Abstract
The fundamental process of protein self-assembly is governed by protein-protein interactions between subunits, which combine to form structures that are often on the nano-scale. The nano-cage protein, bacterioferritin from Escherichia coli, a maxi-ferritin made up of 24 subunits, was chosen as the basis for an alanine-shaving mutagenesis study to discover key amino acid residues at symmetry-related protein-protein interfaces that control protein stability and self-assembly. By inspection of these interfaces and “virtual alanine scanning,” nine mutants were designed, expressed, purified, and characterized using transmission electron microscopy, size exclusion chromatography, dynamic light scattering, native PAGE, and temperature-dependent CD. Many of the selected amino acids act as hot spot residues. Four of these (Arg-30, which is located at the two-fold axis, and Arg-61, Tyr-114, and Glu-128, which are located at the three-fold axis), when individually mutated to alanine, completely shut down detectable solution formation of 24-mer, favoring a cooperatively folded dimer, suggesting that they may be oligomerization “switch residues.” Furthermore, two residues, Arg-30 and Arg-61, when changed to alanine form mutants that are more thermodynamically stable than the native protein. This investigation into the structure and energetics of this self-assembling nano-cage protein not only can act as a jumping off point for the eventual design of novel protein nano-structures but can also help to understand the role that structure plays on the function of this important class of proteins.
Keywords: Protein Folding, Protein Self-assembly, Protein Stability, Protein Structure, Protein-Protein Interactions, Alanine Shaving, Ferritin, Nano-cage, Nano-structure, Self-assembly
Introduction
Mistakes can occur in the folding of proteins. These errors, in turn, can cause cellular disorders due either to the loss of function of the misfolded protein or to new and pathogenic activities attributed to the altered fold. Diseases associated with protein misfolding are wide ranging (Alzheimer disease, bovine spongiform encephalopathy, Huntington disease, Parkinson disease, Type II diabetes, etc.) and have been discovered in many different tissues and organs (1). Thus, how proteins fold is not only one of the “big unanswered scientific questions” (2); understanding the fundamentals of protein folding could also identify potential errors in the processes leading to misfolding and could therefore shed significant light on all of these diseases in general.
Protein-protein interactions have grown in interest within the pharmaceutical industry following the discovery that many, although not all, proteins, whereas possessing interfaces with large surface areas, have binding energy that is focused onto a small number of amino acid residues or “hot spots” (3–5), thereby suggesting that it could be possible to inhibit these medicinally attractive interactions with small molecule drugs (6). An understanding of the fundamentals of protein-protein interactions would therefore provide insight into drug design in addition to facilitating a multitude of other applications such as the engineering of interfaces for the development of antibodies to new or non-immunogenic antigens or the engineering of single-celled organisms for bioremediation.
A sufficient amount of work on the structure of biomacromolecules has been performed in the last 50 years to warrant the awarding of ∼25% of the chemistry Nobel prizes (7), and the field has advanced to the stage where it is now possible to design proteins de novo (8). However, the majority of these designs either have involved small, monomeric proteins or were based on α-helix bundles (9). Only recently has the rational design of multimeric protein structures begun to be studied (10–12). Engineering the quaternary structure of a protein with a novel sequence is the ultimate test of our understanding of the processes of folding and self-assembly.
Many of the ferritin proteins and their structural homologues assemble into multisubunit, hollow, nano-scale cages (13). Ferritins and other cage proteins have been the focus of much recent attention as part of bioorthogonal chemical reaction methodology development (14), drug delivery studies (15, 16), as platforms to generate nano-structured materials (17), and as pharmacological models for ion channels (18). Due to the current interest in their application and the fact that they are composed of monomers folded into a four-helix bundle motif, a fold well studied by the protein de novo design community, ferritins could act as important model systems for developing the fundamentals of how to expand the dimensionality of rational protein engineering into the realm of self-assembly.
Genetic and biochemical evidence suggests a direct role for ferritins in human diseases including those associated with aging and protein misfolding. One of the roles of ferritins in the cell is to sequester iron in a mineralized form, thereby protecting the cell from iron-induced oxidative damage. Human ferritin is made up of a heavy chain (H) and a light chain (L). Although it is possible for homo-oligomeric nano-cages of these proteins to form, the activity of the two monomers is complementary. The H-chain possesses ferroxidase activity, facilitating rapid iron uptake, whereas the L-chain stabilizes the mineralized iron core (19). Iron homeostasis is important for normal cellular function, and iron usage is especially enhanced in the brain due to expanded respiration demands, biosynthesis of neurotransmitters, and myelinogenesis (20). Ferritin up-regulation has been shown to play a role in a number of autoimmune diseases such as lupus and rheumatoid arthritis. In addition, the symptoms of multiple sclerosis, as a disease caused by the demyelination of neurons, can be reversed in mice by the injection of apoferritin (21). Neuroferritinopathy generates motor defects and tremors, possibly causing it to be misdiagnosed as Huntington or Parkinson disease. The condition seems to be caused by single nucleotide missense mutations in the ferritin L-chain gene resulting in a mistranslated and extended C terminus. These mutations destabilize the folded protein and result in the formation of brain inclusions made up of iron and misfolded, aggregated protein (20, 22, 23).
