Abstract
The members of group III hybrid histidine kinases (HHK) are ubiquitous in fungi. Group III HHK have been implicated to function as osmosensors in the high osmolarity glycerol (HOG) pathway that is essential for fungal survival under high osmolarity stress. Recent literature suggests that group III HHK are also involved in conidia formation, virulence in several filamentous fungi, and are an excellent molecular target for antifungal agents. Thus, group III HHK constitute a very important group of sensor kinases. Structurally, group III HHK are distinct from Sln1p, the osmosensing HHK that regulates the HOG pathway in Saccharomyces cerevisiae. Group III HHK lack any transmembrane domain and typically contain HAMP domain repeats at the N terminus. Until now, it is not clear how group III HHK function as an osmosensor to regulate the HOG pathway. To investigate this, we undertook molecular characterization of DhNIK1, an ortholog from osmotolerant yeast Debaryomyces hansenii. We show here that DhNIK1 could complement sln1 mutation in S. cerevisiae thereby confirming its role as a bona fide osmosensor. We further investigated the role of HAMP domains by deleting them systematically. Our results clearly indicate that the HAMP4 domain is crucial for osmosensing by DhNik1p. Most importantly, we also show that the alternative interaction among the HAMP domains regulates the activity of DhNik1p like an “on-off switch” and thus provides, for the first time, an insight into the molecular mechanism of osmosensing by this group of HHKs.
Keywords: Organisms/Fungi, Organisms/Yeast, Signal Transduction/Protein Kinases, Signal Transduction/Protein Kinases/Histidine, Signal Transduction, Debaryomyces hansenii, HAMP Domain, Osmosensing, Two-component Histidine Kinase
Introduction
Sensing the changes in their surroundings is pivotal for the survival of fungi. For this, fungi utilize a number of molecular sensors that are linked to various downstream signaling pathways to elicit appropriate responses. Many of these sensors are two-component histidine kinases that are known to regulate various aspects of cellular physiology, e.g. osmo-adaptation, hyphal development, virulence, sporulation, etc. (1–5). Available genome sequence data suggest that fungi, in general, have a large repertoire of two-component histidine kinases. Nearly all the fungal two-component histidine kinases are hybrid histidine kinases (HHK)3 because the sensor histidine kinase and the receiver domains are present in a single polypeptide. Based on the domain architecture, these kinases have been classified into 11 groups (6). Sln1p was the first HHK to be discovered in fungi, and surprisingly, it is the sole HHK in Saccharomyces cerevisiae (7, 8). It functions as an osmosensor in one of the two upstream branches of the high osmolarity glycerol (HOG) pathway. The HOG pathway, one of the best studied MAPK pathways, is essential for adaptation to high osmolarity stress (9). Under ambient osmolarity conditions, Sln1p activates a multistep phosphorelay involving the phosphor-transfer protein Ypd1p and response regulator Ssk1p and therefore maintains Ssk1p in the phosphorylated form, which is inactive. Exposure to hyper-osmolarity conditions inactivates Sln1p, and the absence of the phosphorelay causes dephosphorylation of Ssk1p (10). The dephosphorylated Ssk1p activates the redundant MAPK kinase kinase Ssk2p/Ssk22p that ultimately causes phosphorylation of Hog1p via MAPK kinase kinase Pbs2p. Thus, Sln1p negatively regulates the HOG pathway. In S. cerevisiae, the HOG pathway can also be activated under high osmolarity stress by another independently acting upstream branch that utilizes the promiscuous MAPK kinase kinase kinase Ste11p to activate Pbs2p. This branch is called SHO1 branch (11). Recent studies showed that two mucin-like proteins Hkr1p and Msb2p function upstream of Sho1p and act as osmosensor in this branch (12).
The N-terminal region of Sln1p has two transmembrane domains that are responsible for its localization in the membrane (13, 14). This architecture made it a typical member of group VI HHK. The membrane localization of Sln1p is essential for its function as it monitors the changes in turgor through the extracellular domain (13, 14). The loss of turgor due to high osmolarity stress appeared to act as a stimulus for its function. Recent studies suggested that besides Sln1p, the members of group III HHK also regulate the HOG pathway in several fungi. These putative osmosensors appear to be ubiquitous in fungi (6). Nik1p of Neurospora crassa (also known as os-1) (15, 16), Daf1p of Botryotinia fuckeliana (Botrytis cinerea) (17), Hik1p of Magnaporthe grisea (18), and CaNik1p of Candida albicans (19–21) are few well known examples of this group. Group III HHKs are also shown to be involved in the hyphal development and virulence in few fungi. Moreover, they appear to be the target of different fungicidal agents (e.g. phenylpyrroles, dicarboximides, and aromatic hydrocarbons) as the cells harboring mutations in these kinases exhibit resistance to these fungicides (4, 6, 22–25). Thus, Nik1 orthologs are a very important class of HHK in fungi.
Structurally, these HHKs are very different from Sln1p orthologs. They lack the transmembrane domains and therefore have been assumed to be soluble proteins. Moreover, they have a unique N-terminal region consisting of HAMP domain repeats. The analysis of the fungicide-resistant field or laboratory isolates of N. crassa, B. cinerea, and Cochliobolus heterostrophus showed that many of these mutants harbor point mutations in the HAMP domain repeats of the respective Nik1 orthologs and therefore suggested that these repeats are functionally very important (17, 26, 27). HAMP domains are so named because of their frequent occurrence in proteins associated with signal transduction in prokaryotic and lower eukaryotic organisms, such as histidine kinases, adenyl cyclases, methyl accepting chemotaxis proteins and phosphatases (28). Nearly 11,750 proteins with HAMP domains are present in the SMART data base. These domains are composed of ∼50 amino acid residues with two amphipathic helices (29). HAMP domains have been shown to play an active role in the intramolecular signal transduction in prokaryotic sensor kinases, and this is achieved through the helix rotation as elucidated recently from NMR-based structure (30, 31).
The precise mechanism by which Nik1 or its orthologs sense and respond to higher osmolarity stress for regulating the downstream HOG pathway remains an enigma. To probe their role in this process, we have cloned an ortholog (DhNIK1) from Debaryomyces hansenii, which is one of the most osmotolerant and halotolerant yeast (32, 33). We report here that DhNIK1 could complement the sln1 mutation and function as an osmosensor in S. cerevisiae. We show that the interactions among HAMP domains constitute an osmosensing molecular switch in DhNik1p. We also present evidence about the existence of a similar mechanism in other orthologs. Together, these results not only illuminate, for the first time, the mechanistic insight in the functioning of these HHKs as osmosensors in fungi but also provide a novel paradigm about the function of HAMP domain as a sensing module.
EXPERIMENTAL PROCEDURES
Yeast Strains and Plasmids
S. cerevisiae strains RJ1428 (MATa sln1Δ::LEU2 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 with pRS-PTP2 URA3) (34) and TM229 (MATa ura3 leu2 his3 sln1-ts4) (8) were used in this study. NM2 (MATa ura3 leu2 his3 sln1-ts4 ste11Δ::hph) was constructed by disrupting the STE11 gene by hygromycin-resistant gene cassette in TM229. S. cerevisiae strain EGY48 (MATα trp1 his3 ura3 6ops-LEU2) was used as host for two-hybrid analysis (35).
Construction of Plasmids Carrying DhNIK1 or Its Different Mutant Forms
D. hansenii DhNIK1 gene was cloned by PCR amplification from genomic DNA of D. hansenii strain CBS767. A 616-bp fragment corresponding to the region upstream to the ATG codon was amplified using forward Dnik1F-BamHI and reverse DnikP-NcoIR primers. Another fragment (3351 bp) comprising the entire ORF was PCR-amplified using forward Dnik-NcoIF and reverse Dnik-XhoIR primers. Following double digestion by BamHI-NcoI and NcoI-XhoI, respectively, the two fragments were ligated to plasmid pRS423 (36) at BamHI-XhoI sites to obtain pDhNIK1. The NcoI site was introduced by silent mutation at the 5′-end of the ORF to conveniently construct different mutants. Similarly, DhNIK1 was also subcloned into low copy vector pRS313. For the expression of the DhNik1-green fluorescent protein (GFP) fusion, the entire ORF encoding DhNik1p was amplified with forward Dnik-NcoIF and reverse DnikR XhoI-GFP primer. Cloning of the amplified fragment in plasmid pMA26 (33) at the SmaI-XhoI site resulted in the plasmid pDhNIK1-GFP. To construct different mutants of DhNIK1 carrying point mutation(s), deletion in HAMP domain(s), or re-shuffled HAMP domains, a PCR-based overlap extension method was followed (37). All the mutant constructs were confirmed by DNA sequencing. For the mutant H500Q, two PCR amplifications were carried out using primer pairs forward Dnik-NcoIF/reverse DnikH500QR1 and forward DnikH500QF1/reverse HAMPR-EcoRI. The two fragments were then combined in a third PCR using Dnik-NcoIF/HAMPR-EcoRI, and the amplified fragment was cloned at the NcoI-EcoRI site of the plasmid pDhNIK1 to replace 1.9-kb fragment. For D915N, a 1.2-kb fragment carrying the desired mutation was created by following a similar strategy, and this was used to replace the corresponding wild type sequences flanked by EcoRI and XhoI restriction sites in the plasmid pDhNIK1. The mutants H13452 and H12435 were made by shuffling the HAMP domains in DhNik1p. For constructing H13452, four PCRs were carried out using primer pairs Dnik-NcoIF/1CR, 1CF/5BR, 5BF/2fR, and 2fF/HAMPR-EcoRI. The PCR product of the first two reactions were recombined by PCR using primer pairs Dnik-NcoIF/5BR, and the other two amplified products were recombined using primer pair 5BF/HAMPR-EcoRI. Finally, the products of these two reactions were recombined by PCR using Dnik-NcoIF/HAMPR-EcoRI, and the cloning of this fragment at NcoI-EcoRI site of the plasmid pDhNIK1 resulted in H13452. The mutant H12435 was constructed following a similar strategy using primers listed in supplemental Table 1.
