Abstract
Arabidopsis thaliana sucrose nonfermenting 1-related protein kinase 1 complexes belong to the SNF1/AMPK/SnRK1 protein kinase family that shares an ancestral function as central regulators of metabolism. In A. thaliana, the products of AtSnAK1 and AtSnAK2, orthologous to yeast genes, have been shown to autophosphorylate and to phosphorylate/activate the AtSnRK1.1 catalytic subunit on Thr175. The phosphorylation of these kinases has been investigated by site-directed mutagenesis and tandem mass spectrometry. The autophosphorylation site of AtSnAK2 was identified as Thr154, and it was shown to be required for AtSnAK catalytic activity. Interestingly, activated AtSnRK1 exerted a negative feedback phosphorylation on AtSnAK2 at Ser261 (Ser260 of AtSnAK1) that was dependent on AtSnAK autophosphorylation. The dynamics of these reciprocal phosphorylation events on the different kinases was established, and structural modeling allowed clarification of the topography of the AtSnAK phosphorylation sites. A mechanism is proposed to explain the observed changes in the enzymatic properties of each kinase triggered by these phosphorylation events.
Keywords: Arabidopsis, Carbohydrate/Plant, Methods/Site-directed Mutagenesis, Phosphorylation/Kinases/Serine-Threonine, Protein/Posttranslational Modification, Signal Transduction/Protein Kinases/Serine/Threonine, AMP-activated Protein Kinase, SNF1-related Protein Kinase 1
Introduction
The SNF14 (sucrose nonfermenting 1)/AMPK (AMP-activated protein kinase)/SnRK1 (SNF1-related protein kinase 1) protein kinase family is a major component of cell energy homeostasis maintenance in Eukaryotes. The conservation of this broad physiological function is accompanied by structural similarities in yeast, mammals, and plants (1–3). In all Eukaryotes, these complexes are composed of three subunits, α, β, and γ, which are the catalytic and the two regulatory subunits, respectively. In Arabidopsis thaliana, two catalytic α-subunits (AtSnRK1.1, AtSnRK1.2) have been shown to interact with the three β-subunits and the two γ-subunits (2). In plants, SnRK1 is involved in the broad regulation of gene expression (4) and the post-translational regulation of key enzymes, e.g. sucrose phosphate synthase and nitrate reductase (5), and it is considered to be a central regulator of metabolism and energy balance (4).
In Saccharomyces cerevisiae, three Ser/Thr protein kinases TOS3, PAK1, and ELM1, are known to activate SNF1 via the phosphorylation of Thr210 located in the canonical phosphorylation domain called T-loop of the α subunit (6). Likewise, in mammals three upstream kinases LKB1 (7), CaMKKβ (8) and TAK1 (9) phosphorylate AMPKα subunits on their Thr172 (10). Moreover, mammalian LKB1 can phosphorylate/activate plant SnRK1, thus indicating that elements of the SnRK1/AMPK pathway may have been conserved during evolution (11).
Indeed, plant SnRK1α subunits contain the T-loop with the conserved threonine residue at position 175 for AtSnRK1.1 and 176 for AtSnRK1.2 (12). Two Arabidopsis protein kinases, AtSnAK1 and 2 (Arabidopsis SnRK1-activating kinases) (13), also named GRIK2 and 1 (geminivirus rep-interacting kinases), respectively (14), have been shown to rescue the yeast tos3 pak1 elm1 triple mutant by restoring SNF1 upstream kinase activity. Furthermore, it has been demonstrated recently that they can phosphorylate truncated AtSnRK1 catalytic subunits in vitro (15), making them good candidates as physiological upstream kinases of AtSnRK1. In addition, it was reported that both AtSnAKs have the capacity to autophosphorylate. However, both the site and the role of this autophosphorylation process were not defined. Inactive AtSnAK2 (mutated in the ATP binding site) showed a substantial decrease in its capacity to interact with AtSnRK1 (15). This suggested that the autophosphorylation of AtSnAK and/or the AtSnRK1 trans-phosphorylation sites were important for kinase interactions (15).
In this work, by using recombinant protein technology combined with tandem mass spectrometry (MS/MS), it is shown that AtSnAK can phosphorylate/activate in vitro nontruncated AtSnRK1 catalytic subunits (AtSnRK1.1 and AtSnRK1.2). The function of AtSnAK autophosphorylation was examined, and it was found that this process is required for AtSnAK catalytic activity. Furthermore, the kinetics and function of a reciprocal phosphorylation process exerted by phosphorylated/activated AtSnRK1.1 that inhibits AtSnAK activity were unraveled. Structural modeling allowed us to suggest a model to explain the mechanisms involved in AtSnAK regulation by phosphorylation. Our biochemical analysis of the reciprocal phosphorylations of AtSnRK1 and AtSnAK kinases highlights novel regulatory mechanisms in the AtSnRK1/AtSnAK pathway that could be maintained in their yeast and animal (LKB1/AMPK) counterparts.