Structural studies have revealed that of the ferritins that naturally self-assemble into nano-capsules, there are two types. Mini-ferritins form hollow spheres composed of 12 monomers, and maxi-ferritins are composed of 24 monomers. Although their monomers share similar four-helix bundle tertiary structure, mini-ferritins such as DNA-binding protein from starved cells (DPS) (24, 25) have tetrahedral symmetry, whereas maxi-ferritins such as Escherichia coli bacterioferritin (BFR)5 (26) have octahedral symmetry (see Fig. 1). Structural comparisons have been made, and yet it is still an open question as to what controls the differing nano-architecture of these proteins. Little modern research has been undertaken to understand how the “structural energetics” of these systems define their quaternary structure (27). Mutagenesis and design studies targeting the ferritins will not only help to understand the fundamentals of protein folding, quaternary structure formation, self-assembly, and misfolded disease states, they may also provide insight into helping to establish dynamic drug delivery systems (28) or the development of new nano-materials using these proteins.6
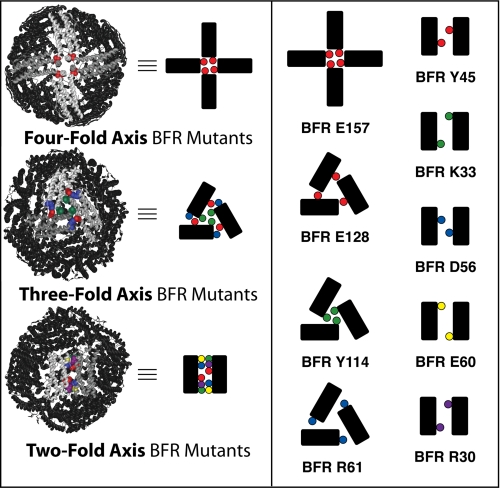
FIGURE 1.
Position of mutated residues with respect to symmetry axes of the nano-cage protein. Left, the BFR crystal structure with mutated residues highlighted with respect to the three axes of symmetry and schematized diagrams representing the protein-protein interactions defining these symmetries. Right, the residues of BFR mutated in this study and their schematized position at the symmetry-related protein-protein interfaces. This schematic convention will be conserved throughout this report.
To determine the importance of specific key residues residing between the monomer subunits, we employed alanine-shaving mutagenesis (3, 30) to explore whether hot spots exist on the protein-protein interfaces and how these affect self-assembly. Through inspection of the crystal structure of BFR (24–26), we focused our attention on interfacial salt bridges, due to their straightforward identification, and aromatic amino acids, because of their hot spot propensity (4, 31). In addition, we performed virtual alanine scanning (32, 33) to confirm the predicted importance of these residues. The selected amino acids were then subjected to mutagenesis to alanine, and the resulting mutants were probed using transmission electron microscopy (TEM), size exclusion chromatography (SEC), dynamic light scattering (DLS), native PAGE, and temperature-dependent CD to determine the role that these individual interfacial interactions play in the stability and structure of the ferritin.
EXPERIMENTAL PROCEDURES
Virtual Alanine Scanning
Computational prediction of energetically important amino acid residues on the protein-protein interfaces followed standard protocol using web tools developed by the Baker laboratory (32). The input consisted of the symmetry-related protein-protein interfaces described by the coordinates for the crystal structure of BFR (Protein Data Bank (PDB) ID: 1bfr). From these coordinates, two monomers on the two-fold axis, three monomers on the three-fold axis, and two monomers that are 90 degrees adjacent at the four-fold axis were used. The output predicted hot spot residues if the ΔΔG for mutagenesis was greater than 1 kcal/mol. The results are displayed in table form in supplemental Figs. S13–S15.
Cloning of the Wild-type and Mutant Genes
Cloning and protein preparation was modified from Fan et al. (34). The oligonucleotide design, cloning characterization, and expression analyses are available in the supporting information (supplemental Figs. S1–S4).
Short oligonucleotides (synthesized by solid phase phosphoramidite technology by 1st Base Pte. Ltd., Singapore) were designed based on the amino acid sequences of the native ferritin so that the 5′ strands overlapped by 12 bp, whereas the 3′ strands had a 15-bp overlap. The overlap and the C/G content were modified so that the oligonucleotides all had similar melting temperatures.
Assembly PCR
The recombinant genes were synthesized in a two-step, total gene synthesis method combining dual asymmetrical PCR and overlap extension PCR as per Young and Dong (35). In the dual asymmetrical PCR, every four consecutive oligonucleotides were used as templates, with the outer two oligonucleotides in four times molar excess to the inner ones. The gene synthesis reactions were performed by adding each of the outer two (2.0 μl of a 10 μm solution) and each of the inner oligonucleotides (0.5 μl of a 10 μm solution) to a solution made up of a dNTP mix (Fermentas, 2.0 μl of a 2 mm solution), Pfu buffer (Fermentas, 2.0 μl, 10×), Pfu DNA polymerase (Fermentas, 0.2 μl, 2.5 units/μl), and deionized water to bring the total volume to 20 μl. An initial denaturation (95 °C, 5 min) was followed by 28 cycles of denaturation (95 °C, 0.5 min), annealing (53 °C, 0.75 min), and extension (72 °C, 0.75 min) followed by a final extension (72 °C, 7 min).
In the subsequent overlap extension PCR, the gene synthesis reactions were performed by adding the reaction product solutions (1.0 μl) from each dual asymmetrical PCR reaction to a solution made up of dNTP mix (Fermentas, 2.0 μl of a 2 mm solution), Pfu buffer (Fermentas, 3.0 μl, 10×), Pfu DNA polymerase (Fermentas, 0.3 μl, 2.5 units/μl), and deionized water to bring the total volume to 30 μl. An initial denaturation (95 °C, 5 min) was followed by 10 cycles of denaturation (95 °C, 1 min), annealing (43 °C, 1 min), and extension (72 °C, 1 min) followed by a final extension (72 °C, 5 min).
Amplification PCR
The assembled gene was amplified by PCR. The reaction was performed by adding Pfu buffer (Fermentas, 2.0 μl, 10×), Pfu polymerase (Fermentas, 0.2 μl, 2.5 units/μl), and dNTP mix (Fermentas, 2.0 μl of a 2 mm solution), the reaction product from the overlap extension PCR (2.0 μl), forward and reverse primers (i.e. the end primers from gene synthesis, 1.0 μl of a 10 μm solution of each), and deionized water to bring the total reaction volume to 20 μl. An initial denaturation (95 °C, 5 min) was followed by 28 cycles of denaturation (95 °C, 5 min), annealing (53 °C, 0.75 min), and extension (72 °C, 0.75 min) followed by a final extension (72 °C, 7 min). Products were visualized and analyzed by 2% agarose gel electrophoresis to ensure that the assembled gene was of the correct size.
