(1A3) and consists of dimer (2×), trimer (3×), hexamer (6×), and several major bands larger than the hexamer (6×). These oligomer differences of SP-A1 and SP-A2 from the TG mice are similar to those observed previously by in vitro expressed SP-As (29, 35).
FIGURE 3.
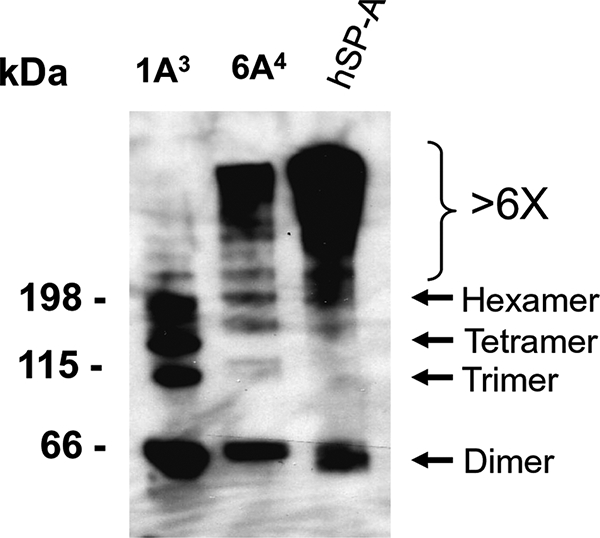
Oligomerization pattern of SP-A1 and SP-A2 in hTG mice as assessed by non-reducing immunoblot analysis. BAL fluid was prepared from hTG mice, SP-A2(1A3) and SP-A1(6A4), and then heated at 95 °C for 10 min in SDS-containing non-reducing buffer. About 15 μg of protein (per lane) was subjected to gel electrophoresis in a polyacrylamide gradient 4–15% gel under non-reducing conditions. The protein in the gel was transferred onto a polyvinylidene difluoride membrane, and SP-A bands on the membrane were detected using a primary rabbit antibody (IgG) to human SP-A and then a horseradish peroxidase-conjugated secondary antibody (goat anti-rabbit IgG). The blot was exposed to XAR film following enhanced ECL chemiluminescent detection. The oligomer pattern between SP-A1 and SP-A2 variants differed, as shown previously by in vitro expressed variants (29, 35). hSP-A protein from the BAL of an alveolar proteinosis patient was used as positive control. Molecular weight markers of protein sizes are shown on the left, and oligomeric forms are pointed out with arrows on the right.
Lung Morphology and Immunohistochemical Analysis of SP-A1 and SP-A2 Expression
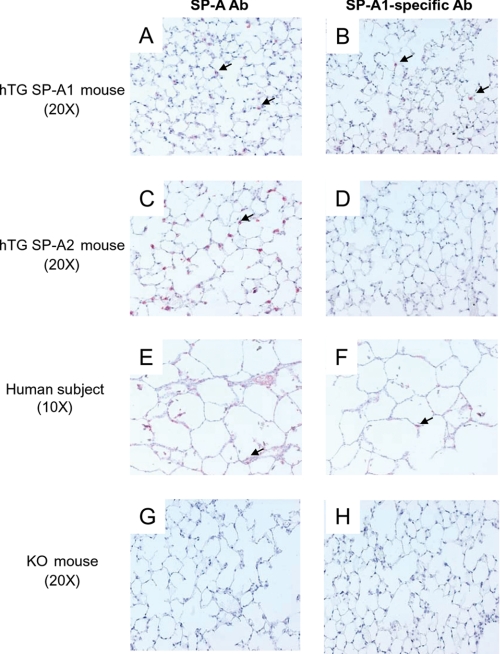
Lung morphology was examined in the hTG mice, SP-A1(6A4) and SP-A2(1A3), and control WT and SP-A KO mice. No obvious significant difference was observed in the alveolar structures of lung morphology, such as septal thickness or size of alveoli, among these mice. We further assessed the expression of human SP-A1 and SP-A2 in the hTG mice by immunohistochemical analysis using hSP-A- and SP-A1-specific antibodies. The results (Fig. 4) showed that alveolar type II cells were positive in tissues from hTG SP-A1 and SP-A2 mice with hSP-A antibody (A and C). With the SP-A1-specifc antibody, as expected, only tissue from the hTG SP-A1(6A4) mouse was positive (Fig. 4B); tissue from the hTG SP-A2(1A3) mouse was negative (D). Serving as a positive control, type II cells from human lung tissue were positive with either human SP-A antibody or SP-A1-specific antibody (Fig. 4, E and F) as well as an antibody to SP-C (type II cell-specific marker) (not shown). No positive signal was observed in SP-A KO mice by either SP-A antibody or SP-A1-specific antibody (Fig. 4, G and H). Furthermore, no SP-A1 and SP-A2 expression was detected in the epithelial Clara cells and airway of the hTG lung (data not shown). These data indicate that the 3.7-kb human SP-C drives alveolar type II cell-specific expression of human SP-A1 and SP-A2, consistent with similar observations made when this promoter was used to drive expression of other genes in TG mice (4, 53, 56).
FIGURE 4.
Immunohistochemical analysis with hSP-A- and SP-A1-specific Abs. Lung tissues from each hTG mouse, SP-A1(6A4) and SP-A2(1A3), SP-A KO mouse, and a human subject were fixed with Bouin's solution (Sigma) for 24 h. About 5-μm thick sections were prepared from these fixed lung tissues. Immunohistochemical analysis using an ABC kit (Vector Laboratories, Inc., Burlingame CA) was performed with human SP-A Ab (A, C, E, and G) and an SP-A1-specific Ab (B, D, F, and H). The results indicate that the human SP-A Ab stains alveolar type II cells of all three types of lung tissues (A, C, and E), but the SP-A1-specific Ab detects alveolar type II cells only of lung tissues from the hTG SP-A1(6A4) mouse (B) and the human subject (F) but not from the SP-A2 mouse (D). As expected, no positive alveolar type II cells were detected in the lung of the SP-A KO mouse with either SP-A Ab (G) or SP-A1-specific Ab (H). The arrows point to positive type II cells.
