Abstract
Bacterial flagellin is important for intestinal immune homeostasis. Flagellins from most species activate Toll-like receptor 5 (TLR5). The principal bacterial food-borne pathogen Campylobacter jejuni escapes TLR5 recognition, probably due to an alternate flagellin subunit structure. We investigated the molecular basis of TLR5 evasion by aiming to reconstitute TLR5 stimulating activity in live C. jejuni. Both native glycosylated C. jejuni flagellins (FlaA and FlaB) and recombinant proteins purified from Escherichia coli failed to activate NF-κB in HEK293 cells expressing TLR5. Introduction of multiple defined regions from Salmonella flagellin into C. jejuni FlaA via a recombinatorial approach revealed three regions critical for the activation of human and mouse TLR5, including a β-hairpin structure not previously implicated in TLR5 recognition. Surprisingly, this domain was not required for the activation of chicken TLR5, indicating a selective requirement for the β-hairpin in the recognition of mammalian TLR5. Expression of the active chimeric protein in C. jejuni resulted in secreted glycosylated flagellin that induced a potent TLR5 response. Overall, our results reveal a novel structural requirement for TLR5 recognition of bacterial flagellin and exclude flagellin glycosylation as an additional mechanism of bacterial evasion of the TLR5 response.
Keywords: Immunology/Innate Immunity, Immunology/Toll Receptors, Organisms/Bacteria, Protein/Protein-Protein Interactions, Protein/Chimeras, Receptors, Receptors/Toll-like, Campylobacter
Introduction
Flagellin, the monomeric subunit of the bacterial motility apparatus, is the natural ligand of the innate immune sensor Toll-like receptor 5 (TLR5)3 (1). Activation of TLR5 by flagellin initiates a powerful host response that provides crucial signals for maintaining intestinal immune homeostasis (2, 3). The immunostimulatory properties make flagellin an attractive vaccine carrier protein and potent vaccine adjuvant. Its intrinsic adjuvant activity is currently being employed in experimental recombinant vaccines against human influenza, West Nile fever, malaria, tuberculosis, and plague (4–9). In addition, flagellin-induced immune activation protects the intestine and other tissues against lethal irradiation due to potent TLR5-mediated anti-apoptotic effects (10, 11).
The immunological impact of flagellin stimulation has driven several bacterial pathogens to evolve mechanisms to escape the effective TLR5-mediated host defense. In Salmonella enterica serotype Typhi, this is achieved by repression of flagellin expression and secretion (12), whereas Listeria shuts off flagellin expression at the host temperature of 37 °C (13). The flagellins of the α- and ϵ-proteobacteria, which include the major food-borne pathogen Campylobacter jejuni and the gastric pathogen Helicobacter pylori, fail to activate TLR5 altogether (14–18). For these organisms the consequences of TLR5 evasion for infection are currently unknown. It has been demonstrated that purified H. pylori flagellin induces severely impaired adaptive immune responses in comparison to TLR5-activating flagellins (19).
The flagellin protein of C. jejuni clearly differs from bacterial flagellins that do activate TLR5. Electron microscopy shows that the C. jejuni flagellar filament comprises seven longitudinal helical arrays of stacked flagellin subunits (protofilaments) instead of the 11 present in e.g. S. enterica serotype typhimurium, suggesting differences in flagellin polymerization between these species (20). Consistent with this hypothesis, the amino acid regions of flagellin involved in filament assembly in Salmonella have diverged in Campylobacter. These changes may contribute to the TLR5 evasion in C. jejuni (18, 21). An additional difference between C. jejuni flagellin and most TLR5-activating flagellins is the presence of pseudaminic acid derivatives that cover the putative surface-exposed region of C. jejuni flagellin and that may comprise up to 10% of their total weight (22). The contribution of the post-translational modification of Campylobacter flagellin to evasion of the TLR5 response is currently unknown.
Considering the important role of TLR5 in intestinal biology and the potential of bacterial flagellin as a vaccine adjuvant, we sought to better define the molecular basis of the TLR5 evasion by C. jejuni flagellin by attempting to restore TLR5-stimulating activity. Mutagenesis or replacement of larger amino acid regions between flagellins has previously been instrumental in defining residues critical for TLR5 recognition (18, 21, 23–26). Using a series of recombinant chimeric flagellins, we succeeded in reconstituting a Campylobacter flagellin with TLR5 activating ability and in the engineering of C. jejuni that processes, glycosylates, and secretes flagellins that yield a potent TLR5 response. Activation of human TLR5 by Campylobacter was not influenced by flagellin glycosylation but required introduction of three defined domains of Salmonella flagellin. Differential activation of mammalian and chicken TLR5 by recombinant flagellins led to the discovery of a β-hairpin structure not previously implicated in mammalian TLR5 recognition.
EXPERIMENTAL PROCEDURES
Cell Lines and Bacterial Strains
HeLa 57A cells stably transfected with a NF-κB luciferase reporter construct (27), HEK293 cells, and HT-29 intestinal epithelial cells were maintained in Dulbecco's modified Eagle's medium (DMEM, Invitrogen) supplemented with 5% fetal calf serum at 37 °C under 5% CO2. NHEK human primary keratinocytes were propagated under the same conditions in KGM-2 Keratinocyte Growth Medium-2 (Lonza).
C. jejuni strains 81116 (NCTC11828) (28), NCTC 11168H1 (29), and their derivatives were grown at 37 °C under microaerobic conditions (5% O2, 10% CO2, and 85% N2) on saponin agar medium containing 4% lysed horse blood with the appropriate antibiotics. S. enterica ssp. enteritidis 90-13-706 and S. enteritidis 90-13-706ΔfliC (30) and Escherichia coli BL21(DE3) Star (Invitrogen), used to express recombinant flagellins, were grown in Luria Bertani broth at 37 °C.
Purification of Native Flagellin
Native flagellin of S. enteritidis was purified overnight cultures as described (31), with minor modifications. Briefly, bacteria were resuspended in 10 mm Tris·HCl, 145 mm NaCl, pH 7.4, homogenized (2 min), and centrifuged twice (10,000 × g, 20 min, 4 °C), discarding the pellet after each centrifugation. Flagella were collected from the supernatant by centrifugation (100,000 × g, 60 min, 4 °C) and depolymerized in 0.2 m glycine, pH 2 (30 min, 20 °C, with stirring). After centrifugation (100,000 × g, 60 min, 4 °C), the supernatant containing monomeric flagellin was adjusted to pH 7.2 with 1 m NaOH, and ammonium sulfate was added to a final concentration of 2.67 m. After overnight incubation (20 °C), repolymerized flagellins were collected by centrifugation (14,000 × g, 15 min, 4 °C), dissolved in H2O, and dialyzed against H2O (24 h, 4 °C). Native flagellin of C. jejuni strain 81116 was purified as described (32) and stored at −20 °C.