Extension PCR
The product of amplification PCR (3.0 μl) was added to a solution of sense and antisense primers (2.5 μl of a 10 μm solution of each), Pfu buffer (Fermentas, 5.0 μl of 10×), Pfu polymerase (Fermentas, 0.5 μl, 2.5 units/μl) and dNTP mix (Fermentas, 5.0 μl of a 2 mm solution), and deionized water to bring the total reaction volume to 50 μl. An initial denaturation (95 °C, 5 min) was followed by 28 cycles of denaturation (95 °C, 5 min), annealing (53 °C, 0.75 min), and extension (72 °C, 0.75 min) followed by a final extension (72 °C, 7 min). Products were gel-purified with extraction from agarose using Perfectprep gel cleanup kit (Eppendorf).
T4 DNA Polymerase Treatment of Target Insert and Annealing to Ek/LIC Vector
The purified product from extension PCR (17.5 μl) was treated with T4 DNA polymerase (Novagen, 0.5 μl, 2.5 units/μl) in a solution of dATP (Novagen, 2.5 μl of a 25 mm solution), NEB Buffer 2 (Novagen, 2.5 μl, 10×), bovine serum albumin (Novagen, 0.25 μl, 100×), and deionized water to bring the total reaction volume to 25 μl. The solution was incubated (22 °C, 30 min), and the enzyme was subsequently inactivated (75 °C, 20 min). Annealing was performed by incubating the polymerase-treated insert (5.0 μl) with pET-32 Ek/LIC vector (Novagen, 2.0 μl, 22 °C, 30 min). EDTA (Novagen Chemicals, 2.3 μl of a 25 mm solution) was subsequently added, and the solution was further incubated (22 °C, 1 h).
The plasmid was then transformed by electroporation into NovaBlue electro-competent cells (Novagen). The resulting carbenicillin-resistant colonies were screened by colony PCR, and the plasmids were verified by sequencing (1st Base Pte. Ltd.) after miniprep (QIAprep spin miniprep kit) “Sequencing Results” in the supplemental material. Mutants BFR R30A and BFR R61A were constructed using the QuikChange site-directed mutagenesis kit (Stratagene) with the BFR vector as the template using the primers in supplemental Fig. S2.
Gene Expression and Purification
Plasmids harboring the desired genes were transformed into E. coli BL21(DE3) (Novagen) cells. Expression culture in 500 ml of LB was induced with isopropyl-β-d-thiogalactopyranoside (Fermentas, 2 ml of a 0.1 m solution), and the culture was further incubated (3 h, 30 °C). The cells were isolated by centrifugation and lysed by sonication. The soluble protein was applied to nickel-nitrilotriacetic acid resin (Qiagen) and eluted by affinity tag cleavage following incubation with enterokinase (New England Biolabs, 8 μl, 2 μg ml−1, 36 h, 4 °C). The protein was concentrated via ultrafiltration (Sartorius Vivaspin 6), and SDS-PAGE was carried out to verify the purity of the protein. Matrix-assisted laser desorption/ionization-time of flight mass spectroscopy of the monomers confirmed the identity of the purified proteins (supplemental Fig. S4).
TEM
A drop of a solution of purified protein (5 μl of a ∼200 μg ml−1 solution) was placed on a piece of Parafilm, and a circular carbon-coated copper grid was placed on top of the drop (1 min). Excess sample was removed by wicking with a piece of filter paper, the grid was placed on a drop of either uranyl acetate or phosphotungstic acid (5 μl of a 1% w/v solution, 1 min), and excess solution was again removed by wicking with filter paper. TEM micrographs were analyzed using ImageJ (National Institutes of Health) to measure the longest diameter of the protein cages. For each protein, 100 particles were measured.
SEC
Chromatography experiments were performed on an ÄKTAFPLCTM (GE Healthcare Life Sciences) system equipped with a UV detector (λ = 280 nm) using a Superdex 200 10/300 GL gel filtration column at a flow rate of 0.5 ml min−1 (running buffer: 50 mm phosphate, 0.15 m NaCl, pH 7.0) at 4 °C. The column was previously calibrated using six well characterized proteins as standards (GE Biosystems calibration kit) (34). Oligomerization state was determined from the calculated molecular weight and the predicted monomer molecular weight.
DLS
DLS analysis was performed on a 90Plus particle sizer instrument (Brookhaven Instruments Ltd.) using a 0.5-cm path length quartz cell. Samples were filtered (0.2 μm) prior to measurement. For each protein (0.6 ml, ∼200 μg ml−1) (with the exception of BFR R61A and BFR R30A (0.6 ml, ∼800 μg ml−1)), three scans (2 min for each scan, 25 °C) were collected. The average diameter of the particle was calculated from three replicates. The polydispersity was less than 20% for all samples.
Temperature-dependent CD Analysis
CD spectra were collected on a Jasco J-710 spectropolarimeter fitted with a Peltier temperature controller in the wavelength range 200–250 nm using a 1-mm quartz cell. The concentration of the protein solutions was determined by BCA (Novagen) and diluted (100 μg ml−1) with phosphate buffer (50 mm NaH2PO4, pH 7.2). The temperature was increased from 20 to 90 °C in 5 °C-steps. Samples were allowed to equilibrate (8 min) at each temperature. After melting, the protein solutions were cooled slowly to 20 °C, and the spectra were compared with those obtained before unfolding. At least three replicates were performed.
Native PAGE
A 7% gel was run using 20 μg of all the proteins (with the exception of BFR E128A where 10 μg was used) and was stained with Coomassie Blue.
Chemical Denaturation
Samples for chemical denaturation were prepared with 100 μg ml−1 protein and GdnHCl or urea (0–8 m) varying in phosphate buffer (50 mm NaH2PO4, pH 7.2). The samples were mixed and incubated with no agitation (room temperature, 16 h). The tryptophan emission at 355 nm was recorded using a Varian Cary Eclipse spectrofluorometer with excitation at 295 nm.