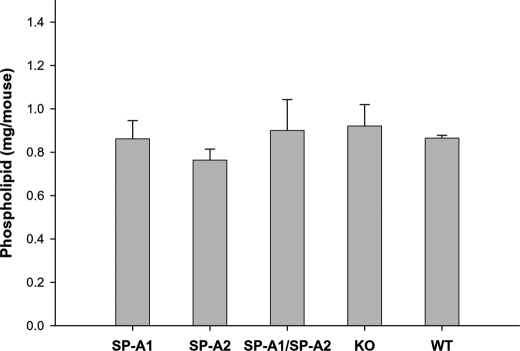
In addition, the content of total phospholipids in mouse BAL fluid was determined quantitatively. The data showed that there were no significant differences among hTG mice, SP-A1(6A4), SP-A2(1A3), SP-A1/SP-A2(6A4/1A3), WT, and KO mice (Fig. 5).
FIGURE 5.
Comparison of total phospholipids in the BAL fluid of hTG, KO, and WT mice. hTG mice (SP-A1(6A4), SP-A2(1A3), and SP-A1/SP-A2(6A4/1A3)), KO mice, and WT mice were killed, and the lung was lavaged three times, each time with 0.5 ml of saline buffer. The BAL was centrifuged at 150 × g to remove cells, and the supernatant was used for analysis. Each group contains five mice. The content of total phospholipids in the BAL fluid from each mouse was then determined by an in vitro phospholipids assay kit (Wako Diagnostics, Richmond, VA). The data were analyzed by a one-way analysis of variance test (significant differences were considered when p was <0.05). The results indicated no significant differences in the levels of total phospholipids of each group among hTG, KO, and WT mice.
Analysis of Integration of hSP-A1 and hSP-A in the Genome of hTG Mice
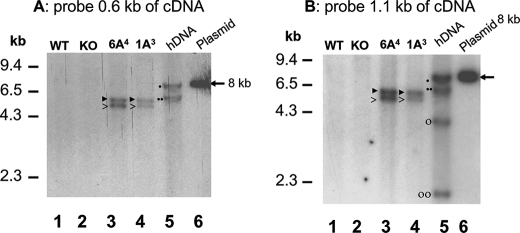
To determine how many copies of SP-A1 and SP-A2 were integrated into the genome of hTG mice, we used F8 hTG mice from two founder lines, SP-A1(6A4)-T1 and the SP-A2(1A3)-T2, for Southern blot and RFLP analyses. These two founder lines, as noted above, exhibited SP-A expression comparable with that in human lung. Total DNA extracted from the tails of hTG or control mice was digested (8 μg) by restriction enzyme EcoRI and subjected to Southern blot analysis. DNA fragments were detected with two cDNA probes (0.6 and 1.1 kb of human SP-A cDNA). When either probe was used, two bands at positions of 5.1 and 5.8 kb were detected (Fig. 6) in both hTG mice, SP-A1(6A4)-T1 and SP-A2(1A3)-T2, whereas no signal was detected in the Southern blot with either probe in DNAs from SP-A KO or WT mice. However, genomic DNA (8 μg) from human lung showed two bands with the 0.6-kb probe (Fig. 6A) and four bands with the 1.1-kb probe (Fig. 6B) on the Southern blot. The two probes used were from the cDNA sequence of human SP-A2(1A3), and as such, they could hybridize with several exons of human genomic SP-A1 and SP-A2 sequences, hence the multiple bands with human genomic DNA. The 0.6-kb probe is located within the coding region (exons I–IV) of SP-A cDNA. In this region, no EcoRI cleavage site exists in either SP-A cDNA or human genomic DNA. The 1.1-kb probe contains the coding region (exons I–IV) and a partial 3′-UTR sequence (present in exon IV) of SP-A cDNA and contains one EcoRI cleavage site. This site is present in the 3′-UTR of SP-A cDNA and genomic DNA.
FIGURE 6.
Southern blot analysis of transgene integration in hTG mice. Genomic DNAs were prepared from hTG mice (SP-A1(6A4) and SP-A2(1A3)), SP-A KO mice, and WT mice as well as from human lung tissue. The DNA samples were digested with EcoRI. The digested DNAs were subjected to agarose-gel electrophoresis and then transferred to a nylon membrane. The hSP-A1 or hSP-A2 DNA bands were detected with 32P-labeled DNA probes of 0.6-kb or 1.1-kb fragments of human SP-A cDNA. The linearized plasmid DNA (about 8 kb, pointed out by an arrow) carrying the hSP-A cDNA (described in the legend to Fig. 1) was used as a control. After hybridization with either probe, the blot membrane was washed three times (see “Materials and Methods”) and then exposed to XAR film. A and B depict the RFLP pattern of hSP-A as detected by the 0.6- and 1.1-kb probes from SP-A cDNA, respectively. The 0.6-kb probe is located within the coding region (exons I–IV) of SP-A cDNA. This region contains no EcoRI cleavage site in either SP-A cDNA or human genomic DNA. The 1.1-kb probe contains the coding region (exons I–IV) and a partial 3′-UTR sequence (present in exon IV) of SP-A cDNA with one EcoRI cleavage site. This site is in the 3′-UTR of SP-A cDNA and human genomic DNA. The data from A indicate that there are two DNA bands (5.8 and 5.1 kb, marked by ▶ and >, respectively) in each of the hTG mice, SP-A1(6A4) and SP-A2(1A3). The human genomic DNA (hDNA) also shows two DNA bands (7.7 and 6.3 kb, marked by ● and ●●, respectively); the 6.3-kb fragment is from SP-A1, and the 7.7-kb fragment is from SP-A2. The RFLP pattern of B with the 1.1-kb probe shows that the hTG mice (SP-A1(6A4) and SP-A2(1A3)) also contained two DNA bands (5.8 and 5.1 kb, marked by ▶ and >, respectively), as shown in A. Human genomic DNA contains four bands. Of these, the 6.3 kb and 4.3 kb bands (marked by ●● and ○, respectively) are from SP-A1, and the 7.7 kb and 2.2 kb bands (marked by ● and ○○, respectively) are from SP-A2.
