Abstract
Inhibition of tumor necrosis factor α (TNFα) is a favorable way of treating several important diseases such as rheumatoid arthritis, Crohn disease, and psoriasis. Therefore, an extensive range of TNFα inhibitory proteins, most of them based upon an antibody scaffold, has been developed and used with variable success as therapeutics. We have developed a novel technology platform using C-type lectins as a vehicle for the creation of novel trimeric therapeutic proteins with increased avidity and unique properties as compared with current protein therapeutics. We chose human TNFα as a test target to validate this new technology because of the extensive experience available with protein-based TNFα antagonists. Here, we present a novel and highly specific TNFα antagonist developed using this technology. Furthermore, we have solved the three-dimensional structure of the antagonist-TNFα complex by x-ray crystallography, and this structure is presented here. The structure has given us a unique insight into how the selection procedure works at a molecular level. Surprisingly little change is observed in the C-type lectin-like domain structure outside of the randomized regions, whereas a substantial change is observed within the randomized loops. Thus, the overall integrity of the C-type lectin-like domain is maintained, whereas specificity and binding affinity are changed by the introduction of a number of specific contacts with TNFα.
Keywords: Antibodies, Cytokines/Tumor Necrosis Factor, Methods/X-ray Crystallography, Protein/Protein-Protein Interactions, Protein/Structure, In Vitro Selection
Introduction
Tetranectin belongs to the large class of C-type lectins characterized by a common fold known as the C-type lectin-like domain (CTLD)3 (1). Tetranectin is a homotrimeric human protein found in both plasma and tissue. This protein binds the lysine-binding kringle domains from apolipoprotein A (2), plasminogen (3), and angiostatin (4). Tetranectin is a 60-kDa protein built from a structural unit composed of three identical chains, each with a CTLD domain located C-terminally to a trimerizing coil-coil region (5). The CTLD domains retain their structural integrity as separate protein domains, (6, 7) and, moreover, it was shown that their binding to the known tetranectin ligand, plasminogen kringle-4, exhibits the same thermodynamic parameters, irrespective of whether it was analyzed in the form of free monomeric domains or as tethered domains in the complete homotrimeric protein (3). In addition, the thermodynamic analysis showed that the formation of the trimer led to an apparent 100-fold affinity increase, which most likely is due to the avidity effect caused by the three-fold clustering of CTLD domains in the complete protein. Comparison of ensembles of natural CTLD domains for which the known structure and ligand specificity are known shows that the ligand-binding site can accommodate a diverse range of ligands. We therefore concluded that the tetranectin CTLD might be a useful scaffold for designing novel protein therapeutics. We could change the sequence of loops within the CTLD scaffold in the monomeric version without perturbing the overall structure. Thus, CTLD serves as an efficient starting point for the in vitro selection of a high affinity antagonist with a low immunogenicity. In this procedure, we use the monomeric CTLD domain during the in vitro selection procedure but use the naturally occurring trimeric version in downstream applications.
Borean Pharma established an extended and coherent technology platform to use C-type lectins as a vehicle for the creation of novel trimeric therapeutic proteins with increased avidity and unique properties as compared with current protein therapeutics, such as antibodies and small protein scaffolds. Human tetranectin may be readily tailored to meet specific therapeutic needs by “reprogramming” CTLD. Each CTLD has five loop regions, each 6–9 amino acids in length, which determine the binding specificities. Reprogramming is performed by creating phage libraries displaying CTLD, where specific loops are randomized, followed by selection. Randomization can be repeated either sequentially or iteratively. Furthermore, the use of the CTLD platform ensures selected protein candidates, which are highly homologous to a native human secreted protein and thus of low immunogenicity. Initial validation of this novel scaffold technology was achieved by selecting an antagonist of hTNFα, as described herein. Subsequently, the platform has been effectively used on a number of diverse and therapeutically relevant targets. Current potency of the tetranectin-derived hTNFα antagonist was obtained through carefully managed in vitro evolution steps (Fig. 1). After each step, the binding kinetics were measured, as well as the ability of the trimeric version to inhibit hTNFα-induced apoptosis in L929 cells (see Table 1).
