Abstract
The voltage-gated proton channel Hv1 has a voltage sensor domain but lacks a pore domain. Although the C-terminal domain of Hv1 is known to be responsible for dimeric architecture of the channel, its role and structure are not known. We report that the full-length Hv1 is mainly localized in intracellular compartment membranes rather than the plasma membrane. Truncation of either the N or C terminus alone or both together revealed that the N-terminal deletion did not alter localization, but deletion of the C terminus either alone or together with the N terminus resulted in expression throughout the cell. These results indicate that the C terminus is essential for Hv1 localization but not the N terminus. In the 2.0 Å structure of the C-terminal domain, the two monomers form a dimer via a parallel α-helical coiled-coil, in which one chloride ion binds with the Nη atom of Arg264. A pH-dependent structural change of the protein has been observed, but it remains a dimer irrespective of pH value.
Keywords: Biophysics, Channels/Ion, Membrane/Proteins, Protein, Cell pH, Crystal Structure, Localization, pH-dependent
Introduction
Voltage-gated proton channel (Hv channel) currents were observed first in snail neurons (1) and later found in many mammalian cells, such as alveolar epithelium cells of the lung, microglia of the brain, skeletal muscle, and many blood cells, including macrophages, neutrophils, and eosinophils (2–5). The Hv channels in mammalian phagocytes were originally proposed to be responsible for the proton-transporting pathway, which regulates intracellular pH during oxygen consumption associated with phagocytosis, called “respiratory burst” (5, 6). Hv channels are activated by depolarization and intracellular acidification, whose activities maintain intracellular pH neutral to keep reactive oxygen species generation (7, 8). Hv channels not only regulate pH in cytoplasm but could also provide protons in the phagosome, a closed membrane compartment for killing and digestion of a pathogen (5). Hv channels are extremely selective for H+, with no detectable permeability to other cations (2, 9, 10). The voltage activation relationship of Hv channels depends strongly on both the intracellular pH (pHi) and extracellular pH (pHo). Increasing pHo or lowering pHi promotes H+ channel opening by shifting the activation threshold to more negative potentials (4, 5). Furthermore, Hv currents are known to be inhibited by submillimolar concentrations of Zn2+ and Cd2+ and other divalent cations (11).
Recently, the human and mouse voltage-gated proton channels, called Hv1 and mVSOP, respectively, were identified using bioinformatics searches based on known cation channels (Hv1) and the voltage sensor domain of Ciona intestinalis VSP (mVSOP) (12, 13). Hv1/mVSOP currents are activated under depolarizing voltage, sensitive to the membrane pH gradient, H+-selective, and Zn2+-sensitive. Hv1/mVSOP is a predicted 273/269 amino acids in length and contains three predicted domains: N-terminal acid and proline-rich domain, transmembrane voltage sensor domain, and C-terminal domain. Voltage-gated K+ channels are composed of four subunits, each of which has a pore domain and a voltage sensor domain. The four pore domains come together to form one single central pore, and four peripheral voltage sensor domains control the gate of the pore (14). In contrast to the voltage-gated K+ channels, the Hv1/mVSOP contains a voltage sensor domain but lacks the pore domain or the phosphatase domain.
Recent studies showed that Hv1/mVSOP functions as a dimer in which the intracellular C-terminal domain is responsible for the dimeric architecture of the proteins, and each subunit contains its own pore (15–17). However, the role and structure of the C terminus of Hv1 are not well known. In this present study, we found that the Hv1 is expressed in intracellular compartment membrane in HeLa cells. The C terminus but not the N terminus is essential for Hv1 localization. In the 2.0 Å structure, two monomers form a dimer via parallel α-helical coiled-coil interaction. Furthermore, a pH-dependent structural change of the protein has been observed, but its dimeric state is irrespective of pH value. These results implicate that the C terminus might regulate the H+ permeability of Hv1 through its structural changes dependent on intracellular pH.
EXPERIMENTAL PROCEDURES
Expression Vectors
Two different kinds of Hv1 expression plasmids were constructed. The Hv1 cDNAs that were full-length (residues 1–273), N terminus-deleted (residues 101–273), C terminus-deleted (residues 1–220), and both N and C terminus-deleted (residues 101–220) (Fig. 1) were amplified by PCR from an expressed sequence tag clone (IMAGE 6424182). The PCR products were subcloned into pEGFP-N1 (Clontech) and pEGFP-C1 (Clontech) between the EcoRI and BamHI sites to create the protein expression plasmids, pHv1-EGFP, pΔN-Hv1-EGFP, pHv1-ΔC-EGFP, and pΔN-Hv1-ΔC-EGFP, fused with the enhanced green fluorescent protein (EGFP)3 moiety attached to the C terminus of Hv1, and pEGFP-Hv1, pEGFP-ΔN-Hv1, pEGFP-Hv1-ΔC, and pEGFP-ΔN-Hv1-ΔC, fused with the EGFP moiety to the N terminus of Hv1.