To further understand the differences in RFLP pattern between the human genomic DNA and that from the hTG mice, human genomic sequences of SP-A1 and SP-A2 were downloaded from the NIH human genome data base (see “Materials and Methods”), and their EcoRI-digested pattern was determined. The results indicated that the bands of 6.3 and 7.7 kb detected by the 0.6-kb probe (Fig. 6A lane 5) were, respectively, from the SP-A1 and SP-A2 genomic sequences. The four bands detected in the human genomic EcoRI digestion with the 1.1-kb probe (Fig. 6B, lane 5) were from SP-A1 (bands 6.3 kb and 4.3 kb) and from SP-A2 (bands 7.7 kb and 2.2 kb).
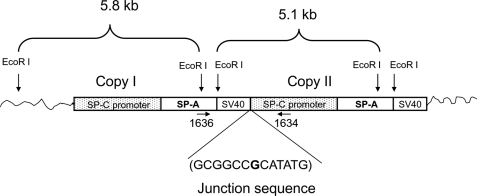
Because each of the hTG mice, SP-A2(1A3) and SP-A1(6A4), carried the cDNA sequence of human SP-A1 and SP-A2, respectively, as shown in Fig. 1, only one band should have been detected in the blot if the hTG mice contained one copy of the transgene of either SP-A2(1A3) or SP-A1(6A4). However, two bands (5.1 and 5.8 kb) were found in the hTG mice with both probes, raising the possibility that two copies of the transgene were integrated into the genome of the hTG mice. This could occur either via the formation of a concatemer of the transgene or integration of the transgene in two independent loci in the genome on the same or different chromosomes. The mice analyzed had been backcrossed with SP-A KO mice for 8 generations; therefore, it is unlikely that the transgene is integrated in two different chromosomes. The most likely scenario is that two copies of the transgene formed a concatemer structure or that the two copies integrated into the same chromosome but at different loci. To address this issue further, we first examined the possibility that there is a concatemer of SP-A2(1A3) or SP-A1(6A4) in the hTG mice. PCR fragments were produced using genomic DNA from the TG mice as template and two primers, one (sense oligonucleotide) located on the hSP-A 3′-UTR and the other (antisense oligonucleotide) on the 3.7-kb SP-C promoter region (Fig. 7). With these two primers, only concatemeric forms of each SP-A1 and SP-A2 transgene in the genome could be amplified to generate a PCR product of about 0.9 kb. DNAs from both TG mice, SP-A2(1A3) and SP-A1(6A4), exhibited an about 0.9-kb PCR product. Furthermore, we purified and sequenced these PCR products and found the junction sequence (GCGGCCGCATATG) of a two-copy transgene, which is the exact junction sequence at the end of the first copy and the start of the second copy (Fig. 7). The results from this specific PCR amplification and DNA sequencing support the conclusion that two copies of either transgene (SP-A1 or SP-A2) have integrated as a concatemer in each of the hTG mice.
FIGURE 7.
Analysis of concatemeric structure of the transgene and the junction sequence of transgenes in hTG mice. Southern blot analysis with either the 0.6-kb or the 1.1-kb probe indicated (as shown in Fig. 6) two DNA bands of about 5.8 and 5.1 kb in the hTG mice SP-A1(6A4) and SP-A2(1A3). These two bands are probably the result of a concatemer structure of the transgene. One 0.9-kb PCR DNA fragment could be generated with primers 1636 (sense) and 1634 (antisense) (data not shown). Because the sense primer 1636 is located in the 3′-UTR of the human SP-A gene and the antisense primer 1634 is on the flanking region of the human SP-C promoter, only a concatemer structure can produce the 0.9-kb PCR product. Sequencing analysis of the 0.9-kb PCR product revealed that two copies of the transgene form a concatemer structure. The junction sequence of the two copies of the transgene consists of a 3′-sequence (GCGGCC) of the first copy and a 5′-sequence (CATATG) of the second copy shown here. One additional base (G, shown in boldface type) was found between the two copies of the transgene.
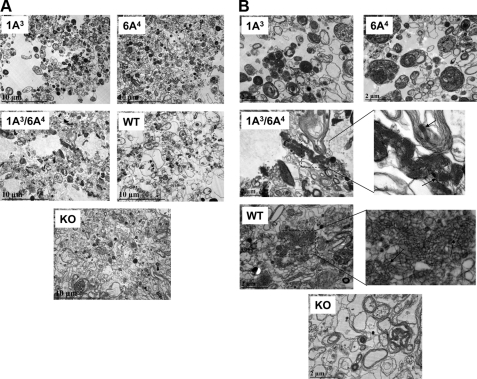
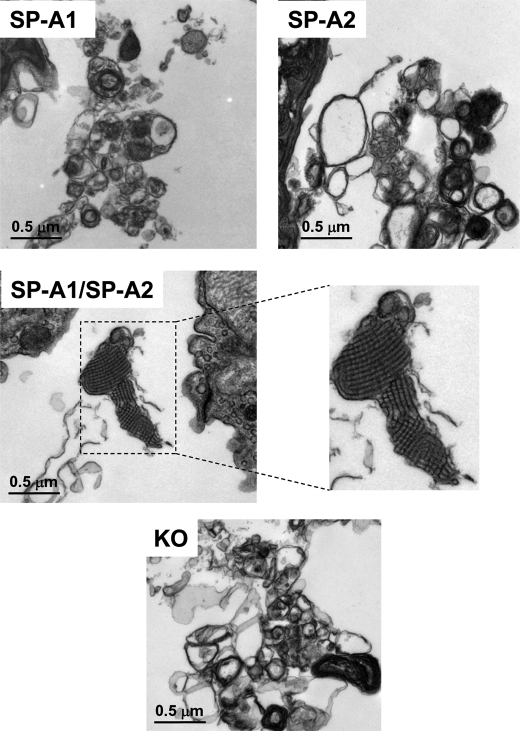
Ultrastructural Analysis of Surfactant Large Aggregates from BAL of hTG Mice; Both hSP-A Gene Products Are Necessary for Tubular Myelin Formation
SP-A KO mice cannot form TM, an extracellular form of surfactant (3, 4). To study whether a single gene product or both gene products of human SP-A1 and SP-A2 are necessary for the formation of TM, surfactant large aggregates isolated from BAL of hTG were studied by transmission electron microscopy for the presence of TM, a lattice-like ultrastructure. Large aggregates from BAL fluid of five mice from each of the five groups (i.e. hTG mice with a single SP-A cDNA, SP-A2(1A3) or SP-A1(6A4); hTG mice with two SP-A cDNAs, SP-A2/SP-A1(1A3/6A4); and control WT and KO mice) were prepared as described under “Materials and Methods,” fixed, and examined by transmission electron microscopy. Fig. 8 shows representative images at two different magnifications (×10,000 in A and ×30,000 in B). The results in Fig. 8A indicate that 1) hTG mice carrying both SP-A2/SP-A1(1A3/6A4) transgenes exhibit a similar pattern of surfactant large aggregates as WT mice, and 2) hTG mice with a single SP-A gene, SP-A2(1A3) or SP-A1(6A4), although they formed compact lamellar bodies, their pattern of surfactant large aggregates (Fig. 8B) differed from that of WT mice and hTG mice of SP-A2/SP-A1(1A3/6A4). SP-A1(6A4) hTG mice appeared to exhibit larger size lamellar bodies than SP-A2(1A3) hTG mice (Fig. 8B). The lattice-like structures of TM (Fig. 8B) were observed only in hTG mice of SP-A2/SP-A1(1A3/6A4) and WT mice (TM is shown in the enlargement and is marked by arrows) and not in hTG mice, expressing a single gene product, SP-A2(1A3) or SP-A1(6A4), or in SP-A KO mice, indicating that both SP-A1 and SP-A2 gene products are required for the formation of lattice-like forms of TM. TM appeared to be more regular in hTG mice of SP-A2/SP-A1(1A3/6A4) than in WT mice, as suggested previously (4).
