Abstract
Lung cancer is the leading cause of cancer-related deaths worldwide. Here we show for the first time that HtrA3 is a mitochondrial stress-response factor that promotes cytotoxicity to etoposide and cisplatin in lung cancer cell lines. Exogenous expression of wild type HtrA3 domain variants significantly attenuated cell survival with etoposide and cisplatin treatment in lung cancer cell lines H157 and A549 compared with expression of protease inactive mutants (S305A) or vector control. Conversely, HtrA3 suppression promoted cell survival with etoposide and cisplatin treatment in lung cancer cell lines Hop62 and HCC827. Survival was attenuated by re-expression of wild type HtrA3 variants during treatment but not by protease inactive mutants or vector control. HtrA3 also co-fractionated and co-localized with mitochondrial markers with both endogenous and exogenous expression in normal lung and lung cancer cell lines but was translocated from mitochondria following etoposide treatment. Moreover, HtrA3 translocation from mitochondria correlated with an increase in cell death that was attenuated by either HtrA3 suppression or Bcl-2 overexpression. Taken together, these results suggest that HtrA3 may be a previously uncharacterized mitochondrial cell death effector whose serine protease function may be crucial to modulating etoposide- and cisplatin-induced cytotoxicity in lung cancer cell lines.
Keywords: Cancer Therapy, Cell Death, Mitochondrial Apoptosis, Serine Protease, Subcellular Fractionation, Chemoresistance, Chemotherapy, Cytotoxicity, HtrA3, Lung Cancer
Introduction
Lung cancer is the leading cause of cancer-related deaths in the United States and worldwide (1, 2). Standard first line treatment for small cell lung cancer includes platinum-based chemotherapy, generally with the DNA and protein cross-linking agent cis-platinum-(II)-diamine dichloride (cisplatin) or carboplatin plus etoposide, a topoisomerase II inhibitor, with or without radiation (3–6). Treatment of non-small cell lung cancer may include surgical resection, radiotherapy, and/or platinum-based chemotherapy (7–10). Despite a promising initial response to therapy, however, 86% of lung cancer patients die within five years of diagnosis from advanced or chemoresistant disease (2, 10–14).
Chemotherapeutic agents function by inhibiting vital cellular processes or by activating programmed cell death pathways (15). Mechanisms of chemoresistance include pharmacokinetic, tumor microenvironment, and cancer cell-specific changes that alter intracellular active drug concentrations, drug-target interactions, target-mediated cell damage, damage-induced cell death signaling, or the cell death effector machinery (15–17). That apoptotic cells can be found in cancers suggests that tumors in vivo may arrive at a unique equilibrium between high proapoptotic signals and high antiapoptotic activity (15). Thus, shifts in a finely tuned balance between survival and death pathways may be pivotal first steps in the etiology of chemoresistant disease (18).
HtrA3 (high temperature requirement A3) belongs to the HtrA family of stress-related serine proteases that is well conserved from bacteria to humans (19). We previously reported that HtrA1 binds microtubules and modulates cisplatin- and paclitaxel-induced cytotoxicity in ovarian and gastric cancers (18, 20–22). HtrA2 is localized to the mitochondrial intermembrane space and is an effector of the apoptotic pathway that has also been implicated in modulating cisplatin-induced cytotoxicity in renal cells (23–28). HtrA3 was first characterized as an up-regulated factor during mouse placental development (29).
In the present study, we show that HtrA3 is a nuclear encoded mitochondrial protease that modulates etoposide- and cisplatin-induced cytotoxicity in lung cancer cell lines by promoting cell death in a manner dependent on serine protease function. We show that both endogenously and exogenously expressed HtrA3 attenuate cell survival with etoposide and cisplatin treatment. Further, we show that HtrA3 co-fractionates and co-localizes with integral mitochondrial markers and is translocated from mitochondria, with HtrA2 and cytochrome c, following etoposide treatment. HtrA3 translocation corresponds to a statistically significant increase in cell death that is attenuated by either HtrA3 suppression or Bcl-2 overexpression. Collectively, our data suggest a crucial role for HtrA3 in modulating etoposide- and cisplatin-induced cytotoxicity in lung cancer cell lines, in part through a protease-dependent role in the apoptotic cell death pathway.
EXPERIMENTAL PROCEDURES
Cell Culture
The cell lines were cultured according to ATCC guidelines.
Cloning
The long HtrA3 isoform open reading frame was obtained from the Invitrogen Full-Length Mammalian Gene Collection and subcloned into pcDNA3.1(+)myc-His A and pEGFP-N1 (Clontech). Protease inactive mutants were generated by site-directed mutagenesis using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The oligonucleotides used for cloning are given in supplemental Table S1. The S-tagged Bcl-2 construct cloned into pSPN was obtained from Scott H. Kaufmann (30).
Immunoblotting
Immunoblotting was performed as previously described (18). The anti-HtrA3 monoclonal antibody was developed at the Mayo Antibody Core Facility Rochester and raised against a polypeptide corresponding to amino acids 144–453 of HtrA3. A polyclonal antibody (Abcam, Cambridge, MA) was used only for detection of endogenous HtrA3 by immunocytochemistry and for detection of PDZ-deleted HtrA3 variants. The S-tag antibody used for detection of exogenous Bcl-2 was provided by Scott H. Kaufmann (30). The antibody to human frataxin was provided by Grazia Isaya. The antibodies used for immunoblotting are listed in supplemental Table S2.
Densitometric Analysis
The immunoblots were analyzed by Bio-Rad Quantity One 4.40 software. The arbitrary intensity values were normalized to β-actin loading controls and expressed as fold changes over controls.
Immunostaining and Fluorescence Microscopy
Immunostaining was performed as previously described (31). The antibodies used for immunocytochemistry are given in supplemental Table S2.
Subcellular Fractionations
Whole cell lysates were collected in Laemmli buffer as previously described (18). Mitochondria-enhanced and postmitochondrial cytoplasmic fractions from which nuclear and mitochondrial materials were depleted were prepared by Dounce homogenization using the Pierce mitochondria isolation kit for cultured cells. Cytoplasmic fractions were concentrated using Millipore (Billerica, MA) Microcon NMWL 10,000 centrifugal filter devices.
Stable Small Hairpin RNA-mediated Down-regulation of HtrA3 and HtrA3 Re-expression
Lentiviral supernatants were generated by transfecting shRNA3 transfer vectors into 293T with both vesicular stomatitis virus G and gag/pol vectors (Sigma) using Lipofectamine LTX (Invitrogen). Supernatants containing lentiviral particles were collected 48 h after transfection. The transductions were carried out 24 h after seeding. The efficiency of knockdown was determined by immunoblot after 48 h. Stable batch clones were selected by puromycin 2 days after transduction. Rescue experiments were done as previously described (32).
Drug Treatments
Chemotherapeutic drug treatment was performed as previously described (18). Etoposide, cisplatin, and 4-(2-aminoethyl)benzenesulfonyl fluoride were purchased from Sigma-Aldrich, dissolved in Me2SO, and added to the culture medium. Z-VAD-FMK was purchased from Calbiochem.
Cell Survival Assays and Hoechst Staining
MTT and clonogenic survival assays and Hoechst staining were done as previously described (18).
IC50 Values
MTT assay results were expressed as log values and fit to a dose-response curve with a standard slope using nonlinear regression curve plots.
Statistics
Two-tailed Student's t test, one-way ANOVA, two-sided χ2 analyses, and nonlinear regression were performed using Prism 3.0 (Graphpad Software). p < 0.05 and α = 0.05 were considered statistically significant.
Annexin V Labeling and Flow Cytometry
Annexin V (Pharmingen) labeling and flow cytometry were performed according to the supplier's instructions and as previously described (33).