Construction and Purification of Chimeric Flagellins
The construction of recombinant Salmonella FliC has been described (33). Recombinant FlaA and FlaB of C. jejuni strain 81116 were obtained after amplification of the corresponding genes with pfu polymerase (Promega) using primer pairs His6-FlaA-F and His6-FlaA-R and pairs His6-FlaB-F and His6-FlaB-R, respectively (Table 1). Products were ligated into expression vector pT7-7 (34) using restriction enzymes BamHI and ClaI. Chimeric flagellins were constructed by overlap extension PCR using primers and a template as depicted in Table 1, unless stated otherwise. For construction of chimera FlaAN, the template was a mixture of FlaA-(1–52), FliC(N), and FlaA-(123–576); for chimera FlaAC, the template was a mixture of FlaA-(1–491), FliC(C), and FlaA-(527–576); for FlaANC, the template was FlaA-(1–491) (prepared with template flaAN), FliC(C), and FlaA(527–576); for FlaANVC, the template was a mixture of FlaA-(1–52), FliC(NVC), and FlaA-(527–576); for FlaAH, the template was a mixture of FlaA-(1–122), FliC(H), and FlaA-(177–576); for FlaANH, the template was FlaA-(1–122) (prepared with template flaAN), FliC(H), and FlaA-(177–576); for FlaAHC, the template was a mixture of FlaA-(1–122), FliC(H), and FlaA(177–576) (prepared with template flaAC); for FlaANHC, the template was FlaA(1–122) (prepared with template flaAN), FliC(H), and FlaA-(177–576) (prepared with template flaAC). The obtained chimeric flagellin genes were amplified using primers FlaA-topoF and FlaA-topoR and ligated in pET101/D-TOPO (Invitrogen). All plasmid constructs were propagated in E. coli DH5α and transformed into E. coli BL21(DE3) Star for protein expression.
TABLE 1.
Primers used in this study
| Product/primer | Primer sequencea | Template DNA |
|---|---|---|
| His6-FlaA-F | Forward, 5′-GGATCCCACCACCACCACCACCACATGGGATTTCGT-3′ | C. jejuni 81116 |
| His6-FlaA-R | Reverse, 5′-ATCGATCTATTGTAATAATCTTAAAACATTTTGCTG-3′ | |
| His6-FlaB-F | Forward, 5′-GGATCCCACCACCACCACCACCACATGGGTTTTAGG-3′ | C. jejuni 81116 |
| His6-FlaB-R | Reverse, 5′-ATCGATTTATTGTAATAGTTTTAAAACATTTTGCTG-3′ | |
| FlaA-(1–52) | Forward, 5′-CACCATGGGATTTCGTATTAACAC-3′ | C. jejuni 81116 |
| Reverse, 5′-GATATTAGAAGTGAAGCGATCTGCTATCGCCATCCC-3′ | ||
| FliC(N) | Forward, 5′-GGGATGGCGATAGCAGATCGCTTCACTTCTAATATC-3′ | S. enteritidis 706 |
| Reverse, 5′-GTATTAGCGATATTATCAAGTTCTTCCAGACGTTGCTGAA-3′ | ||
| FlaA-(123–576) | Forward, 5′-TTCAGCAACGTCTGGAAGAACTTGATAATATCGCTAATAC-3′ | C. jejuni 81116 |
| Reverse, 5′-TTGTAATAATCTTAAAACATTTTGC-3′ | ||
| FlaA-(1–491) | Forward, 5′-CACCATGGGATTTCGTATTAACAC-3′ | C. jejuni 81116 |
| Reverse, 3′-ACAATGCAGAATCAATTGAATCCATAACCGCCATTGC-3′ | ||
| FliC(C) | Forward, 5′-GCAATGGCGGTTATGGATTCAATTGATTCTGCATTGT-3′ | S. enteritidis 706 |
| Reverse, 5′-ATTCTGCTGCTTTAACATTGGTTACCGTATTGCCAAG-3′ | ||
| FlaA-(527–576) | Forward, 5′-CTTGGCAATACGGTAACCAATGTTAAAGCAGCAGAAT-3′ | C. jejuni 81116 |
| Reverse, 5′-TTGTAATAATCTTAAAACATTTTGC-3′ | ||
| FlaA-(1–122) | Forward, 5′-CACCATGGGATTTCGTATTAACAC-3′ | C. jejuni 81116 |
| Reverse, 5′-TTAGAAACGCGATCGATATCTGCTATCGCCATCC-3′ | ||
| FliC(H) | Forward, 5′-GGATGGCGATAGCAGATATCGATCGCGTTTCTAA-3′ | S. enteritidis 706 |
| Reverse, 5′-GTAAAACTTTGAGCACCAACATTGAACCCATCAA-3′ | ||
| FlaA-(177–576) | Forward, 5′-TTGATGGGTTCAATGTTGGTGCTCAAAGTTTTAC-3′ | C. jejuni 81116 |
| Reverse, 5′-TTGTAATAATCTTAAAACATTTTGC-3′ | ||
| FliC(NVC) | Forward, 5′-GGGATGGCGATAGCAGATCGCTTCACTTCTAATATC-3′ | S. enteritidis 706 |
| Reverse, 5′-ATTCTGCTGCTTTAACATTGGTTACCGTATTGCCAAG-3′ | ||
| FlaA3-topoF | 5′-CACCATGGGATTTCGTATTAACAC-3′ | |
| FlaA6-topoR | 5′-TTGTAATAATCTTAAAACATTTTGC-3′ | |
| FlaAB-mutant-R | 5′-AAAGCTATTATTCCCTTACAGGATGAG-3′ | C. jejuni 81116 |
| FlaAFSphI | 5′-GCATGCTAGTAAAATTGAAGATGAAAGAGAG-3′ | C. jejuni 81116 |
| FlaARNsiI | 5′-ATGCATTTTAAATCCTTTAAATAATTTC-3′ | |
| Flagellin-pMA3-F | Forward, 5′-CCGAGCTCAAAAGGATTTAAAATGGGATTTCGTATTAACACAAATGT-3′ | |
| Flagellin-pMA3-R | Reverse, 5′-CCCCGCGGCTATTGTAATAATCTTAAAACATTTTGCTG-3′ |
a Underlines indicate restriction sites used for cloning.
Recombinant His6-tagged proteins were obtained by incubating pellets of isopropyl-1-thio-β-d-galactopyranoside-induced (1 mm, 5 h, 37 °C) bacterial cultures in 8 m urea for 17 h (20 °C). After sonication and centrifugation (5000 × g, 15 min, 20 °C) to remove debris, His6-tagged flagellin was purified with nickel-nitrilotriacetic acid-agarose (Qiagen). After washing with 8 m urea, pH 6.4, flagellin was eluted in steps with 8 m urea, pH 5.3, and 8 m urea, pH 4.5. For recombinant FliC, FlaA, and FlaB, fractions containing flagellin were pooled, dialyzed (24 h, 4 °C) against 10 mm of Tris·HCl (pH 9.0), and centrifuged (100,000 × g, 60 min) to remove protein aggregates. Chimeric flagellins and control recombinant FlaA and FliC were kept at −20 °C in 8 m urea, pH 4.5, at a concentration of 500 μg ml−1. Proteins were analyzed on SDS-PAGE, and concentrations were determined by using the BCA protein assay kit (Thermo Scientific Pierce).