FIGURE 1.
Schematic domain structures of Hv1 and its deletion mutants. The numbers indicate the boundaries of these domains. TM, transmembrane domain.
Cell Culture and Transfections
For this study, HeLa cells were used. The HeLa cells were cultured on glass coverslips in 6-well plates in Dulbecco's modified Eagle's medium (Invitrogen) with 10% fetal bovine serum plus antibiotics (100 units/ml penicillin and 100 μg/ml streptomycin; Invitrogen) in a 5% CO2 incubator at 37 °C. Cells were transiently transfected at 50–70% confluence with the protein expression plasmids by using FuGENE6 (Roche Applied Science), following the manufacturer's protocol. Cells were used for experiments 36–48 h after transfection.
Confocal Laser-scanning Microscopy
Confocal images were acquired on an FV1000S-IX81 microscope (Olympus, Japan) with the fluorescein isothiocyanate filter set for EGFP and the 4′,6-diamidino-2-phenylindole filter set for the nuclear 4′,6-diamidino-2-phenylindole dye. The images were processed by Olympus FLUOVIEW Version 1.6 Viewer software and Adobe Photoshop software.
Fluorescence of EGFP and Rhodamine Phalloidin
HeLa cells on glass coverslips were transfected with the protein expression plasmids for 48 h, washed with serum-free Dulbecco's modified Eagle's medium, and subsequently incubated with rhodamine phalloidin (Invitrogen) that binds to F-actin to delineate the cell's edges, in serum-free Dulbecco's modified Eagle's medium at 37 °C for 30 min, following the manufacturer's protocol. The cells preloaded with rhodamine phalloidin were rinsed with PBS for 5 min for three times and added to Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. The cells on coverslips were directly viewed and photographed under DM RXA2 microscopy (Leica, Germany) with the fluorescein isothiocyanate filter set (excitation at 488 nm) for EGFP fluorescence and rhodamine (excitation at 540 nm) for rhodamine phalloidin fluorescence. The images were recorded immediately to avoid photobleaching.
Expression, Purification, and Crystallization of C Terminus
Expression, purification, and crystallization of recombinant C-terminal Hv1 (residues 221–273) were carried out as described previously (18), with the exception of all buffers for ion exchange containing 5 mm β-mercaptoethanol to protect the forming of the disulfide bond from one free cysteine in the protein. Crystals were grown in 3.0–3.4 m NaCl, 0.1 m sodium citrate, pH 5.0, at 16 °C by hanging drop vapor diffusion. Equal volumes (2 μl) of frozen stock protein (5 mg/ml) were mixed with reservoir solution. Crystals appeared after 1 month. Heavy atom soaks were prepared by soaking crystals in 1 mm HgCl2 solution containing 3.0 m NaCl, 0.1 m sodium citrate (pH 5.0) for 12 h.
Data Collection, Phasing, and Refinement
Crystals were cryoprotected with 20% glycerol containing 0.1 m citrate buffer, pH 5.0, 3 m NaCl, harvested with a cryoloop (Hampton Research, Laguna Niguel, CA), plunged into liquid nitrogen, and stored at 100 K. Native and derivative diffraction data were collected at 100 K on beamline BL 17A (λ = 1.0 Å) at the Photon Factory (KEK, Tsukuba, Japan). All data were processed using MOSFLM and SCALA (19). The crystals belong to the space group P43 (a = b = 37.65 Å, c = 137.04 Å, α = β = γ = 90°) with 4 molecules/asymmetric unit (solvent content 32.2%). The C terminus structure was solved with native and mercury derivative crystals using the single isomorphous replacement with anomalous scattering method. Heavy atom derivative data were collected from a crystal soaked in cryosolution containing 1 mm HgCl2 for 12 h. Initial phases were determined using SOLVE (19). Phase improvement by density modification was performed using RESOLVE (19). The structure was built using ARP/warp (20) and COOT (21) and refined using REFMAC5 (22) with 5% of data set aside as a free set. During subsequent refinement, twin refinement was conducted using REFMAC5 (22). Residues 221–225 and 267–273 in chains A and B were disordered and were not visible in the electron density map. Almost all of the main-chain hydrogen bonding groups were involved in intramolecular α-helical hydrogen bonding. The stereochemistry of the final model was verified using PROCHECK (23). Data collection and refinement statistics are summarized in Table 1.
TABLE 1.