EXPERIMENTAL PROCEDURES
Materials
Oligonucleotides used for PCR amplification and direct site mutagenesis of cDNAs are summarized in supplemental Table S1. AtSnAK1 (At5g60550, clone U82608) and AtSnAK2 (At3g45240, clone U09417) cDNAs were obtained from the TAIR resource center. AtSnAK, AtSnRK1.1 (At3g01090), and AtSnRK1.2 (At3g29160) cDNAs (2) were amplified using specific primers containing the attB1 (in 5′) and attB2 (in 3′) adapter sequences allowing BP recombination within the Gateway® system against the pDONR201 plasmid (Invitrogen). After selection (16), the cDNAs were subcloned into pDEST15 (Invitrogen) using LR recombination. Plasmids were used to transform Escherichia coli BL21 cells (17) for recombinant protein expression.
The mutated coding sequences were obtained by two-strand PCR amplification. Two strands were amplified with forward/midantisense primers and midsense/reverse primers, respectively, and then this two-strand mixture was taken as a template to amplify specific mutations using their corresponding forward/reverse primers. Both midsense and midantisense primers used for mutations are listed in supplemental Table S1.
Production and Purification of Recombinant Proteins
Recombinant proteins were purified from E. coli extracts using glutathione-agarose affinity chromatography (G4510; Sigma-Aldrich) according to the manufacturer's instructions. After SDS-PAGE (18), mass spectrometry (MALDI-TOF) was performed to identify protein bands (Unité de Biochimie Bactérienne, PAPPSO, Bâtiment 526, Domaine de Vilvert, 78352 Jouy en Josas Cedex, France).
Western Blotting
After SDS-PAGE, semidry protein transfer onto polyvinylidene difluoride membranes (P2938; Sigma-Aldrich) using a Trans-blot semidry (Bio-Rad) was performed for 40 min at 20 V in 25 mm Tris, 192 mm glycine, 0.1% SDS, 20% ethanol. For hybridization, SNAP i.d. was used as recommended by Millipore. Anti-GST antibodies from Calbiochem (OB03) and anti-mouse secondary antibodies coupled to horse radish peroxidase (Pierce) were used at 10 μl/3 ml and 0.5 μl/3 ml, respectively. ECL (NEL103001EA; PerkinElmer Life Sciences) was performed to detect GST-containing proteins.
Autoradiography and MS/MS
After SDS-PAGE of in vitro 32P-labeled proteins, signals were revealed using a PhosphorImager (Bio-Rad) following the manufacturer's instructions. Proteins and corresponding peptides were identified using MS/MS as described in the supplemental Experimental Procedures.
Kinase Activity Assay
Kinase activity was measured in the presence of 2 μCi of [γ-32P]ATP (500 μCi/mmol) and 90 μm AMARA peptide (AMARAASAAALARRR) (19) in kinase activity buffer containing 0.1 m HEPES-NaOH, pH 7.3; 5 mm dithiothreitol; 10 mm MgCl2; 0.5 mm EGTA; 20, 30, or 1000 μm ATP; 1 μl of antiprotease mixture (P9599; Sigma-Aldrich) for 1 ml of buffer; 1 μl of each antiphosphatase mixture (P2850 and P5726; Sigma-Aldrich) for 1 ml of buffer, at 30 °C in a total volume of 50 μl. Following 30–60 min of incubation, an aliquot of 10 μl (two replicates) was spotted onto 1 cm2 of Whatman P81 cation-exchange paper. The papers were immediately washed once for 10 min and then twice for 5 min in 200 ml of 1% H3PO4 and dried by a quick wash in acetone. 32P incorporation into the AMARA peptide was counted using a scintillation spectrometer (LS6500; Beckman-Coulter) in 3 ml of scintillation liquid (Ready GelTM P/N596601; Beckman-Coulter).
Modeling the Three-dimensional Structure of AtSnAK1
The AtSnAK1 protein sequence (At5g60550, Q5HZ38) between residues 104 and 374 was modeled using the ROCK1 crystal structure (Protein Data Bank (PDB) code 2ESM chain B) as template by Swiss Model (20). All work with this model, including surface modeling, was done using PyMOL from DeLano Scientific LLC. The AtSnAK1 model structure was compared successfully with the crystal structure of TAK1 (PDB code 2EVA) and the SNF1 kinase domain (PDB code 3HYH). The ATP molecule was added to the AtSnAK1 model from the MEK1 structure (PDB code 3E8N) after superposition of their kinase domains.
