Abstract
We have revealed that in Caenorhabditis elegans, non-sulfated chondroitin is required for normal cell division and cytokinesis at an early developmental stage, whereas heparan sulfate is essential for embryonic morphogenesis in the later stages of development. To clarify the roles of chondroitin sulfate and heparan sulfate in early embryogenesis in mammals, we generated glucuronyltransferase-I (GlcAT-I) knock-out mice by gene targeting. GlcAT-I is an enzyme required for the synthesis of both chondroitin sulfate and heparan sulfate. Here we report that mice with a deletion of GlcAT-I showed remarkable reduction of the synthesis of chondroitin sulfate and heparan sulfate and embryonic lethality before the 8-cell stage because of failed cytokinesis. In addition, treatment of wild-type 2-cell embryos with chondroitinase ABC had marked effects on cell division, although many heparitinase-treated embryos normally developed to blastocysts. Taken together, these results suggest that chondroitin sulfate in mammals, as with non-sulfated chondroitin in C. elegans, is indispensable for embryonic cell division.
Keywords: Cell Division, Chondroitin Sulfate, Gene Knockout, Glycosaminoglycan, Proteoglycan Synthesis
Introduction
Chondroitin sulfate (CS),3 dermatan sulfate (DS), heparan sulfate (HS), and heparin (Hep) are a class of glycosaminoglycans (GAGs) that distribute on the surfaces of virtually all cells and in the extracellular matrices. CS/DS and HS/Hep are covalently linked to a specific serine residue in the core protein and occur as CS/DS proteoglycans (PGs) and HS-PGs. Many of the physiological roles of CS/DS-PGs and HS-PGs are thought to be attributable to CS/DS and HS side chains, with core proteins largely playing the role of a scaffold to make CS/DS and HS functionally available for binding to a variety of ligands. In fact, gene-targeting technology in vertebrates and invertebrates has led to elucidation of the physiological functions of HS in the developmental process and morphogenesis in addition to the regulation of signaling molecules. In contrast to the series of model organisms deficient in HS, we have generated a model lacking CS backbone biosynthesis in only Caenorhabditis elegans (C. elegans) so far (1). Study of these worms revealed that non-sulfated chondroitin is required for normal cell division and cytokinesis at an early developmental stage (2, 3), whereas HS is essential for embryonic morphogenesis in the later stages of development (4). These observations suggested that, whereas the structure of chondroitin is similar to that of HS, the function of chondroitin is different from that of HS in C. elegans (4). In mice, although deficiency of an enzyme that synthesizes HS backbones leads to neonatal lethality not only with abnormal organogenesis but also with the aberration of signaling pathways of morphogens and growth factors (5, 6), little is known about the roles of CS, mainly because of the unexpected redundancy of CS-synthesizing enzymes, thereby making the functional analysis of CS more difficult (1).
CS/DS and HS/Hep chains are synthesized onto a common carbohydrate-protein linkage region structure, GlcUAβ1–3Galβ1–3Galβ1–4Xylβ1-O-Ser (7). The linkage region tetrasaccharide is formed by sequential stepwise addition of monosaccharide residues by the respective specific glycosyltransferases, xylosyltransferase, galactosyltransferase-I, galactosyltransferase-II, and glucuronyltransferase-I (GlcAT-I) (8). The repeating disaccharide region [(-4GlcUAβ1–4GlcNAcα1-)n] of HS/Hep is synthesized on the linkage region by the HS co-polymerase complex of EXT1 and EXT2 (9, 10). In contrast, the repeating disaccharide region [(-4GlcUAβ1–3GalNAcβ1-)n] of CS/DS is formed on the linkage region by any two combinations of chondroitin synthases-1 (11), -2 (12), -3 (13), and chondroitin polymerizing factor (14). Also, the functionally redundant, multiple glycosyltransferases involved in CS/DS have been cloned (15, 16). Thus, as mentioned, this redundancy makes it difficult to investigate the specific role of CS/DS in mammalian early embryogenesis.
In this study, to clarify the functions of CS/DS in mammalian early embryogenesis, we focused on GlcAT-I. Because GlcAT-I transfers GlcUA from UDP-GlcUA to the trisaccharide-serine, Galβ1–3Galβ1–4Xylβ1-O-Ser, finalizing the formation of the common linkage region (17, 18), GlcAT-I knock-out mice would result in the complete elimination of CS/DS as well as HS/Hep. Thus, we generated GlcAT-I knock-out mice and attempted to analyze in vivo functions of CS/DS at an early developmental stage. Here, we demonstrated that most homozygous null mice die by embryonic day 2.5 because of failure of cytokinesis. In addition, almost all embryos treated with chondroitinase ABC died from 2-cell to 8-cell stages, whereas many heparitinase-treated embryos developed normally to blastocysts. These results suggest that CS is indispensable for early embryonic cell division in mammals.