To determine the role of HAMP domains in the functionality of DhNik1p, we made different constructs carrying deletion in HAMP domains. The mutant constructs ΔH1, ΔH2, ΔH3, ΔH4, and ΔH5 carrying different single HAMP domain deletions were made by overlap extension PCR. For ΔH1, a construct designed to produce a DhNik1p mutant lacking first HAMP domain (53–108 amino acid residues), two PCRs were carried out utilizing primer pairs Dnik-NcoIF/ΔH1delR and ΔH1delF/HAMPR-EcoRI. The PCR products were combined in a third PCR using Dnik-NcoIF and HAMPR-EcoRI primers. The resultant ∼1.779-kb fragment was cloned in pDhNIK1 replacing the ∼1.9-kb wild type sequence flanked by NcoI and EcoRI restriction sites. Similarly, ΔH2 (Δ147–199 aa), ΔH3 (Δ239–291 aa), ΔH4 (Δ331–383 aa), and ΔH5 (Δ423–475 aa), devoid of second, third, fourth, and fifth HAMP domains, respectively, were made using the primers listed in the supplemental Table 1. Another set of mutants (ΔH1–2, ΔH1–3, ΔH1–4, and ΔH1–5) was generated, wherein the HAMP domains were deleted sequentially such that the number of these domains was serially reduced. Thus, ΔH1–2 lacked HAMP1 and HAMP2, whereas ΔH1–5 lacked all five HAMP domains. For ΔH1–2, two PCRs were carried out utilizing primer pairs Dnik-NcoIF/ΔH1–2delR and ΔH1–2delF/HAMPR-EcoRI. The PCR products were combined in a third PCR using Dnik-NcoIF and HAMPR-EcoRI primers and cloned in pDhNIK1 replacing ∼1.9-kb NcoI-EcoRI fragment to obtain ΔH1–2 (Δ53–199 aa). Similarly, ΔH1–3 (Δ53–291 aa), ΔH1–4 (Δ53–383 aa), and ΔH1–5 (Δ53–475 aa) were made using the primers listed in the supplemental Table 1. For adding the hemagglutinin (HA) tag at the C terminus of DhNik1p or different HAMP-deleted mutants, an ∼1.4-kb fragment was PCR-amplified by utilizing custom-made forward DNik1F2-EcoRI and reverse DNIK1_ORF_R_2HA primers. The reverse primer was designed to incorporate two HA tag sequences. Following digestion with EcoRI and XhoI, the PCR product was cloned at EcoRI-XhoI site of the plasmid pDhNIK1 to replace the 1.4-kb fragment. Similarly, the 1.4-kb amplified fragment was used to replace the EcoRI-XhoI fragments in different HAMP-deleted mutants for HA tagging. To express DhNik1p carrying the His6 tag at the C terminus, the ORF encoding DhNik1p was PCR-amplified using forward Dnik-NcoIF and reverse DNik1R-XhoI primers. Following double digestion by NcoI-XhoI, the fragment was cloned in pET28c vector at NcoI and XhoI sites to obtain pET-DhNik1. For making pET-ΔH4 and pET-ΔH1–4 constructs, the ∼1.9-kb NcoI-EcoRI fragment of pET-DhNIk1 was replaced by the NcoI-EcoRI fragment of ΔH4 and ΔH1–4, respectively.
Western Blotting
Levels of dually phosphorylated Hog1p in S. cerevisiae strain NM2 (sln1-ts ste11Δ) expressing DhNik1p or its mutants were detected by Western blotting. Cells were grown in SD medium at 28 °C until an A600 of 0.5–0.7 and then shifted to either 37 °C or 0.4 m NaCl or both conditions for different times. Cells were collected by centrifugation, and the resulting cell pellets were immediately frozen in liquid nitrogen. The total cell extract (∼20 μg of protein) from each sample was blotted onto nitrocellulose membrane, and dually phosphorylated Hog1p was detected using anti-dually phosphorylated p38 antibody (Cell Signaling Inc.) as described earlier (38). The level of Hog1p was detected in the same blot after re-probing with anti Hog1p antibody (Y-215; Santa Cruz Biotechnology).
For down-shock experiments, S. cerevisiae strain sln1-ts ste11Δ (NM2) expressing DhNik1p was grown in SD medium at 28 °C until an A600 of 0.5–0.7 and then shifted to 37 °C. After 1 h, cells were exposed to 0.4 m NaCl at 37 °C for 5 min, and subsequently they were collected by centrifugation at 37 °C. The centrifugation was carried out at this temperature to prevent the reactivation of sln1-ts by lowering the temperature. Following resuspension in SD medium that was maintained at 37 °C, cells were incubated further at 37 °C for different times. Cells were harvested by centrifugation, and the resulting cell pellets were immediately frozen in liquid nitrogen. The level of phosphorylated Hog1p in the total cell extract was detected by Western blotting.
Expression and Purification of Recombinant Proteins
The wild type construct (pET-DhNik1) and different mutant constructs were transformed into Escherichia coli strain BL21(DE3) for overexpression and subsequent purification of C-terminally histidine-tagged fusion proteins. Overnight cultures of these clones (∼15 h at 37 °C in LB broth containing 50 μg/ml kanamycin) were re-inoculated and grown to an A600 of 0.5. Cells were then induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside and incubated overnight at 12 °C. Cells were harvested and resuspended in lysis buffer (50 mm Tris, pH 7.5, 200 mm NaCl, 5 mm dithiothreitol, 0.1% Triton X-100, and protease inhibitor mixture, Roche Applied Science). After addition of lysozyme (0.1 mg/ml), the cells were disrupted by sonication. Recombinant DhNik1p protein from the supernatant fraction was then purified under native conditions, using nickel-nitrilotriacetic acid matrix according to the manufacturer's protocol. Concentration of the protein in the purified fractions was determined by the Bradford method.
Kinase Assay
The kinase activity of the purified, recombinant DhNik1p, ΔH4, and ΔH1–4 proteins were determined in vitro using Kinase-Glo Max kit (Promega) (3). The kinase reaction was performed in a microtiter plate with different amounts of DhNik1p, ΔH4, and ΔH1–4 in 50 μl of buffer (50 mm Tris-Cl, pH 7.5, 300 mm NaCl, 5 mm MgCl2) with 10 μm ATP. The reaction was carried out at 30 °C for 30 min. Subsequently, 50 μl of Kinase-Glo reagent was added to each well and incubated at room temperature for 20 min in dark. For each reaction, a separate blank was set up without addition of any protein. The amount of luminescence as relative light units in each well was recorded using a GloMax 96 microplate luminometer (Promega). This assay essentially measures the activity of a kinase by quantitating the amount of ATP remaining in the solution following a kinase reaction. With an increase in kinase activity, the ATP concentration decreases in the reaction, which is detected as a decrease in the luminescence. The activity of DhNik1 and its mutants was plotted as percentage of luminescence with respect to the corresponding blanks.