FIGURE 8.
Ultrastructures of surfactant large aggregates from BAL of hTG mice. BAL fluid was obtained from hTG mice with a single gene, either SP-A1(6A4) or SP-A2(1A3), and both genes SP-A1/SP-A2(6A4/1A3) as well as from the same background SP-A KO and WT mice. To generate a sufficient amount of the large aggregate fraction for this assay, BAL fluids from five mice were pooled for each type of samples. Large aggregates were prepared from BAL using sucrose gradient centrifugation. The large aggregate fraction of surfactant was collected from the interface following centrifugation. Pellets of large aggregates were fixed with 2.5% paraformaldehyde and 4% glutaraldehyde. After 24 h, the fixed pellets were used for analysis by electron microscopy. A and B represent ultrastructures of each sample at magnifications of ×10,000 (A) and ×30,000 (B), respectively. In the single gene-containing hTG mice, SP-A1(6A4) and SP-A2(1A3), the lamellar bodies in the large aggregates appear more dense compared with KO mice (A), and tubular myelin figures are absent compared with WT mice (B). hTG mice SP-A1/SP-A2(6A4/1A3) carrying both genes have a similar pattern of ultrastructures of large aggregates as WT mice, including formation of a significant amount of tubular myelin figures marked by arrows (B) in the enlargement of the 1A3/6A4 and WT.
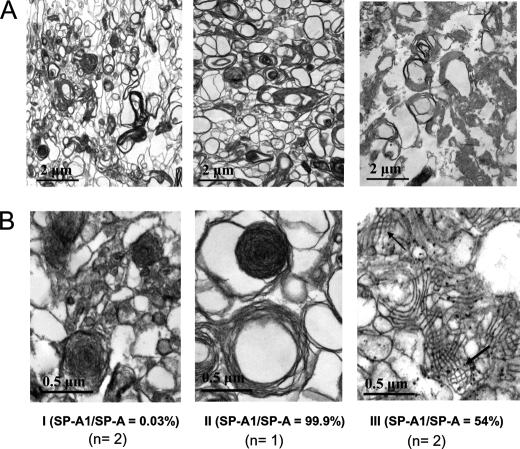
Ultrastructural Analysis of Surfactant Large Aggregates from Human BAL
To further confirm that TM formation in humans required both SP-A1 and SP-A2 gene products, we studied surfactant large aggregates from BAL fluid of human subjects by transmission electron microscopy for the presence of TM. Five human BAL samples were studied as described under “Materials and Methods.” These were selected based on the SP-A1/total SP-A ratio, which was assessed by an enzyme-linked immunosorbent assay in our previous study (25). Two of them had a very low SP-A1/SP-A = 0.003 ratio, one had a very high SP-A1/SP-A = 0.999 ratio, and the other two had a ratio of SP-A1/SP-A2 = 0.588 and 0.546, respectively. The ratio was used as a tool to identify BALs that could potentially have an overabundance of one gene product over the other and samples where both gene products were well represented. The results (Fig. 9) showed that TM lattice-like ultrastructures were observed only in samples containing both SP-A1 and SP-A2 gene products (TM structure is marked by an arrow in Fig. 9B, III). Samples that may contain predominantly either SP-A1 or SP-A2 products lacked TM formation, although large lamellar bodies as well as large size of loose lipid arrays were present in the large aggregates (Fig. 9). These results confirm the observation made with BALs from the hTG mice and together indicate that the in vivo formation of TM structure requires the presence of both human SP-A1 and SP-A2 proteins. This is the first evidence that shows a functional divergence between SP-A1 and SP-A2.
FIGURE 9.
Ultrastructures of surfactant large aggregates from BAL of human subjects. BAL fluid was obtained from human subjects where the ratio of SP-A1 to total SP-A was assessed in our previous work (25). Large aggregates of surfactant were prepared from BAL fluid of three types of individuals: with mostly SP-A2 protein present in the BAL fluid (i.e. SP-A1/SP-A = 0.003) (n = 2) (I); with mostly SP-A1 protein present in the BAL fluid (i.e. SP-A1/SP-A = 0.999) (n = 1) (II); and with both SP-A1 and SP-A2 present in the BAL fluid (n = 2) (III). Large aggregates were prepared from previously frozen BAL fluid using sucrose gradient centrifugation. The large aggregate fraction of surfactant was collected from the interface following centrifugation. Pellets of large aggregates were fixed with 2.5% paraformaldehyde and 4% glutaraldehyde. After 24 h, the fixed pellets were used for analysis by electron microscopy. A and B, ultrastructures of each sample at a magnification of ×10,000 (A) and ×30,000 (B), respectively. Tubular myelin figures were observed only in the samples containing both SP-A1 and SP-A2 gene products (III in B) and not in the ones containing either SP-A1 or SP-A2.