FIGURE 1.
Schematic overview of the selection and maturation process. The red X indicates the amino acid position that was randomized in a given library. The terms Primary lib and mat lib refer to primary and maturation libraries, respectively. The sequences of CTLD libraries used as a basis for the next round of selection are shown in bold. a, amino acids maintained from the primary clone to TN-2-B-1-C31. b, amino acids maintained from the first round maturation clone to TN-2-B-1-C31. c, amino acids maintained from the second round maturation clone to TN-2-B1-C31.
TABLE 1.
Binding kinetics of monomeric CTLD toward hTNFα and inhibitory constant of the trimeric CTLD
| Maturation round | Clone | KDa | kon | koff | Ki | 95% confidence intervals |
|---|---|---|---|---|---|---|
| m | m−1s−1 | s−1 | ng/ml | |||
| Primary | TN2 | >5.0 × 10−5 | NDb | NDb | NDb | |
| First | TN2-B | 7.3 × 10−8 | 1.3 × 107 | 9.2 × 10−1 | >10 × 106 | |
| Second | TN2-B1 | 1.2 × 10−8 | 2.5 × 105 | 2.9 × 10−3 | 1440 | 1026–2017 |
| Third | TN2-B1-C31 | 3.4 × 10−10 | 2.6 × 106 | 8.9 × 10−4 | 2.8 | 1.6–6.6 |
a Primary, first, and second clones were analyzed with ∼500 RU ligand immobilized, and the third clone was analyzed with ∼50 RU. T-values > 100. RU designates Biacore response unit; T-value is defined as parameter value divided by S.E.
b ND, not detectable.
EXPERIMENTAL PROCEDURES
Development and Construction of a Truncated TNFα Antagonist Gene
TN-2-B-1-C31 was developed by the use of phage display selection technology. A phage library displaying the monomeric scaffold structure of the C-type lectin domain of human tetranectin containing random amino acid residues in positions 116–122 (loop 1) and 145–150 (loop 4) was constructed and used for the selection of hTNF-binding CTLDs. A candidate from the primary selection, TN-2, was taken through three rounds of affinity maturation using general phage selection techniques. A construct expressing the full-length protein of TN-2-B-1-C31 was created by cloning the coding region of TN-2-B-1-C31 into the BamHI and EcoRI sites of the vector pT7CIIH6-AmpR (8).
CTLD Purification
Escherichia coli (strain BL21 DE3) cells were harvested by centrifugation and lysed in 100 ml of lysis buffer by sonication followed by the addition of detergent buffer and inclusion body recovery for refolding in the present first generation procedure. Inclusion bodies were washed with 0.5% Triton X-100, 1 mm EDTA, pH 8, and resuspended in 6 m guanidine, 50 mm dithiothreitol, 50 mm Tris-HCl, pH 8. After inclusion body solubilization, the buffer was changed to 8 m urea, 1 m NaCl, 5 mm β-mercaptoethanol, 50 mm Tris-HCl, pH 8, and the fusion CTLD was captured on a nickel-nitrilotriacetic acid-agarose column (Qiagen). After elution with 8 m urea, 1 m NaCl, 5 mm β-mercaptoethanol, 20 mm EDTA, 50 mm Tris-HCl, pH 8, the fusion CTLD was refolded by dilution with 3 m urea, 250 mm NaCl, 50 mm glycine, pH 9.5, 2 mm CaCl2, 0.5/0.3 mm GSH/GSSG overnight at a rate of 0.15 ml/min to a final dilution of 10× sample volume at +7 °C with mixing. After refolding, the CTLD was purified further on SP-Sepharose FF in 8 m urea, 50 mm NaAc, pH 4.5, 0–1 m NaCl gradient. The fusion tag was cleaved by recombinant human granzyme B in 1 m urea, 20 mm Tris-HCl, pH 7.3, 150 mm NaCl, 3 mm CaCl2 overnight before a final purification on a SOURCE 15S column in 8 m urea, 50 mm NaAc, pH 4.5, 0–1 m NaCl gradient and finally buffer-exchanged into 50 mm NaAc, pH 5.0, buffer.