RESULTS
AtSnAK Phosphorylates/Activates AtSnRK1.1
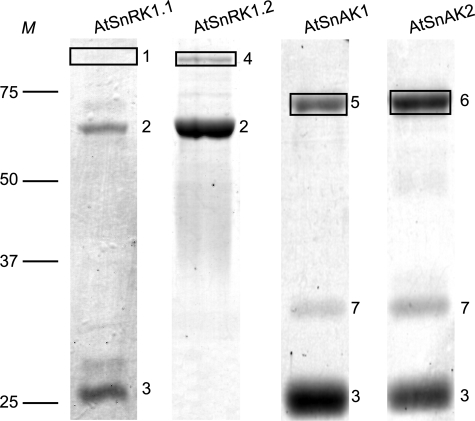
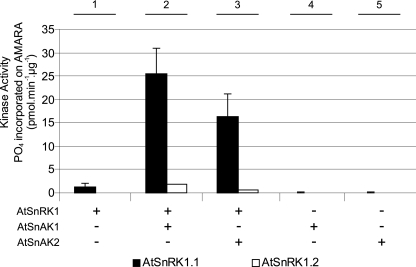
After production in E. coli and affinity purification using glutathione-agarose, SDS-PAGE coupled to mass spectrometry (MALDI-TOF) analysis of the bands confirmed the production of AtSnRK1.1, AtSnRK1.2, AtSnAK1, and AtSnAK2 fused to a GST tag (Fig. 1). Recombinant AtSnRK1.1 and 1.2 alone displayed very little, or no in vitro activity (Fig. 2, condition 1) using the classical phosphorylation assay with the AMARA peptide (19), but they were activated, especially AtSnRK1.1, following incubation with either AtSnAK1 or AtSnAK2 (Fig. 2, conditions 2 and 3). No phosphorylation of the AMARA peptide was observed with AtSnAK alone (Fig. 2, conditions 4 and 5). This is in good agreement with recent data reported by Shen et al. (15) on truncated AtSnRK1. Considering the higher activity displayed by AtSnRK1.1 compared with AtSnRK1.2, as previously described in plant extracts (21), it was decided to use AtSnRK1.1 to study AtSnRK1 phosphorylation and AtSnAK function.
FIGURE 1.
Purification and identification of AtSnRK1 and AtSnAK proteins. Recombinant proteins (5 μg of preparation) produced in E. coli and purified by GSH affinity chromatography were submitted to SDS-PAGE and stained with Coomassie Blue. Boxed bands are for proteins expected to be produced, and numbers indicate major bands. Most of them were identified by mass spectrometry (MALDI-TOF); 1, AtSnRK1.1-GST (84 kDa); 2, groEL (60 kDa), which might appear because of a possible SnRK1 protein toxicity; 3, GST (25 kDa); 4, AtSnRK1.2-GST (84 kDa); 5, AtSnAK1-GST (70 kDa); 6, AtSnAK2-GST (70 kDa); 7, unknown nonspecific contaminant protein. M, protein molecular mass markers in kDa.
FIGURE 2.
Activation of AtSnRK1 catalytic subunits by AtSnAK. AtSnRK1.1-GST and AtSnRK1.2-GST (1 μg of each preparation) were incubated with each AtSnAK-GST (0.5 μg of each preparation) in the reconstituted assay containing 2 μCi of [γ-32P]ATP, 30 μm ATP, and 90 μm AMARA peptide, at 30 °C for 30 min. Radioactivity incorporated onto the AMARA peptide was counted. The results are the average of three independent experiments ± S.D. (error bars).
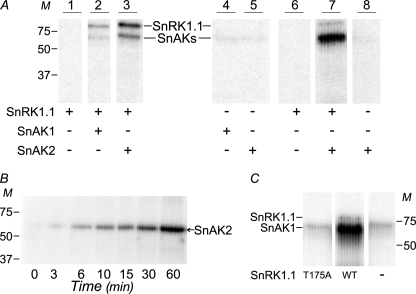
To test whether AtSnRK1 activation involved AtSnRK1 phosphorylation by AtSnAK, the recombinant proteins were subjected to radiolabeling, separated by SDS-PAGE, and autoradiographed (Fig. 3A). This showed that recombinant AtSnRK1.1 was indeed phosphorylated (Fig. 3A, lanes 2 and 3) under activating conditions, whereas no labeling was detected in the absence of AtSnAK (Fig. 3A, lane 1) or AtSnRK1 (Fig. 3A, lanes 4 and 5).
FIGURE 3.
Phosphorylations of AtSnRK1 by AtSnAK. A, purified proteins (0.5 μg of AtSnAKs-GST and 10 μg of AtSnRK1.1-GST preparations) were incubated in kinase activity buffer (containing 30 μm total ATP) supplemented with 2 μCi of [γ-32P]ATP at 30 °C for 60 min and then submitted to SDS-PAGE and autoradiography. In lanes 6–8, assays contained 1 μg of AtSnAK2-GST and/or 3 μg of AtSnRK1.1-GST preparations. The positions of AtSnRK1.1-GST (84 kDa, named SnRK1.1) and AtSnAKs-GST (70 kDa, named SnAKs) are indicated. Lanes 1-5 are from the same autoradiograph whereas lanes 6-8 are from a different autoradiograph. B, purified AtSnAK2 (5 μg of protein preparation) was incubated for the indicated times in the reconstituted assay as described above and analyzed by SDS-PAGE and autoradiography. C, AtSnAK1 was incubated with mutated (T175A) or wild-type (WT) AtSnRK1.1 (5 μg of each protein preparation), in the reconstituted assay and analyzed as described above. All lanes are from the same autoradiograph. M, molecular mass in kDa. All results are representative of three independent experiments.
MS/MS analyses showed that AtSnAK phosphorylated AtSnRK1.1 on Thr175 located in the canonical T-loop sequence: FLKT(175)SCG (supplemental Fig. S1). This process led to the activation of AtSnRK1, as shown previously using a truncated protein (15) and as expected from the phosphorylation site on the animal and yeast homologs (6–9). These data definitely establish that the AtSnAK target on AtSnRK1.1 is the conserved threonine residue in the T-loop and that its phosphorylation confers AtSnRK1 kinase activity.