Construction of C. jejuni Mutants
The complete flaA-flaB region of C. jejuni 81116 was amplified by PCR with primers FlaA3-topoF and FlaAB-mutant-R (Table 1) and ligated into pGEM-T easy (Promega). EcoRV was used to remove the last 714 nucleotides of flaA and the N-terminal 1017 nucleotides of flaB. This fragment was replaced with a chloramphenicol resistance cassette obtained by digestion of pAV35 (35) with PvuII (resulting in plasmid pMR108). C. jejuni mutant strain 11168H1ΔflaAB was constructed by homologue recombination through electroporation using the pMR108 deletion plasmid and strain C. jejuni 11168H1, as described (36). The σ28 flaA promoter region was amplified from C. jejuni 81116 with primers FlaAFSphI and FlaARNsiI and cloned into pMA1 (37) using SphI and NsiI, resulting in pMA3. For the expression of flagellin proteins in C. jejuni, flaA, flaANC, and flaANHC were PCR-amplified with primers flagellin-pMA3-F and flagellin-pMA3-R using the His-tagged expression constructs as template, digested with SacI and SacII, and cloned into the multiple cloning site of pMA3. Conjugation to C. jejuni was performed as described (37).
Transient Transfection
HEK293 and HeLa 57A cells (∼70% confluent) kept in 48-well plates were transiently transfected with 50 μl of a mixture of plasmid DNA and FuGENE 6 (Roche Diagnostics) in DMEM at a lipid to DNA ratio of 3 to 1. HEK293 cells were transfected with 50 ng of NF-κB-luc plasmid and 70 ng of pTK-LacZ, which was used for normalization of transfection efficiency. HeLa 57A cells were transfected with 125 ng of pFLAG-human-TLR5, pFLAG-mouse-TLR5, pFlAG-chicken-TLR5 (33), or pFLAG-CMV1 empty vector (Sigma) together with 125 ng of pTK-LacZ. Cells were used in TLR5 stimulation assays at 48 h after transfection.
Toll-like Receptor 5 Stimulation Assays
Transfected cells were placed in 0.5 ml of fresh DMEM with 5% fetal calf serum before stimulation with bacteria or purified flagellin. For cell stimulation, S. enteritidis and C. jejuni were grown (17 h) in Luria Bertani broth and heart infusion broth, respectively, collected by centrifugation (5000 × g, 10 min, 22 °C), and resuspended in Dulbecco's phosphate-buffered saline. Bacteria were added to the transfected cells at an m.o.i. of 1:100. After 3.5 h of stimulation, cells were rinsed 3 times with DMEM-5% fetal calf serum to prevent bacterial overgrowth and further incubated in fresh DMEM-5% fetal calf serum. Bacterial culture supernatants were collected (5000 × g, 10 min, 22 °C) after 16 h of growth, filtered (0.22 μm, Millipore), and added to transfected cells (10 μl per well). Native flagellin was depolymerized at 70 °C for 20 min before stimulation and added to the cells at the indicated concentrations (ng ml−1). Recombinant FlaA, FliC, and chimeric flagellins, stored in 8 m urea, pH 4.5, at a concentration of 500 μg ml−1 (see above) were instantly diluted 500-fold by adding 1 μl of protein solution per well. All flagellin stimulations were stopped after 5 h by rinsing the cells twice with Dulbecco's phosphate-buffered saline, lysis in 0.1 ml of reporter lysis buffer (Promega), and freezing at −80 °C. Luciferase activity was measured in a luminometer (TD-20/20, Turner Designs) after mixing 20 μl of thawed cell lysate with 0.1 ml of luciferase reagent (Promega). For normalization of transfection efficiency, luciferase values were adjusted to β-galactosidase values determined with the β-galactosidase assay (Promega). Results were expressed in relative light units and represent the means of duplicate values of three independent experiments.
Reverse Transcription-PCR
RNA from HT-29 and NHEK cells stimulated for 2 h with 1 μg ml−1 of recombinant flagellins was isolated using RNA-Bee (Bio-Connect). Subsequent DNase I treatment and reverse transcription-PCR analysis for actin and IL-8 mRNA was performed as described previously (38).
Detection of Flagellins Produced by C. jejuni
Whole bacterial lysates and culture supernatant were prepared from 17 h of C. jejuni cultures. After centrifugation (5000 × g, 10 min, 22 °C), the supernatant was collected, and the pellet was resuspended in an equal amount of Dulbecco's phosphate-buffered saline for SDS-PAGE analysis. Secreted and intracellular flagellins were detected by Western blot analysis using anti-FlaA antibody CF1 (1:500 dilution) (39) and horseradish peroxidase-conjugated goat-anti mouse IgG (Sigma). Reactive bands were visualized using SuperSignal West Pico Chemiluminescent Substrate (Thermal Scientific Pierce). CF1 recognizes an epitope in a 193-amino acid stretch in the variable domain of C. jejuni 81116 FlaA. In chimera FlaANHC, the first 30 amino acids of this stretch has been replaced by the corresponding S. enteritidis sequence, which did not influence CF1 recognition. For two-dimensional electrophoresis, culture supernatant was concentrated 20 times using Centricon YM-30 filters (Millipore) and mixed with rehydration solution (7 m urea, 2 m thiourea, 4% (w/v) CHAPS, 0.5% IPG buffer, pH 4–7 (Amersham Biosciences), and 0.3% (w/v) dithiothreitol). First-dimension isoelectric focusing was performed on an IPGphor (Amersham Biosciences) with immobilized nonlinear pH (3–10) gradient strips (Amersham Biosciences) using the following isoelectric focusing parameters: 12 h at 30 V, 30 min at 500 V, 30 min at 100 V, 1 h at 40 min 6000 V, and 2 h at 500 V. Isoelectric focusing strips were equilibrated for 15 min in 50 mm Tris-HCl, pH 8.8, 6 m urea, 2% SDS, 30% glycerol, and 10 mg ml−1 dithiothreitol followed by 15 min in 50 mm Tris-HCl, pH 8.8, 6 m urea, 2% SDS, 30% glycerol, and 25 mg ml−1 iodoacetamide. Second-dimension SDS-PAGE was performed using 10% polyacrylamide gels. Flagellins were detected as described above.
RESULTS
TLR5-activating Properties of Campylobacter Flagellin
The inability of C. jejuni flagellin to activate TLR5 has been demonstrated for native and recombinant flagellin from strain 81–176 (14, 15, 18). As C. jejuni flagellins show considerable sequence variation between different strains, are present in two differentially regulated isoforms (FlaA and FlaB) (40), and show variable glycosylation (41–43), we first examined the efficacy of C. jejuni strain 81116 to signal via TLR5. Flagellin activity was measured in HEK293 cells expressing TLR5 with an NF-κB-luciferase reporter as a read-out system. In this system, both S. enteritidis and its culture supernatant (containing secreted flagellin), but not the flagellin deficient (ΔfliC) strain, induced a robust TLR5 response (Fig. 1, A and B). In contrast, neither C. jejuni strain 81116 nor its culture supernatant activated NF-κB in the TLR5-expressing cells (Fig. 1, A and B). Analysis of 10 additional clinical C. jejuni isolates confirmed the evolutionary conservation of this trait (data not shown). To exclude limited monomeric flagellin release as a cause of the inability to activate TLR5, native C. jejuni flagellin was purified. Isolated C. jejuni 81116 flagellin was also unable to activate TLR5, even at concentrations 10,000-fold higher than native flagellin of S. enteritidis (Fig. 1C). Experiments with HeLa 57A cells transfected with TLR5 instead of HEK293 cells yielded similar results (Fig. 1D). Competition assays showed that an excess of native C. jejuni 81116 flagellin did not antagonize HEK293 activation by flagellin of S. enteritidis (data not shown).