Two-hybrid Assay
The two-hybrid assay was essentially carried out according to Golemis and Brent (35) using pJG4-5 and pEG202 as the activation domain plasmid (prey) and the LexA DNA binding domain plasmid (bait), respectively. PCR-amplified fragments corresponding to each of the five HAMP domains were cloned into plasmid pJG4-5 at the EcoRI-XhoI site. The amplified fragments encoding HAMP4 and HAMP5 were also cloned in pEG202 at the EcoRI-XhoI site to obtain bait constructs. The primer sequences are listed in supplemental Table 1. Authenticity of all the constructs was confirmed by nucleotide sequencing. S. cerevisiae strain EGY48 was co-transformed with bait and prey constructs and selected on minimal media with 2% glucose and without tryptophan and histidine. Co-transformed cells were grown up to A600 ∼1.0, and 5 μl of the cultures (normalized to A600 1.0) were spotted onto minimal media with 2% galactose, 1% raffinose and without tryptophan, histidine, and leucine. Plates were incubated for 4 days at 30 °C. For quantitative β-galactosidase activity assays, HAMP4 in bait vector was co-transformed with its respective prey plasmid and the reporter plasmid pJK103 (2ops-lacZ) into the yeast strain EGY48. Saturated cultures of the transformants grown in minimal media with 2% raffinose (without tryptophan, histidine, and uracil) were diluted with minimal medium containing 1% raffinose and 2% galactose (without tryptophan, histidine, and uracil) and grown further until the late exponential phase. Cells were harvested by centrifugation, and the β-galactosidase activity in these cells was determined as described earlier (39). Experiments were repeated with two independent pools of six transformants each. For liquid growth assays, EGY48 harboring HAMP4 in bait vector and different prey combinations were grown overnight in minimal media with 2% raffinose (without tryptophan and histidine). The cultures were re-inoculated in minimal media with 1% raffinose and 2% galactose (without tryptophan, histidine, and leucine) at A600 ∼0.10 and grown further. The A600 of the cultures was measured after 40 h.
RESULTS
DhNIK1 Complements sln1 Mutation in S. cerevisiae and Localizes in the Cytoplasm
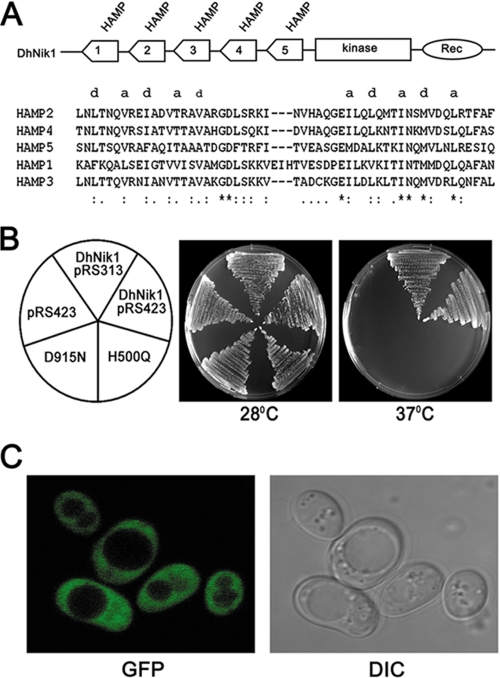
Blast analysis of D. hansenii genome data base revealed the presence of a putative NIK1 ortholog (DhNIK1, accession number XP_462511) encoding a protein of 1116 amino acid residues on the chromosome G. This protein shows more than 80% identity with Nik1 orthologs from yeast, e.g. C. albicans (88%), Pichia stipitis (87%), Candida lusitaniae (83%), and ∼60% identity with those from filamentous fungi like N. crassa and B. fuckeliana. Analysis of the protein sequences by SMART showed the presence of domains that are typically found in group III HHK. The central region contains His kinase A (490–555 aa) and histidine kinase-like ATPase (602–721 aa) domains forming the catalytic core of histidine kinase, including the presumptive autophosphorylated histidine residue (His-500). The canonical receiver domain (865–986 aa) containing a phosphorylatable aspartate residue (Asp-915) was observed in the C-terminal region of DhNik1p. The N terminus of DhNik1p is composed of five HAMP domain repeats (Fig. 1A). The first HAMP domain contains 56 amino acid residues, whereas all others have 53 amino acids. It is apparent from their primary amino acid sequences that they, like a typical HAMP domain, have two segments of hydrophobic amino acid residues in a heptameric arrangement characteristic of the amphipathic α-helix that are joined by a flexible connector (Fig. 1A). To determine the function of DhNIK1, it was introduced into two different S. cerevisiae strains (RJ1428 and TM229) carrying different sln1 mutations. In S. cerevisiae, the sln1 deletion causes inappropriate activation of the HOG pathway, which is lethal for the cell. In RJ1428, the lethality of sln1 deletion was suppressed by expressing a phosphatase, PTP2, from a URA3 based multiple copy plasmid (34). Thus, this strain cannot grow in plates containing 5-fluoroorotic acid as the presence of the plasmid expressing PTP2 is essential for its growth. In contrast, RJ1428 transformed with plasmid harboring the DhNIK1 gene could grow on a 5-fluoroorotic acid plate (data not shown). These results indicated that DhNIK1 could suppress the lethality of the sln1 mutation. To confirm this further, we utilized S. cerevisiae strain TM229, which carries a temperature-sensitive allele of SLN1 (sln1-ts), and therefore it does not grow at 37 °C (8). TM229 transformed with DhNIK1 that was cloned either in a multiple or single copy plasmid showed growth at a nonpermissive temperature (37 °C). This result thus clearly ruled out that the phenotypic complementation was due to multiple copy suppression (Fig. 1B). Next to check whether DhNik1p is an authentic HHK, we have mutated the putative histidine or aspartic acid residues involved in phosphorelay mechanism. The mutants were transformed into TM229, and growth of the transformants was checked at 37 °C. As evident from our results, both the mutants H500Q and D915N failed to complement the sln1-ts mutation (Fig. 1B). These results thus suggested that DhNik1p functioned as an HHK in vivo.
FIGURE 1.
Complementation of sln1 mutation in S. cerevisiae by DhNIK1. A, diagrammatic representation of the domain architecture of DhNik1p. HAMP domains (numbered serially according to their position from the N terminus), histidine kinase (Kinase), and receiver (Rec) domains are shown. Multiple alignments are shown of amino acid residues of the individual HAMP domains using ClustalX1.8. Hydrophobic residues of the two amphipathic helices are denoted by letter a and d to show their heptad periodicity. B, growth of TM229 transformed with only vector pRS423, DhNIK1 in multicopy vector pRS423 or single copy vector pRS313, mutants of DhNIK1 H500Q or D915N on SD agar plate without histidine at 28 or 37 °C after 3 days of incubation. C, subcellular localization of DhNik1p by fluorescence microscopy. Strain TM229 expressing the DhNik1-GFP fusion protein was grown to logarithmic phase in SD minimal medium, and the GFP fluorescence was viewed under an LSM510 META laser scanning confocal microscope (Carl Zeiss). GFP fluorescence (GFP) and differential interference contrast (DIC) image of the same cells are shown.
Bioinformatics analysis of the DhNik1p sequence suggested that as a typical member of a group III HHK, it lacks any transmembrane domain and therefore is unlikely to be present in the membrane. Thus, it was of interest to determine the subcellular localization of DhNik1p in S. cerevisiae. For this purpose, the plasmid pDhNIK1-GFP was made to express DhNik1p as a GFP fusion protein (see “Experimental Procedures”). Transformation of TM229 with this plasmid showed that the fusion protein could complement the sln1-ts mutation in this strain (data not shown). This result indicated that the fusion protein has preserved the biological function of DhNik1p. To determine the subcellular localization of DhNik1p, the transformants were grown to logarithmic phase and examined by fluorescence microscopy. In the cells expressing the DhNik1p-GFP fusion protein, GFP fluorescence was visible throughout the cytoplasm (Fig. 1C). The pattern of GFP fluorescence remained unaltered even after exposing the cells to high osmolarity stress (data not shown). DhNik1p also showed similar subcellular localization in D. hansenii.4
DhNik1p Is a Bona Fide Cytoplasmic Osmosensor
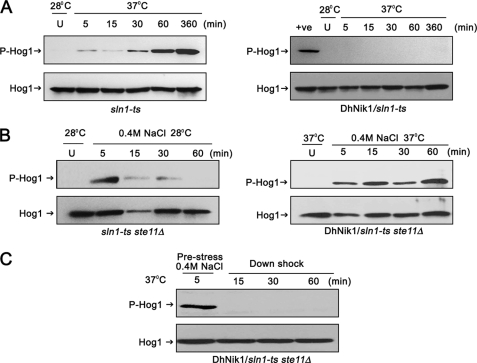
Sln1p is a negative regulator of the HOG pathway in S. cerevisiae. Therefore, the exposure of TM229 (sln1-ts) to nonpermissive temperatures causes constitutive activation of Hog1p, which is lethal for the cell. Therefore, we determined whether DhNik1p could inhibit this activation of Hog1p at nonpermissive temperatures. For this, TM229 with or without DhNIK1, was grown to early logarithmic phase (∼0.7 A600) and then exposed to 37 °C for different periods. The levels of phosphorylated Hog1p in these cells were determined by Western blotting using a commercially available anti-phospho-p38 antibody. To detect the level of Hog1p, membranes were stripped and blotted with a polyclonal antibody against Hog1p (Y-215; Santa Cruz Biotechnology). In sln1-ts cells grown at 28 °C, no phosphorylated Hog1p was detected. However, the phosphorylated Hog1p appeared in these cells within 5 min of exposure to 37 °C. The amount of phosphorylated Hog1p was very pronounced after an hour that possibly indicated the inactivation of Sln1p-ts by this time (Fig. 2A, left panel). In comparison, no phosphorylated Hog1p could be detected in sln1-ts cells expressing DhNik1p even after 6 h of exposure at 37 °C. These results thus indicated that DhNik1p prevented the activation of Hog1p at 37 °C in sln1-ts genetic background (Fig. 2A, right panel).