Summary of data collection and refinement statistics
Values in parentheses are for reflections in the highest resolution shell.
| Parameters | Values |
|
|---|---|---|
| Native | Mercury | |
| Data collection and processing statistics | ||
| Space group | P43 | P43 |
| a = b (Å) | 37.65 | 37.72 |
| c (Å) | 137.04 | 137.44 |
| α = β = γ (degrees) | 90 | 90 |
| Wavelength (Å) | 1.0 | 1.0 |
| Resolution (Å) | 37.65-2.00 (2.11-2.00) | 37.72-2.00 (2.11-2.00) |
| Measured | 80,077 | 131,655 |
| I/σI | 12.0 (4.5) | 10.7 (4.6) |
| Redundancy | 6.2 (6.2) | 10.1 (10.2) |
| Completeness (%) | 100 (100) | 100 (100) |
| Rmergea (%) | 8.1 (32.3) | 11.8 (42.6) |
| Refinement statistics | ||
| Resolution | 37.65-2.00 | |
| Protein atoms | 1430 | |
| Ligand atoms | 2 | |
| Water molecule | 68 | |
| Rwork/Rfreeb (%) | 25.2/26.6 | |
| Root mean square deviations | ||
| Bond lengths (Å) | 0.030 | |
| Bond angles (degrees) | 2.314 | |
a Rmerge = Σ|Ii − 〈I〉|/Σ|Ii, where Ii is the intensity of the ith obvervation and 〈I〉 is the average intensity of multiple observations of symmetry-related reflections.
b R = Σhkl‖Fo| − k|Fc‖/Σhkl|Fo|, where Fo and Fc are the observed and calculated structure factors, respectively. Rfree was calculated for a randomly chosen 5% of reflections, and Rwork was calculated for the remaining 95% of reflections.
Circular Dichroism Spectroscopy
CD data at far-UV (200–250 nm) were collected at 20 °C with 0.1-nm intervals at a rate of 20 nm/min and a bandwidth of 1.0 nm by using a light path length 1-mm cuvette on a Jasco-720 spectropolarimeter. The purified C terminus was dialyzed overnight against various pH value buffers containing 0.2 m NaCl and 0.5 mm dithioerythritol. The following buffers were used: for pH 4.0 and 5.0, 20 mm sodium acetate buffer; for pH 6.0, 20 mm MES buffer; for pH 7.0 and 8.0, 20 mm sodium phosphate buffer; for pH 9.0, 20 mm Tris-HCl buffer. The protein concentration of samples was 0.3 mg/ml. The mean residue weight was 115.5 for calculation of the mean residue ellipticity, [θ]. All CD spectra of the protein were subtracted by the corresponding base-line spectra of buffers used.
Size Exclusion Chromatography
The purified C terminus was dialyzed overnight against various pH value buffers containing 0.2 m NaCl and 1 mm dithioerythritol, as described above. The size exclusion chromatography experiments were carried out on a gel filtration column, Superdex 75 10/300GL (Amersham Biosciences), pre-equilibrated with the corresponding buffers, monitored by an ÄKTAexplorer work station (Amersham Biosciences). 100 μl of dialyzed samples, as described above, with a protein concentration of 2 mg/ml were eluted at a flow rate of 0.8 ml/min. Elution profiles were monitored with an in-line UV-900 detector (Amersham Biosciences) at 230 nm. Peak fractions were analyzed by SDS-PAGE.
Analytical Ultracentrifugation
The purified C terminus was dialyzed overnight against various pH value buffers containing 0.2 m NaCl and 0.5 mm dithioerythritol, as described above. Samples and dialysis buffers were loaded into the Kel-F two-sector cells (cell length 12 mm). Sedimentation equilibrium experiments were performed by using a Beckman Optima XL-A analytical ultracentrifugation with a Ti rotor at 30,000 rpm at 20 °C. Sedimentation equilibrium data were collected at a wavelength of 230 nm in scans every 5 min. The partial specific volume calculated from amino acid composition of the C terminus was 0.750. The sedimentation equilibrium data were analyzed by using the XL-A data analysis software (Beckman, version 4.01).
RESULTS
Hv1 Is Mainly Located in Intracellular Sites, and the C Terminus Is Essential for Its Localization
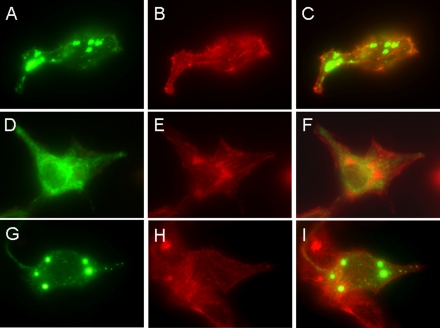
HeLa cells grown on glass coverslips were transiently transfected with the protein expression plasmids as described under “Experimental Procedures,” respectively, and observed by EGFP fluorescence under DM RXA2 microscopy. The cytoskeleton of the cells was labeled with rhodamine phalloidin that binds to F-actin to delineate the cell's edges. As shown in Fig. 2, Hv1 that was full-length (Fig. 2C) and N terminus-deleted (Fig. 2I) was expressed in the intracellular compartment membrane in HeLa cells, whereas the C terminus-deleted Hv1 was present over entire cell (Fig. 2F), demonstrating that the C terminus is essential for Hv1 localization. In nontransfected or EGFP only-transfected cells, the distinctive fluorescence was never seen.