FIGURE 1.
C. jejuni fails to activate TLR5. A and B, NF-κB activation was measured for TLR5-expressing HEK293 cells stimulated with live bacteria (A) or culture supernatant (B) of wild type (wt) C. jejuni, C. jejuni ΔflaAB, wt S. enteritidis, or S. enteritidis ΔfliC. C, NF-κB activation was measured for TLR5-expressing HEK293 cells stimulated for 5 h with purified native C. jejuni flagellin or S. enteritidis flagellin at the indicated concentrations (ng ml−1). D, NF-κB activation was measured for human TLR5 (hTLR5)-transfected or control HeLa 57A cells stimulated for 5 h with purified native C. jejuni and S. enteritidis (S. E) flagellin at the indicated concentrations (ng ml−1). Values represent the increase of NF-κB-induced luciferase activity in stimulated cells compared with non-stimulated cells and are the mean ± S.E. of three independent experiments.
TLR5-stimulating Activity of Recombinant C. jejuni FlaA and FlaB
TLR5 activation by Salmonella flagellin requires the amino acids 89–96 at the bridge of the α-helices ND1a and ND1b in the N-terminal conserved domain (Fig. 2, black box) (18, 21). An additional region, located in the center of the conserved C-terminal CD1 α-helix (Fig. 2, gray box), appears critical for stability of the N-terminal TLR5 binding domain (18, 21, 24). Sequence analysis of C. jejuni FlaA shows considerable deviation of both the relevant N- and C-terminal regions from the corresponding regions of Salmonella FliC (Ref. 18 and Fig. 2). The independently expressed C. jejuni FlaB subunit is identical to FlaA in its N-terminal TLR5 binding site but differs at several amino acids in the center of the CD1 α-helix (Fig. 2, indicated by asterisks). To examine the potential relevance of these changes in amino acid composition for TLR5 activation, we expressed both FlaA and FlaB of C. jejuni strain 81116 as polyhistidine-tagged proteins in E. coli and purified them by Ni2+-affinity chromatography. SDS-PAGE analysis of the native and recombinant C. jejuni flagellins demonstrated a markedly lower apparent molecular mass for the recombinant proteins compared with native C. jejuni flagellin, consistent with the absence of attached glycan moieties (Fig. 3A). The difference in electrophoretic mobility was not observed for recombinant and native S. enteritidis flagellin (FliC), in agreement with the lack of flagellin glycosylation in this species. Functional assays using TLR5-expressing HEK293 cells showed that both recombinant FlaA and FlaB failed to activate NF-κB (Fig. 3B), whereas purified recombinant Salmonella FliC induced a potent response. These results demonstrate that the non-glycosylated forms of both C. jejuni FlaA and FlaB lack TLR5-stimulating activity. As glycosylation of FlaA and FlaB is needed for flagella assembly (44) and, thus, possibly for appropriate folding of the protein, we also tested native flagellins isolated from Campylobacter 81116 FlaA and FlaB mutant strains. These proteins also failed to activate TLR5 (data not shown). Together, the data indicate that neither of the C. jejuni flagellins is able to activate TLR5 irrespective of their state of glycosylation.
FIGURE 2.
ClustalW alignment of C. jejuni 81116 FlaA and FlaB and S. enteritidis 706 FliC. The stretch of amino acids proposed to bind TLR5 in the N-terminal conserved domain are boxed in black, and the crucial residues for TLR5 activation in the C-terminal conserved domain are boxed in gray. Asterisks indicate differences in amino acid sequence between FlaA and FlaB in the C-terminal domain.
FIGURE 3.
Recombinant, non-glycosylated C. jejuni FlaA and FlaB fail to activate TLR5. A, SDS-PAGE was performed to examine differences in electrophoretic mobility between recombinant non-glycosylated C. jejuni flagellins (rFlaA and rFlaB) and native glycosylated C. jejuni flagellin (FlaAB). As a control, recombinant and native S. enteritidis flagellin (rFliC and FliC, respectively) were analyzed. B, NF-κB translocation was measured in TLR5-expressing HEK293 cells after stimulation (5 h) with recombinant non-glycosylated C. jejuni FlaA and FlaB at the indicated concentrations (ng ml−1). S. enteritidis FliC (10 ng ml−1) was used as a positive control. Values represent the increase of NF-κB-induced luciferase activity in stimulated cells compared with non-stimulated cells and are the mean ± S.E. of three independent experiments.
Construction and Function of Chimeric Flagellins
In an attempt to restore the ability of C. jejuni flagellin to bind and activate TLR5, we replaced a part of its ND1 α-helix region with the corresponding region of S. enteritidis FliC that contains the putative TLR5 binding site (chimera FlaAN, Fig. 4). Comparative modeling of FlaA on the structure of S. enteritidis flagellin was used to select amino acids regions that were predicted to yield minimal changes in the overall protein configuration. A second chimeric flagellin was constructed in which the CD1 α-helix region was replaced (chimera FlaAC), and a third was constructed by exchanging both the α-helices ND1 and CD1 (chimera FlaANC). Finally, a control chimera was constructed that contained both S. enteritidis α-helices ND1 and CD1 together with the entire central variable region (chimera FlaANVC, Fig. 4). All recombinant proteins were expressed in E. coli, purified, and tested for their ability to activated TLR5 in HEK293 cells. The control FlaANVC chimera was fully able to induce NF-κB translocation (Fig. 5A), confirming data that the structurally disordered extreme N- and C-terminal regions of flagellin are not involved in TLR5 engagement (23) and excluding the possibility these regions inhibit receptor activation. Functional analysis of the other three chimeric proteins unexpectedly showed that none of the chimeric flagellins had regained the ability to activate NF-κB (Fig. 5A) regardless the presence of both conserved regions critical for Salmonella flagellin to activate TLR5.
FIGURE 4.
Characteristics of chimeric flagellin proteins. A, schematic overview of the constructed chimeric flagellins. Numbers refer to the start and end amino acid positions of the exchanged FlaA domains N, H, V, and C. hTLR5, human TLR5. B, the structure of S. typhimurium flagellin (PDB code 1UCU) shows the different flagellin domains as well as the N- and C-terminal regions proposed to be involved in TLR5 activation (in red and blue, respectively) (left), and the location of the exchanged N (red), H (yellow), and C (blue) domains (right). The potential of the recombinant flagellins to activate human TLR5 is indicated on the right as −, +, ++, or +++.
FIGURE 5.