FIGURE 2.
Regulation of HOG pathway in S. cerevisiae by DhNIK1. A, immunoblots showing the level of Hog1p phosphorylation in S. cerevisiae strain TM229 (sln1-ts) or TM229 expressing DhNik1p at nonpermissive temperature (37 °C). Cells were grown on SD medium with the required nutrient supplement at 28 °C until exponential phase (A600 ∼0.7) before exposing them to 37 °C for indicated time. The level of dually phosphorylated Hog1p (P-Hog1) was detected by immunoblotting the total cell extracts with anti-phospho-p38 antibody (Cell Signaling Inc). The total extract from cells grown at 28 °C was loaded as control (U). The total cell extracts from TM229 exposed to 37 °C for 1 h are included in the right panel as positive control (+ve). Blots were re-probed with anti Hog1 antibody (Y215; Santa Cruz Biotechnology) to detect total Hog1. P-Hog1 or Hog1 indicate phosphorylated or total Hog1p in the blot. B, immunoblots showing high osmolarity-induced Hog1p phosphorylation in S. cerevisiae strain NM2 (sln1-ts ste11Δ) at 28 °C (left panel) and NM2 expressing DhNik1p at 37 °C (right panel). Cells at A600 ∼0.7 were exposed to 0.4 m NaCl for different lengths of time as indicated, and total cell extract from these samples was used for blotting. Total cell extracts from respective cultures not exposed to osmotic stress were loaded as control (U). C, level of dually phosphorylated Hog1p after down-shifting to low osmolarity. NM2 carrying DhNIK1 was grown at 28 °C up to A600 ∼0.7. Cells were incubated at 37 °C for another hour before exposing to 0.4 m NaCl for 5 min at 37 °C. Following centrifugation at 37 °C (to prevent the reactivation of sln1-ts), the cell pellet was resuspended in SD media without salt (maintained at 37 °C) and incubated further at 37 °C for different times as indicated. Total protein extract made from these samples were immunoblotted. All the blots shown above are representative of at least three different experiments.
We next argued that if DhNik1p, like Sln1p, also functions as osmosensor in S. cerevisiae, then exposure to high osmolarity stress should modulate its function to activate the HOG pathway. To check this, we introduced DhNIK1 into S. cerevisiae strain NM2 (sln1-ts ste11Δ). In this strain, the HOG pathway is solely regulated by SLN1 branch as the activation through the other branch was prevented by deleting STE11 (11). NM2 cells transformed with DhNIK1 were grown at 28 °C up to logarithmic phase, and then the culture was shifted to 37 °C. After 1 h of incubation at 37 °C, which would deplete endogenous Sln1p activity (Fig. 2A, left panel), cells were exposed to 0.4 m NaCl for different times. The levels of phosphorylated Hog1p in these cells were detected by immunoblotting. Prior to the application of osmostress, no phosphorylated Hog1p was detected indicating that in the absence of Sln1p, the Hog1p was maintained in the inactive form by DhNik1p at 37 °C. However, within 5 min of the application of the osmostress, phosphorylated Hog1p appeared, and it remained so even after 60 min (Fig. 2B, right panel). Therefore, it was apparent that DhNik1p could activate the HOG pathway under high osmolarity stress. Exposure of sln1-ts ste11Δ to 0.4 m NaCl at permissive temperature (a condition where the endogenous Sln1p remained active) also resulted in the activation of Hog1p within 5 min, but it was transient as phosphorylated Hog1p was barely detectable after 30 min (Fig. 2B, left panel). This discrepancy could be due to the application of higher temperature and osmolarity simultaneously in the earlier experiment that resulted in severe stress to the cells. It is known that severe osmotic stress causes sustained Hog1p activation in S. cerevisiae (40). To be a bona fide osmosensor, DhNik1p should be reversibly activated upon withdrawal of the stress conditions. To test this, after the exposure to 0.4 m NaCl for 5 min at 37 °C, the cells (sln1-ts ste11Δ/DhNIK1) were collected by centrifugation at 37 °C to prevent any reactivation of sln1-ts by lowering the temperature. Following resuspension in SD media that was maintained at 37 °C, cells were incubated further at 37 °C. The disappearance of phosphorylated Hog1p in these cells within 15 min of downshifting to SD medium was a clear indication of transition of DhNik1p back to the active form (Fig. 2C). The results of the down-shock experiment demonstrated that the activity of DhNik1p was modulated by the external osmolarity and thereby clearly establishing its role as an osmosensing HHK.
Essential Role of HAMP Domains in the Functionality of DhNik1p
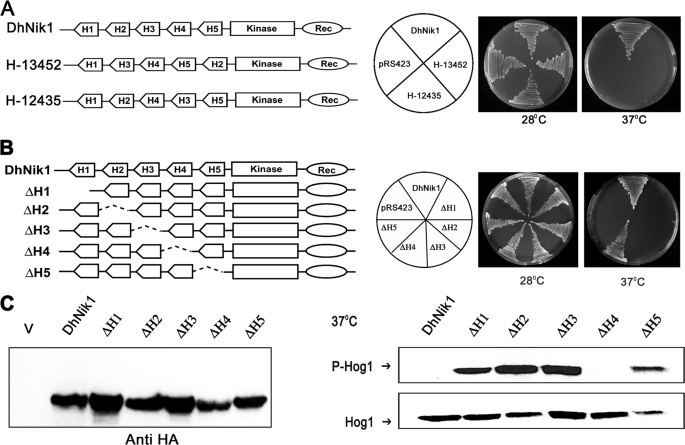
DhNik1p possess five HAMP domains in the N-terminal half of the protein. The presence of HAMP domain repeats is a characteristic feature of the group III HHK. Recently, it has been shown that the mutations in HAMP domain repeats of Nik1p orthologs from B. cinerea (4, 23), C. heterostrophus (27), and N. crassa (41) conferred osmo-sensitive and fungicide resistance phenotypes. These findings clearly suggested that the HAMP domain repeats are very important for the functionality of group III HHKs. In prokaryotic sensor histidine kinases, HAMP domain has been shown to play an active role in the intramolecular signal transduction from the transmembrane to the cytoplasmic kinase domain. Interestingly, in few of these sensor kinases, the HAMP domain could be functionally exchanged indicating their functional similarity (42). The primary amino acid sequences of HAMP domain repeats present in DhNik1p also suggest that each one consists of two amphipathic α-helices flanked by a linker region (Fig. 1A). Therefore, we asked whether the HAMP domain repeats in DhNik1p are functionally alike and whether their arrangements or the positions could be dispensable. For this, we made two different constructs H13452 and H12435 using overlap extension PCR. Both the constructs retained the same five HAMP domains as in DhNik1p, but the positions of the HAMP domains were scrambled (Fig. 3A). These constructs were transformed into sln1-ts, and the transformants were tested for their ability to grow at nonpermissive temperatures. It was apparent from our result that H13452 and H12435 were nonfunctional as both of them failed to complement sln1-ts (Fig. 3A). Thus, the positions and the arrangement of the HAMP domain repeats in DhNik1p are crucial for its function. Furthermore, these results also argue that individual domains might be playing distinct roles.
FIGURE 3.