AtSnAK Proteins Are Subjected to Both Autophosphorylation and Feedback Phosphorylation Processes
The occurrence of an autophosphorylation (AP) of AtSnAK proteins was suggested by the presence of a radiolabeling signal when the recombinant proteins were incubated alone (Fig. 3A, lanes 4 and 5). This point was verified in kinetic experiments with an increased amount of AtSnAK2 (5-fold). Fig. 3B clearly shows substantial phosphate incorporation on AtSnAK2 with time. These data (Fig. 3, A and B) together with previous observations of Shen et al. (15) strengthen the occurrence of an AtSnAK autophosphorylation process. Moreover, in Fig. 3A, radiolabeling was increased substantially when AtSnRK1.1 was also present in the assay (Fig. 3A, lanes 2 and 3). Increasing the amount of AtSnAK2 led to a stronger signal (Fig. 3A, lane 7), whereas the signal was absent or low in control assays (Fig. 3A, lanes 6 and 8). Such observations strongly suggested that a feedback phosphorylation (FP) of AtSnAK2 by activated AtSnRK1.1 occurred in the reconstituted assay.
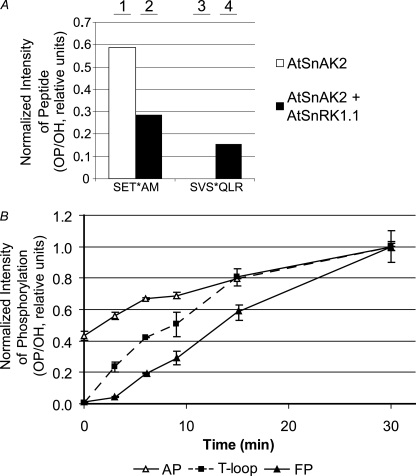
MS/MS was performed to identify these specific phosphorylation sites on recombinant AtSnAK2 and to obtain an estimation of the amounts of the corresponding phosphorylated and nonphosphorylated peptides (Fig. 4A). Consistently, two different phosphorylation sites were found on AtSnAK2 (supplemental Fig. S1). When AtSnAK2 was incubated alone (Fig. 4A, bars 1 and 3) or in the presence of AtSnRK1.1 (Fig. 4A, bar 2) Thr154 in the motif SET(154)AM (called thereafter, the AP site) was phosphorylated. Because no other kinase is expected to be present in the reconstituted assay, it appears to result from the autophosphorylation process detected previously (Fig. 3B). A second phosphorylation site appeared on AtSnAK2 only during incubation with AtSnRK1.1, as clearly indicated by the phosphorylation ratios (Fig. 4A, bar 3 compared with bar 4). This site was identified as SVS(261)QVF with residue 261 as a phosphoserine (named the FP site). The AP (Thr153) and FP (Ser260) sites are conserved in AtSnAK1 (supplemental Fig. S2). Because AtSnAK1 32P-labeling patterns on SDS-PAGE autoradiographs are similar to those of AtSnAK2, it is assumed that both AP and FP occur on AtSnAK1.
FIGURE 4.
MS/MS analysis of AtSnAK2 and AtSnRK1.1 phosphorylation sites. A, proteins (20 μg of AtSnAK2 and 40 μg of AtSnRK1.1 preparations) were incubated in kinase activity buffer (containing 1 mm ATP) at 30 °C for 60 min and then submitted to SDS-PAGE. Ratios of phosphopeptides to unmodified peptides (OP/OH) from the corresponding protein bands were determined by MS/MS. SET*AM and SVS*QLR (asterisk marking the phosphoresidue) were the two phosphorylated sites found on AtSnAK2. B, time course of the phosphopeptide to unmodified peptide ratio (OP/OH) for each phosphorylation site normalized to the maximum value of each reaction (after 30 min). Proteins (10 μg of each preparation) were prepared as described in A. Corresponding bands were analyzed by MS/MS and peptides containing the T-loop site of AtSnRK1.1 (squares), AP (SET*AM, open triangles), and FP (SVS*QLR, closed triangles) sites of AtSnAK2 were quantified. Results are the average of three independent experiments ± S.D. (error bars).
To check whether the FP process was dependent on AtSnRK1.1 activity, a recombinant, constitutively inactive AtSnRK1.1 (T175A) was produced. After validation by Western blotting (using an anti-GST antibody) and a functional test with the AMARA peptide (showing no activity), it was incubated with the wild-type form of AtSnAK1. The data in Fig. 3C clearly suggest that inactive AtSnRK1.1 does not allow any FP to occur (Fig. 3C, lane T175A) as compared with the wild-type AtSnRK1.1 (Fig. 3C, lane WT). These data establish that AtSnRK1.1 activity is required for the FP process.