RESULTS
Exogenously Expressed HtrA3 Modulates Cytotoxicity to Etoposide and Cisplatin
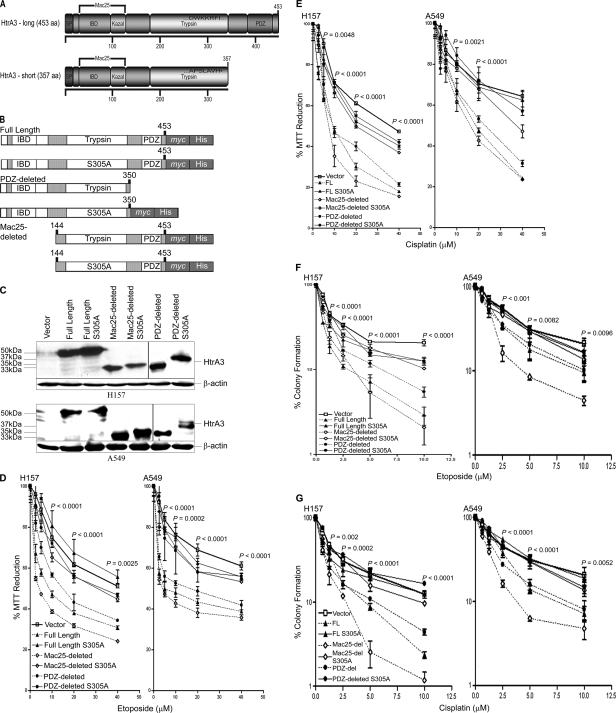
To address whether exogenous HtrA3 expression restores cytotoxicity to etoposide or cisplatin in resistant lung cancer cell lines, the long (full length) and short (PDZ-deleted) HtrA3 isoforms (Fig. 1A) as well as a domain variant lacking the N-terminal Mac25 domain (Mac25-deleted) and protease inactive mutants of each variant were subcloned (Fig. 1B). We previously reported that overexpression of a Mac25 domain-deleted HtrA1 variant induced cell death with activation of caspase 3/7 during the cell death process (18). WT and protease inactive variants and vector control were transiently expressed in HtrA3-deficient lung cancer cell lines H157 and A549 (Fig. 1C) and treated with increasing concentrations either drug. Exogenous expression of WT HtrA3 variants significantly attenuated cell survival with either etoposide or cisplatin treatment in both cell lines compared with expression of protease inactive mutants or vector control (p < 0.05) (Fig. 1, D and E). Further analysis revealed that IC50 for both drugs with expression of WT HtrA3 variants was significantly lower in both cell lines compared with protease inactive mutants or vector control (supplemental Fig. S1, A and B, and Table S3). Exogenous expression of WT variants also resulted in a significant decrease in clonogenic survival with either etoposide or cisplatin treatment in both cell lines compared with expression of protease inactive mutants or vector control (p < 0.05) (Fig. 1, F and G). Collectively, these data support a role for HtrA3 in modulating etoposide- and cisplatin-induced cytotoxicity in a manner dependent on serine protease function in lung cancer cell lines.
FIGURE 1.
WT HtrA3 variants modulate cytotoxicity to etoposide and cisplatin with exogenous expression in lung cancer cell lines. A, domain structural schematics of the long and short HtrA3 splice variants. The long variant is 453 amino acids in length and has an N-terminal signal peptide (SP, amino acids 1–17), an insulin/insulin-like growth factor-binding domain (amino acids 29–94), a Kazal-type serine protease-inhibitor domain (amino acids 89–126), a trypsin protease domain (amino acids 176–341), and one C-terminal PDZ domain (amino acids 384–440). The insulin/insulin-like growth factor-binding domain (IBD) and Kazal domains are homologous to Mac25. The short isoform is 357 amino acids in length and is identical to the long isoform with two exceptions: the short isoform lacks the PDZ domain, and the last seven C-terminal residues in the short form (APSLAVH) differ from the corresponding residues in the long isoform (DWKKRFI). B, domain structural schematics of WT and protease inactive full length, PDZ-deleted, and Mac25-deleted HtrA3 variants that are C-terminally myc- and polyhistidine-tagged. The WT PDZ-deleted variant was not tagged. Protease inactive HtrA3 variants contain a serine to alanine point mutation at residue 305 (S305A) within the conserved trypsin catalytic triad. C, immunoblots showing expression of HtrA3 variants, vector control, and β-actin loading controls in HtrA3-deficient lung cancer cell lines H157 and A549. PDZ-deleted variants were detected using an HtrA3 polyclonal antibody. D and E, MTT survival assays showing statistically significant attenuation of MTT reduction (p < 0.05) with exogenous expression of WT HtrA3 variants during etoposide (D) or cisplatin (E) treatment. The data were expressed as the means ± S.E. and represented at least two independent trials performed in quadruplicate. The p values were calculated using unpaired two-tailed Student's t test for two groups and ANOVA. F and G, clonogenic survival assays showing decreased clonogenic survival with exogenous expression of WT HtrA3 variants following etoposide (F) or cisplatin (G) treatment. The data are expressed as the means ± S.E. and represent independent trials performed in triplicate. The p values were calculated using unpaired two-tailed Student's t test for two groups and ANOVA.
HtrA3 Down-regulation Attenuates Etoposide- and Cisplatin-induced Cytotoxicity
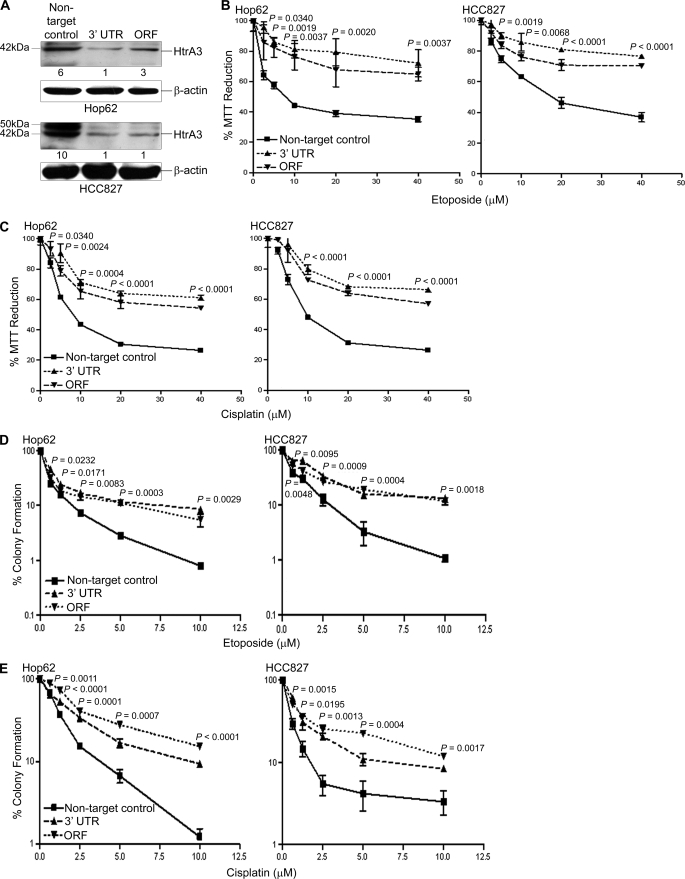
Whether HtrA3 down-regulation results in resistance to etoposide or cisplatin was next addressed. In lung cancer cell lines Hop62 and HCC827, a 2–6-fold decrease in HtrA3 protein expression was observed in pooled clonal lines stably targeted for HtrA3 down-regulation with shRNAs targeting either the open reading frame or the 3′-UTR compared with endogenous HtrA3 expression in nontargeted control lines (Fig. 2A). A significant decrease in cell survival was observed with either etoposide or cisplatin treatment in clonal lines with normal endogenous expression compared with clonal lines in which HtrA3 was stably down-regulated (p < 0.05) (Fig. 2, B and C). Further analysis revealed that the IC50 values for both drugs with endogenous HtrA3 expression were significantly lower than with HtrA3 down-regulation (supplemental Fig. S1, C and D, and Table S1). HtrA3 down-regulation also resulted in significantly greater clonogenic survival with treatment compared with clonogenic survival with normal endogenous HtrA3 expression (p < 0.05) (Fig. 2, D and E).
FIGURE 2.
Down-regulation of endogenous HtrA3 expression attenuates etoposide- and cisplatin-induced cytotoxicity. A, immunoblots showing endogenous HtrA3 expression in pooled clonal lung cancer cell lines stably transduced with a nontargeted control vector and HtrA3 down-regulation in pooled clonal lines stably transduced with shRNA targeting either the open reading frame (ORF) or 3′-UTR. For each cell line, densitometric analysis of fold changes in HtrA3 intensity normalized by β-actin loading controls is given below each band. B and C, MTT survival assays showing statistically significant decrease in MTT reduction with endogenous HtrA3 expression with either etoposide (B) or cisplatin (C) treatment compared with decrease in MTT reduction with HtrA3 down-regulation (p < 0.05). The data are expressed as the means ± S.E. and represent at least two independent trials performed in quadruplicate. The p values were calculated using unpaired two-tailed Student's t test for two groups and ANOVA. D and E, clonogenic survival assays showing significantly greater clonogenic survival with HtrA3 down-regulation following etoposide (D) or cisplatin (E) treatment compared with survival with normal endogenous HtrA3 expression (p < 0.05). The data are expressed as the means ± S.E. and represent independent trials performed in triplicate. The p values were calculated using unpaired two-tailed Student's t test for two groups and ANOVA.