Functional analysis of reshuffled and individually deleted HAMP domain mutants of DhNik1p. A, diagrammatic representations of DhNik1, HAMP domain reshuffled mutants H13452, or H12435 are shown in the left panel. HAMP domains are named H1, H2, H3, H4, and H5 according to their positions in DhNik1. To determine the growth patterns of TM229 harboring vector (pRS423), DhNik1, H13452, or H12435, cells were streaked on minimal SD agar plate without histidine and incubated at 28 or 37 °C for 3 days. Photographs of the respective plates are shown. B, diagrammatic representations of single HAMP domain deletion mutants and their growth patterns on minimal SD agar plate without histidine at 28 or 37 °C are shown. Experiments were repeated three times with similar results. C, left panel, immunoblot showing the level of expression of DhNik1p and HAMP-deleted mutant ΔH1, ΔH2, ΔH3, ΔH4, and ΔH5. Total cell extracts from TM229 expressing HA-tagged DhNik1 and different mutants were immunoblotted with anti-HA antibody (Cell Signaling). Total cell extract from TM229 transformed with pRS423 were loaded as control (V). Right panel, immunoblots showing the phosphorylated Hog1p in cells with different HAMP domain deletion mutants. S. cerevisiae strain TM229 (sln1-ts) carrying DhNik1 or different mutants (ΔH1, ΔH2, ΔH3, ΔH4, and ΔH5) were grown on SD minimal media without histidine at 28 °C until an A600 ∼0.7 and exposed to 37 °C for 1 h. Total protein extract from these cells was analyzed by Western blotting. P-Hog1 or Hog1 indicate phosphorylated or total Hog1p in the blot. Representative blots of three different experiments are shown.
To investigate the role of each HAMP domain, we constructed a set of DhNIK1 mutants (ΔH1, ΔH2, ΔH3, ΔH4, and ΔH5) where individual HAMP domains were deleted (Fig. 3B). These mutants were introduced into S. cerevisiae strain TM229 carrying sln1-ts, and the growth pattern of the transformants was checked at 37 °C. The deletion of HAMP1, HAMP2, HAMP3, or HAMP5 domain resulted in a nonfunctional allele as the cells carrying these mutants could not grow at 37 °C (Fig. 3B). In contrast, the growth pattern of ΔH4 at nonpermissive temperature was quite similar to DhNIK1, which was used as control (Fig. 3B). To check the level of expression of DhNik1p and the mutants, we expressed them as HA-tagged proteins. As evident from the immunoblot analysis using anti-HA antibody, the expression of different HAMP deletion mutants was quite comparable (Fig. 3C, left panel). Moreover, HA-tagged DhNik1p or its mutants exhibited a phenotypic pattern similar to the untagged protein (data not shown). Thus, the phenotypic differences shown by ΔH1, ΔH2, ΔH3, ΔH4, and ΔH5 did not arise from the artifacts related to the expression of the mutant proteins in vivo. We next determined the extent of Hog1p phosphorylation in TM229 carrying ΔH1, ΔH2, ΔH3, ΔH4, and ΔH5. Cells were grown to early logarithmic phase (∼0.7 A600) at 28 °C and then exposed to 37 °C for 1 h. The total extracts from these cells were immunoblotted to determine the levels of phosphorylated Hog1p in these cells. Exposure to 37 °C for an hour resulted in the accumulation of the phosphorylated Hog1p in the cells harboring ΔH1, ΔH2, ΔH3, and ΔH5. However, in the case of ΔH4, Hog1p phosphorylation was not observed (Fig. 3C, right panel). Thus, the mutant allele ΔH4 was as efficient as DhNik1p in suppressing the activation of HOG pathway at a nonpermissive temperature. These results indicated that the HAMP4 domain could be dispensable for kinase function of DhNik1p; on the contrary, HAMP1, HAMP2, HAMP3, or HAMP5 appeared to be essential for this. These mutants also exhibited similar phenotypic pattern in RJ1428 genetic background (data not shown).
HAMP4 Domain Is Crucial for Osmosensing by DhNik1p
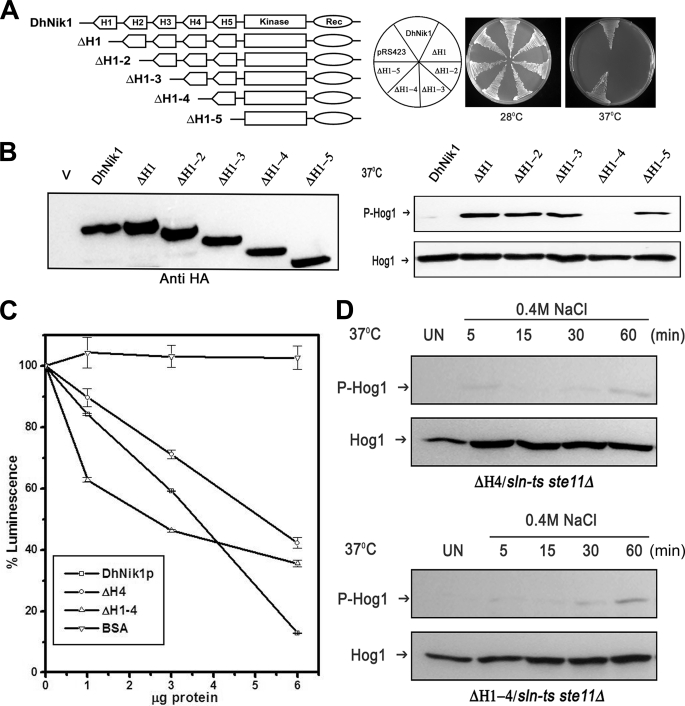
To define the role of the HAMP domains further, we made another set of mutants (ΔH1–2, ΔH1–3, ΔH1–4, and ΔH1–5) by sequential deletion of HAMP domains. Thus, in ΔH1–2, HAMP1 and HAMP2 were deleted, whereas ΔH1–5 was devoid of all the HAMP domains (Fig. 4A). To examine the functional consequences of these deletions, these constructs were transformed into TM229, and the growth pattern of the transformants was examined at 28 and 37 °C. The mutants carrying deletion in first one, two, or three HAMP domains did not grow at nonpermissive temperatures, which indicated that these mutants were nonfunctional (Fig. 4A). To our surprise, ΔH1–4 carrying only HAMP5 domain showed phenotypic complementation. Further removal of the HAMP5 domain (ΔH1–5) also resulted in a nonfunctional allele (Fig. 4A). This series of mutants also displayed comparable levels of expression in vivo as evident from the immunoblot with epitope-tagged constructs (Fig. 4B, left panel). The addition of the HA tag also did not influence the phenotypic pattern (data not shown). The functional consequences of these deletion mutations were further corroborated by testing their ability to suppress inappropriate activation of Hog1p at nonpermissive temperatures (Fig. 4B, right panel). The phenotypic pattern exhibited by these mutants was also confirmed in RJ1428 genetic background (data not shown).
FIGURE 4.
Effect of serial deletions of HAMP domains in DhNIK1. A, diagrammatic representations of serially deleted HAMP domain mutants and growth patterns of TM229 carrying vector pRS423, DhNik1, or different HAMP domain deleted mutants on minimal SD agar plate without histidine at 28 or 37 °C are shown. Experiments were repeated three times with similar results. B, left panel, immunoblot showing the level of expression of DhNik1p, HAMP-deleted mutants ΔH1, ΔH1–2, ΔH1–3, ΔH1–4, and ΔH1–5 after expressing them as HA-tagged proteins. Total cell extract from TM229 transformed with pRS423 were loaded as control (V). Right panel, immunoblots showing the phosphorylated Hog1p in cells with different HAMP domain deletion mutants. S. cerevisiae strain TM229 (sln1-ts) carrying DhNik1 or different mutants (ΔH1, ΔH1–2, ΔH1–3, ΔH1–4, and ΔH1–5) were grown on SD minimal media without histidine at 28 °C until A600 ∼0.7 and exposed to 37 °C for 1 h. The level of phosphorylated Hog1p in these cells was detected by immunoblotting as described previously. Experiments were repeated three times with similar results. C, kinase activities of DhNik1p, ΔH4, and ΔH1–4 were measured by a luminescence assay. Assay was done without any protein or with 1, 3, and 6 μg of purified, recombinant proteins (DhNik1p, ΔH4, and ΔH1–4). Decreasing luminescence indicated increasing kinase activity. Bovine serum albumin (BSA) was used as a negative control. The luminescence observed with different protein concentrations was expressed as percentage of that obtained with the corresponding reaction without any protein. Data are the means ± S.D. of three experiments. D, high osmolarity induced Hog1p phosphorylation in S. cerevisiae strain NM2 (sln1-ts ste11Δ) expressing HAMP domain deletion mutant ΔH4 and mutant ΔH1–4. Cells were grown on SD minimal media without histidine at 28 °C until A600 ∼0.7 and exposed to 37 °C for 1 h. Osmotic stress was given to these cells by exposing them to 0.4 m NaCl at 37 °C for different lengths of time as indicated, and total cell extract from these samples was used for blotting. Total cell extract from respective culture not exposed to osmostress (Un) was loaded as control. Blots were re-probed with anti Hog1 antibody. P-Hog1 or Hog1 indicate phosphorylated or total Hog1p in the blot.