RESULTS
Selection of Key Interfacial Residues for Mutagenesis
Nine mutants of BFR were designed (Fig. 1 and supplemental Fig. S12). The only potential salt bridge identifiable by inspection between the BFR monomers around the four-fold axis involves Glu-157 and Lys-38 (supplemental Fig. S16). However, in the crystal structure, their side chains are pointing slightly away from each other, and virtual alanine scanning (32, 33) (supplemental Figs. S13–S15) suggests that this interaction is subtle. As there are no other even partially promising interactions involving either salt bridges or aromatics, residue Glu-157 was nevertheless selected for mutagenesis. This residue is part of a C-terminal domain adjacent to the E-helix, which is outside of the monomer four-helix bundle but defines a channel at the 24-mer four-fold axis. In a previous study (34), we have determined the necessity of the E-helix for oligomerization; however, the importance of this C-terminal domain has not been established. Along the three-fold symmetry axis, two possible interactions were clearly evident. One is a salt bridge between Glu-128, Arg-61, and Arg-102 (supplemental Fig. S17); the other involves contacts between Tyr-114 and a hydrophobic pocket in the other subunits. These residues were among those predicted to be energetically important by virtual alanine scanning. The calculations indicated that Arg-102 is less significant than Arg-61 and Glu-128; therefore, Arg-102 is not included among the mutants. It has been shown that in some ferritins, the regions located at or near the pores along a three-fold symmetry axis are dynamic and fold independently from the rest of the protein. Moreover, they may be involved in not only iron uptake but also release (36, 37). Of these three-fold-associated residues, Tyr-114 is in the pore, whereas the other two are nearby. One potential interaction along the two-fold symmetry axis involves residue Tyr-45 and a hydrophobic pocket, so it was therefore selected for mutagenesis. The salt bridges on the two-fold symmetry axis are complicated by the fact that four residues appear to be interacting with each other. In the crystal structure, Asp-56 and Glu-60 of one subunit make contact with Arg-30 and Lys-33 of another (supplemental Fig. S18), and virtual alanine scanning reinforces our prediction of the energetic importance of these interactions (supplemental Figs. S13–S15). Although the side chain of Asp-56 is aligned with Arg-30 and that of Glu-60 is aligned with Lys-33, all four are proximal in space and may form an extended hydrogen-bonding “hot stretch” (38) as opposed to a more canonical hot spot. Therefore, conceptually isolating these into simple binary interactions is not possible. Hence, it was decided to mutate Lys-33, Asp-56, and Arg-30 as single mutations and Asp-56 and Glu-60 as a double mutation. Although this is beyond the scope of this current study, we think clustered residues of this type are especially interesting as they exemplify the complexity of many real biological interactions (39). The eventual investigation into this complexity could broaden the understanding outside current reductionist views and help explain the non-additivity (40) that leads to cooperative folding.
Cloning and Gene Expression
The proteins were produced through recombinant gene overexpression in E. coli followed by affinity purification. The genes for expression were generated through the assembly of synthetic oligonucleotides by means of a double PCR procedure (25) (supplemental Figs. S1 and S2). This method proved general and high yielding. These genes were conjugated to a pET-32 Ek/LIC expression vector (Novagen). This expression vector provides proteins fused to a series of affinity tags to simplify purification, and these tags can be removed through cleavage at an enterokinase protease site that abuts the natural start site of the protein, leaving no additional residues, an essential requirement for self-assembly studies. The protein was purified through immobilization on affinity resin followed by on-resin enterokinase cleavage and protein release. The advantage of this strategy is that because the monomer is bound to the resin, it only self-assembles upon release (the large affinity tags presumably prevent self-assembly); thus, it would conceivably be relatively simple to load for drug delivery studies. This purification method proved general and robust (supplemental Figs. S3 and S4).
TEM Analysis of Nano-cage Assembly
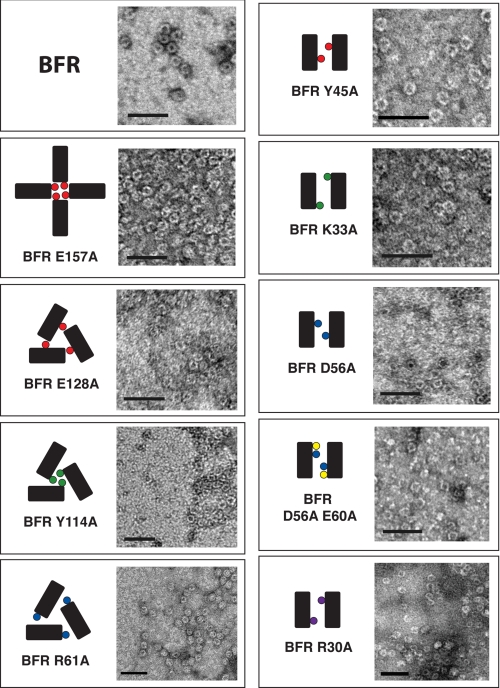
The proteins were analyzed as to whether each possessed the ability to assemble into a nano-cage. Samples of the proteins were prepared and treated with uranyl acetate negative stain. Electron micrographs indicated that all proteins form nano-cages under these conditions (Fig. 2 and supplemental Figs. S5 and S6). Image analysis of these micrographs indicates that the native proteins form structures of sizes on par with those described in the literature (25). Interestingly, the mutants assemble into cages that have the same size as or slightly smaller size than the native protein; none are larger.
FIGURE 2.
All nano-cage protein mutants can form nano-cages under TEM conditions. Negatively stained TEM micrographs indicate that all of the BFR-derived proteins can form nano-cages. Scale bars indicate 50 nm. Schematics follow the convention in Fig. 1.
