Abstract
Sortilin acts as a cell surface receptor for pro-neurotrophins (pro-NT) that upon complex formation with the p75 neurotrophin receptor (p75NTR) is able to signal neuronal cell death. Here we screened a sortilin peptide library comprising 16-mer overlapping sequences for binding of the pro-domains of nerve growth factor and brain-derived neurotrophic factor. We find that a linear surface-exposed sequence, 163RIFRSSDFAKNF174, constitutes an important pro-NT binding epitope in sortilin. Systematic mutational analysis revealed residues Arg163, Phe165, Arg166, and Phe170 to be critical for the interaction. Expression of a sortilin mutant in which these four amino acids were substituted by alanines disrupted pro-NT binding without affecting receptor heterodimerization with p75NTR or binding of ligands that selectively engages the centrally located tunnel in the β-propeller of sortilin. We furthermore demonstrate that a peptide comprising the ligand-binding epitope can prevent pro-NT-induced apoptosis in RN22 schwannoma cells.
Keywords: Growth Factors, Peptides/Neuropeptide, Protein/Protein-Protein Interactions, Protein/Ligand Binding, Cell Surface Receptor, VPS10p Receptor, Pro-neurotrophin, Sortilin
Introduction
Sortilin is one of five members of the mammalian VPS10p-domain receptor family of neuronal type-1 receptors (1). In addition to sortilin, the family comprises sorLA, the sorting receptor for the amyloid precursor protein and major risk factor for late-onset Alzheimer disease (2–4), and three homologous receptors denoted sorCS1, -2, and -3 that have been genetically linked to type-2 diabetes and bipolar disorder (5, 6). Neurotrophins (NTs)3 such as nerve growth factor (NGF) and brain-derived neurotrophic factor (BDNF) are trophic factors essential for development and maintenance of the nervous system (7–11). They are produced as precursors, denoted pro-neurotrophins (pro-NTs), that can be processed to their mature form in the secretory pathway as well as outside the cell (12–14). It is well appreciated that pro-NTs may also have functions in their own right, because they elicit activities opposite to those of their mature counterparts (15–20). Although mature NTs can stimulate neuronal survival and long term potentiation and synaptic strengthening, pro-NTs may induce apoptosis and long term depression (21–23). Sortilin is required for the death signaling pathway, because it forms a high affinity binding site together with the p75NTR neurotrophin receptor (24). Inhibiting binding of the growth factor pro-domains to sortilin prevents apoptosis by pro-NTs from occurring in cultured cells as well as in vivo, e.g. following spinal cord injury (25–32).
Recently, the structure of sortilin, the archetype receptor of the VPS10p-domain family, was solved at the atomic level in complex with the 13-amino acid-long neuropeptide, neurotensin (33). The crystal structure revealed a unique 10-bladed β-propeller structure containing a centrally located tunnel with a diameter exceeding 25 Å that harbors the binding site for neurotensin. Although pro-NT binding to sortilin is partly inhibited by neurotensin (25) the size of the pro-NT pro-domain (∼100–110 amino acids) likely precludes entry of the entire domain into the tunnel. We therefore set out to study if a linear surface exposed amino acid sequence in sortilin might assist in pro-NT binding.
EXPERIMENTAL PROCEDURES
Materials and Proteins
To obtain tagged forms of the neurotrophin pro-domains readily for detection in SPOT analysis, we prepared constructs for each protein allowing for addition of N-terminal S-peptide and polyhistidine tags. Template cDNA for human NGF and BDNF consisted of ATCC clones used for generation of fragments spanning residues Glu1–Arg102 of NGF and Ala1–Arg110 of BDNF using the primer pairs 5′-GGTATTGAGGGTCGCGAACCACACTCAGAGAGCAATGTCCC-3′ and 3′-GGGGGAAGTTGTCCTGAGTGTCCTCGTTCGCCACTCCGAGATTGAGAGGAGA-5′, and 5′-GGTATTGAGGGTCGCGCCCCCATGAAAGAAGCAAACATCCGAGG-3′ and 3′-CACGTTTGTACAGGTACTCCCAGGCCGCGACTCCGAGATTGAGAGGAGA-5′, with compatible overhangs for ligation-independent cloning into the pET-30 Xa/LIC vector from Novagen (catalog no. 70073-3) and amplification using Phusion DNA polymerase and following the protocol as provided by the manufacturer. Proteins were expressed in the BL21/DE3 strain of Escherichia coli, efficiently extracted from bacterial inclusion bodies using Bugbuster reagent from Novagen (catalog no. 70921) with added benzonase (Novagen, catalog no. 70750), and purified by standard nickel-nitrilotriacetic acid affinity chromatography in 500 mm NaCl, 5 mm Imidazole, and 20 mm Tris-HCl, pH 8.0. Protein elution was done in buffer supplemented with 20 mm EDTA. Verification of the intact tagged versions of HisS-NGFpro and HisS-BDNFpro was carried out by SDS-PAGE analysis followed by Coomassie staining or Western blotting using either antibody against the histidine tag from Sigma (H-1029) and secondary HRP-conjugated anti-mouse antibody from Calbiochem (catalog no. 401207), or alternatively by direct binding of HRP-conjugated S-protein from Novagen (catalog no. 69047-3).