Phenotypic analysis of the different HAMP domain deletion mutants suggested that both ΔH4 and ΔH1–4 mutants are functional in vivo. To check their activity in vitro, we therefore expressed them in E. coli as C-terminal His-tagged proteins (see “Experimental Procedures”). The kinase activity of the purified recombinant DhNik1p, ΔH4, and ΔH1–4 proteins was determined in the presence of 10 μm ATP using a commercially available kit (see “Experimental Procedures”) (3). From our results, it was apparent that with the increase in the amount of recombinant DhNik1p, ΔH4, and ΔH1–4 proteins, the luminescence was reduced significantly (Fig. 4C). In contrast, with bovine serum albumin as control, no significant change in the luminescence was observed. Thus, our results clearly showed that both ΔH4 and ΔH1–4 retained the kinase activity, and the phenotypic complementation exhibited by them did not arise due to adventitious functionality. However, it was not clear whether they were proficient in regulating the HOG pathway upon exposure to high osmolarity conditions. To test this, we transformed ΔH4 and ΔH1–4 into S. cerevisiae strain NM2 (sln1-ts ste11Δ), and the transformants were exposed to high osmolarity stress (0.4 m NaCl) for different time intervals at 37 °C. The amount of phosphorylated Hog1p in these cells was measured by immunoblotting. As shown in the Fig. 4D, neither ΔH4 nor ΔH1–4 could activate Hog1p upon exposure to high osmolarity stress even up to 60 min. This was in contrast to our previous experiment with cells expressing DhNik1p, where phosphorylated Hog1p was detected within 5 min (Fig. 2B, right panel). We presume, like Sln1p, DhNik1p also regulated the HOG pathway in S. cerevisiae negatively. The absence of the HOG pathway activation in response to the high osmolarity stress in ΔH4 or ΔH1–4 clearly indicated that they could possibly be a “constitutive kinase” mutant. The mutant ΔH1–4 retaining only one HAMP domain, i.e. HAMP5, was a functional kinase and an addition of HAMP4 domain to it, as in the mutant ΔH1–3, completely inhibited its activity. The obvious explanation could be the negative regulation of the kinase activity by HAMP4 domain. This raises an interesting possibility that in response to high osmolarity stress, the HAMP4 domain can negatively regulate the activity of the wild type DhNik1p. Our results with mutant ΔH4 strongly support this view, as the absence of HAMP4 domain in this construct resulted in a kinase that remained active under high osmolarity stress.
Selective Interactions among HAMP Domains Create an On-Off Switch
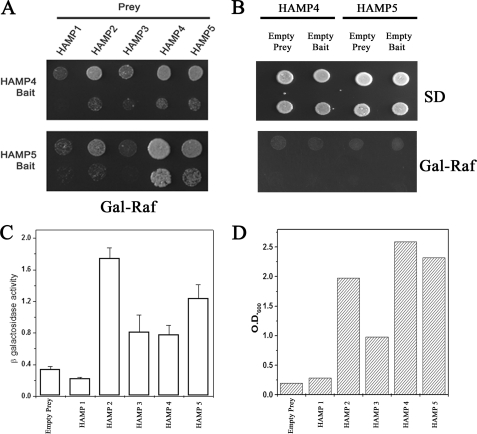
HAMP domains have been shown to interact with each other to form a dimer (30, 31). Therefore, it is plausible that HAMP4 interacts with HAMP5 in ΔH1–3 to inhibit the kinase function. To determine whether the HAMP domains present in DhNik1p are capable of interacting with each other, we performed the yeast two-hybrid assay. As shown in Fig. 5A, HAMP4 strongly interacted with HAMP5, HAMP4, and HAMP2, but its interaction with HAMP3 and HAMP1 was much lower. Like HAMP4, HAMP5 showed no interaction with HAMP1 and HAMP3 but strong interaction with HAMP4 and HAMP5. With HAMP2, it showed moderate interaction as the sequence of HAMP2 has highest homology with HAMP4. In contrast, both HAMP4 and HAMP5 domains in combination with either empty prey or bait did not grow in the selection media (Fig. 5B) thereby confirming that the interactions mentioned above were specific. To obtain a quantitative comparison of the two-hybrid interactions of HAMP4 domain with other HAMP domains, we conducted both LacZ reporter and liquid growth assays (43). Both these assays showed that the HAMP4 domain was capable of strongly interacting with HAMP5 and HAMP2 domains (Fig. 5, C and D). These results indicate that selective interactions could occur among HAMP domains intramolecularly to regulate the activity of DhNik1p in vivo.
FIGURE 5.
Interactions between HAMP domains of DhNik1p. A, two-hybrid assay for interactions between HAMP domains. HAMP4 or HAMP5 cloned in pEG202 vector was used as bait, and five HAMP domains were cloned individually in plasmid pJG4-5 for prey construction. Both bait and prey constructs (in pairs as indicated) were transformed into S. cerevisiae strain EGY48. Growth of the transformants on Gal-Raf minimal medium is shown after dilution spotting. Results are representative of three different experiments. B, two-hybrid interactions in HAMP4 and HAMP5 with empty bait or prey combinations. S. cerevisiae strain EGY48 transformed with HAMP4 bait/empty prey; HAMP4 prey/empty bait; HAMP5 bait/empty prey, and HAMP5 prey/empty bait were grown in minimal SD medium, and serial dilution of the culture was spotted onto minimal SD and Gal-Raf plate. C, β-galactosidase activity in yeast strain EGY48 harboring lacZ reporter plasmid, HAMP4 bait along with empty prey or different HAMP domains in prey vector. Experiments were repeated with two independent pools of six transformants each. β-Galactosidase activity is expressed as nanomoles of o-nitrophenyl β-d-galactopyranoside utilized per min by 1 ml of culture after normalizing its A600 to 1.0. D, EGY48 harboring HAMP4 bait along with empty prey or different HAMP domains in prey vector were grown overnight in minimal media with 2% raffinose (without tryptophan and histidine). The cultures were re-inoculated in minimal media with 1% raffinose and 2% galactose (without tryptophan, histidine and leucine) at A600 ∼0.10 and grown further for 40 h. Representative data of two independent experiments are shown here.
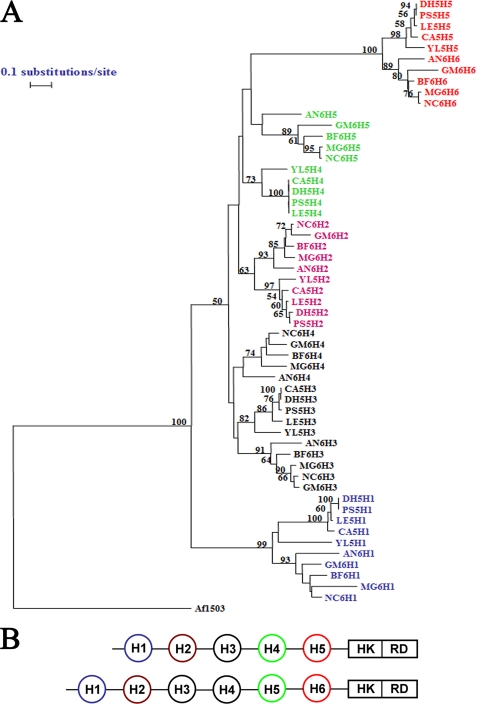
It is evident from our results that the individual HAMP domains play distinct roles in osmosensing by DhNik1p, and their functional uniqueness was dependent on both the position and the sequence of the domain. Sequence analysis of various Nik1 orthologs showed that the orthologs from filamentous fungi have six HAMP domains whereas those from yeast (e.g. DhNik1p) have five HAMP domains. We therefore studied the phylogenetic relationships among HAMP domains from different orthologs to see whether the “functional uniqueness” is preserved across different species. For this, the individual HAMP domain sequences from different yeast and filamentous fungi were aligned using ClustalX program (44), and the multiple alignment was used to draw a neighbor joining tree utilizing TREECON software package (45). From the phylogenetic tree, it was apparent that the N- or C-terminal HAMP domains formed separate groups. Similarly, the HAMP domains that were penultimate from either the N- or C-terminal side also formed a separate clade, irrespective of whether they were from orthologs with five or six HAMP domains (Fig. 6). This clustering of HAMP domains according to their positions in the respective molecules suggested that each might have a separate evolutionary history. Possibly, they had evolved independently and came together by insertion events rather than by duplication of domains intramolecularly. Most importantly, such clustering also suggested conservation of their function toward regulating the activity of the orthologs from other species through selective interactions.
FIGURE 6.