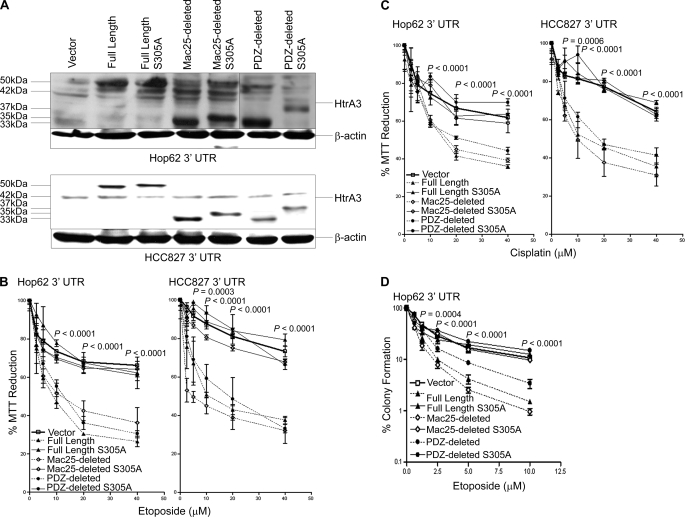
Re-expression of WT HtrA3 variants in clonal Hop62 and HCC827 cell lines stably targeted for HtrA3 down-regulation by the 3′-UTR (Fig. 3A) significantly attenuated MTT reduction with either etoposide or cisplatin treatment compared with expression of protease inactive variants or vector control (p < 0.05) (Fig. 3, B and C). Further analysis revealed that IC50 for both drugs with re-expression of WT HtrA3 variants was significantly lower in both clonal lines than with expression of protease inactive mutants or vector control (supplemental Fig. S1, E and F, and Table S1). Re-expression of WT HtrA3 variants also significantly attenuated clonogenic survival with etoposide treatment in Hop62 3′-UTR compared with expression of protease inactive variants or vector control (p < 0.05) (Fig. 3D). Collectively, these results suggest that the loss of endogenous HtrA3 expression promotes resistance to etoposide- and cisplatin-mediated cytotoxicity and that re-expression of proteolytically active HtrA3 variants restores sensitivity to both drugs in lung cancer cell lines.
FIGURE 3.
HtrA3 re-expression promotes etoposide and cisplatin cytotoxicity in lung cancer cell lines. A, immunoblots showing exogenous expression of WT and protease inactive full length, Mac25-deleted, and PDZ-deleted HtrA3 variants, vector control, and β-actin loading controls in Hop62 and HCC827 clonal cell lines stably down-regulated for HtrA3 expression through shRNA-mediated targeting of the 3′-UTR. Endogenous HtrA3 is expressed at 42 kDa. B and C, MTT survival assays showing statistically significant attenuation of MTT reduction with re-expression of WT HtrA3 variants during etoposide (B) or cisplatin (C) treatment compared with expression of protease inactive variants or vector control (p < 0.05). The data are expressed as the means ± S.E. and represent at least two independent trials performed in quadruplicate. p values were calculated using unpaired two-tailed Student's t test for two groups and ANOVA. D, clonogenic survival assay showing significant decrease in clonogenic survival in Hop62 with exogenous expression of WT HtrA3 variants following etoposide treatment compared with expression of protease inactive mutants or vector control. The data are expressed as the means ± S.E. and represent independent trials performed in triplicate. The p values were calculated using unpaired two-tailed Student's t test for two groups and ANOVA.
Endogenous HtrA3 Is Localized to Mitochondria and Is Translocated from Mitochondria by Etoposide-induced Cytotoxic Stress
The effect of chemotherapy-induced cytotoxic stress on endogenous HtrA3 expression and localization was next assessed. HtrA3 expression was induced by both etoposide and cisplatin in the human bronchial cell line BEAS-2B and in Hop62 and HCC827 (supplemental Fig. S2, A–C). Further analysis revealed that, like HtrA1 (18) and HtrA2 (23, 25, 27, 28, 34, 35), HtrA3 may also be a substrate for limited autoproteolysis. In lung cancer cell line H526, endogenous HtrA3 was up-regulated and cleaved with etoposide and cisplatin treatment, yielding a 35-kDa cleavage product that may be homologous to the unrestricted, proteolytically active Mac25-deleted cleavage product previously identified for HtrA1 (18) (supplemental Fig. S3, A–C). HtrA3 cleavage was uninhibited by the broad spectrum caspase inhibitor Z-VAD-FMK but was attenuated by the serine protease inhibitor 4-(2-aminoethyl)benzenesulfonyl fluoride (supplemental Fig. S2, D and E). Interestingly, HtrA3 cleavage appeared to correlate with a greater number of cells showing the early (17.1%) and late (25.7%) stages of apoptosis with etoposide treatment that was attenuated by HtrA3 down-regulation (supplemental Fig. S3F).
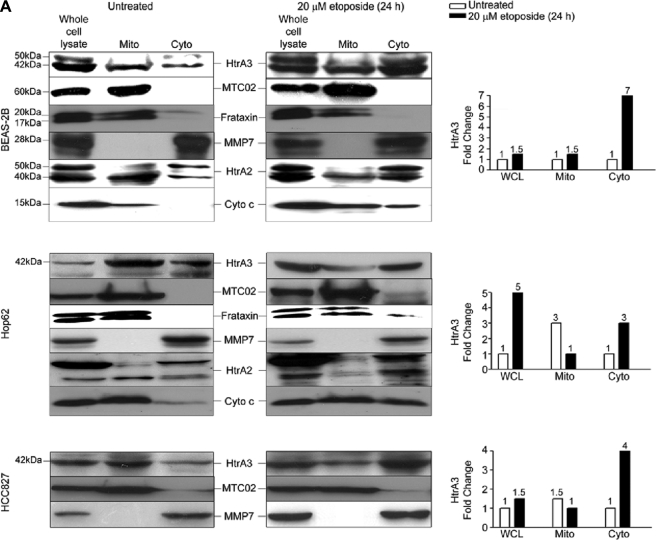
In assessing HtrA3 localization, endogenous MTC02, a 60-kDa nonglycosylated protein component of human mitochondria (36–41), and human frataxin, a nuclear encoded mitochondrial matrix protein thought to be involved in iron metabolism (42–46), were used as markers of mitochondrial localization (Fig. 4A). Immunoblot analysis of whole cell lysates and of mitochondria-enriched and postmitochondrial cytoplasmic fractions revealed that HtrA3, MTC02, frataxin, and MMP7 were each expressed in whole cell lysates, whereas HtrA3 and MTC02 and frataxin were also co-expressed in mitochondria-enriched fractions, and MMP7 was expressed in postmitochondrial cytoplasmic fractions. In contrast, immunoblot analysis following etoposide treatment revealed that HtrA3 was up-regulated and co-expressed with MMP7 in both whole cell lysates and postmitochondrial cytoplasmic fractions, where expression was increased by 3–7-fold, whereas MTC02 and frataxin were expressed in mitochondria-enriched fractions. Assessment of endogenous HtrA2 and cytochrome c expression revealed that in BEAS-2B and Hop62, cytochrome c and the mature form of HtrA2 (40 kDa) were more highly expressed in mitochondria-enriched fractions prior to etoposide treatment and in postmitochondrial cytoplasmic fractions after treatment. Cytochrome c up-regulation in mitochondria-enriched fractions following etoposide treatment may have been due to increases in protein production and mitochondrial translocation in response to cytotoxic stress (47).
FIGURE 4.