hTNFα Purification
E. coli strain BL-21 (Invitrogen) was transformed with a pT7-CIIH6-TNFα expression vector encoding human TNFα and grown to an A600 of 0.5 in standard tryptone-yeast extract medium with 100 μg/ml ampicillin and 10 mm MgSO4 before induction with λCE46 phage and rifampicin addition to 200 μg/ml. Cells from 6 liters of culture were harvested by centrifugation, washed (0.1 m Tris-HCl, pH 8, 1 m NaCl), sonicated, and centrifuged. The supernatant was loaded on a nickel-nitrilotriacetic acid column (Qiagen), and the fusion TNFα was captured, eluted, and further purified on an SP-Sepharose FF column (GE Healthcare). Digestion with factor Xa at +5 °C overnight was applied afterward. The protease and cleaved fusion tag were captured on soybean trypsin inhibitor and nickel-nitrilotriacetic columns, respectively. A final Q-Sepharose FF step yielded a pure hTNFα batch.
Biacore Assay
Surface plasmon resonance binding analysis was performed on a Biacore 3000 instrument. The hTNFα capture antibody (AHC3712) to be immobilized was dissolved in 10 mm NaAc, pH 5.0, and then immobilized on a CM5 Biacore sensor chip using the amine coupling kit (Biacore). Binding analysis was performed at a flow rate of 5 μl/min. Before loading of the protein sample, the chip was equilibrated in 10 mm Tris, pH 8.0, 150 mm NaCl, 2 mm CaCl2, 50 μm EDTA, and 0.005% surfactant P20. Recombinant human TNFα was dissolved in 10 mm Tris, pH 8.0, 150 mm NaCl, 2 mm CaCl2 at 10 μg/ml, and 10 μl was injected. Aliquots (20 μl) of CTLD C31 were injected using the KINJECT option. 5 minutes of dissociation were allowed before the chip was regenerated with sequential injection of 0.05% SDS and 10 mm glycine, pH 2.5. Binding of CTLD C31 was analyzed at six different protein concentrations, ranging from 1.5 to 50 nm. Binding data were evaluated using the BIAevaluation program version 3.2 (Biacore).
Inhibition of hTNFα-induced Apoptosis
The effect of inhibition of hTNFα by trimeric TN-2-B1-C31 was analyzed in a standard cell assay for the antagonistic effect of hTNFα binders on the cytotoxic effect of hTNFα on the murine L929 cell line as described in Ref. 9.
Crystallization, Data Collection, and Structure Determination
The complex was formed by mixing hTNFα with CTLD at a 1:1 molar ratio. Initial crystallization screening was performed with the Crystal ScreenTM system (Hampton Research). Optimal crystals were grown by mixing equal volumes of protein solution with reservoir solution containing 0.1 m Tris-HCl, pH 8.0, 0.35 m MgAc, and 20% 2-propanol. The crystals were frozen directly in the crystallization solution at the I911-3 beam line at the National Laboratory for Synchrotron Radiation in Lund, Sweden, MAX-lab (10) or the X12 beam line at European Molecular Biology Laboratory (EMBL) Hamburg. The crystals belonged to the P6322 space group with cell dimensions of a = b = 84 Å, c = 150 Å.
The structure of the CTLD-hTNFα complex was solved by molecular replacement using Phaser (11). We used a search model composed of one wild type hTNFα monomer (Protein Data Bank (PDB) code 1TNF) (12) and the wild type CTLD (6) (PDB code 1TN3), excluding the loops, which were changed during the in vitro evolution procedure. The resulting model was used as a starting point for wARP/ARP (13), which did build 98% of the complex (including the maturation loops). The model was finalized with Coot (14) and refined in REFMAC5 (15). The final model consists of residues 9–157 for hTNFα, residues 46–180 for CTLD excluding the loop region 52–56, for which no electron density was seen, 244 water molecules, and one magnesium ion. 99.1% of the residues are in the allowed regions of the Ramachandran plot, with 0.4% (1 residue) in the generously allowed region and Arg-120 from CTLD in the disallowed region. Figures were made using the program PyMOL (19).