The TLR5 stimulatory activity of the recombinant chimeric flagellins. A and B, NF-κB activation was measured for TLR5-expressing HEK293 cells after 5 h of stimulation with the indicated flagellins (1 μg ml−1). C, TLR5-expressing HEK293 cells stimulated with increasing concentrations (ng ml−1) of recombinant FlaANHC or FliC show the dose-response relationship. D, NF-κB activation in HeLa 57A cells transfected with either human TLR5 (hTLR5) or empty vector after stimulation (5 h) with 1 μg ml−1 of the indicated recombinant flagellins. Values represent the increase of NF-κB-induced luciferase activity in stimulated cells compared with non-stimulated cells and are the mean ± S.E. of three independent experiments.
Reconstitution of Human TLR5 Recognition in C. jejuni FlaA Requires a Variable β-Hairpin Region of S. enteritidis FliC
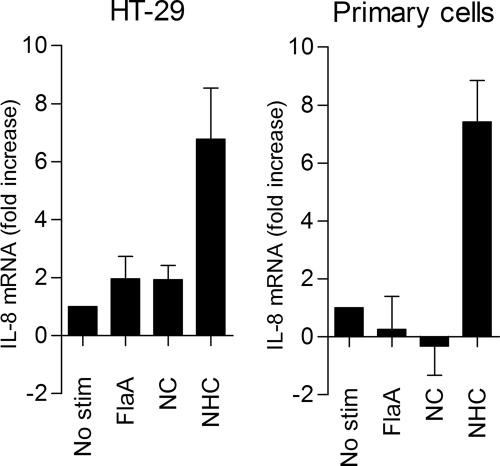
In the search for additional regions required for restoration of TLR5 activation in a Campylobacter flagellin backbone, we focused on the β-hairpin region after the ND1b helix in Salmonella flagellin. This hairpin structure is involved in multimerization of flagellin subunits and may further stabilize the intramolecular structure formed by the highly conserved α-helices (45). Due to low sequence homology between bacterial species, the β-hairpin region has thus far been ignored as part of a direct TLR5 binding site. To assess the role of the 56-amino acid β-hairpin, we constructed a second series of chimeric flagellins (Fig. 4). Replacement of the β-hairpin region of C. jejuni FlaA with the β-hairpin from S. enteritidis FliC (FlaAH) was not sufficient to induce TLR5 activation (Fig. 5B). Similarly, chimeras consisting of C. jejuni flagellin with two of three Salmonella regions (chimera FlaANH and chimera FlaACH) were inactive (Fig. 5B). However, a chimeric flagellin containing the conserved ND1 and CD1 regions together with the variable β-hairpin region (chimera FlaANHC) strongly activated TLR5 in both HEK293 cells (Fig. 5, B and C) and HeLa 57A carrying human TLR5 but not empty vector (Fig. 5D). To further verify that the β-hairpin plays a role in TLR5 stimulation, we tested the activity of the chimeric flagellins FlaANC and FlaANHC in the non-transfected human intestinal epithelial cell-line HT-29 and in non-transformed primary human epithelial cells, which both endogenously express TLR5. FlaANHC, but not FlaA or FlaANC, enhanced IL-8 transcript levels in both cell types (Fig. 6). Together, these results indicate that at least three distinct sections of Salmonella flagellin are required to reconstitute human TLR5-stimulating activity in Campylobacter flagellin.
FIGURE 6.
IL-8 mRNA induction by FlaANHC in human non-transfected intestinal and primary epithelial cells. HT-29 cells (A) and non-transformed primary human cells (B) were stimulated (2 h) with 1 μg ml−1 recombinant FlaA, FlaANC, or FlaANHC. IL-8 transcripts were analyzed by reverse transcription-PCR and are presented as -fold increase in mRNA levels in stimulated versus non-stimulated cells. Values are the mean ± S.E. of three independent experiments.
The β-Hairpin Region of Flagellin Determines TLR5 Species Specificity
To further explore the importance of the β-hairpin region in TLR5 recognition, we tested the abilities of constructed chimeric flagellins to activate TLR5 from different species. This approach has previously been instrumental in dissecting ligand properties required for TLR activations (33, 46, 47). All constructed chimeras that failed to activate human TLR5 were unable to activate mouse TLR5, except for FlaANHC (Fig. 7, A and B). FlaANHC induced lower levels of NF-κB activation in mouse TLR5 than in human TLR5-transfected cells. This effect was also observed for Salmonella FliC (data not shown) and is likely caused by intrinsic differences in TLR5, different expression levels, and/or the expression of mTLR5 in a heterologous (human) background. Chimeric flagellin FlaANHC was also able to activate chicken TLR5 (Fig. 7C). Unexpectedly, however, chicken TLR5 responded also to chimeric flagellin FlaANC, in clear contrast to human and mouse TLR5. As the only difference between flagellin FlaANC and FlaANHC is the presence of the S. enteritidis β-hairpin, these results indicate that this hairpin structure is not merely needed for proper folding of the flagellin but, rather, is essential for activation of mammalian TLR5 but not chicken TLR5.
FIGURE 7.
Species-specific activation of TLR5 by FlaANHC. A, B, and C, HeLa 57A cells were transfected with either human (A), mouse (B), or chicken (C) TLR5. NF-κB translocation was measured after stimulation with 1 μg ml−1 of the indicated recombinant flagellins. Values represent the increase of NF-κB-induced luciferase activity in stimulated cells compared with non-stimulated cells and are the mean ± S.E. of three independent experiments.
Glycosylation and Secretion of Biologically Active FlaANHC by C. jejuni
The TLR5-activating Campylobacter flagellins used above were overexpressed in E. coli, purified under denaturing conditions, and refolded in vitro. In Campylobacter, flagellins are only successfully secreted after post-translational modification. To engineer C. jejuni that express TLR5-activating flagellins, we expressed the genes encoding wild type FlaA, the chimeric flagellin FlaANC, and TLR5-activating chimeric flagellin FlaANHC in C. jejuni strain 81116. The genes were cloned into plasmid pMA3, a shuttle vector suitable for protein expression in C. jejuni under the control of the endogenous flaA σ28 promoter, and transformed into a flagella-deficient and non-motile C. jejuni 11168H1ΔflaAB mutant. Introduction of the plasmid encoding FlaA but not FlaANC or FlaANHC flagellin restored flagella formation and bacterial motility in a ΔflaAB background (data not shown). Western blot analysis of whole cell lysates using anti-FlaA antibody CF1 as a probe yielded reactive proteins for both FlaANC and FlaANHC (Fig. 8A). Analysis of the bacterial culture supernatants also yielded reactive flagellin bands for both strains that were larger in size than the non-secreted intracellular proteins, consistent with the attachment of glycan moieties during protein export (Fig. 8A). Two-dimensional gel electrophoresis followed by immunoblotting with anti-FlaA antibodies demonstrated that both wild type flagellin produced by 11168H1ΔflaAB+FlaA and secreted FlaANHC appeared as an array of similarly sized proteins of different isoelectric points, a pattern shown by mass spectrometry to be typical for variable glycosylation of the protein (42, 48) (Fig. 8, B and C). Overall, these results indicate that the chimeric flagellins were expressed, processed, and secreted in the C. jejuni native background.
FIGURE 8.