EXPERIMENTAL PROCEDURES
Materials
Proteus vulgaris chondroitinase ABC (CSase) (EC 4.2.2.4), Flavobacterium heparinum heparitinase and heparinase, and the monoclonal antibodies (LY111, Hepss-1, and 3G10) were purchased from Seikagaku Corp. (Tokyo, Japan).
Targeting Vector Construction
The mouse GlcAT-I gene isolated from a 129/SvJ genome library (Stratagene) was used to construct the targeting vector (see Fig. 1A). The PGKneobpA cassette (19), in which the neomycin resistance gene was ligated under the phosphoglycerate kinase I promoter and the polyadenylation site from bovine growth hormone was ligated downstream of the neo gene, was inserted in reverse transcriptional orientation into the EcoRV site in GlcAT-I exon 2 for positive selection. The DT-A cassette (20), in which the diphtheria toxin A fragment gene was ligated under the MC1 promotor, was ligated as the 3′-end of the targeting vector for negative selection.
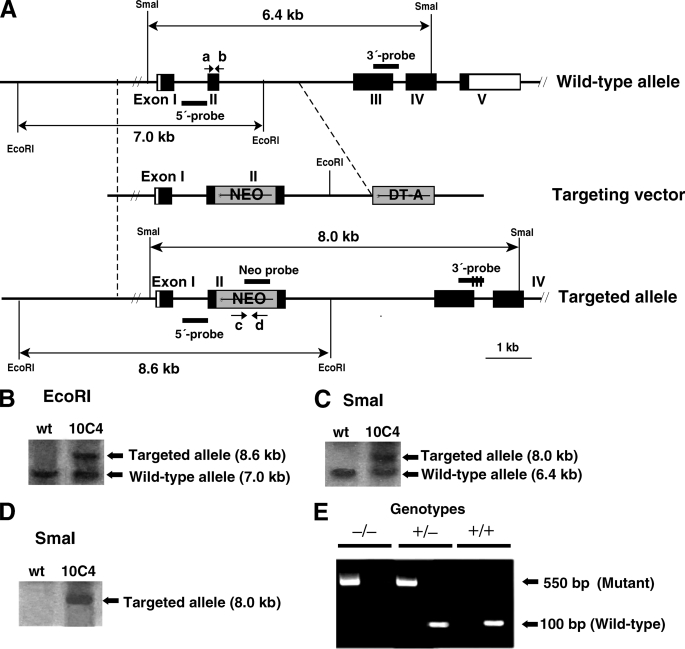
FIGURE 1.
Targeted disruption of mouse GlcAT-I gene. A, generation of GlcAT-I-deficient mice. The neomycin resistance cassette was inserted into exon II of GlcAT-I gene. Coding and non-coding exons of GlcAT-I gene are shown by closed and open boxes, respectively, and the PGKneobpA cassette (NEO) and diphtheria toxin A fragment gene cassette (DT-A) are represented by gray boxes. PCR primers (a, b, c, and d) used for genotyping are shown by arrows. B–D, Southern blot analysis of wild-type (wt) and targeted (10C4) ES cells demonstrated homologous recombination in GlcAT-I gene. Genomic DNA (10 μg) from wild-type and targeted ES cells was digested with EcoRI and hybridized with the 5′ probe (B). Genomic DNA (10 μg) from wild-type and targeted ES cells was digested with SmaI and hybridized with the 3′ probe (C), or with the Neo probe (D). The expected DNA fragments of the wild-type allele and mutant allele are shown in B–D. E, genotypes were determined by tail DNA PCR using wild-type allele-specific primers (primers a and b, left lanes) or mutant allele-specific primers for the neo gene (primers c and d, right lanes), respectively. The mutant allele was detected as a 550-bp band using primers c and d and the wild-type allele was detected as a 100-bp band using primers a and b.
Generation of GlcAT-I−/− Mice
The linearized targeting vector (20 μg) was electroporated (250 V, 500 μF) into 107 E14–1 mouse embryonic stem (ES) cells (21) and selected with 250 μg (active form)/ml G418 (Invitrogen) for 7–10 days. Homologous recombinants were screened by PCR and confirmed by Southern blot hybridization with an external 5′ probe, 3′ probe, and Neo probe (see Fig. 1). Chimeric mice were generated by the aggregation method (22) with some modifications. Chimeras were mated with C57BL/6J females, and homozygous mutant mice were generated by intercrossing of heterozygotes. Genotypes were determined by tail DNA PCR using wild-type allele-specific primers (primers a: 5′-CTGAGGATATCCCAGTTGC-3′ and b: 5′-ACATAGATAGTAGGCAGGGCC-3′) or mutant allele-specific primers for the neo gene (primers c: 5′-GGAAGGGCGAAGTCACTGTTG-3′ and d: 5′-GAAGAACTCGTCAAGAAGGCGATAG-3′), respectively. Mice were kept under specific pathogen-free conditions in an environmentally controlled clean room at the Institute of Laboratory Animals, Kobe Pharmaceutical University. The experiments were conducted according to institutional ethics guidelines for animal experiments and safety guidelines for gene manipulation experiments.