RESULTS AND DISCUSSION
A primary phage display library was generated by randomizing loops 1 and 4 in the CTLD domain of tetranectin, and selection for binding to hTNFα was performed by panning. This resulted in a candidate molecule of high specificity but suboptimal affinity (Clone TN-2). This clone was then subjected to three consecutive steps of affinity maturation. First, loops 1 and 4 were randomized individually to provide full sequence coverage of each loop, and phage display selection was performed to select for higher affinity. This did not result in any changes in loop 1 (the loop 1 sequence was reselected), but 3 out of 4 amino acids were changed in loop 4 (only the 1st proline was conserved and would remain conserved throughout the maturation). An additional unexpected change occurred outside the randomized area, where DCTLD157 was changed to Gly. The significance of this change, although not fully clear, may have helped stabilize the CTLD structure and resulted in substantially improved affinity (Table 1). However, the TN-2-B clone was incapable of inhibiting hTNFα-induced apoptosis. In the second maturation step, loop 1 was randomized in increments of 3 amino acids, and the selection procedure was changed to favor clones with low off-rates. This was done by including prolonged washing steps and competition by free hTNFα. The resulting TN-B-1 clone showed a 300-fold reduction in off-rate and improved bioactivity. During the third and final stage of maturation, loop 3 was randomized together with two positions in loop 4. This only led to a marginal improvement in the binding kinetics (35-fold in affinity and 6-fold in off-rate for the monomeric CTLD). However, the bioactivity improved by almost 200-fold for the trimeric tetranectin derivative.
The improved bioactivity of clones obtained by applying selective pressure for “better” dissociation kinetics demonstrated the potential of this strategy to improve the efficacy of tetranectin-based therapeutics. It should be noted that the selection as well as the binding data originate from the monomeric version of CTLD, whereas the trimeric version was used for measuring the biological activity. As compared with the moderate improvement in binding of the monomeric CTLD after the third round of maturation, the 200-fold improvement in biological activity of the trimeric tetranectin derivative might well be explained by a large increase in avidity of the trimeric tetranectin derivative.
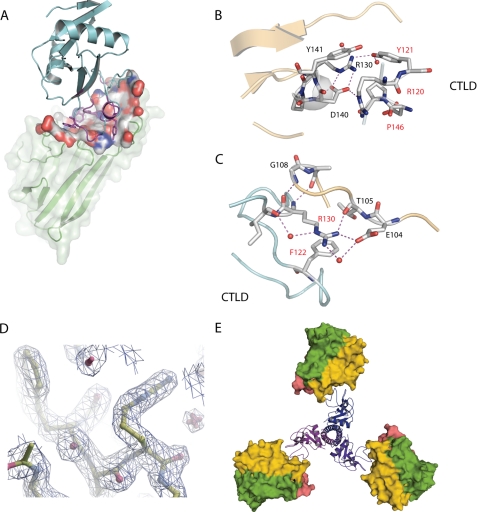
To further understand the three-dimensional interactions responsible for the high affinity binding and to assure that the overall structure of tetranectin was maintained, we determined the crystal structure of the CTLD-hTNFα complex by molecular replacement at 2.1 Å resolution (Fig. 2A). The final model consists of residues 9–157 for hTNFα and residues 46–180 for CTLD, excluding residues 52–56, for which no electron density was observed (Table 2). The secondary structure of hTNFα bound to CTLD closely resembles that of free hTNFα, with a root mean square deviation of 1.2 Å for Cα atoms (12, 16). Likewise, the non-randomized part of the CTLD scaffold exhibited little change in structure (root mean square deviation for Cα atoms with free native CTLD is less than 1 Å), which validates the use of human tetranectin as a scaffold or platform for engineering new biologics. As for the CTLD loops, the structure has changed substantially in the randomized loop 3 and loop 4 (supplemental Fig. 1). In contrast, the randomized loop 1 backbone resembles that of native CTLD despite a complete change of sequence. The side chains of the randomized maturation loops display a systematic directionality toward hTNFα, creating a visibly depressed surface in the previously rather flat hTNFα area. The buried, accessible surface area at the interface between hTNFα and the CTLD is 865 Å2, and the interaction surface features a mixture of polar, van der Waals, and hydrophobic interactions. Hydrophobic interactions are often the driving force in tight protein-protein interactions. However, we observed a more diverse set of interactions, which in this case is most likely found because the CTLD must adapt to the hTNFα surface, including neutralizing the charge present.