For the production of the sortilin ectodomain, a construct encompassing the entire coding region of the N-terminal part of human sortilin, including the endogenous signal peptide and followed by a C-terminal polyhistidine tag inserted in the pCEP-Pu vector was kindly provided by D. Militz, Max-Delbrueck-Center for Molecular Medicine, Berlin. The DNA was transferred into EBNA 293 cells that were selected by G418 (Invitrogen catalog no. 10131-027, 300 μg/ml) and Puromycin from Sigma (catalog no. P8833, 1 μg/ml) before proteins were collected from medium conditioned for 48 h, and used for purification by applying to nickel-nitrilotriacetic acid-Sepharose. The secreted recombinant sortilin polypeptide chain spanning the intact extracellular domain of human sortilin thus ended at Ser725 (+AMIEGRGVGHHHHHH containing the fXa site and polyhistidine tag). The quality of the protein was tested by silver staining of SDS-PAGE analysis.
Peptides for binding and competition studies were synthesized in-house at the Charite (Berlin) or purchased from Eurogentec. GST-NGFpro was prepared as previously described and unprocessed precursor pro-NGF was purchased from Millipore (catalog no. GF210).
Sortilin Mutants
Single amino acid substitutions of ligand binding residues within the extracellular domain of sortilin were introduced using the QuikChange II XL site-directed mutagenesis kit from Stratagene (catalog no. 200521) and the wild-type sortilin pCEP-Pu expression construct as template. Mutagenesis primers were from BioTez (Berlin). Multiple rounds of mutagenesis allowed for generation of double, triple, and quadruple mutant constructs knocking out several ligand binding residues in the sortilin domain.
To generate the quadruple mutations in a sortilin construct encoding the entire sortilin receptor, including the transmembrane and cytoplasmic domains, we took advantage of internal HpaI and BsrGI (New England Biolabs catalog nos. R0105S and R0575S, respectively) restriction recognition sites in the sortilin sequence. Using both enzymes we cut out the sortilin fragment containing the 4A mutations from the pCEP-Pu expression vector for the soluble sortilin and exchanged that for the corresponding fragment from a full-length sortilin expression vector in pcDNA3.1/zeo(+) thereby generating a mutant full-length (fl)-sortilin-4A construct. In parallel, the same fragment was also introduced into an endocytosis-deficient mutant of sortilin (34) to generate a version of full-length sortilin-4A that resides at the cell surface (fl-sortilin-4AΔendo).
CD Spectroscopy
The purity of sortilin proteins was assessed by SDS-PAGE, and the purified proteins were dialyzed against 20 mm NaH2PO4, pH 7.4. Protein concentrations were determined using the absorbance at 280 nm and an estimated extinction coefficient of 133,240 m−1cm−1 (from the ExPaSy proteomics server tools). Ten CD spectra were recorded at 25 °C for each protein on a Jasco J-810 spectropolarimeter (Jasco Spectroscopic, Japan) using a polypeptide concentration of ∼0.1 mg/ml and a cuvette of 2-mm path length. CD data were obtained in the range from 260 to 200 nm at a resolution of 1 nm using a bandwidth of 2.0 nm. The scan speed was 100 nm/min, and the response time was 1 s.
Surface Plasmon Resonance Analysis
Determination of direct binding of ligand to immobilized protein was performed on a biacore2000 instrument (Biacore, Sweden) using CaHBS as standard running buffer (10 mm HEPES, pH 7.4, 140 mm NaCl, 2 mm CaCl2, 1 mm EGTA, and 0.005% Tween 20). A biosensor chip from Biacore (CM5, catalog no. BR-1000-14) was activated using the NHS/EDC method as described by the supplier followed by coating with sortilin to a protein density of 79 fmol/mm2 and used for affinity measurements of the recombinant pro-domains of NGF and BDNF. Preparation of a biosensor surface with pro-sortilin followed an equal procedure. Regeneration of the flow cell after each cycle of ligand binding experiment was done by two 10-μl pulses of regeneration buffer (10 mm glycine-HCl, pH 4.0, 500 mm NaCl, 20 mm EDTA, and 0.005% Tween 20) and a single injection of 0.001% SDS. Fitting of sensorgrams for affinity estimations was done using BIAevaluation version 3.1.