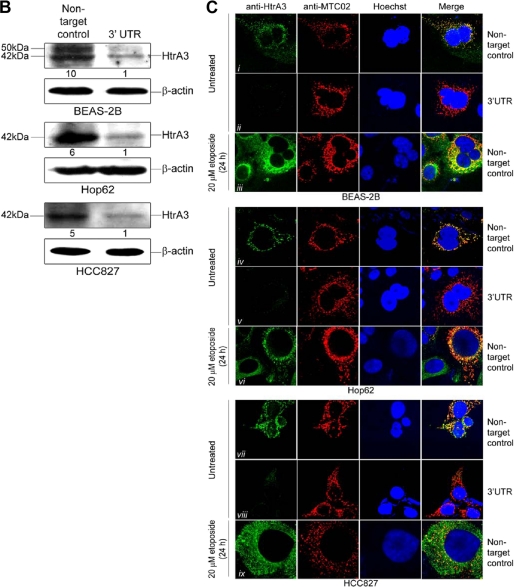
Endogenous HtrA3 is localized to mitochondria and is translocated from mitochondria by etoposide-induced cytotoxic stress. A, immunoblots showing endogenous expression of HtrA3, mitochondrial marker MTC02, human frataxin, MMP7, HtrA2, and cytochrome c in whole cell lysates, mitochondria-enriched fractions (Mito), and postmitochondrial cytoplasmic fractions (Cyto) before and after etoposide treatment in the human bronchial cell line BEAS-2B and in lung cancer cell lines Hop62 and HCC827. Both the mature (40 kDa) and immature (50 kDa) HtrA2 forms are shown. Densitometric analysis shows fold changes in HtrA3 intensity for each cell line. B, immunoblots showing endogenous HtrA3 expression, stable HtrA3 down-regulation by 3′ UTR-targeted shRNA, and β-actin loading controls in pooled stable clonal cell lines. A polyclonal antibody was used for detection of endogenous HtrA3. Densitometric analysis of fold changes in HtrA3 intensity normalized by β-actin loading controls is given for each cell line. C, immunocytochemistry using an HtrA3 polyclonal antibody showing HtrA3 and MTC02 co-localization before and after etoposide-mediated apoptotic stimulation. Hoechst stain binds nuclear material. Original magnification, ×100.
Immunocytochemical analysis of endogenous HtrA3 expression in clonal cell lines stably targeted for HtrA3 down-regulation and nontargeted control lines (Fig. 4B) showed that HtrA3 co-localized with MTC02 in untreated clonal cell lines with endogenous HtrA3 expression (Fig. 4C, panels i, iii, iv, vi, vii, and ix) but did not co-localize with MTC02 following etoposide treatment. Interestingly, HtrA3 mislocalization was inhibited by overexpression of Bcl-2, an antiapoptotic factor that inhibits apoptosis in part by suppressing cytochrome c release from the mitochondrial intermembrane space (supplemental Fig. S4, A and B) (48, 49). HtrA3 expression was undetected in clonal lines down-regulated for HtrA3 expression, indicating that the polyclonal antibody used in this assay was specific for HtrA3 expression (Fig. 3C, panels ii, v, and viii). Collectively, these results confirm that HtrA3 is a mitochondrial protease that is translocated from mitochondria in a manner similar to HtrA2 and cytochrome c by etoposide-induced cytotoxic stress in BEAS-2B and in lung cancer cell lines.
HtrA3 Localization Is Regulated by Domain Composition and Protease Function with Etoposide-induced Cytotoxic Stress
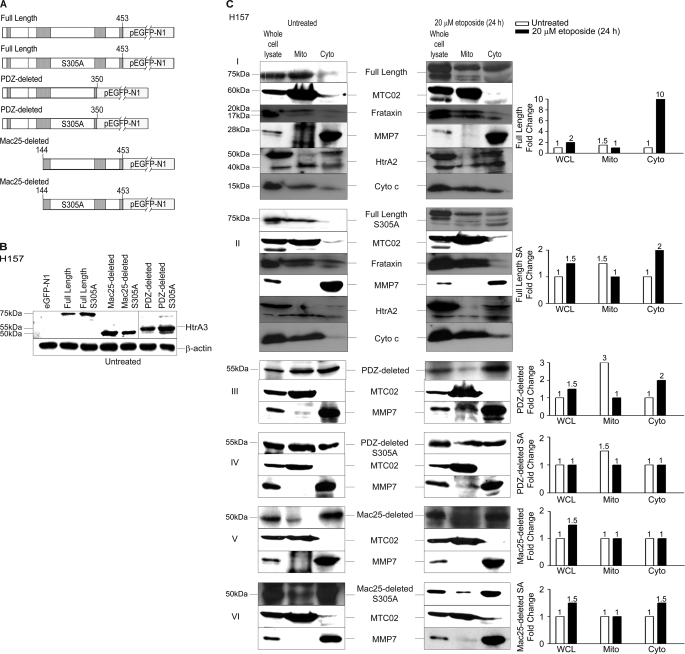
The long HtrA3 isoform (full length), the short isoform (PDZ-deleted), and a Mac25 deletion variant (Mac25-deleted) as well as proteolytically inactive mutants of each (S305A) were each N-terminally GFP-tagged and expressed in lung cancer cell line H157 (Fig. 5, A and B). Immunoblot analysis showed that although both WT and inactive full-length HtrA3 variants were expressed with MTC02, frataxin, and MMP7 in whole cell lysates, they co-fractionated with MTC02 and frataxin in mitochondria-enriched fractions but not with MMP7 (Fig. 5C, panels I and II). With etoposide treatment, the WT full-length variant co-fractionated with MMP7 in postmitochondrial cytoplasmic fractions, where expression was increased by 10-fold, whereas the inactive full-length protease mutant co-fractionated with MTC02 and frataxin in mitochondria-enriched fractions. Analysis of endogenous HtrA2 and cytochrome c expression showed that both cytochrome c and the mature HtrA2 isoform (40 kDa) were more highly expressed in mitochondria-enriched fractions prior to etoposide treatment and in postmitochondrial cytoplasmic fractions after treatment.
FIGURE 5.
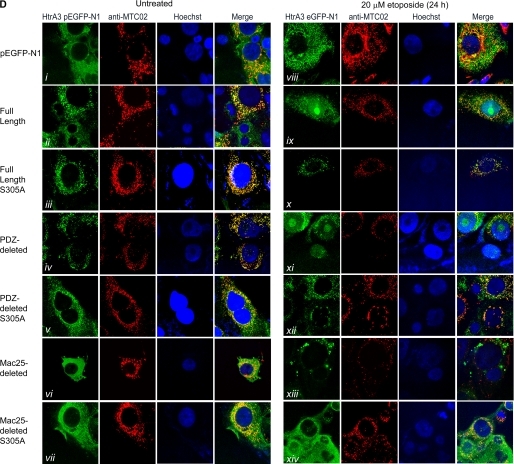
HtrA3 localization is regulated by domain composition and protease function with etoposide-induced cytotoxic stress. A, domain structural schematics of GFP-tagged WT and protease inactive HtrA3 variants. Inactive variants contain a single serine to alanine point mutation at amino acid residue 305 (S305A) within the conserved trypsin catalytic triad. B, immunoblots showing expression of GFP-tagged HtrA3 variants in lung cancer cell line H157. PDZ-deleted variants were detected using an HtrA3 polyclonal antibody. β-actin shows equal loading. C, panels I–VI, immunoblots showing expression of HtrA3 variants, mitochondrial marker MTC02 and MMP7 in whole cell lysates (WCL) and in mitochondria-enriched (Mito) and postmitochondrial cytoplasmic fractions (Cyto) before and after etoposide treatment. Endogenous expression of human frataxin, HtrA2, and cytochrome c are given in panels I and II. Both the mature (40 kDa) and immature (50 kDa) HtrA2 forms are shown. Densitometric analysis shows fold changes in HtrA3 intensity for each HtrA3 variant. D, immunocytochemistry showing subcellular localization of MTC02 and GFP-tagged HtrA3 variants before and after etoposide treatment. Hoechst stain binds nuclear material. Original magnification, ×100.
For both WT and inactive PDZ-deleted variants, expression was detected in whole cell lysates and evenly detected in mitochondria-enriched and postmitochondrial cytoplasmic fractions in untreated samples, whereas only the WT variant showed a 2-fold increase in expression in the postmitochondrial cytoplasmic fraction with etoposide treatment (Fig. 5C, panels III and IV). Interestingly, both WT and inactive Mac25-deleted variants were expressed with MTC02 and MMP7 in whole cell lysates but co-fractionated with MMP7 and not with MTC02 in both untreated and treated samples (Fig. 5C, panels V and VI).
Immunocytochemical analysis of HtrA3 expression revealed that both WT and inactive full-length and PDZ-deleted variants co-localized with MTC02 in untreated cell populations, whereas only protease inactive variants were localized with MTC02 following etoposide treatment (Fig. 5D, panels ii–v and ix–xii). Interestingly, mislocalization of the full-length HtrA3 variant was inhibited by overexpression of Bcl-2 (supplemental Fig. S4, C and D). Neither WT nor inactive Mac25-deleted variants nor vector control co-localized with MTC02 in either untreated or treated populations (Fig. 5D, panels i, vi, vii, viii, xiii, and xiv). Collectively, these data suggest that the Mac25 domain may be essential to mitochondrial localization and that HtrA3 is translocated from mitochondria by etoposide-induced cytotoxic stress in a manner dependent on protease function.