FIGURE 2.
Localizations of full-length, C terminus-deleted, and N terminus-deleted Hv1. HeLa cells grown on glass coverslips were transfected with the expression plasmids, pHv1-EGFP, pΔC-Hv1-EGFP, and pΔN-Hv1-EGFP, and observed under a DM RXA2 microscope. The localizations of full-length (A), C terminus-deleted (D), and N terminus-deleted (G) Hv1 are shown in green. The cytoskeleton is labeled with rhodamine phalloidin (shown in red; B, E, and H). Images merging actin (red) with the full-length Hv1 (C), C terminus-deleted Hv1 (F), and N terminus-deleted Hv1 (I) (green) show that the full-length, C terminus-deleted, and N terminus-deleted Hv1 all localize in intracellular sites, in which the C terminus-deleted Hv1 is present over the entire cell.
To further confirm the localizations of Hv1 that was full-length, C terminus-deleted, and N terminus-deleted in HeLa cells, the cells transfected with the plasmids pHv1-EGFP pΔC-Hv1-EGFP and pΔN-Hv1-EGFP were observed by focal microscopy with the fluorescein isothiocyanate filter set for EGFP. As shown in Fig. 3, the full-length Hv1 is mainly expressed in intracellular sites in HeLa cells, but truncation of the C or N terminus alone alters the localization.
FIGURE 3.
The full-length Hv1 is mainly expressed in intracellular sites in HeLa cells, but truncation of the C or N terminus alone alters localization. HeLa cells grown on glass coverslips were transfected with the expression plasmids pHv1-EGFP, pΔC-Hv1-EGFP, and pΔN-Hv1-EGFP and observed by an FV1000S-IX81 focal microscope. The localizations of with full-length (A–C), C terminus-deleted (D–F), and N terminus-deleted (G–I) Hv1 are shown in green. B, E, and H, the phase-contrast images of the same fields corresponding to A, D, and G, respectively. C, F, and I, images that merge A and B, D and E, and G and H, respectively.
C Terminus Forms a Well Defined Coiled-coil Structure
To obtain the atomic structure of the C terminus of the human voltage-gated proton channel Hv1, we purified and crystallized the protein after expression in Escherichia coli (18) (Fig. 4). The SDS-PAGE of the purified protein under both reducing and non-reducing conditions showed that the cysteine (Cys249) in the protein does not form a disulfide bond (Fig. 4A), which is consistent with the result of Lee et al. (17). However, in the crystals of the protein, the disulfide bridge was formed in a time-dependent manner, as shown in Fig. 4B.
FIGURE 4.
Analysis of the formation of the disulfide bond in the C terminus by 12.5% SDS-PAGE. The SDS-PAGE was stained by Coomassie Brilliant Blue. MW, positions of the migrations of standard proteins and their molecular masses (in kDa) presented on the left. A, the freshly purified C terminus under both reducing (line 1) and non-reducing (line 2) conditions; B, the C terminus crystals under both reducing (line 1) and non-reducing (line 2) conditions, showing that the two cysteines in the dimeric C terminus form a disulfide bond in crystals.
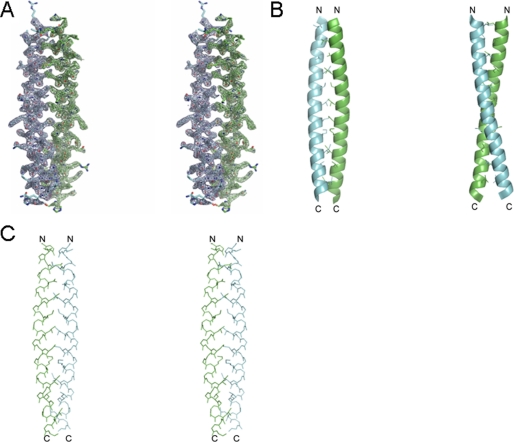
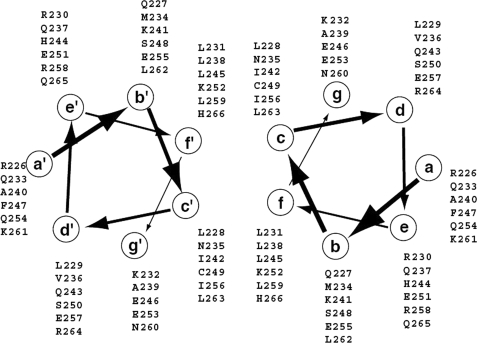
The crystallographic studies on the crystals of the protein showed that there are two antiparallel dimers in the crystallographic asymmetric unit. The final model consists of residues 226–266 with good stereochemistry and 70 solvent molecules with an Rfree of 26.6% and Rwork of 25.2% (Table 1). Residues 221–225 and 267–273 are not visible in the electron density map and must be disordered. The experimental electron density map at 2.0 Å resolution based on phases from a mercury derivative (Table 1) reveals a dimeric parallel-oriented coiled-coil quaternary structure (Fig. 5A). As seen in Fig. 5, B and C, the two helices form more or less a classic coiled-coil arrangement, each helix having a slight negative superhelical twist in order to maintain the interface along the length of the dimer. Following the conventional nomenclature for coiled-coil structures, the positions within each heptad repeat are named a–g, with the key residues of the interface occupying positions c and f. Assignment of the residues of Hv1 to positions a–g is straightforward based on the calculated structure, and the result is summarized in a schematic helical wheel representation in Fig. 6.