Phosphorylation Dynamics
To determine whether the phosphorylation events involving the two different kinases were hierarchical, kinetic experiments were performed. Aliquots were taken at different time points during a reconstituted phosphorylation assay containing AtSnRK1.1 and AtSnAK2. After protein separation by SDS-PAGE, the amounts of each phosphorylated peptide were determined by MS/MS. The results are shown as the normalized ratio of detected phosphorylated (OP) and nonphosphorylated (OH) peptides (Fig. 4B). These phosphorylation events appeared in three steps that are in favor of the following sequence: AtSnAK2 AP (Thr154) followed by AtSnRK1.1 phosphorylation (Thr175, activating), and finally AtSnAK2 FP (Ser261). Two different mechanisms can be put forward to account for such kinase interactions. First, it can be sequential, AtSnRK1.1 phosphorylation/activation by autophosphorylated AtSnAK followed by FP. Second, it could be simultaneous, the two kinases cross-phosphorylating in an AtSnAK·AtSnRK1 complex. In the first case, the increase in AtSnRK1.1 activity should be delayed and cooperative in nature. This was not observed in our kinetic experiments (Fig. 5), thus suggesting the occurrence of the second mechanism.
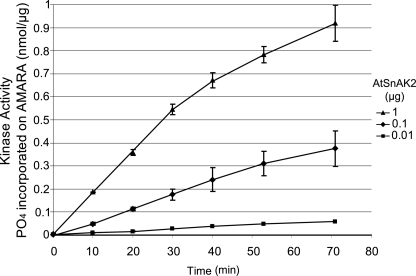
FIGURE 5.
Kinetics of AtSnRK1.1 assays in the presence of increasing amounts of AtSnAK2. AtSnRK1.1-GST (2 μg of the preparation) was incubated with AtSnAK2-GST (0.01 (■), 0.1 (♦), or 1 μg (▲) of the preparation) in a reconstituted assay containing 2 μCi of [γ-32P]ATP, 20 μm ATP, and 90 μm AMARA peptide, at 30 °C for 0, 10, 20, 30, 40, 50, 60, or 70 min. Radioactivity incorporated onto the AMARA peptide was counted. The results are the average of two independent experiments ± S.D. (error bars).
Mutagenesis Reveals the Functional Aspects of AtSnAK1 Phosphorylation
To determine the role of each phosphorylation event with respect to AtSnRK1, site-directed mutagenesis (T153A, T153D, S260A, and S260D) of AtSnAK1 was carried out. Mutation to alanine or aspartate can mimic a nonphosphorylated or a fully phosphorylated kinase, respectively. After PCR-based mutagenesis of the cDNA, recombinant mutated proteins were produced, purified, and identified by Western blot analyses using anti-GST antibodies. Similar amounts of each recombinant protein were used to compare the catalytic activity of wild-type and mutated forms in a coupled enzymatic assay with recombinant AtSnRK1.1 and the AMARA peptide (Fig. 6A). For AtSnAK1 mutated to an alanine on Thr153, a dramatic loss of activity was observed (Fig. 6A, bar 1) compared with wild-type AtSnAK1 (Fig. 6A, bar 5). In a marked contrast, when this residue was changed to an aspartate, introducing a negative charge at this position and thus mimicking a phosphorylated protein, the corresponding enzyme displayed a substantial catalytic activity (similar to the wild-type form) toward AtSnRK1.1 (Fig. 6A, bar 2). Interestingly, these data establish that AtSnAK autophosphorylation is required for full catalytic activity. Thus, the AP is an activating process.
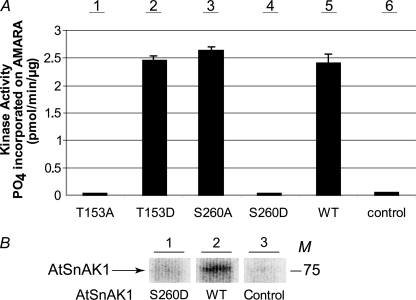
FIGURE 6.
Coupled kinase assays of mutated forms of AtSnAK1 and confirmation of the FP site. A, AtSnRK1.1-GST (2 μg of protein preparation) was incubated with each AtSnAK1-GST form (2 μg of protein preparation) in the reconstituted assay containing 2 μCi of[γ-32P]ATP, 20 μm ATP, and 90 μm AMARA peptide, at 30 °C for 60 min. Radioactivity incorporated onto the AMARA peptide was measured. The results are the average of three independent experiments ± S.D. (error bars). B, AtSnAK1 WT or S260D (2 μg of protein preparation) was incubated with AtSnRK1.1 T175D (2 μg) in the same reconstituted assay. After SDS-PAGE, the proteins were submitted to autoradiography. All three lanes are from the same exposure. The results are representative of three independent experiments. WT, wild-type; Control, AtSnRK1.1 without AtSnAK; M, molecular mass in kDa.
Site-directed mutagenesis was also used to investigate the Ser260 FP site. To this end, a constitutively active, T175D-mutated AtSnRK1.1 was produced and validated (Western blotting and kinase assay) as described above for AtSnRK1.1 T175A. Incubation of this mutated enzyme form with wild-type AtSnAK1 led to phosphate incorporation (as seen on the autoradiograph of Fig. 6B), as expected, whereas the S260D mutated AtSnAK1 form did not show any labeling. Thus, these results are in good agreement with previous mass spectrometry data (Fig. 4A) and show that Ser260 is the unique residue involved in the FP process.
In addition, the S260A mutation introduced in AtSnAK1 had no effect on the enzyme activity (Fig. 6A, bar 3), whereas the S260D mutation led to a dramatic reduction in the ability of AtSnAK1 to phosphorylate/activate AtSnRK1.1 (Fig. 6A, bar 4). Thus, the FP is an inhibiting process.