HtrA3 Promotes Cell Death in Response to Etoposide- and Cisplatin-induced Cytotoxicity
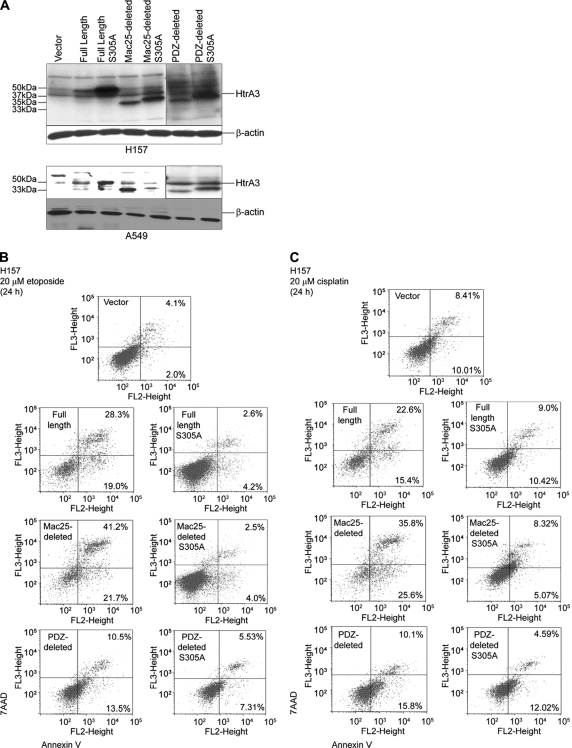
Whether HtrA3 responds to chemotherapy-induced cytotoxic stress through a mechanism that promotes activation of the canonical, mitochondrial apoptotic pathway was next assessed. WT and protease inactive HtrA3 variants were expressed in H157 and A549 (Fig. 6A), treated with 20 μm etoposide or cisplatin for 24 h, and assayed for cell death. Annexin V labeling followed by flow microfluorometry showed that more cells showed early and late stages of cell death with overexpression of WT HtrA3 variants during either etoposide (Fig. 6B) or cisplatin (Fig. 6C) treatment than with overexpression of protease inactive mutants or vector control.
FIGURE 6.
HtrA3 promotes cell death in response to etoposide- and cisplatin-induced cytotoxic stress. A, immunoblots showing expression of exogenous WT and inactive HtrA3 variants in HtrA3-deficient lung cancer cell lines H157 and A549. PDZ-deleted variants were detected using an HtrA3 polyclonal antibody. β-actin shows equal loading. B and C, analysis of apoptotic activity with exogenous overexpression of WT and protease inactive HtrA3 variants with etoposide (B) or cisplatin (C) treatment in lung cancer cell line H157. Apoptosis activity was assessed by annexin V labeling followed by flow microfluorimetry. The percentage of cells in early (bottom right quarters) and late (top right quarters) stages of apoptosis is given. The results show greater cell death with overexpression of WT variants than with expression of protease mutants or vector control. D, Hoechst nuclear staining contrasting the normal nuclear morphology of untreated cells with nuclear condensation and fragmentation characteristic of apoptosis with transient expression of WT HtrA3 variants during either etoposide or cisplatin treatment. Original magnification, ×100. E and F, quantitative analyses of Hoechst staining showing more apoptotic nuclei with exogenous expression of WT HtrA3 variants in lung cancer cell lines H157 (E) and A549 (F) with either etoposide or cisplatin treatment than with exogenous expression of protease inactive mutants or vector control. The error bars represent ± S.D. of three independent trials performed at least in triplicate. G, immunoblots showing up-regulation of apoptosis effectors with exogenous expression of WT HtrA3 variants during etoposide treatment in H157. An HtrA3 polyclonal antibody was used to detect PDZ-deleted variants. Densitometric analysis showing fold changes in intensity normalized by β-actin loading controls is given below each blot.
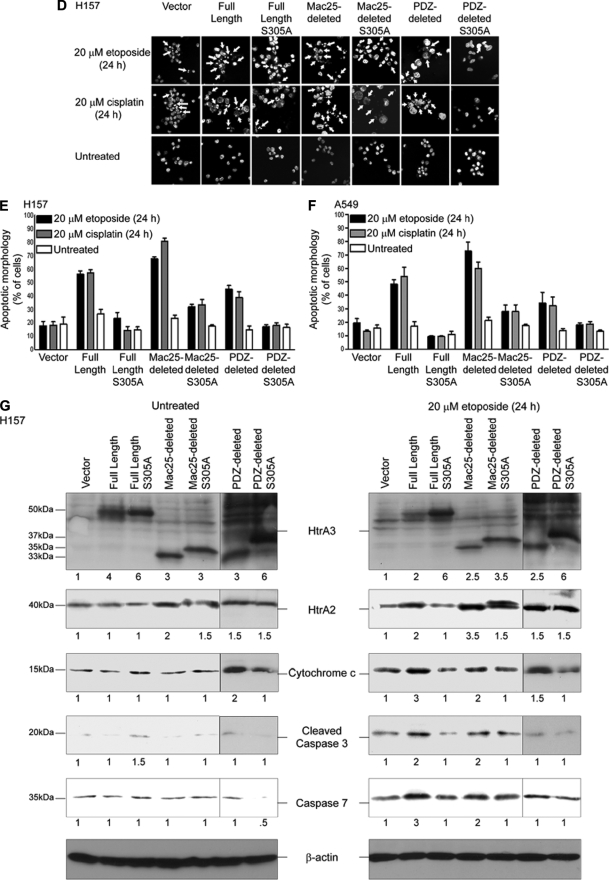
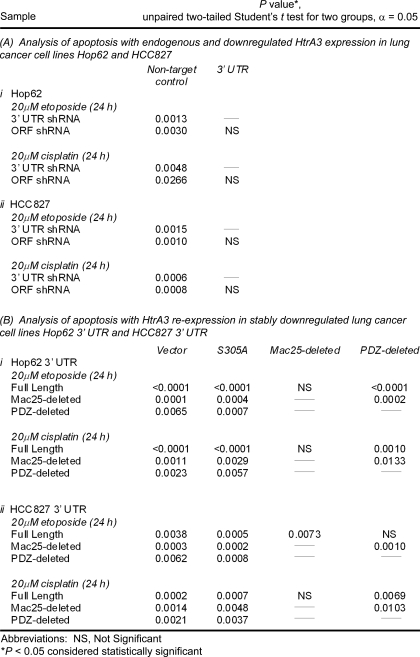
In assaying for morphologic changes in nuclear structure consistent with an apoptotic cell death, microscopic inspection revealed a significantly greater number of condensed and fragmented nuclei in H157 and A549 cells expressing WT full length or Mac25-deleted HtrA3 variants with etoposide or cisplatin treatment than in untreated cells or those expressing the PDZ-deleted variant, protease mutants, or vector control (p < 0.05) (Fig. 6, D–F, and Table 1, parts A and B). Along these lines, immunoblot analysis of untreated and etoposide-treated H157 transfectants showed 2–3-fold up-regulation of apoptosis effectors HtrA2, cytochrome c, and caspase 7 as well as caspase 3 cleavage products with exogenous expression of WT HtrA3 variants compared with the expression of protease mutants or vector control (Fig. 6G).
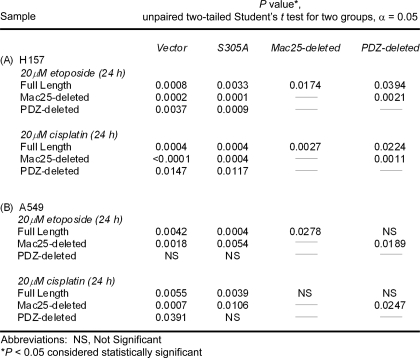
TABLE 1.