FIGURE 5.
Crystal structure of the C terminus. A, a stereoview of the 2.0 Å resolution 2|Fo| − |Fc| electron density map (contoured at 1.5σ) from residues 226–266 of Hv1. B, the backbone as a ribbon with side chains of interfacial residues added. C, a stereoview of the backbone and interfacial residues. C and N termini are indicated with the letters C and N, respectively.
FIGURE 6.
Helical wheel representation of the assignment of residues in the C terminus to positions within heptad repeats.
In the dimer of the C terminus, the parallel two-stranded coiled-coil is stabilized by complementary hydrophobic and polar interactions between the helices (Figs. 5 and 6). The residues that are involved in the interactions at positions c and f of the heptad repeats are leucine, isoleucine, asparagine, cysteine, lysine, and histidine. In addition, charged side chains and polar residues in positions b and g may help to stabilize the coiled-coil structure by local charge compensation. These interactions involve mainly basic side chains (Lys232-Lys241-Ser248) and acidic side chains (Glu246-Glu253-Glu255).
In ideal coiled-coils, the heptad motif a-b-c-d-e-f-g is repeated n times, and the a and d residues make up the hydrophobic core of the two helix bundles (24). In our case, the hydrophobic core is composed of the c and f residues, not the a and d residues. The buried surface consists of the alternation of three and four residues in the heptad repeat, but compared with other known coiled-coil structures, this assembly has a higher content of polar residues. The high frequency of favorable electrostatic interactions has not been observed previously in coiled-coil structures. It might underlie the pH dependence of the association/dissociation of dimers or structural changes.
The most remarkable feature of the structure is the presence of the asparagine, histidine, and cysteine residues (Asn235, Cys249, and His266) at the c and f positions. Asparagine, histidine, and cysteine residues are extremely rare at c or f positions in other coiled-coil structures,4 these interfacial positions being occupied almost universally by hydrophobic residues. The other interfacial positions in the structure are occupied predominantly by leucine and isoleucine residues, so that the interfacial asparagine, histidine, and cysteine residues pack extensively against these side chains.
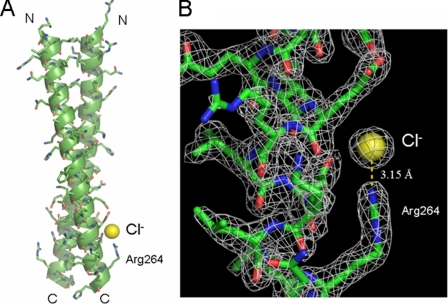
There is a “solvent molecule” with low temperature factor near the Nη atom of Arg264 of one monomer of the dimer. The temperature factor of the atom became lower when further refinements were done as a water molecule. The low temperature factor of the atom and high positive peaks in both |Fo| − |Fc| and 2|Fo| − |Fc| Fourier electron density maps indicated that the atom should be a Cl− ion. The Cl− ion with a temperature factor of 23.7 Å2 only binds with the Nη atom of Arg264 at a distance of 3.15 Å via a hydrogen interaction, and no water molecules interacting with the Cl− ion have been observed (Fig. 7).
FIGURE 7.