C. jejuni expresses, glycosylates, and secretes chimeric flagellins FlaANC and FlaANHC. A, Western blotting was performed to examine the electrophoretic mobility of bacteria-associated (B) and secreted (S) chimeric flagellins FlaANC and FlaANHC produced by C. jejuni. Blots were probed with the flagellin-specific antibody CF1. As controls, recombinant FlaA (rFlaA) and native C. jejuni flagellin (FlaAB) were used. B and C, two-dimensional electrophoresis followed by Western blotting using antibody CF1 was performed to visualize the flagellin glycosylation pattern on C. jejuni produced and secreted (B) FlaA and (C) FlaANHC.
Infection of TLR5-expressing HEK293 cells with live C. jejuni that secrete chimeric glycosylated FlaANHC flagellin yielded a potent NF-κB response, whereas no activation was observed for C. jejuni producing the chimera FlaANC and wild type FlaA (Fig. 9, A and B). Similar results were obtained with sterile culture supernatants of C. jejuni secreting FlaANHC. These results indicate that viable C. jejuni strains can be engineered that activate TLR5 and that glycosylation of flagellin does not interfere with TLR5 receptor recognition.
FIGURE 9.
C. jejuni expressing FlaANHC induces NF-κB activation in HEK293 cells. NF-κB activation in TLR5-expressing HEK293 cells was measured after stimulation (5 h) with live bacteria (A) or culture supernatant of C. jejuni ΔflaAflaB expressing FlaA, FlaANC or FlaANHC (B). As a control, C. jejuni ΔflaAflaB carrying the empty expression vector pMA3 was used. Values represent the increase of NF-κB-induced luciferase activity in stimulated cells compared with non-stimulated cells and are the mean ± S.E. of three independent experiments.
DISCUSSION
Knowledge of the molecular basis of TLR recognition is important to understand bacteria-host interactions and to exploit bacterial components for targeted modulation of the immune system. In the present study we took advantage of the inability of C. jejuni flagellin to activate TLR5 to better define the molecular requirements for TLR5 recognition. Using a reverse-engineering approach, we reconstituted TLR5-stimulating activity in C. jejuni and discovered that besides the conserved N-terminal ND1a and ND1b α-helices and the C-terminal CD1 α-helix in flagellin, an adjacent β-hairpin structure is required for activation of mammalian TLR5. This β-hairpin was not required for activation of chicken TLR5 (Fig. 7C), indicating species-specific interaction of the flagellin with TLR5. C. jejuni O-linked glycosylation of flagellin did not interfere with TLR5 activation, which may hold promise for modification of flagellin to alter its physical properties when used e.g. as a vaccine adjuvant.
Previous studies on the ability of Campylobacter to avoid TLR5 recognition were performed with C. jejuni strain 81–176 (14, 15, 18). However, C. jejuni flagellins are known to show high sequence variability between strains and are present in two differentially regulated isoforms, FlaA and FlaB, that consistently differ at 12 amino acid positions in their conserved N- and C-terminal regions (49). Our results demonstrate that the evasion of the TLR5 response is a conserved trait among the C. jejuni species and holds for both FlaA and FlaB (Fig. 3B and data not shown) irrespective of the composition of the flagella (FlaA/FlaB subunit ratio) or the variable glycan modification. These results lend support to the notion that this species and other α- and ϵ-Proteobacteria have evolved an alternate class of flagellins that may provide a selective advantage in the host by evading the TLR5 innate immune response (18). The different flagellin structure may explain why the Campylobacter flagella fiber is formed by 7 instead of 11 subunit helices (28). Although its contribution to pathogenesis is unknown, the widespread evolutionary conservation of TLR5 evasion suggests that this trait adds a valuable selective advantage during colonization or infection.
The evasion of the mammalian TLR5 sensing machinery by C. jejuni has thus far been mostly attributed to deviations in the proposed TLR5 binding region, a stretch of eight amino acids located in the N-terminal conserved domain flagellin and crucial for flagella formation in Salmonella (18). The successful engineering of a recombinant Campylobacter flagellin that activates human TLR5 required, besides the known N- and C-terminal regions, the presence of the β-hairpin domain from Salmonella FliC (Fig. 5). This domain has previously been discarded as a potential binding region for TLR5 due to low sequence homology among bacteria, although disruption of the β-hairpin domain by transposon insertion of a 31-amino acid polypeptide resulted in a significant decrease in TLR5 activation (21). As the construction of chimeric proteins brings the possibility of incorrect protein folding, it could be argued that proper flagellin folding and subsequent TLR5 activation in humans is only achieved with the presence of three distinct flagellin domains from the same origin. Indeed, in Salmonella flagellin, the ND1 and CD1 helices form multiple intramolecular domain-domain interactions that provide structural strength in the flagellin protein. The absence of the interactions in FlaAN and FlaAC, which contain one helix of Salmonella and one of Campylobacter, may explain the biological inactivity of these chimeras. Evidence that the β-hairpin structure likely confers more than protein folding and stability is that the Salmonella β-hairpin proved necessary for activation of human and mouse TLR5 but not chicken TLR5. This receptor was activated by both the chimeras FlaANC and FlaANHC (Fig. 7C). The activation of chicken TLR5 by FlaNC (but not FlaN or FlaC) indicates that the protein is folded into a TLR5 activating state.
Amino acid sequence analysis suggests that C. jejuni flagellin contains a β-hairpin structure at grossly the same position as in Salmonella flagellin. Although several amino acids are conserved between the β-hairpin of Campylobacter and Salmonella, the inactivity of chimera FlaANC toward human and mouse TLR5 shows that the C. jejuni β-hairpin domain cannot substitute for the Salmonella β-hairpin structure in the receptor interaction. Chicken TLR5 is activated by flagellins that contain either the Salmonella or Campylobacter β-hairpin. This may indicate that chicken TLR5 has a more relaxed ligand specificity than mammalian TLR5 with respect to the β-hairpin. Indeed, we previously demonstrated that chicken TLR5 has different flagellin sensing qualities compared with human TLR5 (33). In addition, Smith et al. (21) showed that the disruption of the β-hairpin in flagellin in Salmonella significantly decreased biological activity of flagellin for human TLR5 but not mouse TLR5. Together, these data suggest the hairpin stretch contributes to the species specificity of flagellin recognition by TLR5.
Campylobacter flagellins are heavily decorated with an array of variably modified pseudaminic acid residues, which is needed for flagella formation (44). In Pseudomonas aeruginosa, flagellin glycosylation promotes TLR5 stimulation (50). The glycosylation moieties present on the flagellin of C. jejuni are located on the predicted surface exposed variable domain, mostly in a 200-amino acid hydrophobic patch. Structural modeling of C. jejuni FlaA on the crystal structure of Salmonella flagellin reveals that the sugar moieties are not in close proximity to the predicted TLR5 binding site. Expression of chimeric FlaANHC by live Campylobacter, which resulted in glycosylated and secreted proteins, presented us with the opportunity to, for the first time, directly assess the role of the flagellin-glycosylation on TLR5 activation. Culture supernatants containing glycosylated FlaANHC as well as live C. jejuni secreting glycosylated flagellins showed the ability to strongly activate TLR5, suggesting that the modification of C. jejuni flagellin does not serve as an additional mechanism to prevent or promote activation of TLR5.