Mating and in Vitro Blastocyst Culture
All embryos were generated by natural matings. Heterozygous male and female mice were bred to obtain wild-type, heterozygous, and homozygous mouse embryos. The morning of the day on which a vaginal plug was detected was designated E0.5. Embryos at E1.5 were collected by flushing oviducts with HEPES-buffered medium 2 (M2, Sigma). Culture was done under 5% CO2 in m-KSOM medium.
Immunocytochemistry of Mouse Oocytes Using an Anti-CS and/or Anti-HS Monoclonal Antibody
Embryos at E1.5 were collected by flushing oviducts with HEPES-buffered medium 2 (M2; Sigma). Immediately after collection, eggs were cultured in m-KSOM medium for 1 h, transferred in 50-μl drops of m-KSOM medium containing Cy3-conjugated LY111 (diluted 1:1000) or Alexa Fluor 488-conjugated Hepss-1 (diluted 1:1000), and incubated for 1 h, and then washed three times in 50-μl drops of m-KSOM medium. Fluorescent images were obtained using a fluorescence microscope, Biozero (Keyence, Osaka, Japan).
For staining blastocysts with anti-CS or anti-HS monoclonal antibodies, embryos from heterozygous intercrosses were fixed in cold methanol at −20 °C for 10 min and washed three times with PBS. After blocking with PBS containing 0.1% Triton X-100 and 3% BSA for 1 h at room temperature, embryos were incubated with anti-CS monoclonal antibody LY111 (diluted 1:1000 in 0.1% BSA/PBS) or anti-HS monoclonal antibody Hepss-1 (diluted 1:100 in 0.1% BSA/PBS) at room temperature for 1 h. After washing, the embryos were incubated with an antibody against mouse IgM conjugated to Alexa 488 for 1 h. After three further washes in 0.1% BSA/PBS, embryos were rinsed once in PBS and incubated at 37 °C with 0.1 mg/ml RNase A (Roche) in PBS with propidium iodide (2 μg/ml). To confirm the specificity of staining with these antibodies, blastocysts were pretreated with CSase (2 milliinternational units) or a mixture of heparitinase and heparinase (HSase) (0.5 milliinternational units each) to remove CS or HS, respectively, and then processed for immunostaining as described above.
For double staining of blastocysts with anti-CS and anti-HS monoclonal antibodies, embryos were treated with a mixture of heparitinase and heparinase (HSase) (0.5 milliinternational units each) for 1 h and incubated with anti-CS monoclonal antibody LY111 and anti-proteoglycan ΔHS monoclonal antibody 3G10 (diluted 1:100 in 0.1% BSA/PBS) at room temperature for 1 h. After washing, the embryos were incubated with an antibody against mouse IgM conjugated to Alexa 488 and an antibody against mouse IgG conjugated to Alexa 568 for 1 h. After three further washes in 0.1% BSA/PBS, embryos were rinsed once in PBS. Fluorescent images were obtained using a laser-scanning confocal microscope FLUOVIEW (Olympus, Tokyo, Japan).
Embryonic Culture with CSase or HSase
Embryos at E1.5 were collected by flushing oviducts with HEPES-buffered medium 2 (M2; Sigma). Culturing was performed under 5% CO2 in m-KSOM medium with the addition of CSase (2 milliinternational units), HSase (2 milliinternational units) or heat-inactivated CSase and HSase. To visualize nuclei, embryos were incubated in Hoechst 33342 (Molecular Probes) for 10 min at room temperature. Fluorescent images were obtained using a fluorescence microscope, Biozero (Keyence, Osaka, Japan).
RESULTS AND DISCUSSION
To examine CS/DS functions in mammalian early embryogenesis, we inactivated the GlcAT-I gene via homologous recombination in mouse ES cells. The mouse GlcAT-I gene contains five putative coding exons (Fig. 1A). The targeting vector was constructed by inserting a neomycin resistance cassette into exon 2 (Fig. 1A). The targeting vector was electroporated into mouse ES cells and G418-resistant colonies were picked up. Eleven ES clones were selected out of 586 G418-resistant clones by PCR and among the 11 independent ES clones, six homologous recombinants were found. Correct targeting was confirmed in two of the six ES clones by Southern blot analysis using an external 3′ probe (Fig. 1B), 5′ probe (Fig. 1C), and neo probe (Fig. 1D). Data regarding one of the two targeted ES clones were depicted (Fig. 1, B–D). The two independent ES clones were aggregated with C57BL/6 x BDF1 8-cell-stage embryos to generate a chimera and the target allele was transmitted through a germ line in two independent ES clones. GlcAT-I+/− mice were backcrossed to C57BL/6 mice for 11 generations. The phenotypes described here were identical in the two mouse lines.