A, a Cl− ion near the Nη atom of Arg264 of one monomer of the dimer, in which the backbone of the dimeric coiled-coil structure is depicted as a ribbon with all side chains. B, 2|Fo| − |Fc| electron density map of the Cl− binding site with a contour level of 1.5σ, in which the Cl− ion only binds with the Nη atom of Arg264 at a distance of 3.15 Å via a hydrogen interaction.
pH-dependent Structural Changes
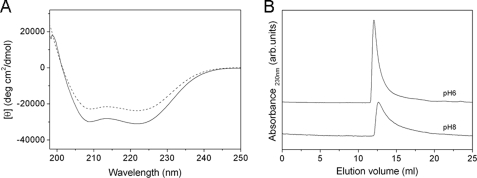
The fact that the H+ permeability of Hv1 depends strongly on the pHi (4, 5) prompted a study of changes in secondary structure and oligomeric state of the C terminus in response to pH. The CD spectra of the C terminus revealed changes in secondary structure in response to change with pH values (Fig. 8A). At pH 6.0, the spectrum of the protein showed two pronounced double minima at 222 and 208 nm, characteristic of α-helical secondary structure (25). The molar ellipticities at 222 and 208 nm were −31010.4 and −29356.1 degrees cm2 dmol−1, respectively. The analysis by the K2d program showed that the C terminus secondary structure at this pH comprised nearly 94% α-helix, 0% β-sheet, and only 6% random coil. At pH 8.0, the characteristic double minima of α-helix was remained; however, the molar ellipticities at 222 and 208 nm were changed from −31,010.4 to −23,775.9 degrees cm2 dmol−1 and from −29,356.1 to −22,405.1 degrees cm2 dmol−1, respectively, in contrast to pH 6.0, indicating that a decrease in secondary structure occurred. The secondary structure of the C terminus at pH 8.0 analyzed by the K2d program was 79% α-helix, 0% β-sheet, and 21% random coil. Furthermore, we also measured the CD spectra of the protein at pH 4.0, pH 5.0, pH 7.0, and pH 9.0. The results showed that the protein at these pH values only contains α-helical secondary structure and no β-sheet secondary structure (data not shown).
FIGURE 8.
A, far-UV CD spectra of the C terminus at pH 6.0 (solid line), and pH 8.0 (dashed line). CD spectra were carried out at a protein concentration of 0.3 mg/ml at 20 °C. For pH 6.0 and pH 8.0, 20 mm MES and 20 mm sodium phosphate were used, respectively, both of which contained 0.2 m NaCl and 0.5 mm dithioerythritol. Base lines measured for the buffers alone were subtracted from both of the original spectra. B, size exclusion chromatography of the C terminus on a Superdex 75 10/300GL gel filtration column eluted with 20 mm MES (pH 6.0) buffer and 20 mm sodium phosphate (pH 8.0) buffer, respectively, both of which contained 0.2 m NaCl and 1 mm dithioerythritol, at a flow rate of 0.8 ml/min and monitored at 230 nm. The concentration of the protein was 2.0 mg/ml.
The far-UV CD spectrum revealed a pH-dependent secondary structural change of the C terminus. To examine the pH-dependent tertiary structural changes and estimate the pH-dependent assembly behavior of the protein, size exclusion chromatography and sedimentation equilibrium were employed. Size exclusion chromatography is a useful method for estimating the hydrodynamic dimensions of a protein and can elucidate the compactness of the protein's tertiary structure (26). The C terminus in various pH ranges was eluted on size exclusion chromatography. Only one elution peak exhibiting asymmetry was observed for each pH value. This result indicated that the C terminus exists as a homogeneous aggregation in different pH solution. Fig. 8B showed the elution patterns of the C terminus on size exclusion chromatography for pH 6.0 and pH 8.0. At pH 6.0, the elution peak of the protein was at 12.1 ml and sharp, whereas at pH 8.0, the elution peak was moved to 12.7 ml and broadened, demonstrating a pH-dependent structural change. The sedimentation equilibrium analysis showed that the protein remains a dimer from pH 4.0 to pH 9.0. The measured molecular masses of the protein at pH 6.0 and pH 8.0 were 13,585 Da and 12,765 Da, which were in good agreement with its dimeric molecular mass of 13,394 Da predicted from its amino acid sequence (Fig. 9). This result also showed that the C terminus remains a dimer, like the whole Hv1 molecule.
FIGURE 9.
Analysis of the oligomeric states of the C terminus at pH 6.0 (A) and pH 8.0 (B) by sedimentation equilibrium analysis. The sedimentation equilibrium experiments were performed at a protein concentration of 1.0 mg/ml at 20 °C in 20 mm MES buffer for pH 6.0 and 20 mm sodium phosphate buffer for pH 8.0, both of which contained 0.2 m NaCl and 0.5 mm dithioerythritol.
DISCUSSION
The results from the expression of the full-length, N terminus- or C terminus-truncated, and both N and C terminus-truncated Hv1, for the first time, indicate that Hv1 is mainly expressed in intracellular sites in HeLa cells, and the presence of the C-terminal domain is essential for Hv1 localization. The crystal structure of the C terminus shows that the C terminus forms a dimer and exhibits a coiled-coil structure. One chloride ion binding to residue Arg264 has been observed in the crystal structure, but no water molecules interacting with the chloride ion have been seen. Furthermore, a pH-dependent structural change of the C terminus has also been observed, but the dimerization of the C terminus is irrespective of pH value.