Phylogenetic analysis of HAMP domains. A, phylogenetic tree. Individual HAMP domain sequences from the NIK1 orthologs having five HAMP domains from Yarrowia lipolytica (Q6C775_YARLI), Lodderomyces elongisporus (A5E5X7_LODEL), P. stipitis (A3LYT9_PICST), D. hansenii (DhNik1p; Q6BH10_DEBHA), and C. albicans (CaNik1p; Q9URL9_CANAL) were obtained from SMART data base. YL5H1, YL5H2, YL5H3, YL5H4, and YL5H5 were HAMP domains from Y. lipolytica ortholog. They were named serially according to their position in the protein from N terminus. Similarly, LE5H1, LE5H2, LE5H3, LE5H4, and LE5H5 were from L. elongisporus; PS5H1, PS5H2, PS5H3, PS5H4, and PS5H5 were from P. stipitis; DH5H1, DH5H2, DH5H3, DH5H4, and DH5H5 were from D. hansenii; CA5H1, CA5H2, CA5H3, CA5H4, and CA5H5 were from C. albicans. Representative orthologs of NIK1 containing six HAMP domains are B. fuckeliana (Q8X1E7_BOTCI), Gibberella moniliformis (Q6SLB2_GIBMO), N. crassa (Nik1p; Q01309_NEUCR), M. grisea (Q9C1U1_MAGGR), and Aspergillus niger (A2QP39_ASPNG). BF6H1, BF6H2, BF6H3, BF6H4, BF6H5, and BF6H6 are individual HAMP domain sequences (named serially according to their position from N terminus) from ortholog in B. fuckeliana. Similarly, GM6H1, GM6H2, GM6H3, GM6H4, GM6H5, and GM6H6 were from G. moniliformis; NC6H1, NC6H2, NC6H3, NC6H4, NC6H5, and NC6H6 were from N. crassa; MG6H1, MG6H2, MG6H3, MG6H4, MG6H5, and MG6H6 were from M. grisea; AN6H1, AN6H2, AN6H3, AN6H4, AN6H5, and AN6H6 were from A. niger. Amino acid sequences of the individual HAMP domains were compared using the Clustal_X program (supplemental Table 2 and supplemental Fig. 1). Aligned sequences were analyzed by using the TREECON software package. A neighbor-joining tree (50% or more boot strap values of 1000 replicates are indicated) is shown. HAMP domain sequence from Archaeoglobus fulgidus protein Af1503 (30) was included as out-group. B, schematic showing Nik1 orthologs carrying five HAMP domains and six HAMP domains. Similarly colored HAMP domains from either group are clustered together in the phylogenetic tree. HK, histidine kinase; RD, receiver domain.
DISCUSSION
Nik1 or its orthologs are highly conserved in euascomycetes. They have been implicated to function as osmosensor in the HOG pathway. Besides this role, recent literature suggests that they are also involved in conidia formation, virulence in several filamentous fungi, and an excellent molecular target for antifungal agents (2, 22, 46, 47). Thus, they constitute a very important group of sensor kinases. To establish their role firmly as an osmosensor in the HOG pathway, we undertook the characterization of DhNik1p, a typical Nik1 ortholog, from a highly osmotolerant yeast D. hansenii. The functional analysis was carried out in a heterologous host S. cerevisiae as the HOG pathway in this species is one of the most well characterized pathways in fungi. The expression of DhNik1p could suppress the lethality of sln1 mutation in S. cerevisiae (Fig. 1). Moreover, we have observed that the HOG pathway could be reversibly activated by DhNik1p in response to the changes in external osmolarity (Fig. 2). Together, these findings unequivocally established that it was functionally similar to Sln1p at least in S. cerevisiae. The N-terminal region of Sln1p has two transmembrane domains that are responsible for its localization in the membrane. This architecture is essential for its function (13, 14). In contrast, DhNik1p lacks any transmembrane domain and therefore is unlikely to be present in the membrane. To determine its cellular localization, we made DhNikp-GFP fusion construct. Fluorescence microscopic examination of the cells expressing the fusion protein indicated that DhNik1p is localized in the cytoplasm (Fig. 1), and it remained there even at high osmolarity stress. Therefore, our results not only provide, for the first time, the experimental evidence regarding the intracellular localization of a Nik1 ortholog but also clearly establish that these HHKs are indeed cytosolic osmosensors.
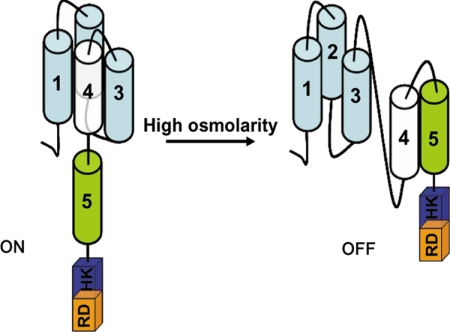
In S. cerevisiae, Hog1p is regulated independently by two distinct branches, and each utilizes a separate class of membrane-bound osmosensors. In the SLN1 branch, Sln1p acts as an osmosensor, and the high osmolarity-induced changes in the turgor appear to act as its stimulus (13, 14). The other branch, i.e. SHO1 branch, utilizes the mucin-like transmembrane proteins Hkr1p and Msb2p as osmosensors. Interestingly, mucin-like proteins have also been implicated as osmosensors in mammals. Mucins are highly hydrated proteins, and therefore osmo-stress-induced changes in their volume appear to function as a trigger for them (12, 48). In contrast to Sln1p or Hkr1p/Msb2p, DhNik1p is not present in the membrane and therefore is unlikely to monitor the external osmolarity directly. What it is sensing as high osmolarity stress and how the change in the external osmolarity regulates its activity thus become interesting but challenging questions. The N-terminal nonkinase region of DhNik1p typically contains HAMP domain repeats. Two lines of evidence led us to believe that they may be importance for osmosensing by DhNik1p. First, several osmosensitive and fungicide-resistant mutants were identified that carried point mutations in HAMP domain repeat in Nik1 orthologs (17, 26, 27). Second, we have observed that a chimeric construct where the N-terminal nonkinase region (which is essential for its membrane localization and osmosensing) of DhSln1p (a Sln1p ortholog in D. hansenii) was swapped with the N-terminal region of DhNik1p containing HAMP repeats could also complement sln1 mutation in S. cerevisiae.4 To delineate the role of HAMP domains in the functionality of DhNik1p, we therefore created several mutants either by shuffling their positions or by deleting these domains systematically. The results obtained from the analysis of these mutants provided a novel insight into the functionality of DhNik1p, which is presented as a model in Fig. 7. Under ambient osmolarity, the HAMP1, HAMP2, and HAMP3 domains hold back HAMP4 from HAMP5 thereby leaving it free to form active kinase possibly upon dimerization. All three domains appear to be essential for this as the deletion of any one of them (ΔH1, ΔH2, and ΔH3) leads to a nonfunctional kinase (Fig. 3B). Yeast two-hybrid assay showed that the interaction of HAMP4 with either HAMP1 or HAMP3 was weaker compared with that with HAMP2 (Fig. 5). Therefore, it is possible that HAMP1 and HAMP3 have an indirect, but essential role by facilitating interaction between HAMP2 and HAMP4. Disruption of these interactions upon exposure to high osmolarity allows HAMP4 to bind to HAMP5 thereby forming an inactive kinase. Our results with the mutant ΔH1–4 and ΔH4 corroborate well with this model as it posits that the absence of HAMP4 will form a kinase unresponsive to the changes in the external osmolarity. Thus, the alternative interaction among the HAMP domains, which are sensitive to changes in external osmolarity, creates an “on-off” switch in DhNik1p, and such a mechanism appeared to be a common theme in this group of kinases as revealed from the phylogenetic analysis of different HAMP domains. NIK1 orthologs from filamentous fungi are directly linked with the resistance to few important fungicidal agents used in agriculture. Point mutations in different HAMP domain repeats in Nik1p or its orthologs from B. fuckeliana and C. heterostrophus could confer varying degrees of drug resistance and osmosensitivity to the host (17, 27, 41). Thus, the structural insights about the HAMP domain repeats will not only help to understand these interesting mutants but also will aid in developing novel fungicides in the future.
FIGURE 7.
Model depicting how the activity of DhNik1 is regulated under high and low osmolarity conditions. Each of the HAMP domains (cylinder) is numbered. The histidine kinase and receiver domain are shown in box labeled as HK and RD. Under high osmolarity stress, HAMP4 interacts with HAMP5 to shut off the kinase activity of DhNik1p.