Statistical comparison of apoptotic response to etoposide or cisplatin treatment with exogenous expression of HtrA3 variants
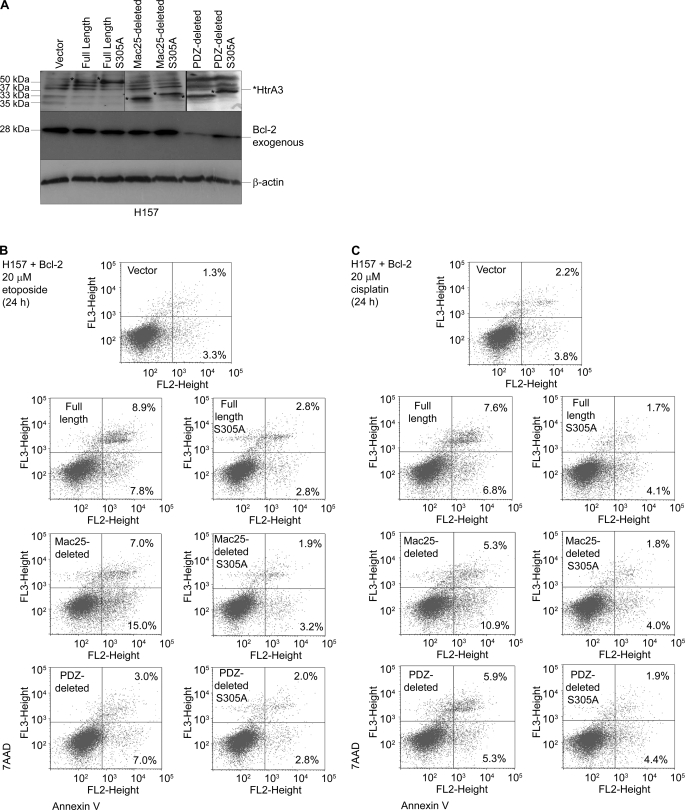
Further analysis showed that Bcl-2 overexpression attenuated HtrA3-induced cell death. WT and protease inactive HtrA3 variants were expressed with Bcl-2 in H157 (Fig. 7A), treated with 20 μm etoposide or cisplatin for 24 h, and assayed for cell death by annexin V labeling followed by flow microfluorimetry. With Bcl-2 overexpression, the results show a statistically insignificant difference in the number of cells showing late and early stages of cell death between cells with exogenous expression of WT HtrA3 variants and those expressing protease inactive mutants or vector control with either etoposide (Fig. 7B) or cisplatin (Fig. 7C) treatment. Taken together, these findings suggest that HtrA3 induces cell death in a manner dependent on serine protease function either through or in cooperation with the intrinsic, mitochondrial cell death pathway in response to etoposide or cisplatin in lung cancer cell lines.
FIGURE 7.
Exogenous Bcl-2 expression attenuates HtrA3-induced cell death in response to etoposide and cisplatin. A, immunoblots showing expression of exogenous WT and inactive HtrA3 variants and exogenous Bcl-2 in HtrA3-deficient lung cancer cell line H157. PDZ-deleted variants were detected using an HtrA3 polyclonal antibody. β-Actin shows equal loading. B and C, analysis of apoptosis activity with exogenous overexpression of WT and protease inactive HtrA3 variants and Bcl-2 with etoposide (B) or cisplatin (C) treatment in lung cancer cell line H157. Apoptosis activity was assessed by annexin V labeling followed by flow microfluorimetry. The percentage of cells in early (bottom right quarters) and late (top right quarters) stages of apoptosis is given.
HtrA3 Down-regulation Attenuates the Apoptotic Response to Both Etoposide and Cisplatin Treatment
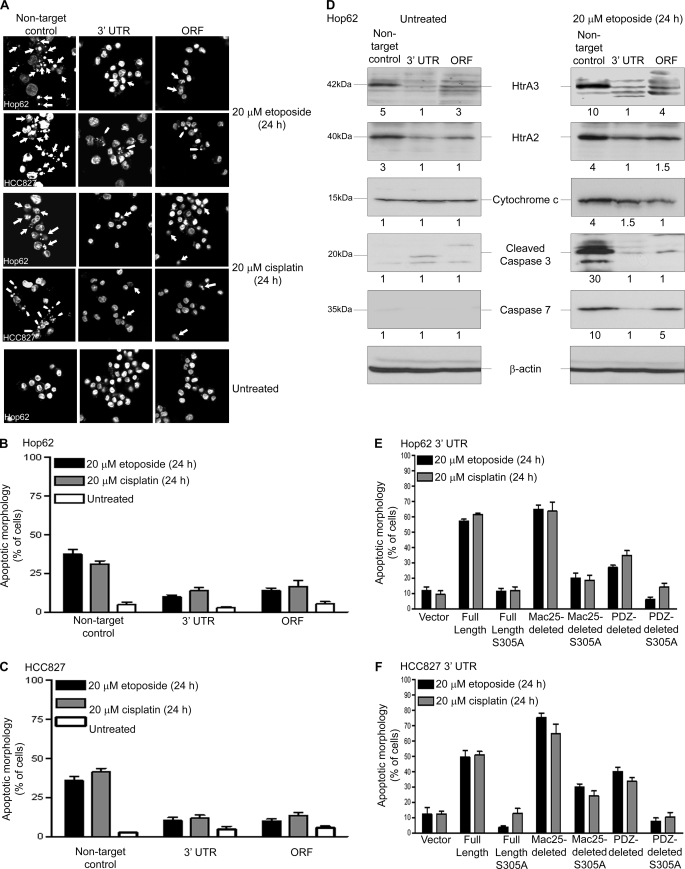
In assessing whether the loss of HtrA3 expression diminishes the apoptotic response, microscopic inspection of clonal lung cancer cell lines Hop62 and HCC827 stably targeted for HtrA3 down-regulation and their nontargeted controls (Fig. 2A) revealed significantly fewer fragmented and condensed nuclei with either etoposide or cisplatin treatment in cells with low HtrA3 expression compared with untreated cells or with those with normal HtrA3 expression (Fig. 8, A–C, and Table 2, part A, i and ii). Moreover, immunoblot analysis showed 4–30-fold up-regulation of apoptosis effectors HtrA2, cytochrome c, and caspase 7 as well as an increase in caspase 3 cleavage products with etoposide treatment in the control Hop62 clonal line with normal HtrA3 expression compared with untreated control (Fig. 8D). In contrast, HtrA3 down-regulation attenuated up-regulation of apoptosis effectors with etoposide treatment. Re-expression of WT HtrA3 variants, but not of protease inactive variants or vector control, significantly increased the appearance of apoptotic nuclei by Hoechst staining with either etoposide or cisplatin treatment (Fig. 8, E and F, and Table 2, part B, i and ii). Collectively, these results suggest that HtrA3 down-regulation abrogates the cell death response to etoposide- and cisplatin-induced cytotoxicity in lung cancer cell lines, whereas HtrA3 re-expression restores toxicity to either etoposide or cisplatin in resistant cell lines.
FIGURE 8.
HtrA3 down-regulation attenuates the apoptotic response to etoposide- and cisplatin-induced cytotoxicity in lung cancer cell lines. A, Hoechst nuclear staining contrasting the normal nuclear morphology of untreated Hop62 and HCC827 cells and of pooled stable clones in which HtrA3 expression was down-regulated with nuclear condensation and fragmentation characteristic of apoptosis with endogenous HtrA3 expression during either etoposide or cisplatin treatment. Original magnification, ×100. B and C, quantitative analysis of Hoechst staining showing fewer apoptotic nuclei with HtrA3 down-regulation in Hop62 (B) and HCC827 (C) with either etoposide or cisplatin treatment than with normal endogenous HtrA3 expression. The error bars represent ± S.D. of three independent trials performed at least in triplicate. D, immunoblots showing decrease in expression of apoptosis effectors with HtrA3 down-regulation during etoposide treatment in Hop62. Densitometric analysis showing fold changes in intensity normalized by β-actin loading controls is given below each blot. E and F, quantitative analyses of Hoechst staining showing a greater number of apoptotic nuclei with transient re-expression of catalytically active HtrA3 variants in a pooled Hop62 (E) and HCC827 (F) clonal lines stably down-regulated for HtrA3 expression with either etoposide or cisplatin treatment. The error bars represent ± S.D. of three independent trials performed at least in triplicate. ORF, open reading frame.
TABLE 2.
Statistical comparison of apoptotic response to etoposide or cisplatin treatment with HtrA3 down-regulation and re-expression in lung cancer cell lines
DISCUSSION
HtrA are widely known as stress-response factors (23, 26, 50). Here we have provided the first evidence that HtrA3 is a mitochondrial protease that promotes etoposide and cisplatin-induced cytotoxicity in lung cancer cell lines through activation of the apoptotic pathway.