Recent studies showed that the voltage-gated proton channel functions as a dimer in the cell membrane, and each subunit has its own conduction pathway, in which the intracellular C-terminal domain is responsible for the dimeric architecture of the protein (15–17). Lee et al. (17) found that Hv1 is a dimer in the cell membrane by DSS cross-linking and defined the dimer interfaces by cysteine cross-linking, in which the C terminus forms the dimer interface on the intracellular side of the membrane. The cysteine (Cys249) in the C terminus may help to stabilize the dimer, but a disulfide bridge at this position is not essential. Koch et al. (15) showed that mVSOP channel is expressed as a dimer, and truncation of the cytoplasmic regions of mVSOP significantly reduced the dimerization. The C terminus-deleted mVSOP and especially both the C and N terminus-deleted mVSOP opened much faster than wild type. Tombola et al. (16) also showed that Hv1 assembles as a dimer. In summary, the recent studies on the proton channel showed that the C terminus is essential to the channel activity. The N-terminal region is largely unstructured and does not affect the oligomeric state of the protein, confirmed by limited proteolysis and preliminary biochemical data analysis (17). In our case, the C terminus-deleted Hv1 is expressed over entire HeLa cells but not the N terminus-deleted Hv1. Our results suggest strongly that the C terminus but not the N terminus is essential for Hv1 localization. Furthermore, we expressed N-terminal domain of Hv1 as an inclusion body in E. coli and attempted to refold it in the presence of the C-terminal domain. However, we could not obtain a complex of the N and C termini (data not be shown).
The C terminus of Hv1 is a homodimer with a parallel coiled-coil structure. The C terminus of many ion channels exhibits a coiled-coil structure that is known to mediate subunit interactions in assembly and functions (27–29). The most remarkable feature of the structure is the presence of two cysteine residues (Cys249) at the hydrophobic core of the two helix bundles. A disulfide bond between the two cysteines has been observed in the crystal structure. The formation of the disulfide bond is due to the oxidation of two hydrosulfide groups of the two cysteines during crystallization. Furthermore, one chloride ion in the dimer has been observed, which binds only with the side chain of Arg264. No water molecules that interact with the chloride ion have been observed. Several mechanisms of pH regulation of voltage-gated proton channels have been proposed (5, 30). Byerly et al. (30) suggested that protonation of specific acidic sites that regulate H+ channel gating must exist and must be accessible to the external and internal solutions. Whether the binding site of the chloride ion to Arg264 is involved in the protonation and the protonation of the site is unknown.
The activity of Hv channels depends strongly on both the pHi and pHo (4, 5), suggesting a pH-dependent structural change and oligomeric state of the C terminus. We used three different techniques to assess pH-dependent structural changes and assembly behavior of the C terminus: CD spectroscopy, size exclusion chromatography, and sedimentation equilibrium analysis. First, we measured the CD spectra of the protein from pH 4.0 to pH 9.0. The protein only contains α-helical content and no β-sheet, irrespective of pH value, and the secondary structure is increased with a pH decrease. Second, we eluted the protein in various pH values from pH 4.0 to 9.0 on a gel filtration column. A structural change occurred at pH 7.0. Below pH 7.0, a sharp elution peak at almost the same position was observed; however, above pH 7.0, the peak became broad and appeared at a larger elution volume in contrast to below pH 7.0. This demonstrates that the structural changes of the protein are dependent on pH value. Although the structural changes of the protein depend on solution pH value, its oligomeric state is independent of pH value, confirmed by sedimentation equilibrium analytical ultracentrifugation.
The Hv channels have been described in the plasma or phagosome membranes of many blood cells, including macrophage, neutrophils, and eosinophils, that undergo phagocytosis. However, the Hv1 described here is localized in intracellular compartment membranes in HeLa cells, in which the C terminus is essential for the protein localization. The differential localization of Hv1 in differential cells might indicate that there is some novel function for the protein. The C terminus is known to regulate the dimerization of Hv1. It is possible that the dimeric form of Hv1 is more stable than the monomeric state of the C terminus-truncated Hv1 in the membranes. Whether the C terminus affects the localization of Hv1 in plasma membrane and phagosome membranes in phagocytes is not clear yet.
In conclusion, we found that Hv1 is mainly confined to localize in the intracellular compartment membrane in HeLa cells but not on the cell surface. The C terminus is essential for the channel localization. The crystallographic studies showed that the C terminus forms a dimer via a parallel α-helical coiled-coil. A pH-dependent structural change of the C terminus has been observed, but it remains a dimer irrespective of pH value. In the intact channel, the exactly dimeric coiled-coil structure and the pH-dependent conformational changes of the C terminus may be different than these of the isolated C terminus.