Modeling the Phosphorylation Sites on AtSnAK1
Sequence comparisons of AtSnAK and AtSnRK1 with related protein kinases having known crystal structures (supplemental Fig. S2) allowed us to propose a three-dimensional model of the AtSnAK protein (Fig. 7) to localize the topography of the newly discovered phosphorylation sites (supplemental Fig. S2, underlined in green and purple). The model (Fig. 7) delineates the classical structure of protein kinases with two lobes forming the active pocket in which ATP is positioned. Conserved residues responsible for binding ATP (22), Thr153 (AP site), and Ser260 (FP site) are represented in stick form (Fig. 7A). These two phosphorylated residues are found in poorly conserved flexible loops (supplemental Fig. S2) and positioned in the upper and lower parts of the catalytic cleft, respectively (Fig. 7, A and B, orange and yellow loops). Surface representation of the AtSnAK1 three-dimensional structure (Fig. 7B) suggested that Thr153 (red spot in the orange domain) is freely accessible to phosphorylation, whereas Ser260 (red spot in the yellow domain), inside the pocket, is not. Because the AP process activates the enzyme, it is tempting to speculate that the catalytic pocket is closed in the nonphosphorylated enzyme and that Thr153 phosphorylation brings about a conformational change that allows substrates to enter the pocket. Consistent with the probable sequence of each phosphorylation event (Fig. 4B), this is also expected to allow the internal Ser260 to become accessible to FP by AtSnRK1.1.
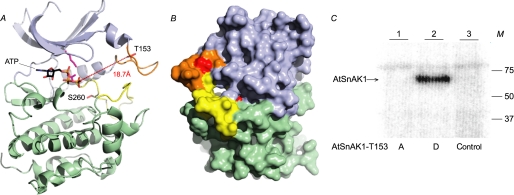
FIGURE 7.
Structural model of AtSnAK1 and accessibility of Ser260. This model was built by using the ROCK1 structure (PDB code: 2ESM, chain B) as template. A, AtSnAK1 protein as a schematic representation. The small lobe is in blue and large lobe in green, autophosphorylation (AP) loop is in orange and feedback phosphorylation (FP) loop in yellow. ATP is positioned according to the structural alignment with MEK1 (PDB code: 3E8N) and shown as lines. The side chains of interesting residues are represented in stick form (Thr153 and Ser260 in red). Distance between Thr153, and the γ-phosphate of ATP is visualized in hatched red and indicated in Å. B, protein surface model (opposite face compared with A). C, mutated AtSnAK1 proteins (2 μg of preparation) (T153A or T153D) incubated in the presence of 2 μg of AtSnRK1.1 T175D (constitutively active form) preparation in the reconstituted assay containing 2 μCi of [γ-32P]ATP and 20 μm ATP, at 30 °C for 60 min. Proteins were submitted to SDS-PAGE and autoradiography. M, molecular mass in kDa. The results are representative of three independent experiments.
To check this hypothesis, the constitutively active AtSnRK1.1 mutant (T175D) was incubated in a reconstituted assays containing mutated forms of AtSnAK1 on the autophosphorylation site (T153D and T153A). The autoradiograph after SDS-PAGE (Fig. 7C) clearly shows that AtSnAK1 T153D (catalytically active) can be phosphorylated on Ser260 by AtSnRK1.1 (Fig. 7C, lane 2), whereas T153A (catalytically inactive) cannot (Fig. 7C, lane 1). These results strongly suggest that autophosphorylation of AtSnAK1 is required for Ser260 accessibility to feedback phosphorylation by AtSnRK1.1.
DISCUSSION
In this work we show that AtSnAK requires autophosphorylation before it can phosphorylate and activate AtSnRK1 in vitro, in good agreement with recent data reported by Shen et al. (15). Examining in more detail the autophosphorylation process by mass spectrometry led to the identification of Thr153 (referenced to AtSnAK1) as the phosphorylated residue. Based on the AtSnAK1 amino acid sequence, Swiss Model was used to generate a model describing the three-dimensional structure of the enzyme (Fig. 7). This model shows the AP site on a flexible loop and located at the entry of the catalytic cleft (Fig. 7A). Interestingly, the distance between the γ-phosphate of bound ATP and Thr153 is very large (18.7 Å), suggesting that AP is probably an intermolecular autophosphorylation of AtSnAK. Sequence comparisons of animal, yeast, and plant enzymes showed that the loop containing the AP site has been poorly conserved throughout evolution (supplemental Fig. S2, underlined in green). This is consistent with the fact that this loop is highly variable in kinase structures (22). However, five of the six upstream kinases of yeast and mammals known to phosphorylate SNF1-related family members display at least one putative phosphorylatable residue in this loop (supplemental Fig. S2, blue boxes in domain underlined in green), suggesting that the AP mechanism might be conserved in Eukaryotes. However, none of the autophosphorylated residues of the animal homolog LKB1 (23) aligns with Thr153 (supplemental Fig. S2, four green boxes on LKB1 sequence). Importantly, site-directed mutagenesis of Thr153 (T153A and T153D) indicated that AtSnAK activity is up-regulated by this posttranslational modification.