The successful engineering of Campylobacter strains, which secrete flagellins that variably activate TLR5, indicates that the modified regions are not critical for transport through the C. jejuni flagellar secretion apparatus. Successful secretion of flagellin through the flagellar basal body requires the ND0 domain, which contains a secretion signal (51), and the CD0 region, which binds chaperone FliS to inhibit cytosolic flagellin polymerization (52, 53). None of the chimeric flagellins constructed in this study have alterations in either the putative secretion signal in domain ND0 or in domain CD0, and the chimeras are, thus, predicted to bind C. jejuni FliS in a similar fashion as the wild type flagellin. Furthermore, as in Campylobacter flagellin glycosylation is essential for secretion (44), all putative glycosylation sites in the constructed chimeric flagellins were left intact. The conservation of the basic Campylobacter flagellin architecture may explain the successful secretion of flagellins with the incorporated foreign domains needed for TLR5 activation. However, despite secretion, none of the chimeric flagellins in C. jejuni assembled into a filament. This may indicate a dysregulation of the flagellar components needed for fiber assembly and/or incompatibility of the chimeric structure with e.g. the Campylobacter filament capping protein FliD, a defective multimerization, or altered axial interactions between the flagellin subunits. Elucidation of the crystal structure of C. jejuni flagellin may resolve this issue.
Recent studies have identified two additional cellular receptors for flagellin, the intracellular Nod-like receptor (NLR) Ipaf and NLR apoptosis inhibitory protein 5 (Naip5) (54, 55). Localized intracellularly, these receptors are involved in sensing flagellin that is injected into the host cell, for instance through the type III secretion system (T3SS) of S. typhimurium, or the type IV secretion system (T4SS) of Legionella pneumophila. Activation of Ipaf and/or Naip5 results in caspase-1-dependent IL-1β and IL-18 secretion. So far, a functional injection machinery like T3SS or T4SS has not been found in Campylobacter. In the case that Campylobacter flagellin gains access to the cytosol, it may activate Ipaf and Naip5, as the C-terminal 35-amino acid flagellin domain that is sensed by these receptors is highly conserved when compared with Legionella FlaA (56). As this region is not altered in FlaANHC, this chimeric flagellin is predicted to activate both TLR5 and Ipaf/Naip5.
In conclusion, we constructed a live C. jejuni secreting glycosylated flagellins with reconstituted TLR5 activity by the introduction of multiple domains from Salmonella flagellin. Through the construction of a series of chimeric flagellins, we identified a previously unknown role for the flagellin β-hairpin domain in the activation of TLR5 and showed that this structure determines TLR5 species specificity in flagellin response. These results provide more insight in the flagellin-TLR5 interaction and contribute to the current knowledge on the application of flagellin for vaccination purposes.
The work was supported by the Dutch Ministry of Agriculture, Nature, and Food Quality.
- TLR5
- Toll-like receptor 5
- FlaA
- C. jejuni flagellin A
- FlaB
- C. jejuni flagellin B
- FliC
- S. enteritidis or S. typhimurium Phase I flagellin
- DMEM
- Dulbecco's modified Eagle's medium
- Naip5
- Nod-like receptor (NLR) Ipaf and NLR apoptosis inhibitory protein 5
- IL
- interleukin
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid.
REFERENCES
- 1.Hayashi F., Smith K. D., Ozinsky A., Hawn T. R., Yi E. C., Goodlett D. R., Eng J. K., Akira S., Underhill D. M., Aderem A. (2001) Nature 410, 1099–1103 [DOI] [PubMed] [Google Scholar]
- 2.Vijay-Kumar M., Aitken J. D., Gewirtz A. T. (2008) Semin. Immunopathol. 30, 11–21 [DOI] [PubMed] [Google Scholar]
- 3.van Aubel R. A., Keestra A. M., Krooshoop D. J., van Eden W., van Putten J. P. M. (2007) Mol. Immunol. 44, 3702–3714 [DOI] [PubMed] [Google Scholar]
- 4.Song L., Nakaar V., Kavita U., Price A., Huleatt J., Tang J., Jacobs A., Liu G., Huang Y., Desai P., Maksymiuk G., Takahashi V., Umlauf S., Reiserova L., Bell R., Li H., Zhang Y., McDonald W. F., Powell T. J., Tussey L. (2008) PLoS ONE 3, e2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bargieri D. Y., Rosa D. S., Braga C. J., Carvalho B. O., Costa F. T., Espíndola N. M., Vaz A. J., Soares I. S., Ferreira L. C., Rodrigues M. M. (2008) Vaccine 26, 6132–6142 [DOI] [PubMed] [Google Scholar]
- 6.Le Moigne V., Robreau G., Mahana W. (2008) Mol. Immunol. 45, 2499–2507 [DOI] [PubMed] [Google Scholar]
- 7.McDonald W. F., Huleatt J. W., Foellmer H. G., Hewitt D., Tang J., Desai P., Price A., Jacobs A., Takahashi V. N., Huang Y., Nakaar V., Alexopoulou L., Fikrig E., Powell T. J. (2007) J. Infect. Dis. 195, 1607–1617 [DOI] [PubMed] [Google Scholar]
- 8.Honko A. N., Sriranganathan N., Lees C. J., Mizel S. B. (2006) Infect. Immun. 74, 1113–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skountzou I., Martin M. D., Wang B., Ye L., Koutsonanos D., Weldon W., Jacob J., Compans R. W. (2009) Vaccine, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burdelya L. G., Krivokrysenko V. I., Tallant T. C., Strom E., Gleiberman A. S., Gupta D., Kurnasov O. V., Fort F. L., Osterman A. L., Didonato J. A., Feinstein E., Gudkov A. V. (2008) Science 320, 226–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neish A. S. (2007) Am. J. Physiol. Gastrointest Liver Physiol. 292, G462–G466 [DOI] [PubMed] [Google Scholar]
- 12.Winter S. E., Raffatellu M., Wilson R. P., Rüssmann H., Bäumler A. J. (2008) Cell Microbiol. 10, 247–261 [DOI] [PubMed] [Google Scholar]
- 13.Way S. S., Thompson L. J., Lopes J. E., Hajjar A. M., Kollmann T. R., Freitag N. E., Wilson C. B. (2004) Cell Microbiol. 6, 235–242 [DOI] [PubMed] [Google Scholar]
- 14.Watson R. O., Galán J. E. (2005) Cell Microbiol. 7, 655–665 [DOI] [PubMed] [Google Scholar]