Size Exclusion Chromatographic Analysis of Assembly in Solution
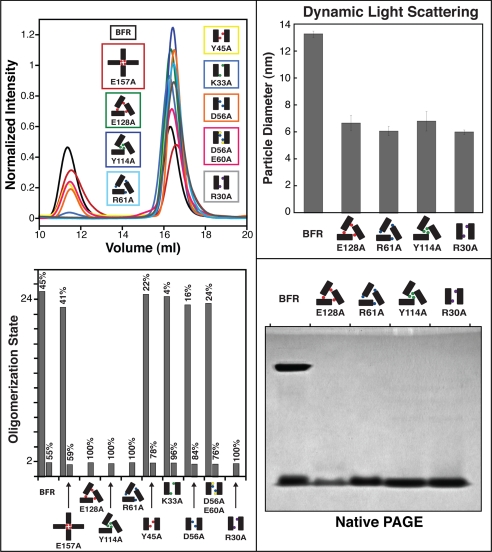
The proteins were analyzed by size exclusion chromatography, which separates different structures by size, to determine the effect the mutations have on the ability to assemble in solution (Fig. 3). The retention volumes of each species were correlated to the molecular weights of protein standards (supplemental Figs. S7 and S8), which were then used to calculate approximate oligomerization states. The native protein BFR, consistent with the literature (34), forms a mixture of 24-mer and dimer. The mutants were also found to form only these states. The mutant BFR E157A exists in a similar ratio to that of BFR, whereas the other mutants form altered ratios of dimer and 24-mer, all of which favor the dimer. Strikingly, BFR R30A, R61A, Y114A, and E128A only assemble into dimer. No 24-mer is observable for all of these four mutants. This is somewhat surprising because these mutants form hollow nano-structures under TEM conditions (see above). As SEC experiments were conducted in neutral conditions and uranyl acetate arguably is acidic, 1% phosphotungstic acid, which is neutral, was used as a negative stain in additional TEM experiments. These results (data not shown) agree with those using uranyl acetate, suggesting that the difference in observed nano-cage formation in SEC and TEM is unrelated to pH. In addition, the 24-mer:dimer ratio was further probed with SEC, and no concentration dependence was determined (data not shown), although it should be noted that the concentration range is limited due to UV detector limits. We are further exploring the fundamentals of this observation with other techniques; however, to resolve these inconsistencies and to confirm these solution data, two other solution techniques, dynamic light scattering and native PAGE, were employed (see below).
FIGURE 3.
All nano-cage protein mutants form 24-mer and/or dimer in solution, and four residues control the oligomerization state upon single point mutation. Top left, SEC chromatograms of all the proteins showing mixtures of two states, which are identified as 24-mer and dimer (bottom left). Dynamic light scattering (top right) and native gel electrophoresis (bottom right) of the four mutants that exhibit no detectable 24-mer agree with SEC solution results confirming that these amino acids act as oligomerization switch residues. Schematics follow the convention in Fig. 1. Error bars indicate S.D.
DLS Analysis of BFR R30A, R61A, Y114A, and E128A Assembly in Solution
To resolve the inconsistencies between the SEC and TEM results, DLS was used to determine whether the four mutants, BFR R30A, R61A, Y114A, and E128A, form 24-mer nano-structures in solution (Fig. 3). Comparison of the DLS data for the four mutants and the native protein, BFR, reinforces the SEC experiments. All four mutants form particles in solution that are much smaller than BFR.
Native Gel Electrophoretic (Native PAGE) Analysis of BFR R30A, R61A, Y114A, and E128A Assembly in Solution
To further resolve the inconsistencies between the solution (SEC and DLS) and the TEM data, native PAGE was performed (Fig. 3). Consistent with the SEC chromatogram, the native protein separates into two bands corresponding to 24-mer and dimer oligomerization states. The lanes corresponding to the four mutants display only single bands that have the same mobility as the dimer state in BFR. With the strength of three solution techniques all supporting each other, we conclude that in solution, these mutants only form dimer with no detectable 24-mer and that the TEM results do not correspond to what is happening in solution.
It is probable that the TEM results are an artifact of the drying step during sample preparation. It is possible that as drying occurs, the concentration of protein reaches a threshold level, driving the subunits together into a cage. Although the TEM does not correlate with the solution data, the micrographs suggest that although these mutants do not form nano-cages in solution, mutation does not completely destroy the potential of the mutants to assemble under “forcing conditions”; it is simply disfavored in solution. In addition, these results emphasize the importance of confirming electron microscopy-based conclusions with data derived from solution experiments.
The Solution Secondary Structure: Its Thermal Stability and Reversibility after Melting, as Monitored by CD
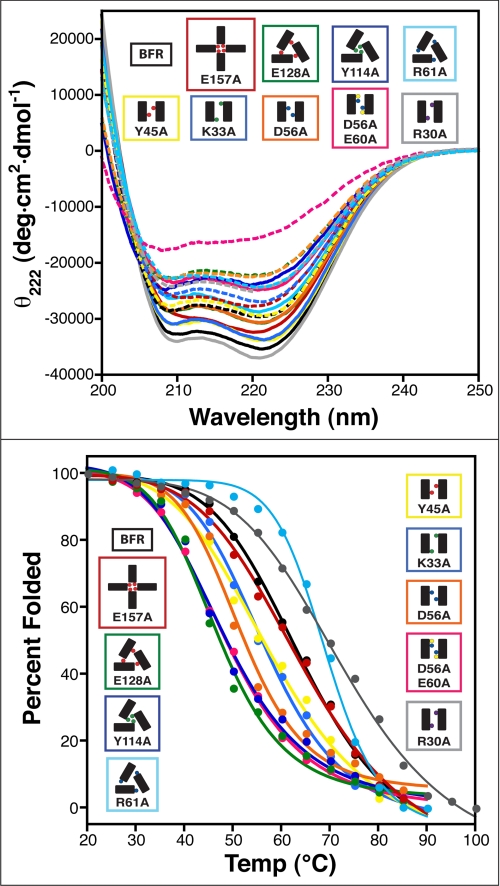
The native protein and all of the mutants fold into helical secondary structure as evidenced by the presence of CD minima at 208 and 222 nm (Fig. 4). Nearly all of the mutants exhibit reduced secondary structure when compared with the parent. Not unexpectedly, the lone exception is BFR R30A, which is the most thermally stable (see below). The double mutant BFR D56A,E60A, which is predictably less stable than the two proteins that eliminate single interactions within the cluster (BFR K33A and BFR D56A), also exhibits the largest reduction in helicity.