GlcAT-I+/− mice had an apparently normal phenotype and were born at largely Mendelian frequency. They were intercrossed, and more than 300 offspring were genotyped by PCR (Fig. 1E). We did not detect any GlcAT-I−/− neonates and embryos after E6.5, indicating that mutant embryos died during early embryogenesis (Table 1). In fact, even at E2.5 (8-cell stage), only 2% GlcAT-I−/− embryos were detected, suggesting that most GlcAT-I−/− mutants died before E2.5 (8-cell stage).
TABLE 1.
Genotype analysis of progeny from GlcAT-I heterozygous intercrosses
Percentages of different genotypes appear in parentheses.
| Day | No. of progeny with genotypea |
No. resorbedb | No. total | ||
|---|---|---|---|---|---|
| +/+ | +/− | −/− | |||
| Neonate | 46 (38%) | 74 (62%) | 0 (0%) | 120 | |
| E8.5 | 22 (39%) | 31 (56%) | 0 (0%) | 3 (5%) | 56 |
| E7.5 | 12 (19%) | 43 (67%) | 0 (0%) | 9 (14%) | 64 |
| E6.5 | 17 (39%) | 18 (41%) | 0 (0%) | 9 (20%) | 44 |
| E2.5 | 19 (45%) | 22 (53%) | 1 (2%) | 42 | |
a Genotyping of each developmental stage was performed by PCR.
b Resorbed embryos were not genotyped.
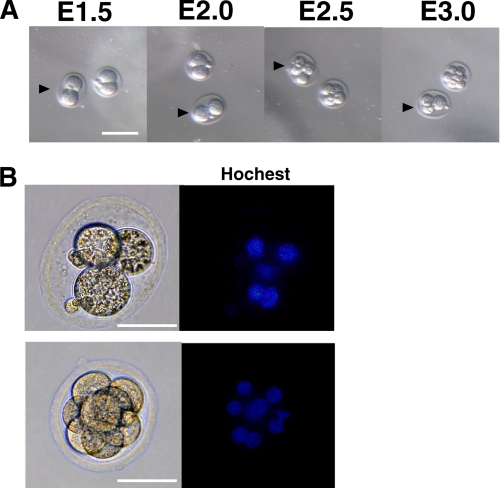
To further analyze the lethal stages and phenotypes of GlcAT-I−/− embryos during early development, 2-cell stage embryos derived from heterozygous intercrosses (GlcAT-I+/− × GlcAT-I+/−) or other matings (GlcAT-I+/− × GlcAT-I+/+ or GlcAT-I+/+ × GlcAT-I+/+) were cultured until blastocyst stages (Table 2). The results of in vitro culture showed that 13% of embryos from GlcAT-I+/− heterozygous intercrosses died between 2-cell and 8-cell stages although all of embryos from GlcAT-I+/− × GlcAT-I+/+ or GlcAT-I+/+ × GlcAT-I+/+ were viable (Table 2). Notably, of embryos from heterozygous intercrosses, we could identify only 7% GlcAT-I−/− embryos at the implantation stage, while the fraction of GlcAT-I+/+ and GlcAT-I+/− embryos was viable within Mendelian expectations (1:2), confirming that GlcAT-I inactivation is lethal before 8-cell stages. Moreover, reversion of cell division was observed in embryos only from GlcAT-I+/− heterozygous intercrosses. Fig. 2 showed a representative example of reversion of cell division in one embryo from GlcAT-I+/− heterozygous intercrosses. The 2-cell (E1.5) embryo divided into a 4-cell embryo, and then insufficient cytoplasmic division seemed to force the embryonic cell compartments to revert to an unusual 3-cell embryo with four nuclei. The unusual embryo eventually died, most likely due to incomplete cytokinesis (Fig. 2, A and B). These results indicate that the GlcAT-I function is essential for embryonic cytokinesis and cell division.
TABLE 2.