- 15.Johanesen P. A., Dwinell M. B. (2006) Infect. Immun. 74, 3437–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gewirtz A. T., Yu Y., Krishna U. S., Israel D. A., Lyons S. L., Peek R. M., Jr. (2004) J. Infect. Dis. 189, 1914–1920 [DOI] [PubMed] [Google Scholar]
- 17.Lee S. K., Stack A., Katzowitsch E., Aizawa S. I., Suerbaum S., Josenhans C. (2003) Microbes Infect. 5, 1345–1356 [DOI] [PubMed] [Google Scholar]
- 18.Andersen-Nissen E., Smith K. D., Strobe K. L., Barrett S. L., Cookson B. T., Logan S. M., Aderem A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 9247–9252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanders C. J., Yu Y., Moore D. A., 3rd, Williams I. R., Gewirtz A. T. (2006) J. Immunol. 177, 2810–2818 [DOI] [PubMed] [Google Scholar]
- 20.Galkin V. E., Yu X., Bielnicki J., Heuser J., Ewing C. P., Guerry P., Egelman E. H. (2008) Science 320, 382–385 [DOI] [PubMed] [Google Scholar]
- 21.Smith K. D., Andersen-Nissen E., Hayashi F., Strobe K., Bergman M. A., Barrett S. L., Cookson B. T., Aderem A. (2003) Nat. Immunol. 4, 1247–1253 [DOI] [PubMed] [Google Scholar]
- 22.Szymanski C. M., Logan S. M., Linton D., Wren B. W. (2003) Trends Microbiol. 11, 233–238 [DOI] [PubMed] [Google Scholar]
- 23.Eaves-Pyles T. D., Wong H. R., Odoms K., Pyles R. B. (2001) J. Immunol. 167, 7009–7016 [DOI] [PubMed] [Google Scholar]
- 24.Murthy K. G., Deb A., Goonesekera S., Szabó C., Salzman A. L. (2004) J. Biol. Chem. 279, 5667–5675 [DOI] [PubMed] [Google Scholar]
- 25.Donnelly M. A., Steiner T. S. (2002) J. Biol. Chem. 277, 40456–40461 [DOI] [PubMed] [Google Scholar]
- 26.Jacchieri S. G., Torquato R., Brentani R. R. (2003) J. Bacteriol. 185, 4243–4247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodriguez M. S., Thompson J., Hay R. T., Dargemont C. (1999) J. Biol. Chem. 274, 9108–9115 [DOI] [PubMed] [Google Scholar]
- 28.Pearson B. M., Gaskin D. J., Segers R. P., Wells J. M., Nuijten P. J., van Vliet A. H. (2007) J. Bacteriol. 189, 8402–8403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karlyshev A. V., Linton D., Gregson N. A., Wren B. W. (2002) Microbiology 148, 473–480 [DOI] [PubMed] [Google Scholar]
- 30.Van Asten F. J., Hendriks H. G., Koninkx J. F., Van der Zeijst B. A., Gaastra W. (2000) FEMS Microbiol. Lett. 185, 175–179 [DOI] [PubMed] [Google Scholar]
- 31.Ibrahim G. F., Fleet G. H., Lyons M. J., Walker R. A. (1985) J. Clin. Microbiol. 22, 1040–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Logan S. M., Trust T. J. (1983) Infect. Immun. 42, 675–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keestra A. M., de Zoete M. R., van Aubel R. A., van Putten J. P. M. (2008) Mol. Immunol. 45, 1298–1307 [DOI] [PubMed] [Google Scholar]
- 34.Tabor S., Richardson C. C. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 4767–4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Vliet A. H., Wooldridge K. G., Ketley J. M. (1998) J. Bacteriol. 180, 5291–5298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wassenaar T. M., Fry B. N., van der Zeijst B. A. (1993) Gene 132, 131–135 [DOI] [PubMed] [Google Scholar]
- 37.van Mourik A., Bleumink-Pluym N. M., van Dijk L., van Putten J. P., Wösten M. M. (2008) Microbiology 154, 584–592 [DOI] [PubMed] [Google Scholar]
- 38.de Zoete M. R., Keestra A. M., Roszczenko P., van Putten J. P. M. (2010) Infect. Immun. 78, 1229–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nuijten P. J., van der Zeijst B. A., Newell D. G. (1991) Infect. Immun. 59, 1100–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wösten M. M., Wagenaar J. A., van Putten J. P. M. (2004) J. Biol. Chem. 279, 16214–16222 [DOI] [PubMed] [Google Scholar]
- 41.Thibault P., Logan S. M., Kelly J. F., Brisson J. R., Ewing C. P., Trust T. J., Guerry P. (2001) J. Biol. Chem. 276, 34862–34870 [DOI] [PubMed] [Google Scholar]
- 42.van Alphen L. B., Wuhrer M., Bleumink-Pluym N. M., Hensbergen P. J., Deelder A. M., van Putten J. P. M. (2008) Microbiology 154, 3385–3397 [DOI] [PubMed] [Google Scholar]
- 43.Logan S. M., Hui J. P., Vinogradov E., Aubry A. J., Melanson J. E., Kelly J. F., Nothaft H., Soo E. C. (2009) FEBS J. 276, 1014–1023 [DOI] [PubMed] [Google Scholar]
- 44.Goon S., Kelly J. F., Logan S. M., Ewing C. P., Guerry P. (2003) Mol. Microbiol. 50, 659–671 [DOI] [PubMed] [Google Scholar]
- 45.Yonekura K., Maki-Yonekura S., Namba K. (2003) Nature 424, 643–650 [DOI] [PubMed] [Google Scholar]
- 46.Steeghs L., Keestra A. M., van Mourik A., Uronen-Hansson H., van der Ley P., Callard R., Klein N., van Putten J. P. M. (2008) Infect. Immun. 76, 3801–3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andersen-Nissen E., Smith K. D., Bonneau R., Strong R. K., Aderem A. (2007) J. Exp. Med. 204, 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Logan S. M., Kelly J. F., Thibault P., Ewing C. P., Guerry P. (2002) Mol. Microbiol. 46, 587–597 [DOI] [PubMed] [Google Scholar]
- 49.Meinersmann R. J., Hiett K. L. (2000) Microbiology 146, 2283–2290 [DOI] [PubMed] [Google Scholar]
- 50.Verma A., Arora S. K., Kuravi S. K., Ramphal R. (2005) Infect. Immun. 73, 8237–8246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Végh B. M., Gál P., Dobó J., Závodszky P., Vonderviszt F. (2006) Biochem. Biophys. Res. Commun. 345, 93–98 [DOI] [PubMed] [Google Scholar]
- 52.Evdokimov A. G., Phan J., Tropea J. E., Routzahn K. M., Peters H. K., Pokross M., Waugh D. S. (2003) Nat. Struct. Biol. 10, 789–793 [DOI] [PubMed] [Google Scholar]
- 53.Auvray F., Thomas J., Fraser G. M., Hughes C. (2001) J. Mol. Biol. 308, 221–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grassl G. A., Finlay B. B. (2008) Curr. Opin. Gastroenterol. 24, 22–26 [DOI] [PubMed] [Google Scholar]
- 55.Miao E. A., Andersen-Nissen E., Warren S. E., Aderem A. (2007) Semin. Immunopathol. 29, 275–288 [DOI] [PubMed] [Google Scholar]
- 56.Lightfield K. L., Persson J., Brubaker S. W., Witte C. E., von Moltke J., Dunipace E. A., Henry T., Sun Y. H., Cado D., Dietrich W. F., Monack D. M., Tsolis R. M., Vance R. E. (2008) Nat. Immunol. 9, 1171–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]