HAMP domains are one of the very common structural modules associated with the signaling molecules from prokaryotes and lower eukaryotes. Our current knowledge about this domain is based on studying few transmembrane receptor histidine kinases of prokaryotic origin that possess a single HAMP domain. In these examples, the HAMP domain mainly serves as a transducing module that inter-converts the mechanical signals from the transmembrane domain to the kinase module. This study shows that the way the HAMP domains function in DhNik1p is quite distinct from this and thus provides a novel paradigm about the functioning of the HAMP domains. Analysis of DhNik1p through the SMART data base revealed that there are ∼400 two-component histidine kinases that contain more than one HAMP domain. The numbers of repeat units vary, and in some kinases there are as many as 16 such units. We speculate these repeat units have evolved as a distinct module in histidine kinase to function as a sensor for different physicochemical stimuli.
Supplementary Material
Acknowledgments
We thank H. Saito and J. Fassler for yeast strains used in this study. We are thankful to R. Sharma and D. Bhatt for excellent technical assistance.
This work was supported in part by a grant from Department of Biotechnology, India.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1 and Tables 1 and 2.
N. Meena and A. K. Mondal, unpublished results.
- HHK
- hybrid histidine kinase
- HOG
- high osmolarity glycerol
- MAPK
- mitogen-activated protein kinase
- ORF
- open reading frame
- aa
- amino acid
- GFP
- green fluorescent protein
- HA
- hemagglutinin.
REFERENCES
- 1.Yamada-Okabe T., Mio T., Ono N., Kashima Y., Matsui M., Arisawa M., Yamada-Okabe H. (1999) J. Bacteriol. 181, 7243–7247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Santos J. L., Shiozaki K. (2001) Sci. STKE 2001, RE1. [DOI] [PubMed] [Google Scholar]
- 3.Nemecek J. C., Wüthrich M., Klein B. S. (2006) Science 312, 583–588 [DOI] [PubMed] [Google Scholar]
- 4.Viaud M., Fillinger S., Liu W., Polepalli J. S., Le Pêcheur P., Kunduru A. R., Leroux P., Legendre L. (2006) Mol. Plant-Microbe Interact. 19, 1042–1050 [DOI] [PubMed] [Google Scholar]
- 5.Bahn Y. S., Xue C., Idnurm A., Rutherford J. C., Heitman J., Cardenas M. E. (2007) Nat. Rev. Microbiol. 5, 57–69 [DOI] [PubMed] [Google Scholar]
- 6.Catlett N. L., Yoder O. C., Turgeon B. G. (2003) Eukaryot. Cell 2, 1151–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ota I. M., Varshavsky A. (1993) Science 262, 566–569 [DOI] [PubMed] [Google Scholar]
- 8.Maeda T., Wurgler-Murphy S. M., Saito H. (1994) Nature 369, 242–245 [DOI] [PubMed] [Google Scholar]
- 9.Hohmann S. (2002) Microbiol. Mol. Biol. Rev. 66, 300–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Posas F., Wurgler-Murphy S. M., Maeda T., Witten E. A., Thai T. C., Saito H. (1996) Cell 86, 865–875 [DOI] [PubMed] [Google Scholar]
- 11.Posas F., Saito H. (1997) Science. 276, 1702–1705 [DOI] [PubMed] [Google Scholar]
- 12.Tatebayashi K., Tanaka K., Yang H. Y., Yamamoto K., Matsushita Y., Tomida T., Imai M., Saito H. (2007) EMBO J. 26, 3521–3533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ostrander D. B., Gorman J. A. (1999) J. Bacteriol. 181, 2527–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reiser V., Raitt D. C., Saito H. (2003) J. Cell Biol. 161, 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alex L. A., Borkovich K. A., Simon M. I. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 3416–3421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schumacher M. M., Enderlin C. S., Selitrennikoff C. P. (1997) Curr. Microbiol. 34, 340–347 [DOI] [PubMed] [Google Scholar]
- 17.Cui W., Beever R. E., Parkes S. L., Weeds P. L., Templeton M. D. (2002) Fungal Genet. Biol. 36, 187–198 [DOI] [PubMed] [Google Scholar]
- 18.Motoyama T., Kadokura K., Ohira T., Ichiishi A., Fujimura M., Yamaguchi I., Kudo T. (2005) Fungal Genet. Biol. 42, 200–212 [DOI] [PubMed] [Google Scholar]
- 19.Alex L. A., Korch C., Selitrennikoff C. P., Simon M. I. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 7069–7073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagahashi S., Mio T., Ono N., Yamada-Okabe T., Arisawa M., Bussey H., Yamada-Okabe H. (1998) Microbiology 144, 425–432 [DOI] [PubMed] [Google Scholar]
- 21.Srikantha T., Tsai L., Daniels K., Enger L., Highley K., Soll D. R. (1998) Microbiology 144, 2715–2729 [DOI] [PubMed] [Google Scholar]
- 22.Avenot H., Simoneau P., Iacomi-Vasilescu B., Bataillé-Simoneau N. (2005) Curr. Genet. 47, 234–243 [DOI] [PubMed] [Google Scholar]
- 23.Oshima M., Banno S., Okada K., Takeuchi T., Kimura M., Ichiishi A., Yamaguchi I., Fujimura M. (2006) J. Gen. Plant. Pathol. 72, 65–73 [Google Scholar]
- 24.Chapeland-Leclerc F., Paccallet P., Ruprich-Robert G., Reboutier D., Chastin C., Papon N. (2007) Eukaryot. Cell 6, 1782–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dongo A., Bataillé-Simoneau N., Campion C., Guillemette T., Hamon B., Iacomi-Vasilescu B., Katz L., Simoneau P. (2009) Appl. Environ. Microbiol. 75, 127–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller T. K., Renault S., Selitrennikoff C. P. (2002) Fungal Genet. Biol. 35, 147–155 [DOI] [PubMed] [Google Scholar]
- 27.Yoshimi A., Tsuda M., Tanaka C. (2004) Mol. Genet. Genomics 271, 228–236 [DOI] [PubMed] [Google Scholar]
- 28.Aravind L., Ponting C. P. (1999) FEMS Microbiol. Lett. 176, 111–116 [DOI] [PubMed] [Google Scholar]
- 29.Appleman J. A., Stewart V. (2003) J. Bacteriol. 185, 89–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hulko M., Berndt F., Gruber M., Linder J. U., Truffault V., Schultz A., Martin J., Schultz J. E., Lupas A. N., Coles M. (2006) Cell 126, 929–940 [DOI] [PubMed] [Google Scholar]
- 31.Swain K. E., Falke J. J. (2007) Biochemistry 46, 13684–13695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prista C., Loureiro-Dias M. C., Montiel V., García R., Ramos J. (2005) FEMS Yeast Res. 5, 693–701 [DOI] [PubMed] [Google Scholar]
- 33.Aggarwal M., Mondal A. K. (2006) Eukaryot. Cell 5, 262–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tao W., Malone C. L., Ault A. D., Deschenes R. J., Fassler J. S. (2002) Mol. Microbiol. 43, 459–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Golemis E. A., Brent R. (1997) in The Yeast Two-hybrid System (Bartel P. L., Fields S. eds) pp. 43–72, Oxford University Press, Oxford, UK [Google Scholar]
- 36.Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P. (1992) Gene 110, 119–122 [DOI] [PubMed] [Google Scholar]
- 37.Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989) Gene 77, 51–59 [DOI] [PubMed] [Google Scholar]
- 38.Sharma P., Meena N., Aggarwal M., Mondal A. K. (2005) Curr. Genet. 48, 162–170 [DOI] [PubMed] [Google Scholar]
- 39.Germino F. J., Moskowitz N. K. (1999) Methods Enzymol. 303, 422–450 [DOI] [PubMed] [Google Scholar]
- 40.Van Wuytswinkel O., Reiser V., Siderius M., Kelders M. C., Ammerer G., Ruis H., Mager W. H. (2000) Mol. Microbiol. 37, 382–397 [DOI] [PubMed] [Google Scholar]
- 41.Ochiai N., Fujimura M., Motoyama T., Ichiishi A., Usami R., Horikoshi K., Yamaguchi I. (2001) Pest. Manag. Sci. 57, 437–442 [DOI] [PubMed] [Google Scholar]
- 42.Appleman J. A., Chen L. L., Stewart V. (2003) J. Bacteriol. 185, 4872–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stephens D. J., Banting G. (2000) Traffic 1, 763–768 [DOI] [PubMed] [Google Scholar]
- 44.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997) Nucleic Acids Res. 24, 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van de Peer Y., De Wachter R. (1994) Comput. Appl. Biosci. 10, 569–570 [DOI] [PubMed] [Google Scholar]
- 46.Bahn Y. S., Kojima K., Cox G. M., Heitman J. (2006) Mol. Biol. Cell 17, 3122–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kruppa M., Calderone R. (2006) FEMS Yeast Res. 6, 149–159 [DOI] [PubMed] [Google Scholar]
- 48.de Nadal E., Real F. X., Posas F. (2007) Trends Cell Biol. 17, 571–574 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.