Our analysis revealed that HtrA3 variants in which domains N-terminal to the trypsin domain were intact localized to mitochondria, whereas the Mac25-deleted variant was cytoplasmic (Figs. 4 and 5). These results suggest that the Mac25 domain, which includes an insulin-like growth factor-binding domain and a Kazal-type serine protease inhibitory domain (Fig. 1A), may be essential to HtrA3 subcellular translocation. However, analysis of the HtrA3 sequence with mitochondrial localization signal prediction programs PSORT II (51) and MitoProt II (52) did not reveal a canonical N-terminal mitochondrial localization signal, suggesting that HtrA3 may contain one of the largely uncharacterized and inconsistent internal targeting signals (50). Alternatively, HtrA3 may translocate to mitochondria by binding nuclear-encoded mitochondrial proteins. Although HtrA family members are structurally defined by one or more C-terminal PDZ protein interaction domain(s) (19), human HtrA also derive substrate specificity from the insulin/insulin-like growth factor-binding domain or Kazal domains (53, 54).
We previously reported that HtrA3 localizes to mitochondria in ovarian cancer cell line OV202 and is released into the cytosol upon induction of apoptosis by the broad kinase inhibitor UCN-01 (21). In support of these findings, we have shown here that HtrA3 was up-regulated in postmitochondrial cytoplasmic fractions (Figs. 4A and 5C) and was translocated from mitochondria (Figs. 4C and 5D) with etoposide-induced cytotoxic stress. Moreover, changes in HtrA3 expression following etoposide treatment were correlated to similar changes in HtrA2 and cytochrome c (Figs. 4A and 5C, panels I and II). These data and evidence that Bcl-2 overexpression inhibited both HtrA3 translocation from mitochondria (supplemental Fig. S4, B and D) and HtrA3-mediated apoptosis (Fig. 7, B and C) imply that, like HtrA2 and cytochrome c, HtrA3 has a role in promoting activation of the intrinsic, mitochondria-mediated apoptotic pathway that is dependent on its release into the cytoplasm. Interestingly, Bcl-2 overexpression attenuated the induction of apoptosis mediated by the cytoplasmically localized Mac25-deleted HtrA3 variant (Fig. 7, B and C), perhaps in support of a role for cytoplasmic HtrA3 in functioning synergistically with other apoptotic effectors in propagating intrinsic apoptotic pathway signaling. Along these lines, the Mac25-deleted variant attenuated cell survival and caused greater induction of cell death than full length or PDZ-deleted variants (Figs. 1, 2, 6, and 8). We have also demonstrated here that HtrA3 attenuates cell survival with either etoposide or cisplatin treatment in a manner dependent on serine protease function (Figs. 1–3). HtrA3 may therefore modulate cell death through a mechanism involving irreversible proteolysis of factors crucial to cell survival. A similar serine protease-dependent mechanism has been proposed for HtrA2 interaction with several substrates, most notably XIAP (X-linked inhibitor of apoptosis protein) (55–59). However, HtrA3 lacks the canonical IAP (inhibitor of apoptosis protein)-binding motif found in HtrA2, and our preliminary studies indicate that HtrA3 overexpression does not result in a loss of XIAP expression by immunoblot (data not shown). Thus, further investigation of HtrA3 binding partners and substrates is clearly needed.
The evidence provided here could ultimately offer a new venue for targeted lung cancer treatment. Because most cancer cells maintain a finely tuned balance between survival and apoptotic signals (15), HtrA3 down-regulation may tip cancer cells toward a clinically unfavorable survival response to anticancer drugs. The identification of potential markers of therapeutic index and treatment response may contribute to the selection of favorable chemotherapeutic strategies, which could in turn alter the target approach from a general to an individual treatment strategy. Thus, the role of HtrA3 in promoting cytotoxicity to anti-cancer drugs clearly warrants further investigation in confirmatory studies and in different tumor types.
Supplementary Material
Acknowledgments
We thank Scott. H. Kaufmann for providing the Bcl-2 construct and the S-tag antibody for its detection and Grazia Isaya for providing the antibody to endogenous human frataxin. We also thank members of the Shridhar Laboratory as well as Julian Molina and Dev Mukhopadhyay for discussions and critical review of the data.
This work was supported by the Edith and Bernard Waterman Foundation and the Mayo Foundation (to V. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Figs. S1–S4.
- shRNA
- small hairpin RNA
- MMP
- matrix metalloproteinase
- WT
- wild type
- Z-VAD-FMK
- benzyloxycarbonyl-Val-Ala-Asp(OMe) fluoromethylketone
- ANOVA
- analysis of variance
- UTR
- untranslated region
- GFP
- green fluorescent protein
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide.
REFERENCES
- 1.Jemal A., Siegel R., Ward E., Hao Y., Xu J., Murray T., Thun M. J. (2008) CA-Cancer J. Clin. 58, 71–96 [DOI] [PubMed] [Google Scholar]
- 2.Minna J. D., Schiller J. H. (2008) Harrison's Principles of Internal Medicine, 17th Ed., McGraw-Hill Book Co., New York [Google Scholar]
- 3.Ciombor K. K., Rocha Lima C. M. (2006) Curr. Treat. Options Oncol. 7, 59–68 [DOI] [PubMed] [Google Scholar]
- 4.Ferraldeschi R., Baka S., Jyoti B., Faivre-Finn C., Thatcher N., Lorigan P. (2007) Drugs 67, 2135–2152 [DOI] [PubMed] [Google Scholar]
- 5.Fischer B., Arcaro A. (2008) Rev. Recent Clin. Trials 3, 40–61 [DOI] [PubMed] [Google Scholar]
- 6.Hann C. L., Rudin C. M. (2008) Oncology 22, 1486–1492 [PMC free article] [PubMed] [Google Scholar]
- 7.Bunn P. A., Jr. (2004) Clin. Lung Cancer 6, 85–98 [DOI] [PubMed] [Google Scholar]
- 8.Machtay M., Lee J. H., Stevenson J. P., Shrager J. B., Algazy K. M., Treat J., Kaiser L. R. (2004) J. Thorac Cardiovasc. Surg. 127, 108–113 [DOI] [PubMed] [Google Scholar]
- 9.Sanborn R. E., Lally B. E. (2008) Thorac Surg. Clin. 18, 423–435 [DOI] [PubMed] [Google Scholar]
- 10.Rosell R., Cecere F., Santarpia M., Reguart N., Taron M. (2006) Curr. Opin. Pharmacol. 6, 323–331 [DOI] [PubMed] [Google Scholar]
- 11.Damiani L. A., Yingling C. M., Leng S., Romo P. E., Nakamura J., Belinsky S. A. (2008) Cancer Res. 68, 9005–9014 [DOI] [PubMed] [Google Scholar]
- 12.Zhang J., Kamdar O., Le W., Rosen G. D., Upadhyay D. (2009) Am. J. Respir. Cell Mol. Biol. 40, 135–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirsch F. R., Lippman S. M. (2005) J. Clin. Oncol. 23, 3186–3197 [DOI] [PubMed] [Google Scholar]
- 14.Gallego M. A., Ballot C., Kluza J., Hajji N., Martoriati A., Castéra L., Cuevas C., Formstecher P., Joseph B., Kroemer G., Bailly C., Marchetti P. (2008) Oncogene 27, 1981–1992 [DOI] [PubMed] [Google Scholar]
- 15.Kaufmann S. H., Vaux D. L. (2003) Oncogene 22, 7414–7430 [DOI] [PubMed] [Google Scholar]
- 16.Agarwal R., Kaye S. B. (2003) Nat. Rev. Cancer 3, 502–516 [DOI] [PubMed] [Google Scholar]