The activation of AtSnAK by the AP process allows the phosphorylation and activation of AtSnRK1 that, once activated, exerts a feedback phosphorylation on the Ser260 of its modifying enzyme, AtSnAK. We checked that this FP process is dependent on AtSnRK1.1 activity (Fig. 3C) and that this residue (Ser260) is the only one involved in the FP (Fig. 6B) as suggested by MS/MS experiments (Fig. 4A). Our site-directed mutagenesis approach established that, contrary to AP, the FP is an inhibitory process. To our knowledge, this is the first time that such a negative FP is shown to occur in the AtSnRK1/AtSnAK pathway, or in their animal and yeast counterparts. In the structural model, this phosphorylated serine is located in the lower part of the catalytic cleft (Fig. 7, A and B). Interestingly, all known yeast and mammalian upstream kinases that phosphorylate SNF1-related kinases display potential phosphorylatable Ser or Thr residues (supplemental Fig. S2, blue boxes) in the loop containing the AtSnAK FP site (supplemental Fig. S2, underlined in purple). Once again, none of the LKB1 residues phosphorylated by other kinases align with Ser260 of AtSnAK1 (supplemental Fig. S2, red boxes in LKB1 sequence). This domain (subdomains VII and VIII, between conserved DFG and APE motifs) is known to contribute to catalytic regulation through phosphorylation in many kinases, examples include T-loop phosphorylation of SNF1-related protein kinases (2), Thr197 of PKA-Cα, Thr183 of Erk2, and Thr160 of Cdk2 (22).
The phosphorylation kinetics of AtSnAK and AtSnRK1 were investigated by mass spectrometry to establish the sequence of the events and to determine the extent of the modification processes. This approach suggested that the sequence of phosphorylation events was the following: AtSnAK AP on Thr153 (activation) and then, simultaneously, AtSnRK1 on the T-loop (activation) and AtSnAK FP on Ser260 (inhibition). Our site-directed mutagenesis experiments confirmed this assumption; indeed, neither T-loop phosphorylation nor FP occurred with inactive mutant forms (AtSnAK; S260D) and (AtSnRK1.1; T175A), respectively.
Because the enzymatic system is reciprocal and thus its components functionally linked, the kinetic behavior observed (Fig. 5) was expected. In vitro, the feedback phosphorylation concerns the entire autophosphorylated/activated AtSnAK population, thereby desensitizing this kinase system. In vivo, it is expected that this kinase system is counteracted by protein phosphatases and reaches a dynamic steady state.
Our kinetic experiments on AtSnRK1 activity suggested that the reciprocal phosphorylation mechanism is simultaneous, involving cross-phosphorylation in the kinase complex. In support of this hypothesis is the fact that both kinases form a stable complex in vitro (15). Metaphorically speaking, this interaction can be viewed as a “mortal embrace” for the modifying AtSnAK protein.
Regarding the underlying mechanism, it was tempting to speculate that access of the protein substrate to the catalytic pocket is strongly limited in the nonautophosphorylated AtSnAK. AP modifies the structural organization of this domain thus opening the pocket and leading to an active enzyme. Phosphorylation/activation of the target AtSnRK1 occurs and triggers the feedback process on AtSnAK at the FP site. The fact that the corresponding serine can only become phosphorylated in the autophosphorylated enzyme form brings strong support to our proposed hypothesis. The down-regulation induced by this process should be due to the introduction of the strong negative charge of the phosphate in this region, possibly impairing access to the substrates. Interestingly, a similar cross-phosphorylation mechanism has been shown to occur in smooth muscle cells between the platelet-derived growth factor receptor-β (PDGFRβ) and G protein-coupled receptor kinase 5 (GRK5) where PDGFRβ triggers its own desensitization by phosphorylating/activating GRK5 (24).
In planta, the feedback phosphorylation process might be a signal for AtSnAK proteolysis via the proteasome pathway that occurs in Arabidopsis (14). Such a mechanism would imply that the kinase activity depends more on the molecular turnover rather than restoration by a phosphatase.
In vivo, a regulatory cross-phosphorylation between the two kinases would provide the system with attenuation properties linked to the extent of the induced AtSnRK1 phosphorylation state/activity. In fine, a highly homeostatic behavior is predicted. This appears to be in agreement with the recent results of Baena-González et al. (4), showing that stresses trigger a rapid response including a modulation of selected gene expression with little or no change in the phosphorylation state of AtSnRK1.1 and 1.2, even though without this phosphorylation, AtSnRK1 is not active. This suggested that the underlying regulatory mechanism apparently not only involved a posttranslational modification of the kinase, but more likely activation by other factors such as metabolites. Indeed, AtSnRK1 is known to be a central regulator of energy homeostasis; during a stress, by promoting catabolism and down-regulating anabolism-related genes its action contributes to energy saving. However, this has to be finely controlled with respect to the extent and the duration of the stress, so as not to be damaging to the cell. In extreme cases, the cell will enter a senescence program and the system becomes irreversible.