FIGURE 4.
The role mutation plays in nano-cage protein thermal stability and ability to refold after denaturation. Top, CD spectra of the BFR derivatives before denaturation (solid lines) and after slow cooling post-thermal denaturation (dashed lines). Bottom, thermal transitions of the BFR derivatives as monitored by CD. Schematics follow the convention in Fig. 1, and the coloring follows that of Fig. 3. deg, degrees.
Increasing the temperature results in the melting of the secondary structure (Fig. 4 and supplemental Figs. S9 and S10). By monitoring the 222 nm band, it is evident that for all the proteins, this unfolding is cooperative, suggesting that they all adopt tight, stable folds. Most of the mutants melt at a lower temperature than native BFR, suggesting that even simple point mutations can affect the protein thermal stability. The exceptions to this trend are BFR E157A, which melts at nearly the same temperature as BFR, and BFR R61A and R30A, both of which exhibit increased thermal stability. As it was surprising that the removal of salt bridges would result in an increase in protein stability, these latter results were confirmed by denaturation with both guanidine and urea (supplemental Fig. S11) and monitoring endogenous tryptophan fluorescence. In both cases, the concentration of denaturant required to unfold the protein is greater than that for the parent protein. These data confirm that R61A and R30A are indeed more stable than native BFR.
Slowly reducing the temperature back to starting conditions after the melting experiments allows for the exploration of folding reversibility (Fig. 4). The most thermally stable protein, BFR R30A, exhibited the least folding reversibility.
DISCUSSION
Many ferritin nano-cage proteins are formed through the homo-oligomerization of relatively small and simple monomers that fold into helix bundles, a motif that has been extensively studied. Therefore, these proteins may develop into a next generation of model systems for the protein engineering and design communities, which are turning their attention from tertiary structure to more complex quaternary structure.
The octahedrally symmetric maxi-ferritin, E. coli bacterioferritin, BFR, was used in this study to help establish an understanding of the role that protein-protein interactions play in the self-assembly of nano-structure. By conceptually shaving to alanine the side chains of nine amino acids, identified by inspection of the crystal structures in concert with virtual alanine scanning, the role of specific interactions at symmetry-related protein-protein interfaces was probed. Although the residues that were chosen for mutagenesis are only a fraction of those located at the various protein-protein interfaces, we predicted that they would be among the most significant. These interfacial amino acids were organized based on their relationship with different symmetry elements within the octahedral BFR nano-cage with the hope that they could help shed light on the importance, if any, these symmetries have on the stability or assembly of the protein. The selected nine amino acid residues were either aromatic or involved in salt-bridging interactions. One of the mutants disrupted interactions at the four-fold axis, three mutants were at the three-fold axis, and five mutants were focused on residues along the two-fold axis. After cloning, expression, and purification, we determined that all nine of the mutants formed cooperatively folded α-helical secondary structures and had the ability to form nano-cages as evidenced by temperature-dependent CD and TEM.
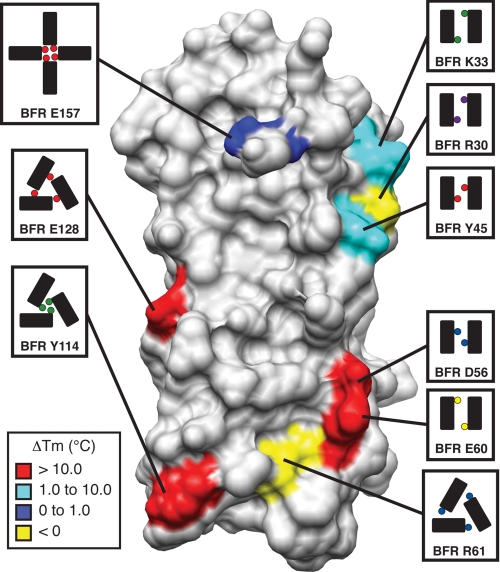
Comparison of the role mutation plays on the thermal stability of the helical structure resulted in a wide range of melting temperatures (Fig. 5 and supplemental Fig. S10). The only residue about the four-fold axis that was mutated was Glu-157. This residue, which is located on a C-terminal strand adjacent to the structurally significant E-helix, appears to be making a hydrogen bond, albeit a poorly aligned one, with Lys-38 supplemental Fig. S16). However virtual alanine scanning predicts that this interaction makes a negligible contribution to the overall stability (see above). The data agree with the computation in that the E157A mutant, which destroys the possibility of forming this hydrogen bond, has a thermal denaturation transition at nearly the same temperature as the native protein. Moreover, this mutation has no effect on the oligomerization state. Interestingly, our previous studies showed that deletion of the entire E-helix resulted in thermal destabilization by more than 10 °C and complete disruption of the 24-mer in favor of the dimer (34). Taken together, these data suggest that Glu-157, and possibly the entire C-terminal strand, is not contributing to the structural energetic importance of the E-helix (41). The point mutants, E128A and Y114A, both located along the three-fold axis, and the double mutant D56A,E60A at the two-fold axis have melting temperatures depressed by more than 15 °C from the native protein. These data suggest that thermodynamic hot spots do indeed exist in this system. In contrast are the thermal stabilities of the three-fold axis-associated mutant, R61A and R30A, which is located at the two-fold axis. These single point mutants exhibit melting temperatures 4 and 8 °C higher than the native protein, and the increased stability over that of BFR was confirmed using the chemical denaturants, urea and guanidine. The R61A result is even more surprising when compared with the destabilization observed upon mutation of Glu-128 with which it interacts. An explanation of these somewhat surprising observations could be reflected in the complexity of interactions that exist in natural systems. The basic amino acids, arginine and lysine, due to the long aliphatic linker between the α-carbon and charged head group often play steric roles in concert with the role of a component in a salt bridge (42, 43). Therefore, it may not be a coincidence that these two residues are arginine, and it could be possible that removal of these large side chains is permitting rearrangement of the interface to maximize stability. Whatever their origin, these results suggest that BFR has not evolved solely to maximize structural stability. This is reasonable considering that BFR exists naturally as a mixture of oligomerization states (34, 44) and has a complex cellular function that is intrinsically linked to its structure.