In Vivo Rescue TM Formation in the Lung of the SP-A KO Mice
To address two questions, 1) whether exogenous SP-A administered into mouse lung is able to restore TM formation in vivo and 2) whether there are different effects on the TM formation in vivo by the three types of SP-A (SP-A1, SP-A2, and SP-A1/SP-A2), SP-A KO mice that lack TM structure were used as the recipient in this in vivo study. SP-A (2.5 or 5 μg/mouse) in 50 μl of saline buffer with 2 mm Ca2+ was administered into the lung of SP-A KO mice intratracheally. Six or 12 h after treatment, the mice were killed, and the lung tissues were fixed and examined for TM formation by electron microscopy. The results indicated that intact lattice-like forms of TM (although not plentiful and rather scarce) were found only in samples at 6 h after treatment with 5 μg of exogenous SP-A consisting of both SP-A1 and SP-A2 and not in those treated with SP-A1 or SP-A2 alone (Fig. 10). As expected, no TM was found in the negative control KO mice, which were treated with only 50 μl of saline buffer.
FIGURE 10.
Ultrastructures of the lung tissues from in vivo TM rescue in SP-A KO mice. SP-A KO mice that lack TM structures were treated with exogenous SP-A protein. Fifty μl of saline buffer containing 2.5 or 5 μg of SP-A (one of the three types of SP-A (SP-A1, SP-A2, or SP-A1/SP-A2) described in the legend to Fig. 9) was administered into the lung of SP-KO mouse intratracheally. One negative control used 50 μl of saline without SP-A (see “Materials and Methods”). The mice were killed at 6 and 12 h after treatment, and the lung tissues were fixed immediately with Karnovsky's fixative solution for 24 h. The fixed lung tissues were used for analysis by electron microscopy. Ultrastructures of each sample (5 μg of SP-A and 6 h after treatment) were shown at a magnification of ×30,000. TM structures were observed in the lung tissues from the SP-A KO mice treated with SP-A containing both SP-A1 and SP-A2 gene products but not in those treated with SP-A1 or SP-A2 alone. As expected, no TM structure was found in the negative control.
DISCUSSION
SP-A plays important roles in surfactant structure and metabolism and in the innate host defense of the lung (1, 57). Human SP-A is encoded by two functional genes, SP-A1 and SP-A2 (13). Although human SP-A1 and SP-A2 share a high level of similarity (∼97.6%) at the amino acid sequences, structural and functional differences between in vitro expressed SP-A1 and SP-A2 variants have been observed (26–29, 31, 32, 34, 35, 58), with amino acid 85, one of the four “core” amino acids residues that distinguish SP-A1 from SP-A2, making a major contribution to these gene-specific differences (35). However, all of the observations from the in vitro studies about SP-A1 and SP-A2 structural and functional properties still lack in vivo documentation. In the present study, we generated and characterized hTG SP-A1 and SP-A2 mice and used them to study in vivo the role of SP-A1 and SP-A2 in surfactant structure. The hTG mice expressed SP-A at levels comparable with those in human lung, and the expressed SP-A1 and SP-A2 exhibited an oligomer size pattern similar to that observed by in vitro expressed SP-A1 and SP-A2 (29). The results further showed that both SP-A1 and SP-A2 gene products are necessary for the formation of TM, one of the extracellular forms of surfactant. The requirement of both gene products for TM formation in the humanized transgenic model was further confirmed with BAL fluid from human subjects in which SP-A consisted primarily of one or the other gene product, or both gene products were presumably well represented. Mouse TM rescue experiments provided further in vivo support that both SP-A gene products are necessary for TM formation. The inability of either single gene product to form TM and the need for both gene products for TM formation provide the first evidence either in vivo or in vitro of divergence of human SP-A1 and SP-A2 function.
Although the strategy of random integration of a foreign gene (DNA fragment) into the genome of fertilized oocytes is widely utilized for generation of transgenic mice (45), the random integration of foreign DNA can alter or damage expression of essential gene(s) of development and/or spermatogenesis, leading to male infertility. In the present study, three of eight founder mice were either infertile (male founder T-2 of SP-A1(6A4)) or could not produce positive transgenic offspring (male founders T-1 and T-3 of SP-A2(1A3)). Chimera transgenic founder mice may not carry the transgene in their spermatocytes, explaining therefore their inability to produce offspring that would be positive for the transgene, as observed in previous studies with different genes (59, 60). Of interest, at the initial stage of this project, we used fertilized oocytes from SP-A KO C57BL/6 mice as host of DNA microinjection but failed to obtain positive transgenic mice in more than 150 potential transgenic mice. The reasons for this are currently unknown. Although SP-A has been shown to play a role in parturition (61), SP-A KO mice do reproduce successfully. On the other hand, when fertilized oocytes from WT C57BL/6 were used, ∼14% of positive transgenic mice were produced, which is within the expected range. The mouse endogenous SP-A gene was subsequently eliminated through backcross with the SP-A KO mouse.
Foreign gene expression in transgenic mice is influenced by several factors, including 1) the type of promoter in the construct utilized, 5′-UTR, and 3′-UTR sequences; 2) the integration position of the transgenic DNA in the transgenic mouse; and 3) the number of copies of the transgene integrated in the genome (44, 45). In the present study, as well as in other studies, the use of the 3.7-kb human SP-C promoter sequence ensured alveolar type II cell-specific expression in lung (4, 50–53, 56). The two founder lines (T1 of SP-A1(6A4) and T2 of SP-A2(1A3)) analyzed in detail each contained two copies of the transgene, and each could express SP-A1 or SP-A2 protein at levels comparable with that of human lung tissues. Therefore, it is tempting to speculate that the presence of two copies is the reason why these mice expressed levels of SP-A similar to those in the human lung. However, an overexpression (7–8-fold higher) of another transgene driven by the same promoter has been observed previously (49). Although it is difficult to compare the levels of human SP-A1 and SP-A2 in hTG mice with the level of the mouse endogenous SP-A due to differences in antibody affinities for human and mouse SP-A, the similarity of the SP-A content in hTG mice and the human lung tissue indicates the appropriateness of these hTG mice for further in vivo study.