Genotype analysis of embryos cultured in vitro from 2-cell-stage embryos to blastocyst implantation stages
Percentages of different genotypes appear in parentheses.
| Parental genotype | No. of progeny with genotypea |
No. of dead embryosb |
No. of total | ||||||
|---|---|---|---|---|---|---|---|---|---|
| +/+ | +/− | −/− | 2-cell to 8-cell | 8-cell to morula | Morula to blastocyst | Blastocyst to hatched blastocyst | Total dead embryos | ||
| GlcAT-I+/− × GlcAT-I+/− | 32 (29%) | 55 (49%) | 8 (7%) | 15 | 0 | 0 | 2 | 17 (15%) | 112 |
| GlcAT-I+/− × GlcAT-I+/+ | 23 (59%) | 14 (36%) | 0 | 0 | 0 | 2 | 2 (5%) | 39 | |
| GlcAT-I+/+ × GlcAT-I+/+ | 42 (98%) | 0 | 1 | 0 | 0 | 1 (2%) | 43 | ||
a Genotyping was performed by PCR.
b Dead embryos were not genotyped, but their lethal stages were determined.
FIGURE 2.
Reversion of cytokinesis in embryos from GlcAT-I heterozygous intercrosses. A, E1.5 embryos were isolated from heterozygous crosses and cultured. Representative features are depicted. Reversal of cytokinesis was observed in one embryo from GlcAT-I heterozygous intercrosses (arrowhead). No reversal of cytokinesis was detected in embryos from crosses of wild-type and heterozygous mice. B, two embryos shown in A were stained with Hoechst 33342. The abnormal embryo (arrowheads in A) failed to complete cytokinesis and a double nucleated cell appeared (upper), whereas cell division of the other embryo continued normally (lower).
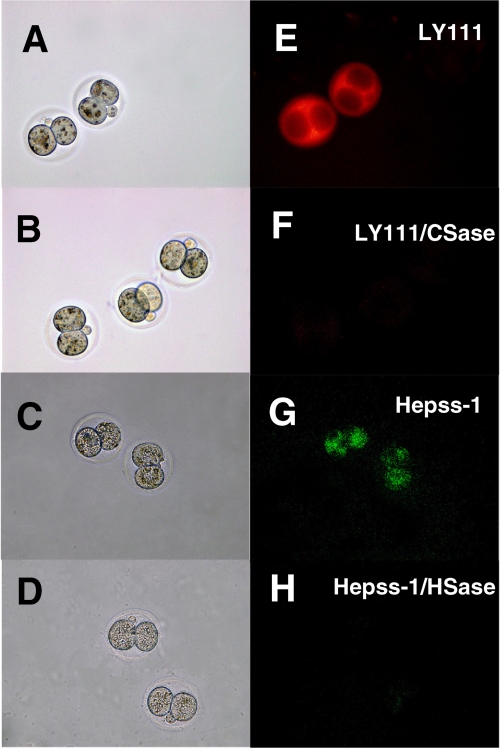
In a previous study, we demonstrated that GlcAT-I transfers GlcUA from UDP-GlcUA to the trisaccharide-serine, Galβ1–3Galβ1–4Xylβ1-O-Ser, finalizing the formation of the common GAG-protein linkage region, GlcUAβ1–3Galβ1–3Galβ1–4Xylβ1-O-Ser (17). Therefore, it was expected that inactivation of GlcAT-I would abolish both CS and HS chains in mouse embryos. We then attempted to determine whether GlcAT-I−/− embryos lack both CS and HS. For this analysis, immunocytochemistry with wild-type mouse 2-cell embryos and blastocysts was first performed using an anti-CS (LY111) or anti-HS (Hepss-1) monoclonal antibody because, to our knowledge, the existence of GAG chains in mouse 2-cell embryos and blastocysts has not been demonstrated. As expected, fluorescent signals were detected in all 2-cell embryos and blastocysts using either of these antibodies (Figs. 3, E, G, and 4, A, and C), and the corresponding signals were eliminated by CSase (Figs. 3F and 4B) or HSase (Figs. 3H and 4D), respectively, indicating that both CS and HS were produced in mouse 2-cell embryos and blastocysts.
FIGURE 3.
Immunocytohistochemistry of mouse 2-cell embryos using an anti-CS or anti-HS monoclonal antibody. Wild-type embryos at E1.5 were collected and stained as described under “Experimental Procedures.” Left panels (A–D) show embryos examined by phase-contrast microscopy. Immunofluorescent staining by anti-CS (LY111) or anti-HS (Hepss-1) antibody is shown in right panels (E–H). Treatment of 2-cell embryos with CSase (LY111/CSase) or HSase (Hepss-1/HSase) eliminated staining by the anti-CS (LY111) or anti-HS (Hepss-1) antibody, respectively.
FIGURE 4.
Immunocytohistochemistry of blastocysts using an anti-CS or anti-HS monoclonal antibody. Wild-type embryos at E3.5 were fixed in cold methanol and stained as described under “Experimental Procedures.” Immunofluorescent staining of CS with LY111 antibody (A, green) or with Hepss-1 antibody (C, green) is shown. Treatment of blastocysts with CSase or HSase eliminated the staining by LY111 antibody (B, green) or by Hepss-1 antibody (D, green), respectively. Nuclei were visualized by PI staining (red).