FIGURE 5.
The role protein-protein interface residues have on thermodynamic stability of the BFR nano-cage protein. The change in thermal stability was mapped onto the BFR crystal structure for interfacial residues. Schematics follow the convention in Fig. 1. The legend in the lower left indicates the color scheme, which reflects the degree to which each residue influences the stability.
Electron microscopy indicates that mutagenesis does not disrupt the ability of the proteins to form nano-cage structures. In solution, however, all the mutants, with the exception of E157A, demonstrate a shift in the ratio of oligomerization to favor the dimer over the 24-mer. Four residues, Glu-128, Tyr-114, Arg-61, and Arg-30, when mutated to alanine, result in proteins that form no detectable 24-mer in solution, suggesting that they are “switch residues” that control the formation of nano-assembly, whereas having little effect on the lower order structure. Although these data present a caveat concerning oligomerization state conclusions based on TEM of semidried samples, when taken together, they suggest that even if the solution composition is altered, mutagenesis may not completely destroy the ability to form specific assembled structure under extreme conditions even if it completely shifts the oligomerization state in solution. The fact that R30A is among these four mutants is especially intriguing as the mutated residue is located at the two-fold axis. Conventional wisdom would dictate that alanine shaving at the dimer interface would result in destruction of interactions that favor dimerization. A possible explanation is that the conformational rearrangement upon removal of the large arginine side chain is transferred to other surfaces of the protein, thereby resulting in disruption of other symmetry-related interfaces. This remodeling could result in an unpacking of the protein in general, which may explain the decreased cooperativity in the thermal denaturation profile (Fig. 4). We are currently exploring this hypothesis by determining the change in solvent-exposed hydrophobic surface area upon mutation. Another possibility is that this dimer is unique and unrelated to the dimer centered around the two-fold axis in the 24-mer. A dimer is a proposed intermediate in the mechanism of assembly of ferritins (29, 45), and the potential for the existence of alternative dimers warrants further investigation. Together these four mutants further emphasize the strength of the model system in its sensitivity to even subtle mutagenesis. In addition, these results are especially intriguing in that they demonstrate that self-assembling nano-structure can be controlled by single point mutation.
The residues in this study were selected by simple inspection and thus an arguably arbitrary selection strategy. Confidence in our approach was enhanced by the fact that many of these residues agreed with virtual alanine scanning calculations. It is a testament to the tractability of this system that of the nine mutants predicted through such a low level algorithm, only one had no effect on either the stability or the oligomerization state. That the residues selected for mutation clearly are energetically and structurally significant is evidenced by the fact that three mutants have depressed melting temperatures by at least 15 °C, and two mutants have melting temperatures elevated by at least four degrees. Moreover, four of the mutants have completely lost the ability to form the nano-cage structure and only form cooperatively folded dimer in solution. Therefore, these are oligomerization switch residues. It is remarkable that multiple properties of the system can be tuned so easily. We are currently exploring the possibility that other computationally predicted residues are significant either structurally or energetically. Moreover, we are actively working to disentangle the relationship between thermal stability and nano-structure, which this current study indicates are linked in perhaps quite complicated ways. In addition, we are applying similar strategies to other nano-cages with similar and yet structurally distinct oligomerization states and symmetries so that the knowledge gleaned can be used to rationally design oligomers with unique structures, which in turn can be used to template unique inorganic nano-materials (17)6 and to aid the fundamental analysis of the evolutionary relationships of key interfacial interactions that control assembly in this class of proteins.
Acknowledgments
We thank Prof. Julien Lescar (Nanyang Technological University School of Biological Sciences) and Prof. Anh Tuan Phan (SPMS Division of Physics and Applied Physics) for the use of the fast protein liquid chromatography machines, Ai Hua Seow for the use of the CBC teaching laboratory CD spectrometer, and Prof. Hongyu Chen's laboratory for help with TEM. We also thank Maziar Ardejani and Tianpei Fu for helpful discussions and K. M. H. Chiew for critical reading of the manuscript.
This work was supported by a School of Physical and Mathematical Sciences (SPMS) start up grant and Singapore Ministry of Education Academic Research Fund Tier 1 Grant RG 53/06.
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S18.
R. Fan, S. W. Chew, V. V. Cheong, and B. P. Orner, manuscript in preparation.
- BFR
- bacterioferritin
- SEC
- size exclusion chromatography
- TEM
- transmission electron microscopy
- DLS
- dynamic light scattering
- Ek
- enterokinase
- LIC
- ligation-independent cloning.