SP-A KO mice have been shown to lack TM, an extracellular form of surfactant (3, 4, 62). Although the physiological role of TM in the lung is not known, it has been postulated that TM is a precursor/reservoir of the surface-active film (63), and it may play a critical role in enhanced lung preservation and surfactant integrity in lung injury (43). The structure and formation of TM in vitro has been studied (37, 38, 64), demonstrating that SP-A is an essential component for this TM lattice-like structure of surfactant in vitro (37, 38). Moreover, the SP-A molecule in its entirety (4) and in the appropriate oligomeric structure (41) is required for the formation of TM, suggesting that the sequence and structure are necessary for this function of SP-A. However, the role (if any) that TM plays in the lung under normal or pathological conditions remains unknown. The present findings indicate that SP-A1 or SP-A2 alone is not sufficient to restore TM structure in vivo in the SP-A KO genetic mouse background and that only hTG mice carrying both SP-A1 and SP-A2 genes could restore TM structures. Furthermore, this observation was confirmed by an in vivo“rescue” study in which three types of SP-A (SP-A1, SP-A2, and SP-A1/SP-A2) were delivered into the lung of SP-A KO mice. Only SP-A containing SP-A1 and SP-A2 could restore TM formation. This may be due in part to the differences in oligomeric structures in human SP-As consisting of both gene products (SP-A1 and SP-A2) and those consisting of either SP-A1 or SP-A2 (29) and the potential requirement of certain SP-A multimeric/oligomeric structures for TM formation.
The need for both SP-A1 and SP-A2 gene products for TM formation was further confirmed in the present study by analysis of BAL fluids from human subjects with a ratio of SP-A1 to total SP-A either very high or very low. Currently, we have no direct knowledge as to what constitutes optimal SP-A1 and SP-A2 content in the lung and how deviation from this putative optimal content can impact surfactant structure and SP-A-related functions. However and of interest, in a recent study, the ratio of SP-A1 to total SP-A in BAL samples from human subjects was found to vary depending on the age and/or health status of individuals (25). A predominance of SP-A2 products in BAL fluid has also been observed in a single patient with alveolar proteinosis (65). Furthermore, it is unknown how a varied SP-A1 and SP-A2 expression affects TM content and its potential significance in lung health. Although several in vitro studies have shown between SP-A1 and SP-A2 variants differences in regulation (54, 66), in function (including their ability to enhance phagocytosis of pathogens by alveolar macrophages) (28–32, 35), and in their oligomerization pattern (26, 29, 35), the present study shows for the first time that SP-A1 and SP-A2 have diverged completely in terms of TM formation because neither one alone can form TM.
In summary, hTG mice have been generated in the SP-A KO background and characterized with each carrying two copies of SP-A1(6A4) or SP-A2(1A3) transgene and each expressing levels of SP-A similar to those in the human lung. The function of SP-A1 and S-A2 gene products has diverged with regard to TM formation. Neither one alone is sufficient to form TM; TM can only be detected if both gene products are present. Similar observations were made with BAL samples from human subjects that were identified predominantly with SP-A1 or SP-A2 or with both gene products being well represented and with mouse in vivo TM rescue experiments. Because alterations in SP-A levels have been associated with several pulmonary diseases, in view of the present in vivo studies and the structural and functional differences between SP-A1 and SP-A2 variants documented in several in vitro observations, it becomes necessary for the focus of study to shift to the study of the individual SP-A1 and SP-A2 levels rather than the total SP-A content. Such an approach provides the opportunity not only to make available markers that could be specific for a given lung disease but also to shed light on their individual contributions to disease pathogenesis. Toward this endeavor, the hTG model described and characterized in the present report provides a useful biological reagent to study in vivo key in vitro findings as well as providing a means that could enable the study of physiological and pathological SP-A-related disease processes that are difficult to investigate with study of human subjects.
Acknowledgments
We thank Drs. J. Whitsett and S. Glasser (Cincinnati Children's Research Foundation) and Dr. S. Hawgood (University of California, San Francisco) for providing the vector of 3.7-hSP-C/SV40 and SP-A KO mice, respectively. We also greatly appreciate Drs. D. Phelps and J. Shenberger for helpful discussions and Sanmei Hu for excellent technical support. We gratefully acknowledge the Gift of Life Donor Program (Philadelphia, PA) and the generosity of the organ donor families for allowing these organs that are not suitable for transplantation to be utilized to advance the understanding of human disease. We also acknowledge the core facilities of the Penn State Hershey College of Medicine for providing animal care, electron microscopy analysis, DNA sequencing, and oligonucleotide synthesis in the present study.
This work was supported, in whole or in part, by National Institutes of Health Grant ES09882.
- SP-A
- -B, -C, and -D, surfactant protein A, B, C, and D, respectively
- hSP-A and -C
- human SP-A and -C, respectively
- BAL
- bronchoalveolar lavage
- TG
- transgenic mouse
- hTG
- humanized transgenic mouse
- KO
- knock-out
- RFLP
- restriction fragment length polymorphism
- TM
- tubular myelin
- WT
- wild-type
- UTR
- untranslated region
- Ab
- antibody.