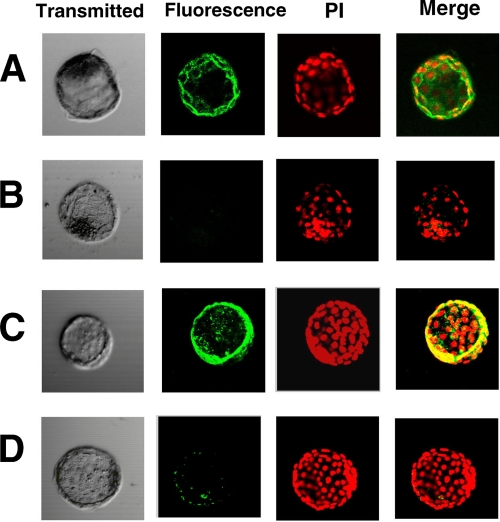
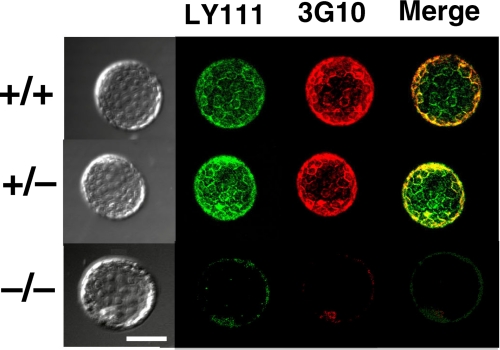
Next, double immunostaining of GlcAT-I−/− and GlcAT-I+/− embryos as well as GlcAT-I+/+ embryos using anti-CS (LY111) and anti-ΔHS (3G10) monoclonal antibodies was carried out. As shown in Fig. 5, GlcAT-I+/+ and GlcAT-I+/− embryos were stained by both anti-CS and anti-ΔHS monoclonal antibodies, whereas the GlcAT-I−/− embryo was not stained. These findings suggest that GlcAT-I−/− embryos seem to lack both CS and HS chains.
FIGURE 5.
Analysis of GAGs in GlcAT-I−/− blastocysts. Blastocysts from heterozygous intercrosses were fixed in cold methanol and stained as described under “Experimental Procedures.” Immunofluorescent staining of CS with LY111 (green) and 3G10 (red) is shown. Wild-type (GlcAT-I+/+) and GlcAT-I+/− embryos (upper and middle panels, respectively) were strongly stained by LY111 and 3G10, whereas the GlcAT-I−/− embryo (lower panels) failed to be stained. Note that some GlcAT-I−/− embryos lacking both CS and HS seem to have progressed through the early cell division stages normally, presumably because of partial rescue by maternal CS. Scale bars: 50 μm.
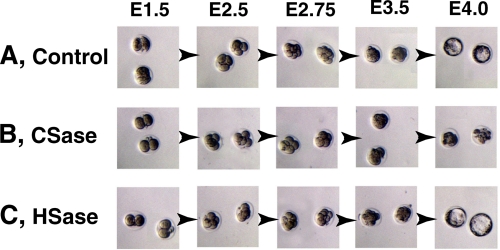
As described above, GlcAT-I−/− embryos showed loss of synthesis of both CS and HS and died before 8-cell stages due to failure of cytokinesis. However, it is unclear that the embryonic cell death observed for GlcAT-I−/− embryos is attributable to deficiency of CS, HS, or both. If CS or HS is indispensable for proper embryonic cytokinesis and cell division, digestion of CS or HS at the embryonic cell surface might also induce abnormal cell division. Treatment of 2-cell embryos with CSase showed that 67% of the treated embryos died between 2-cell and 8-cell stages. In contrast, most embryos treated with heat-inactivated CSase/HSase and 65% of embryos treated with HSase developed normally to blastocysts (Table 3, Fig. 6). These results indicate that CS, but not HS, chains are involved in controlling embryonic cell division and cytokinesis.
TABLE 3.
Analysis of lethal stages of in vitro cultured embryos after treatment with CSase or HSase
| No. of dead embryos |
No. of viable embryos to blastocyst stage | No. total | |||
|---|---|---|---|---|---|
| 2-cell to 8-cell | 8-cell to morula | Morula to blastocyst | |||
| Controla | 0 | 0 | 2 | 29 | 31 |
| CSase treatment | 22 | 5 | 0 | 6 | 33 |
| HSase treatment | 2 | 5 | 1 | 15 | 23 |
a Embryos were treated with heat-inactivated CSase/HSase.
FIGURE 6.