FIGURE 2.
A, overview of hTNFα-CTLD complex structure. The hTNFα secondary structure is shown as green ribbon, combined with a transparent surface representation. The part of the hTNFα surface involved in interactions with CTLD is shown in space-fill coloring according to electrostatic potential. The antagonist is depicted as cyan ribbon, except for the randomized part, which is shown in magenta. B and C, close up of the hTNFα-CTLD interactions. Amino acid labels in red refer to CTLD, and amino acid labels in black refer to hTNFα. D, reconstruction of trimeric version of the TN2-B1-C31 using the structure of natural trimeric tetranectin as a model. This was done by substituting the structure of the CTLD domain found in the structure of natural tetranectin (PDB ID 1HTN) with the structure of the TN2-B1-C31 clone in complex with hTNFα. Next we added the symmetry mates of hTNFα to reconstruct the naturally occurring trimeric hTNFα (panel E).
TABLE 2.
Data collection and refinement statistics
| TN-2B-1-C31:hTNFα | |
|---|---|
| Data collection | |
| Space group | P6322 |
| Cell dimensions | |
| a, b, c (Å) | 84, 84, 149 |
| α, β, γ (°) | 90, 90, 120 |
| Resolution (Å) | 74-2.1 (2.15-2.1) |
| Rsym or Rmerge | 0.08 (0.48) |
| I/σI | 55.1 (9.0) |
| Completeness (%) | 99.9 (99.8) |
| Redundancy | 33 (26.1) |
| Refinement | |
| Resolution (Å) | 2.1 |
| No. of reflections | 17855 |
| Rwork/Rfree | 0.17/0.22 |
| No. of atoms | |
| Protein | 2209 |
| Ligand/ion | 1 |
| Water | 244 |
| B-factors | |
| Protein | 28.2 |
| Ligand/ion | 46.4 |
| Water | 39.185 |
| r.m.s.a deviations | |
| Bond lengths (Å) | 0.016 |
| Bond angles (°) | 1.16 |
a r.m.s., root mean square.
Residues KCTLD116 and SRYFCTLD-(119–122) in loop 1 are conserved throughout the maturation procedure. We were unable to select new loop 1 sequences in the first maturation round by complete randomization of loop 1. In the second round of maturation, where we performed partial randomization of loop 1, KCTLD116 and SRYFCTLD-(119–122) were reselected, and only VRCTLD-(117–118) was changed to RW. This suggests that these residues play an essential role in the binding of CTLD to hTNFα. The structure shows that they interact with a well organized loop structure in hTNFα consisting of residues 138–141 (Fig. 2B). This loop in hTNFα has multiple internal contacts; RTNF138 hydrogen-bonds with DTNF140 and is further prevented from lateral flexibility by close van der Waals packing with YTNF141, thus providing a stable binding epitope for the CTLD. RCTLD120 and YCTLD121 form hydrogen bonds with DTNF140 and RTNF138 of hTNFα, respectively. Considerable strain is put on RCTLD120, making it fit into the tight space at the complex interface, which results in ϕ/ψ angles outside the allowed regions of the Ramachandran plot. The hydroxyl group of SCTLD119 forms multiple hydrogen bonds with the backbone of KCTLD116 and FCTLD122 and thus locks the conformation of the randomized loop 1. Furthermore, SCTLD119 is situated at the center of the CTLD-TNFα interaction surface pointing into the core of the CTLD, and thus there is substantial steric restriction on this residue. KCTLD116 is also conserved in the selection procedure but does not contact hTNFα. However, KCTLD116 forms several contacts with both the backbone and the side chain of QCTLD148 and the side chain of QCTLD151. Thus, the length and charge of lysine are favorable at position 116, stabilizing the structure of the randomized loops. The selection scheme not only produces strong interactions with hTNFα but also ensures the stability of the interacting framework. Both TCTLD121 and FCTLD122 participate in close van der Waals packing with hTNFα. FCTLD122 also stacks closely with RCTLD130 and redirects the side chain of this residue toward hTNFα. RCTLD130 is found well outside the randomized part, but the side chain has adopted a completely different conformation from that found in free CTLD. The guanidine group of RCTLD130 forms hydrogen bonds with the side chain of ETNF104 and the backbone of TTNF105 (Fig. 