REFERENCES
- 1.Wright J. R. (2005) Nat. Rev. Immunol. 5, 58–68 [DOI] [PubMed] [Google Scholar]
- 2.Bates S. R., Dodia C., Tao J. Q., Fisher A. B. (2008) Am. J. Physiol. Lung Cell Mol. Physiol. 294, L325–L333 [DOI] [PubMed] [Google Scholar]
- 3.Korfhagen T. R., Bruno M. D., Ross G. F., Huelsman K. M., Ikegami M., Jobe A. H., Wert S. E., Stripp B. R., Morris R. E., Glasser S. W., Bachurski C. J., Iwamoto H. S., Whitsett J. A. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 9594–9599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ikegami M., Elhalwagi B. M., Palaniyar N., Dienger K., Korfhagen T., Whitsett J. A., McCormack F. X. (2001) J. Biol. Chem. 276, 38542–38548 [DOI] [PubMed] [Google Scholar]
- 5.Jain D., Dodia C., Bates S. R., Hawgood S., Poulain F. R., Fisher A. B. (2003) Am. J. Physiol. Lung Cell Mol. Physiol. 284, L759–L765 [DOI] [PubMed] [Google Scholar]
- 6.Li G., Siddiqui J., Hendry M., Akiyama J., Edmondson J., Brown C., Allen L., Levitt S., Poulain F., Hawgood S. (2002) Am. J. Respir. Cell Mol. Biol. 26, 277–282 [DOI] [PubMed] [Google Scholar]
- 7.LeVine A. M., Bruno M. D., Huelsman K. M., Ross G. F., Whitsett J. A., Korfhagen T. R. (1997) J. Immunol. 158, 4336–4340 [PubMed] [Google Scholar]
- 8.LeVine A. M., Kurak K. E., Bruno M. D., Stark J. M., Whitsett J. A., Korfhagen T. R. (1998) Am. J. Respir. Cell Mol. Biol. 19, 700–708 [DOI] [PubMed] [Google Scholar]
- 9.Giannoni E., Sawa T., Allen L., Wiener-Kronish J., Hawgood S. (2006) Am. J. Respir. Cell Mol. Biol. 34, 704–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mikerov A. N., Haque R., Gan X., Guo X., Phelps D. S., Floros J. (2008) Respir. Res. 9, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haque R., Umstead T. M., Ponnuru P., Guo X., Hawgood S., Phelps D. S., Floros J. (2007) Toxicol. Appl. Pharmacol. 220, 72–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haque R., Umstead T. M., Ahn K., Phelps D., Floros J. (2009) Pneumon. 22, 143–155 [PMC free article] [PubMed] [Google Scholar]
- 13.Floros J., Wang G., Mikerov A. N. (2009) Crit. Rev. Eukaryot. Gene Expr. 19, 125–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phelps D. S., Floros J. (1988) Am. Rev. Respir. Dis. 137, 939–942 [DOI] [PubMed] [Google Scholar]
- 15.Goss K. L., Kumar A. R., Snyder J. M. (1998) Am. J. Respir. Cell Mol. Biol. 19, 613–621 [DOI] [PubMed] [Google Scholar]
- 16.Lin Z., deMello D., Phelps D. S., Koltun W. A., Page M., Floros J. (2001) Pediatr. Pathol. Mol. Med. 20, 367–386 [PubMed] [Google Scholar]
- 17.MacNeill C., Umstead T. M., Phelps D. S., Lin Z., Floros J., Shearer D. A., Weisz J. (2004) Immunology 111, 91–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saitoh H., Okayama H., Shimura S., Fushimi T., Masuda T., Shirato K. (1998) Am. J. Respir. Cell Mol. Biol. 19, 202–209 [DOI] [PubMed] [Google Scholar]
- 19.Auten R. L., Watkins R. H., Shapiro D. L., Horowitz S. (1990) Am. J. Respir. Cell Mol. Biol. 3, 491–496 [DOI] [PubMed] [Google Scholar]
- 20.Ochs M., Johnen G., Müller K. M., Wahlers T., Hawgood S., Richter J., Brasch F. (2002) Am. J. Respir. Cell Mol. Biol. 26, 91–98 [DOI] [PubMed] [Google Scholar]
- 21.Phelps D. S., Floros J. (1991) Exp. Lung Res. 17, 985–995 [DOI] [PubMed] [Google Scholar]
- 22.Akiyama J., Hoffman A., Brown C., Allen L., Edmondson J., Poulain F., Hawgood S. (2002) J. Histochem. Cytochem. 50, 993–996 [DOI] [PubMed] [Google Scholar]
- 23.Voss T., Melchers K., Scheirle G., Schäfer K. P. (1991) Am. J. Respir. Cell Mol. Biol. 4, 88–94 [DOI] [PubMed] [Google Scholar]
- 24.Karinch A. M., deMello D. E., Floros J. (1997) Biochem. J. 321, 39–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tagaram H. R., Wang G., Umstead T. M., Mikerov A. N., Thomas N. J., Graff G. R., Hess J. C., Thomassen M. J., Kavuru M. S., Phelps D. S., Floros J. (2007) Am. J. Physiol. Lung Cell Mol. Physiol. 292, L1052–L1063 [DOI] [PubMed] [Google Scholar]
- 26.Huang W., Wang G., Phelps D. S., Al-Mondhiry H., Floros J. (2004) Am. J. Physiol. Lung Cell Mol. Physiol. 286, L546–L553 [DOI] [PubMed] [Google Scholar]
- 27.Wang G., Phelps D. S., Umstead T. M., Floros J. (2000) Am. J. Physiol. Lung Cell Mol. Physiol. 278, L946–L954 [DOI] [PubMed] [Google Scholar]
- 28.Wang G., Umstead T. M., Phelps D. S., Al-Mondhiry H., Floros J. (2002) Environ. Health Perspect. 110, 79–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang G., Bates-Kenney S. R., Tao J. Q., Phelps D. S., Floros J. (2004) Biochemistry 43, 4227–4239 [DOI] [PubMed] [Google Scholar]
- 30.Mikerov A. N., Umstead T. M., Huang W., Liu W., Phelps D. S., Floros J. (2005) Am. J. Physiol. Lung Cell Mol. Physiol. 288, L150–L158 [DOI] [PubMed] [Google Scholar]
- 31.Mikerov A. N., Wang G., Umstead T. M., Zacharatos M., Thomas N. J., Phelps D. S., Floros J. (2007) Infect. Immun. 75, 1403–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mikerov A. N., Umstead T. M., Gan X., Huang W., Guo X., Wang G., Phelps D. S., Floros J. (2008) Am. J. Physiol. Lung Cell Mol. Physiol. 294, L121–L130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oberley R. E., Snyder J. M. (2003) Am. J. Physiol. Lung Cell Mol. Physiol. 284, L871–L881 [DOI] [PubMed] [Google Scholar]