Depletion of CS results in cytokinesis defects. Wild-type embryos at E1.5 were collected and incubated with heat-inactivated CSase/HSase (A, control), CSase (B) or HSase (C), respectively. Treatment of embryos with CSase showed that these two embryos died from 2-cell to 8-cell stages (B), although control (A) and HSase-treated (C) embryos developed normally to blastocyst (also see Table 3). Representative features are depicted.
Prior studies of EXT1 or EXT2 in mice demonstrated that these genes are essential for HS synthesis and early development (5, 6). Notably, EXT1- or EXT2-null embryos developed normally until around E6.5, when they became growth arrested and failed to gastrulate. In addition, the marked reduction of HS in ES cells from EXT1- or EXT2-deficient mice was reported (5, 6). These findings also support the notion that the abnormal cytokinesis and embryonic cell death observed for mouse GlcAT-I-null mutants are attributable to inhibition of the synthesis of CS but not HS. In addition, it should be noted that some GlcAT-I−/− embryos lacking both CS and HS seem to have progressed through the early cell division stages normally (see Table 2 and Fig. 5), presumably because of partial rescue by maternal CS.
So far, there are no reports on the role of CS or CSPG in cytokinesis and cell division in mice. In C. elegans, Olson et al. (23) reported that simultaneous depletion of two of C. elegans chondroitin proteoglycan core proteins, CPG-1/CEJ-1 and CPG-2, by RNAi results in defective cytokinesis during the first embryonic cell division. This phenotype is identical to that observed when the chondroitin synthase (sqv-5) or chondroitin-polymerizing factor (pfc-1) was silenced by RNAi or by a loss of function mutation (2, 3, 24–26). CPG-1/CEJ-1 and CPG-2 are expressed during embryonic development and bind chitin (23). Therefore, CPG-1/CEJ-1 and CPG-2 could act as structural elements of the eggshell or might bridge chitin polymers in the eggshell with other components of the embryonic plasma membrane that result in transmembrane signaling to cytoskeletal components involved in cytokinesis, as suggested (23). Interestingly, immunocytohistochemical analysis of mouse 2-cell embryos using an anti-CS antibody showed large amounts of CS on the embryonic cell surface (Fig. 3E), which was similar to the findings that there is abundant chondroitin in fertilized eggshells and at the cell surfaces of cleavage stage embryos in C. elegans (2). Thus, although vertebrates lack chitin as a structural component and orthologues of CPG-1/CEJ-1 and CPG-2, it is likely that other structural components, such as hyaluronan and CSPGs, which are not synthesized in C. elegans, are involved in cell division in vertebrates. In fact, hyaluronan is reported to be associated with cell division of human fibroblasts (27) and to be bound by many CSPGs, including aggrecan, versican, brevican, and neurocan (28–30). These observations suggest that the synthesis of extracellular matrices may be an evolutionary highly conserved component of vertebrate and invertebrate cytokinesis. To gain more insight into the role of CSPG in mammalian cell division, future studies on the identification of core proteins modified with CS involved in mouse embryonic cell division are needed.
Acknowledgments
We thank Yuko Tone and Masateru Taoka for help in constructing the targeting vector.
This work was supported in part by Core Research for Evolutional Science and Technology (CREST) of the Japan Science and Technology (JST) Corporation (to H. K.), the Science Research Promotion Fund of the Japan Private School Promotion Foundation, the Naito Foundation (to H. K.), and Grants-in-aid for Scientific Research-B 16390026 (to K. S.) and 21390025 (to H. K.) from MEXT, Japan.
- CS
- chondroitin sulfate
- CSase
- chondroitinase ABC
- GAG
- glycosaminoglycan
- PG
- proteoglycan
- HS
- heparan sulfate
- HSase
- a mixture of heparitinase and heparinase
- GlcAT-I
- glucuronyltransferase-I
- PBS
- phosphate-buffered saline
- BSA
- bovine serum albumin
- E
- embryonic day.