2C). Furthermore, the backbone of RCTLD130 forms hydrogen bonds with the backbone of GTNF108 and ATNF109 of hTNFα. Salt bridges between flexible residues, such as glutamic acid and arginine, are usually rather unstable, but here FCTLD122 restricts the flexibility of RCTLD130, and RCTLD130 is caught in a pocket between FCTLD122 and hTNFα. Thus, not only do the randomized residues contact hTNFα themselves, but they also reprogram part of the non-randomized part of the structure to interact with hTNFα. RCTLD117 and WCTLD118 appear during the second round of maturation, where there is a selection for low off-rate of the antagonist. WCTLD118 increases the buried hydrophobic surface substantially, and it also stacks with the guanidine group of RCTLD117 and stabilizes RCTLD117 hydrogen binding with hTNFα.
Loop 4 has changed conformation considerably as compared with the native tetranectin CTLD structure. However, the loop makes relatively few contacts with hTNFα that are predominantly made by WCTLD149. PCTLD146 appears in the first round of selection and is maintained throughout the maturation. We believe that PCTLD146 dictates a change in the conformation of loop 4, which is required to avoid sterical conflict with hTNFα. Thus, in the first round of selection, loop 1 provides the major part of the binding affinity, and loop 4 changes position to allow this interaction. In the later maturation stages, WCTLD149 appears and provides additional hydrophobic interaction with hTNFα and probably contributes to the lower off-rate.
Loop 3 was randomized during the third and final round of selection. Loop 3 does not make any direct contact with hTNFα. However, the position of the loop has shifted substantially relative to the native CTLD structure. It is likely that the changes in loop 3 help to stabilize the conformation of both loop 3 and loop 4 and provide a more rigid interaction platform. Loop 3 is normally involved in binding of Ca2+, but Mg2+ is found bound in the former Ca2+-binding site, and crystallization is performed in the presence of Mg2+. However, none of the ions are needed for binding to hTNFα, and selections have been performed in the absence of both ions (data not shown).
Several anti-TNFα drugs have been marketed, based either upon antibody technology or upon modified soluble TNFα receptor (17, 18). We compared the CTLD clone selected here with the available drugs. In our in vitro bioassays, the TN2-B1-C31 clone compared favorably to these drugs, both in terms of binding affinity and when comparing the ability to inhibit hTNFα-induced apoptosis (data not shown).
In summary, the data show that the CTLD scaffold can be “reprogrammed” for binding to novel targets through a short series of directed evolutionary steps while maintaining its core scaffold structure. Furthermore, the crystal structure of the CTLD-TNFα complex provided a unique insight into the workings of directed evolution. Initial binding affinity is provided by loop 1, which is accompanied by a change in the structure of loop 4 removing steric hindrance. During later steps of selection, in which a low off-rate was favored, additional buried hydrophobic contacts appeared, represented by WCTLD118 and WCTLD149. The selection also seems to favor additional rigidity of the antagonist, as exemplified by the changes of structure in loops 3 and 4. The successful selection for low off-rates is of particular importance for development of therapeutically useful antagonists. In subsequent selections for candidates against a number of targets, low nm affinity candidates were selected in the first round and then further affinity-matured for low off-rates as described here, thereby rendering solid therapeutic drug candidates with favorable pharmacodynamic profiles.
Supplementary Material
Acknowledgments
Beamline access was funded partly via the Danscatt consortium. We are grateful to the staff of beam lines I911 at MAX-lab Lund and X12 at EMBL Hamburg for help in data collection. We are also grateful to the staff at Borean Pharma A/S.