- 34.García-Verdugo I., Wang G., Floros J., Casals C. (2002) Biochemistry 41, 14041–14053 [DOI] [PubMed] [Google Scholar]
- 35.Wang G., Myers C., Mikerov A., Floros J. (2007) Biochemistry 46, 8425–8435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang G., Taneva S., Keough K. M., Floros J. (2007) Biochim. Biophys. Acta 1768, 2060–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams M. C., Hawgood S., Hamilton R. L. (1991) Am. J. Respir. Cell Mol. Biol. 5, 41–50 [DOI] [PubMed] [Google Scholar]
- 38.Poulain F. R., Allen L., Williams M. C., Hamilton R. L., Hawgood S. (1992) Am. J. Physiol. 262, L730–L739 [DOI] [PubMed] [Google Scholar]
- 39.Poulain F. R., Akiyama J., Allen L., Brown C., Chang R., Goerke J., Dobbs L., Hawgood S. (1999) Am. J. Respir. Cell Mol. Biol. 20, 1049–1058 [DOI] [PubMed] [Google Scholar]
- 40.Hattori A., Kuroki Y., Sohma H., Ogasawara Y., Akino T. (1996) Biochem. J. 317, 939–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hattori A., Kuroki Y., Katoh T., Takahashi H., Shen H. Q., Suzuki Y., Akino T. (1996) Am. J. Respir. Cell Mol. Biol. 14, 608–619 [DOI] [PubMed] [Google Scholar]
- 42.Voorhout W. F., Veenendaal T., Haagsman H. P., Verkleij A. J., van Golde L. M., Geuze H. J. (1991) J. Histochem. Cytochem. 39, 1331–1336 [DOI] [PubMed] [Google Scholar]
- 43.Fehrenbach H., Tews S., Fehrenbach A., Ochs M., Wittwer T., Wahlers T., Richter J. (2005) Respir. Res. 6, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shultz L. D., Ishikawa F., Greiner D. L. (2007) Nat. Rev. Immunol. 7, 118–130 [DOI] [PubMed] [Google Scholar]
- 45.Gonzalez F. J., Yu A. M. (2006) Annu. Rev. Pharmacol. Toxicol. 46, 41–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reaume A. G., Howland D. S., Trusko S. P., Savage M. J., Lang D. M., Greenberg B. D., Siman R., Scott R. W. (1996) J. Biol. Chem. 271, 23380–23388 [DOI] [PubMed] [Google Scholar]
- 47.Taneja V., David C. S. (1998) J. Clin. Invest. 101, 921–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scheer N., Ross J., Rode A., Zevnik B., Niehaves S., Faust N., Wolf C. R. (2008) J. Clin. Invest. 118, 3228–3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elhalwagi B. M., Zhang M., Ikegami M., Iwamoto H. S., Morris R. E., Miller M. L., Dienger K., McCormack F. X. (1999) Am. J. Respir. Cell Mol. Biol. 21, 380–387 [DOI] [PubMed] [Google Scholar]
- 50.Glasser S. W., Burhans M. S., Eszterhas S. K., Bruno M. D., Korfhagen T. R. (2000) Am. J. Physiol. Lung Cell Mol. Physiol. 278, L933–L945 [DOI] [PubMed] [Google Scholar]
- 51.Glasser S. W., Eszterhas S. K., Detmer E. A., Maxfield M. D., Korfhagen T. R. (2005) Am. J. Physiol. Lung Cell Mol. Physiol. 288, L625–L632 [DOI] [PubMed] [Google Scholar]
- 52.Glasser S. W., Korfhagen T. R., Wert S. E., Bruno M. D., McWilliams K. M., Vorbroker D. K., Whitsett J. A. (1991) Am. J. Physiol. 261, L349–L356 [DOI] [PubMed] [Google Scholar]
- 53.Kingma P. S., Zhang L., Ikegami M., Hartshorn K., McCormack F. X., Whitsett J. A. (2006) J. Biol. Chem. 281, 24496–24505 [DOI] [PubMed] [Google Scholar]
- 54.Wang G., Guo X., Floros J. (2005) Am. J. Physiol. Lung Cell Mol. Physiol. 289, L497–L508 [DOI] [PubMed] [Google Scholar]
- 55.Floros J., Steinbrink R., Jacobs K., Phelps D., Kriz R., Recny M., Sultzman L., Jones S., Taeusch H. W., Frank H. A. (1986) J. Biol. Chem. 261, 9029–9033 [PubMed] [Google Scholar]
- 56.Palaniyar N., Zhang L., Kuzmenko A., Ikegami M., Wan S., Wu H., Korfhagen T. R., Whitsett J. A., McCormack F. X. (2002) J. Biol. Chem. 277, 26971–26979 [DOI] [PubMed] [Google Scholar]
- 57.Floros J., Phelps D. S. (2002) in Surfactant-Update of Intensive Care Medicine (Nakos G., Lekka M. E. eds) Vol. 6, pp. 87–102, University of Ioannina, Ioannina, Greece [Google Scholar]
- 58.Mikerov A. N., Umstead T. M., Huang W., Liu W., Phelps D. S., Floros J. (2005) Am. J. Physiol. Lung Cell Mol. Physiol. 288, L150–L158 [DOI] [PubMed] [Google Scholar]
- 59.Fung-Leung W. P., De Sousa-Hitzler J., Ishaque A., Zhou L., Pang J., Ngo K., Panakos J. A., Chourmouzis E., Liu F. T., Lau C. Y. (1996) J. Exp. Med. 183, 49–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Albertelli M. A., Scheller A., Brogley M., Robins D. M. (2006) Mol. Endocrinol. 20, 1248–1260 [DOI] [PubMed] [Google Scholar]
- 61.Condon J. C., Jeyasuria P., Faust J. M., Mendelson C. R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4978–4983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ikegami M., Korfhagen T. R., Whitsett J. A., Bruno M. D., Wert S. E., Wada K., Jobe A. H. (1998) Am. J. Physiol. 275, L247–L254 [DOI] [PubMed] [Google Scholar]
- 63.Schmiedl A., Vieten G., Mühlfeld C., Bernhard W. (2007) Pediatr. Pulmonol. 42, 548–562 [DOI] [PubMed] [Google Scholar]
- 64.Froh D., Ballard P. L., Williams M. C., Gonzales J., Goerke J., Odom M. W., Gonzales L. W. (1990) Biochim. Biophys. Acta 1052, 78–89 [DOI] [PubMed] [Google Scholar]
- 65.Berg T., Leth-Larsen R., Holmskov U., Højrup P. (2000) Biochim. Biophys. Acta 1543, 159–173 [DOI] [PubMed] [Google Scholar]
- 66.Hoover R. R., Floros J. (1999) Am. J. Physiol. 276, L917–L924 [DOI] [PubMed] [Google Scholar]