- 17.Meng X. W., Lee S. H., Kaufmann S. H. (2006) Curr. Opin. Cell Biol. 18, 668–676 [DOI] [PubMed] [Google Scholar]
- 18.Chien J., Aletti G., Baldi A., Catalano V., Muretto P., Keeney G. L., Kalli K. R., Staub J., Ehrmann M., Cliby W. A., Lee Y. K., Bible K. C., Hartmann L. C., Kaufmann S. H., Shridhar V. (2006) J. Clin. Invest. 116, 1994–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clausen T., Southan C., Ehrmann M. (2002) Mol. Cell 10, 443–455 [DOI] [PubMed] [Google Scholar]
- 20.Chien J., He X., Shridhar V. (2009) J. Cell. Biochem. 107, 253–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chien J., Ota T., Aletti G., Shridhar R., Boccellino M., Quagliuolo L., Baldi A., Shridhar V. (2009) Mol. Cell. Biol. 29, 4177–4187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chien J., Campioni M., Shridhar V., Baldi A. (2009) Curr. Cancer Drug Targets 9, 451–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hegde R., Srinivasula S. M., Zhang Z., Wassell R., Mukattash R., Cilenti L., DuBois G., Lazebnik Y., Zervos A. S., Fernandes-Alnemri T., Alnemri E. S. (2002) J. Biol. Chem. 277, 432–438 [DOI] [PubMed] [Google Scholar]
- 24.Cilenti L., Kyriazis G. A., Soundarapandian M. M., Stratico V., Yerkes A., Park K. M., Sheridan A. M., Alnemri E. S., Bonventre J. V., Zervos A. S. (2005) Am. J. Physiol. Renal Physiol. 288, F371–F379 [DOI] [PubMed] [Google Scholar]
- 25.Suzuki Y., Imai Y., Nakayama H., Takahashi K., Takio K., Takahashi R. (2001) Mol. Cell 8, 613–621 [DOI] [PubMed] [Google Scholar]
- 26.Martins L. M., Iaccarino I., Tenev T., Gschmeissner S., Totty N. F., Lemoine N. R., Savopoulos J., Gray C. W., Creasy C. L., Dingwall C., Downward J. (2002) J. Biol. Chem. 277, 439–444 [DOI] [PubMed] [Google Scholar]
- 27.van Loo G., van Gurp M., Depuydt B., Srinivasula S. M., Rodriguez I., Alnemri E. S., Gevaert K., Vandekerckhove J., Declercq W., Vandenabeele P. (2002) Cell Death Differ. 9, 20–26 [DOI] [PubMed] [Google Scholar]
- 28.Verhagen A. M., Silke J., Ekert P. G., Pakusch M., Kaufmann H., Connolly L. M., Day C. L., Tikoo A., Burke R., Wrobel C., Moritz R. L., Simpson R. J., Vaux D. L. (2002) J. Biol. Chem. 277, 445–454 [DOI] [PubMed] [Google Scholar]
- 29.Nie G. Y., Hampton A., Li Y., Findlay J. K., Salamonsen L. A. (2003) Biochem. J. 371, 39–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hackbarth J. S., Lee S. H., Meng X. W., Vroman B. T., Kaufmann S. H., Karnitz L. M. (2004) BioTechniques 37, 835–839 [PubMed] [Google Scholar]
- 31.Lingle W. L., Salisbury J. L. (2000) Curr. Top. Dev. Biol. 49, 313–329 [DOI] [PubMed] [Google Scholar]
- 32.Staub J., Chien J., Pan Y., Qian X., Narita K., Aletti G., Scheerer M., Roberts L. R., Molina J., Shridhar V. (2007) Oncogene 26, 4969–4978 [DOI] [PubMed] [Google Scholar]
- 33.Chien J., Staub J., Hu S. I., Erickson-Johnson M. R., Couch F. J., Smith D. I., Crowl R. M., Kaufmann S. H., Shridhar V. (2004) Oncogene 23, 1636–1644 [DOI] [PubMed] [Google Scholar]
- 34.Martins L. M. (2002) Cell Death Differ. 9, 699–701 [DOI] [PubMed] [Google Scholar]
- 35.Vande Walle L., Lamkanfi M., Vandenabeele P. (2008) Cell Death Differ. 15, 453–460 [DOI] [PubMed] [Google Scholar]
- 36.Dinser R., Pelled G., Müller-Ladner U., Gazit D., Neumann E. (2009) J. Tissue Eng. Regen. Med. 3, 124–128 [DOI] [PubMed] [Google Scholar]
- 37.Li Y., Maedler K., Shu L., Haataja L. (2008) PLoS One 3, e1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li C., Hao M., Cao Z., Ding W., Graves-Deal R., Hu J., Piston D. W., Coffey R. J. (2007) Mol. Biol. Cell 18, 3081–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gandhi S., Muqit M. M., Stanyer L., Healy D. G., Abou-Sleiman P. M., Hargreaves I., Heales S., Ganguly M., Parsons L., Lees A. J., Latchman D. S., Holton J. L., Wood N. W., Revesz T. (2006) Brain 129, 1720–1731 [DOI] [PubMed] [Google Scholar]
- 40.Millar J. K., James R., Christie S., Porteous D. J. (2005) Mol. Cell Neurosci. 30, 477–484 [DOI] [PubMed] [Google Scholar]
- 41.James R., Adams R. R., Christie S., Buchanan S. R., Porteous D. J., Millar J. K. (2004) Mol. Cell Neurosci. 26, 112–122 [DOI] [PubMed] [Google Scholar]
- 42.Campuzano V., Montermini L., Moltò M. D., Pianese L., Cossée M., Cavalcanti F., Monros E., Rodius F., Duclos F., Monticelli A., Zara F., Cañizares J., Koutnikova H., Bidichandani S. I., Gellera C., Brice A., Trouillas P., De Michele G., Filla A., De Frutos R., Palau F., Patel P. I., Di Donato S., Mandel J. L., Cocozza S., Koenig M., Pandolfo M. (1996) Science 271, 1423–1427 [DOI] [PubMed] [Google Scholar]
- 43.Gibson T. J., Koonin E. V., Musco G., Pastore A., Bork P. (1996) Trends Neurosci. 19, 465–468 [DOI] [PubMed] [Google Scholar]
- 44.Babcock M., de Silva D., Oaks R., Davis-Kaplan S., Jiralerspong S., Montermini L., Pandolfo M., Kaplan J. (1997) Science 276, 1709–1712 [DOI] [PubMed] [Google Scholar]
- 45.Koutnikova H., Campuzano V., Foury F., Dollé P., Cazzalini O., Koenig M. (1997) Nat. Genet. 16, 345–351 [DOI] [PubMed] [Google Scholar]
- 46.Campuzano V., Montermini L., Lutz Y., Cova L., Hindelang C., Jiralerspong S., Trottier Y., Kish S. J., Faucheux B., Trouillas P., Authier F. J., Dürr A., Mandel J. L., Vescovi A., Pandolfo M., Koenig M. (1997) Hum. Mol. Genet. 6, 1771–1780 [DOI] [PubMed] [Google Scholar]
- 47.Chandra D., Liu J. W., Tang D. G. (2002) J. Biol. Chem. 277, 50842–50854 [DOI] [PubMed] [Google Scholar]
- 48.Vaux D. L., Cory S., Adams J. M. (1988) Nature 335, 440–442 [DOI] [PubMed] [Google Scholar]
- 49.Kroemer G. (1997) Nat. Med. 3, 614–620 [DOI] [PubMed] [Google Scholar]
- 50.Neupert W., Herrmann J. M. (2007) Annu. Rev. Biochem. 76, 723–749 [DOI] [PubMed] [Google Scholar]
- 51.Nakai K., Horton P. (1999) Trends Biochem. Sci. 24, 34–36 [DOI] [PubMed] [Google Scholar]
- 52.Claros M. G. (1995) Comput. Appl. Biosci. 11, 441–447 [DOI] [PubMed] [Google Scholar]
- 53.Murwantoko, Yano M., Ueta Y., Murasaki A., Kanda H., Oka C., Kawaichi M. (2004) Biochem. J. 381, 895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Runyon S. T., Zhang Y., Appleton B. A., Sazinsky S. L., Wu P., Pan B., Wiesmann C., Skelton N. J., Sidhu S. S. (2007) Protein Sci. 16, 2454–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Srinivasula S. M., Gupta S., Datta P., Zhang Z., Hegde R., Cheong N., Fernandes-Alnemri T., Alnemri E. S. (2003) J. Biol. Chem. 278, 31469–31472 [DOI] [PubMed] [Google Scholar]
- 56.Yang Q. H., Church-Hajduk R., Ren J., Newton M. L., Du C. (2003) Genes Dev. 17, 1487–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bartke T., Pohl C., Pyrowolakis G., Jentsch S. (2004) Mol. Cell 14, 801–811 [DOI] [PubMed] [Google Scholar]
- 58.Seong Y. M., Choi J. Y., Park H. J., Kim K. J., Ahn S. G., Seong G. H., Kim I. K., Kang S., Rhim H. (2004) J. Biol. Chem. 279, 37588–37596 [DOI] [PubMed] [Google Scholar]
- 59.Suzuki Y., Takahashi-Niki K., Akagi T., Hashikawa T., Takahashi R. (2004) Cell Death Differ. 11, 208–216 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.