Furthermore, it was recently seen that the overexpression of AtSnRK1.1 protein led to a glucose hypersensitivity in Arabidopsis (21). Indeed, the addition of glucose produced an increase in both AtSnRK1.1 phosphorylation state and enzymatic activity compared with control plants. The phenotype could reflect a deregulation of the cross-phosphorylation mechanism described in this work due to increased AtSnRK1.1 levels and an activated sugar-signaling pathway by glucose. In vivo, our proposed homeostatic mechanism of cross-phosphorylation should allow such dramatic consequences of deregulating the master kinase, AtSnRK1, to be avoided.
Supplementary Material
Acknowledgments
We thank Dr. Stéphane Lemaire for a critical reading of the manuscript. We thank William Bourbon, Camille Verly, and François Rey for excellent contributions to the production of recombinant AtSnRK1 and AtSnAK.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, an additional reference, Table S1, and Figs. S1 and S2.
- SNF1
- sucrose nonfermenting 1
- AMPK
- AMP-activated protein kinase
- SnRK1
- SNF1-related protein kinase 1
- AtSnRK1
- Arabidopsis thaliana SNF1-related protein kinase 1
- AtSnAK
- AtSnRK1 activating kinase
- MS/MS
- tandem mass spectrometry
- MALDI-TOF
- matrix-assisted laser desorption ionization time-of-flight
- GST
- glutathione S-transferase
- PDB
- Protein Data Bank
- AP
- autophosphorylation
- FP
- feedback phosphorylation.
REFERENCES
- 1.Hedbacker K., Carlson M. (2008) Front. Biosci. 13, 2408–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Polge C., Thomas M. (2007) Trends Plant Sci. 12, 20–28 [DOI] [PubMed] [Google Scholar]
- 3.Hardie D. G. (2007) Nat. Rev. Mol. Cell Biol. 8, 774–785 [DOI] [PubMed] [Google Scholar]
- 4.Baena-González E., Rolland F., Thevelein J. M., Sheen J. (2007) Nature 448, 938–942 [DOI] [PubMed] [Google Scholar]
- 5.Sugden C., Donaghy P. G., Halford N. G., Hardie D. G. (1999) Plant Physiol. 120, 257–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hong S. P., Leiper F. C., Woods A., Carling D., Carlson M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8839–8843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woods A., Johnstone S. R., Dickerson K., Leiper F. C., Fryer L. G., Neumann D., Schlattner U., Wallimann T., Carlson M., Carling D. (2003) Curr. Biol. 13, 2004–2008 [DOI] [PubMed] [Google Scholar]
- 8.Hawley S. A., Pan D. A., Mustard K. J., Ross L., Bain J., Edelman A. M., Frenguelli B. G., Hardie D. G. (2005) Cell Metab. 2, 9–19 [DOI] [PubMed] [Google Scholar]
- 9.Momcilovic M., Hong S. P., Carlson M. (2006) J. Biol. Chem. 281, 25336–25343 [DOI] [PubMed] [Google Scholar]
- 10.Hawley S. A., Davison M., Woods A., Davies S. P., Beri R. K., Carling D., Hardie D. G. (1996) J. Biol. Chem. 271, 27879–27887 [DOI] [PubMed] [Google Scholar]
- 11.Harthill J. E., Meek S. E., Morrice N., Peggie M. W., Borch J., Wong B. H., Mackintosh C. (2006) Plant J. 47, 211–223 [DOI] [PubMed] [Google Scholar]
- 12.Sugden C., Crawford R. M., Halford N. G., Hardie D. G. (1999) Plant J. 19, 433–439 [DOI] [PubMed] [Google Scholar]
- 13.Hey S., Mayerhofer H., Halford N. G., Dickinson J. R. (2007) J. Biol. Chem. 282, 10472–10479 [DOI] [PubMed] [Google Scholar]
- 14.Shen W., Hanley-Bowdoin L. (2006) Plant Physiol. 142, 1642–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen W., Reyes M. I., Hanley-Bowdoin L. (2009) Plant Physiol. 150, 996–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanahan D. (1983) J. Mol. Biol. 166, 557–580 [DOI] [PubMed] [Google Scholar]
- 17.Studier F. W., Moffatt B. A. (1986) J. Mol. Biol. 189, 113–130 [DOI] [PubMed] [Google Scholar]
- 18.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 19.Dale S., Wilson W. A., Edelman A. M., Hardie D. G. (1995) FEBS Lett. 361, 191–195 [DOI] [PubMed] [Google Scholar]
- 20.Arnold K., Bordoli L., Kopp J., Schwede T. (2006) Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 21.Jossier M., Bouly J. P., Meimoun P., Arjmand A., Lessard P., Hawley S., Grahame Hardie D., Thomas M. (2009) Plant J. 59, 316–328 [DOI] [PubMed] [Google Scholar]
- 22.Hanks S. K., Hunter T. (1995) FASEB J. 9, 576–596 [PubMed] [Google Scholar]
- 23.Jansen M., Ten Klooster J. P., Offerhaus G. J., Clevers H. (2009) Physiol. Rev. 89, 777–798 [DOI] [PubMed] [Google Scholar]
- 24.Cai X., Wu J. H., Exum S. T., Oppermann M., Premont R. T., Shenoy S. K., Freedman N. J. (2009) Mol. Pharmacol. 75, 626–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.