REFERENCES
- 1.Dobson C. M. (2002) Nature 418, 729–730 [DOI] [PubMed] [Google Scholar]
- 2.Kennedy D., Norman C. (2005) Science 309, 75–102 [DOI] [PubMed] [Google Scholar]
- 3.Jin L., Wells J. A. (1994) Protein Sci. 3, 2351–2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogan A. A., Thorn K. S. (1998) J. Mol. Biol. 280, 1–9 [DOI] [PubMed] [Google Scholar]
- 5.Moreira I. S., Fernandes P. A., Ramos M. J. (2007) Proteins 68, 803–812 [DOI] [PubMed] [Google Scholar]
- 6.Fletcher S., Hamilton A. D. (2007) Curr. Top. Med. Chem. 7, 922–927 [DOI] [PubMed] [Google Scholar]
- 7.Seringhaus M., Gerstein M. (2007) Science 315, 40–41 [DOI] [PubMed] [Google Scholar]
- 8.Calhoun J. R., Bell C. B., 3rd, Smith T. J., Thamann T. J., DeGrado W. F., Solomon E. I. (2008) J. Am. Chem. Soc. 130, 9188–9189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hill R. B., Raleigh D. P., Lombardi A., DeGrado W. F. (2000) Acc. Chem. Res. 33, 745–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papapostolou D., Smith A. M., Atkins E. D., Oliver S. J., Ryadnov M. G., Serpell L. C., Woolfson D. N. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10853–10858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grueninger D., Treiber N., Ziegler M. O., Koetter J. W., Schulze M. S., Schulz G. E. (2008) Science 319, 206–209 [DOI] [PubMed] [Google Scholar]
- 12.Bromley E. H., Channon K., Moutevelis E., Woolfson D. N. (2008) ACS Chem. Biol. 3, 38–50 [DOI] [PubMed] [Google Scholar]
- 13.Liu X., Theil E. C. (2005) Acc. Chem. Res. 38, 167–175 [DOI] [PubMed] [Google Scholar]
- 14.Wang Q., Lin T., Tang L., Johnson J. E., Finn M. G. (2002) Angew. Chem. Int. Ed. Engl. 41, 459–462 [DOI] [PubMed] [Google Scholar]
- 15.Destito G., Yeh R., Rae C. S., Finn M. G., Manchester M. (2007) Chem. Biol. 14, 1152–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ren Y., Wong S. M., Lim L. Y. (2007) Bioconjug. Chem. 18, 836–843 [DOI] [PubMed] [Google Scholar]
- 17.Uchida M., Klem M. T., Allen M., Suci P., Flenniken M., Gillitzer E., Varpness Z., Liepold L. O., Young M., Douglas T. (2007) Adv. Mater. 19, 1025–1042 [Google Scholar]
- 18.Butts C. A., Xi J., Brannigan G., Saad A. A., Venkatachalan S. P., Pearce R. A., Klein M. L., Eckenhoff R. G., Dmochowski I. J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 6501–6506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Curtis A. R., Fey C., Morris C. M., Bindoff L. A., Ince P. G., Chinnery P. F., Coulthard A., Jackson M. J., Jackson A. P., McHale D. P., Hay D., Barker W. A., Markham A. F., Bates D., Curtis A., Burn J. (2001) Nat. Genet. 28, 350–354 [DOI] [PubMed] [Google Scholar]
- 20.Levi S., Cozzi A., Arosio P. (2005) Best Pract. Res. Clin. Haematol. 18, 265–276 [DOI] [PubMed] [Google Scholar]
- 21.Zandman-Goddard G., Shoenfeld Y. (2007) Autoimmun. Rev. 6, 457–463 [DOI] [PubMed] [Google Scholar]
- 22.Vidal R., Delisle M. B., Rascol O., Ghetti B. (2003) J. Neurol. Sci. 207, 110–111 [DOI] [PubMed] [Google Scholar]
- 23.Ingrassia R., Gerardi G., Biasiotto G., Arosio P. (2006) J. Biochem. 139, 881–885 [DOI] [PubMed] [Google Scholar]
- 24.Almirón M., Link A. J., Furlong D., Kolter R. (1992) Genes Dev. 6, 2646–2654 [DOI] [PubMed] [Google Scholar]
- 25.Grant R. A., Filman D. J., Finkel S. E., Kolter R., Hogle J. M. (1998) Nat. Struct. Biol. 5, 294–303 [DOI] [PubMed] [Google Scholar]
- 26.Dautant A., Meyer J. B., Yariv J., Précigoux G., Sweet R. M., Kalb A. J., Frolow F. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 16–24 [DOI] [PubMed] [Google Scholar]
- 27.Luzzago A., Cesareni G. (1989) EMBO J. 8, 569–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tong G. J., Hsiao S. C., Carrico Z. M., Francis M. B. (2009) J. Am. Chem. Soc. 131, 11174–11178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerl M., Jaenicke R., Smith J. M., Harrison P. M. (1988) Biochemistry 27, 4089–4096 [DOI] [PubMed] [Google Scholar]
- 30.Cunningham B. C., Wells J. A. (1989) Science 244, 1081–1085 [DOI] [PubMed] [Google Scholar]
- 31.Hu Z., Ma B., Wolfson H., Nussinov R. (2000) Proteins 39, 331–342 [PubMed] [Google Scholar]
- 32.Kortemme T., Kim D. E., Baker D. (2004) Sci. STKE 2004, pl2. [DOI] [PubMed] [Google Scholar]
- 33.Kortemme T., Baker D. (2002) Proc. Natl. Acad. U.S.A. 99, 14116–14121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan R., Boyle A. L., Cheong V. V., Ng S. L., Orner B. P. (2009) Biochemistry 48, 5623–5630 [DOI] [PubMed] [Google Scholar]
- 35.Young L., Dong Q. (2004) Nucleic Acids Res. 32, e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X., Jin W., Theil E. C. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3653–3658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bellapadrona G., Stefanini S., Zamparelli C., Theil E. C., Chiancone E. (2009) J. Biol. Chem. 284, 19101–19109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pieraccini S., Saladino G., Cappelletti G., Cartelli D., Francescato P., Speranza G., Manitto P., Sironi M. (2009) Nat. Chem. 1, 642–648 [DOI] [PubMed] [Google Scholar]
- 39.Turk J. A., Smithrud D. B. (2001) J. Org. Chem. 66, 8328–8335 [DOI] [PubMed] [Google Scholar]
- 40.Jencks W. P. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 4046–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levi S., Luzzago A., Franceschinelli F., Santambrogio P., Cesareni G., Arosio P. (1989) Biochem. J. 264, 381–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chamberlain A. K., Lee Y., Kim S., Bowie J. U. (2004) J. Mol. Biol. 339, 471–479 [DOI] [PubMed] [Google Scholar]
- 43.Segrest J. P., Jones M. K., De Loof H., Brouillette C. G., Venkatachalapathi Y. V., Anantharamaiah G. M. (1992) J. Lipid Res. 33, 141–166 [PubMed] [Google Scholar]
- 44.Andrews S. C., Smith J. M., Hawkins C., Williams J. M., Harrison P. M., Guest J. R. (1993) Eur. J. Biochem. 213, 329–338 [DOI] [PubMed] [Google Scholar]
- 45.Stefanini S., Vecchini P., Chiancone E. (1987) Biochemistry 26, 1831–1837 [DOI] [PubMed] [Google Scholar]