REFERENCES
- 1.Uyama T., Kitagawa H., Sugahara K. (2007) in Comprehensive Glycoscience (Kamerling J. P. ed) Vol. 3, pp. 79–104, Elsevier, Amsterdam [Google Scholar]
- 2.Mizuguchi S., Uyama T., Kitagawa H., Nomura K. H., Dejima K., Gengyo-Ando K., Mitani S., Sugahara K., Nomura K. (2003) Nature 423, 443–448 [DOI] [PubMed] [Google Scholar]
- 3.Izumikawa T., Kitagawa H., Mizuguchi S., Nomura K. H., Nomura K., Tamura J., Gengyo-Ando K., Mitani S., Sugahara K. (2004) J. Biol. Chem. 279, 53755–53761 [DOI] [PubMed] [Google Scholar]
- 4.Kitagawa H., Izumikawa T., Mizuguchi S., Dejima K., Nomura K. H., Egusa N., Taniguchi F., Tamura J., Gengyo-Ando K., Mitani S., Nomura K., Sugahara K. (2007) J. Biol. Chem. 282, 8533–8544 [DOI] [PubMed] [Google Scholar]
- 5.Lin X., Wei G., Shi Z., Dryer L., Esko J. D., Wells D. E., Matzuk M. M. (2000) Dev. Biol. 224, 299–311 [DOI] [PubMed] [Google Scholar]
- 6.Stickens D., Zak B. M., Rougier N., Esko J. D., Werb Z. (2005) Development 132, 5055–5068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindahl U., Rodén L. (1972) in Glycoprotein (Gottschalk A. ed) pp. 491–517, Elsevier, Amsterdam [Google Scholar]
- 8.Sugahara K., Kitagawa H. (2000) Curr. Opin. Struct. Biol. 10, 518–527 [DOI] [PubMed] [Google Scholar]
- 9.McCormick C., Leduc Y., Martindale D., Mattison K., Esford L. E., Dyer A. P., Tufaro F. (1998) Nat. Genet. 19, 158–161 [DOI] [PubMed] [Google Scholar]
- 10.Lind T., Tufaro F., McCormick C., Lindahl U., Lidholt K. (1998) J. Biol. Chem. 273, 26265–26268 [DOI] [PubMed] [Google Scholar]
- 11.Kitagawa H., Uyama T., Sugahara K. (2001) J. Biol. Chem. 276, 38721–38726 [DOI] [PubMed] [Google Scholar]
- 12.Izumikawa T., Uyama T., Okuura Y., Sugahara K., Kitagawa H. (2007) Biochem. J. 403, 545–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Izumikawa T., Koike T., Shiozawa S., Sugahara K., Tamura J., Kitagawa H. (2008) J. Biol. Chem. 283, 11396–11406 [DOI] [PubMed] [Google Scholar]
- 14.Kitagawa H., Izumikawa T., Uyama T., Sugahara K. (2003) J. Biol. Chem. 278, 23666–23671 [DOI] [PubMed] [Google Scholar]
- 15.Uyama T., Kitagawa H., Tamura J., Sugahara K. (2002) J. Biol. Chem. 277, 8841–8846 [DOI] [PubMed] [Google Scholar]
- 16.Uyama T., Kitagawa H., Tanaka J., Tamura J., Ogawa T., Sugahara K. (2003) J. Biol. Chem. 278, 3072–3078 [DOI] [PubMed] [Google Scholar]
- 17.Kitagawa H., Tone Y., Tamura J., Neumann K. W., Ogawa T., Oka S., Kawasaki T., Sugahara K. (1998) J. Biol. Chem. 273, 6615–16618 [DOI] [PubMed] [Google Scholar]
- 18.Tone Y., Kitagawa H., Imiya K., Oka S., Kawasaki T., Sugahara K. (1999) FEBS Letters 459, 415–420 [DOI] [PubMed] [Google Scholar]
- 19.Soriano P., Montogomery C., Geske R., Bradley A. (1991) Cell 64, 693–702 [DOI] [PubMed] [Google Scholar]
- 20.Yagi T., Nada S., Watanabe N., Tamemoto H., Kohmura N., Ikawa Y., Aizawa S. (1993) Anal. Biochem. 214, 77–86 [DOI] [PubMed] [Google Scholar]
- 21.Kühn R., Rajewsky K., Müller W. (1991) Science 254, 707–710 [DOI] [PubMed] [Google Scholar]
- 22.Nagy A., Rossant J., Nagy R., Abramow-Newerly W., Roder J. C. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 8424–8428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olson S. K., Bishop J. R., Yates J. R., Oegema K., Esko. J. D. (2006) J. Cell Biol. 173, 985–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herman T., Hartwieg E., Horvitz H. R. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 968–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hwang H. Y., Horvitz H. R. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 14218–14223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hwang H. Y., Olson S. K., Esko J. D., Horvitz H. R. (2003) Nature 423, 439–443 [DOI] [PubMed] [Google Scholar]
- 27.Brecht M., Mayer U., Schlosser E., Prehm P. (1986) Biochem. J. 239, 445–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faltz L. L., Caputo C. B., Kimura J. H., Schrode J., Hascall V. C. (1979) J. Biol. Chem. 254, 1381–1387 [PubMed] [Google Scholar]
- 29.Yamagata M., Yamada K. M., Yoneda M., Suzuki S., Kimata K. (1986) J. Biol. Chem. 261, 13526–13535 [PubMed] [Google Scholar]
- 30.Lesley J., Hyman R. (1998) Front. Biosci. 3, d616–d630 [DOI] [PubMed] [Google Scholar]