Acknowledgments
We are deeply grateful to Prof. Tomitake Tsukihara (Institute for Protein Research, Osaka University) for support and encouragement of the research and many useful discussions on the crystallization and the x-ray crystallographic analysis. We especially thank Prof. Thomas E. DeCoursey (Department of Molecular Biophysics and Physiology, Rush University Medical Center) for many helpful discussions about the manuscript. We also thank Miyo Sakai (Institute for Protein Research, Osaka University) for excellent technical assistance with the analytical ultracentrifugation experiments.
This work was supported by National Natural Science Foundation of China Grants 30840028, 30970579, and 30623005; “863” program from the Chinese Ministry of Science and Technology Grant 2006AA02Z173; Basic Science and Advance Technology Research Program of Tianjin Grant 08JCYBJC25800; and Ph.D. Program Foundation of the Ministry of Education of China Grant 200800551035.
The atomic coordinates and structure factors (code 3A2A) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
M. Suzuki, personal communication.
- EGFP
- enhanced green fluorescent protein
- MES
- 4-morpholineethanesulfonic acid.
REFERENCES
- 1.Thomas R. C., Meech R. W. (1982) Nature 299, 826–828 [DOI] [PubMed] [Google Scholar]
- 2.DeCoursey T. E. (1991) Biophys. J. 60, 1243–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kapus A., Romanek R., Qu A. Y., Rotstein O. D., Grinstein S. (1993) J. Gen. Physiol. 102, 729–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eder C., DeCoursey T. E. (2001) Prog. Neurobiol. 64, 277–305 [DOI] [PubMed] [Google Scholar]
- 5.Decoursey T. E. (2003) Physiol. Rev. 83, 475–579 [DOI] [PubMed] [Google Scholar]
- 6.Henderson L. M., Chappell J. B., Jones O. T. (1987) Biochem. J. 246, 325–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark R. A., Leidal K. G., Pearson D. W., Nauseef W. M. (1987) J. Biol. Chem. 262, 4065–4074 [PubMed] [Google Scholar]
- 8.Morgan D., Cherny V. V., Murphy R., Katz B. Z., DeCoursey T. E. (2005) J. Physiol. 569, 419–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeCoursey T. E., Cherny V. V. (1994) J. Membr. Biol. 141, 203–223 [DOI] [PubMed] [Google Scholar]
- 10.Cherny V. V., Markin V. S., DeCoursey T. E. (1995) J. Gen. Physiol. 105, 861–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cherny V. V., DeCoursey T. E. (1999) J. Gen. Physiol. 114, 819–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramsey I. S., Moran M. M., Chong J. A., Clapham D. E. (2006) Nature 440, 1213–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sasaki M., Takagi M., Okamura Y. (2006) Science 312, 589–592 [DOI] [PubMed] [Google Scholar]
- 14.Long S. B., Campbell E. B., Mackinnon R. (2005) Science 309, 897–903 [DOI] [PubMed] [Google Scholar]
- 15.Koch H. P., Kurokawa T., Okochi Y., Sasaki M., Okamura Y., Larsson H. P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9111–9116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tombola F., Ulbrich M. H., Isacoff E. Y. (2008) Neuron 58, 546–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee S. Y., Letts J. A., Mackinnon R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7692–7695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li S. J., Zhao Q., Zhou Q., Zhai Y. (2009) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65, 279–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collaborative Computational Project 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 20.Morris R. J., Perrakis A., Lamzin V. S. (2003) Methods Enzymol. 374, 229–244 [DOI] [PubMed] [Google Scholar]
- 21.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 22.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 23.Laskowski R. A., Macarthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 24.Lupas A. (1996) Trends Biochem. Sci. 21, 375–382 [PubMed] [Google Scholar]
- 25.Saxena V. P., Wetlaufer D. B. (1971) Proc. Natl. Acad. Sci. U.S.A. 68, 969–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uversky V. N. (1993) Biochemistry 32, 13288–13298 [DOI] [PubMed] [Google Scholar]
- 27.Margeta-Mitrovic M., Jan Y. N., Jan L. Y. (2000) Neuron 27, 97–106 [DOI] [PubMed] [Google Scholar]
- 28.Mei Z. Z., Xia R., Beech D. J., Jiang L. H. (2006) J. Biol. Chem. 281, 38748–38756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiener R., Haitin Y., Shamgar L., Fernández-Alonso M. C., Martos A., Chomsky-Hecht O., Rivas G., Attali B., Hirsch J. A. (2008) J. Biol. Chem. 283, 5815–5830 [DOI] [PubMed] [Google Scholar]
- 30.Byerly L., Meech R., Moody W., Jr. (1984) J. Physiol. 351, 199–216 [DOI] [PMC free article] [PubMed] [Google Scholar]