This work was supported by a Novo Nordisk Foundation senior research grant and by grants from The Danish Medical Research Council (Grant 22-04-0704) and The Carlsberg Foundation and an Arne Hansen stipend (to R. H.).
The atomic coordinates and structure factors (code 3L9J) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- CTLD
- C-type lectin-like domain
- TNFα
- tumor necrosis factor α
- hTNFα
- human TNFα.
REFERENCES
- 1.Zelensky A. N., Gready J. E. (2005) FEBS J. 272, 6179–6217 [DOI] [PubMed] [Google Scholar]
- 2.Caterer N. R., Graversen J. H., Jacobsen C., Moestrup S. K., Sigurskjold B. W., Etzerodt M., Thøgersen H. C. (2002) Biol. Chem. 383, 1743–1750 [DOI] [PubMed] [Google Scholar]
- 3.Graversen J. H., Lorentsen R. H., Jacobsen C., Moestrup S. K., Sigurskjold B. W., Thøgersen H. C., Etzerodt M. (1998) J. Biol. Chem. 273, 29241–29246 [DOI] [PubMed] [Google Scholar]
- 4.Mogues T., Etzerodt M., Hall C., Engelich G., Graversen J. H., Hartshorn K. L. (2004) J. Biomed. Biotechnol. 2004, 73–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nielsen B. B., Kastrup J. S., Rasmussen H., Holtet T. L., Graversen J. H., Etzerodt M., Thøgersen H. C., Larsen I. K. (1997) FEBS Lett. 412, 388–396 [DOI] [PubMed] [Google Scholar]
- 6.Kastrup J. S., Nielsen B. B., Rasmussen H., Holtet T. L., Graversen J. H., Etzerodt M., Thøgersen H. C., Larsen I. K. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 757–766 [DOI] [PubMed] [Google Scholar]
- 7.Nielbo S., Thomsen J. K., Graversen J. H., Jensen P. H., Etzerodt M., Poulsen F. M., Thøgersen H. C. (2004) Biochemistry 43, 8636–8643 [DOI] [PubMed] [Google Scholar]
- 8.Holtet T. L., Graversen J. H., Clemmensen I., Thøgersen H. C., Etzerodt M. (1997) Protein Sci. 6, 1511–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hogan M. M., Vogel S. N. (2001) in Current Protocols in Immunology (Coligan J. E. ed) p. 183, John Wiley & Sons, Inc., New York [Google Scholar]
- 10.Ursby T., C. B. M., Cerenius Y., Svensson C., Sommarin B., Fodje M. N., Kvick Å., Logan D. T., Als-Nielsen J., Thunnissen M. M. G. M., Larsen S., Liljas A. (2004) in AIP Conference Proceedings, Eight International Conference on Synchrotron Radiation Instrumentation, Vol. 705, pp. 1241–1246, American Institute of Physics, College Park, MD [Google Scholar]
- 11.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eck M. J., Sprang S. R. (1989) J. Biol. Chem. 264, 17595–17605 [DOI] [PubMed] [Google Scholar]
- 13.Lamzin V. S., Wilson K. S. (1997) Methods Enzymol. 277, 269–305 [DOI] [PubMed] [Google Scholar]
- 14.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 15.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 16.Eck M. J., Beutler B., Kuo G., Merryweather J. P., Sprang S. R. (1988) J. Biol. Chem. 263, 12816–12819 [PubMed] [Google Scholar]
- 17.Maini R., St Clair E. W., Breedveld F., Furst D., Kalden J., Weisman M., Smolen J., Emery P., Harriman G., Feldmann M., Lipsky P. (1999) Lancet 354, 1932–1939 [DOI] [PubMed] [Google Scholar]
- 18.Moreland L. W., Baumgartner S. W., Schiff M. H., Tindall E. A., Fleischmann R. M., Weaver A. L., Ettlinger R. E., Cohen S., Koopman W. J., Mohler K., Widmer M. B., Blosch C. M. (1997) N. Engl. J. Med. 337, 141–147 [DOI] [PubMed] [Google Scholar]
- 19.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.