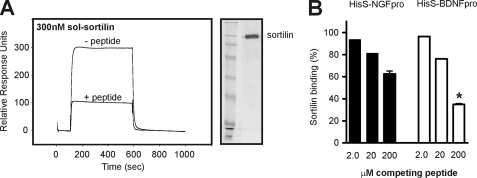
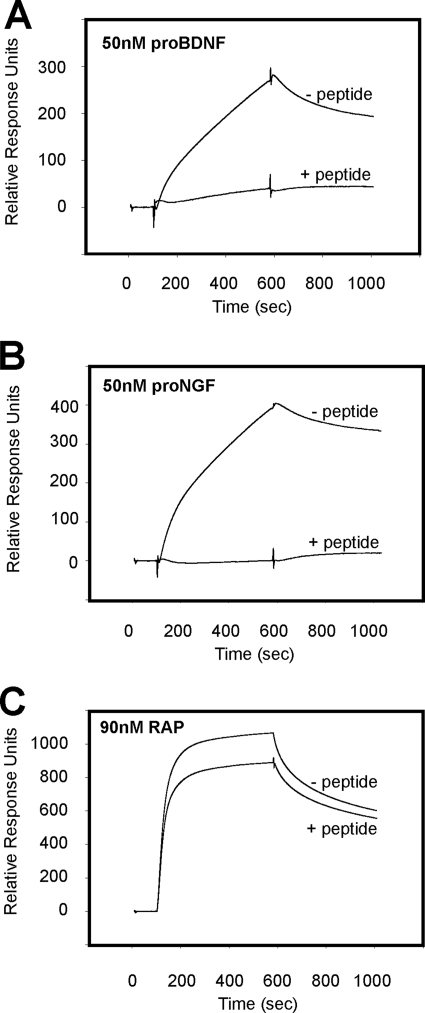
Following similar protocols, immobilization of HisS-NGFpro or HisS-BDNFpro was also done on a CM5 biosensor chip using the NHS/EDC coupling kit according to the manufacturer's instructions (Biacore, Sweden), giving similar surface densities of immobilized protein (∼300 fmol/mm2). Purified peptides were applied to the chip at increasing concentrations to verify the direct binding of pro-neurotrophic domains to linear sortilin peptides. This chip was subsequent used to examine the binding of 390 nm wild-type sortilin domain in CaHBS buffer at a flow of 5 μl/min, in the absence or presence of competing sortilin peptide. In another competition assay, we applied the unprocessed versions of pro-NGF (50 nm) and pro-BDNF (50 nm) as well as receptor-associated protein (RAP, 90 nm) to immobilized sortilin and measured the binding in absence or presence of 200 μm competing peptide.
Cellulose Membrane Preparation
We generated peptide libraries for all members of the VPS10p-domain receptor gene family or specific peptide variations in terms of substitution or length analyses of identified ligand binding peptides. A total of 2181 peptides was used for representation of the sortilin gene family, corresponding to 273 peptides for sortilin (accession code: CAA66904), 734 peptides for sorLA (accession code: NP_003096), 389 peptides for sorCS1 (accession code: NP_001013049), 382 peptides for sorCS2 (accession code: Q96PQ0), and 403 peptides for sorCS3 (accession code: CAI64579), with a 13-amino acid overlap between 16-mer peptides (35).
A cellulose support was prepared as an N-modified cellulose-aminohydroxylpropyl ether membrane, and all rounds of synthesis started with spot definition by 9-fluorenylmethoxycarbonyl-β-alanine-pentafluoophenyl ester that created an alanine linker between peptide and membrane. Then followed an automated linear synthesis of stepwise addition of the different amino acids protected at their N-terminal by 9-fluorenylmethoxycarbonyl and appropriate side-chain protection for the growing peptide chain. The pattern of de-protection, activation, and coupling continued until 16-mer peptides were produced, resulting in an equally distributed array of covalently anchored peptides to the cellulose support at their C-terminal ends with N-terminal free ends (details are given in Ref. 36). Removal of the side protection groups was done in two steps: First the membrane was treated with 90% trifluoroacetic acid (in dichlormethane, containing 3% triisobutylsilane and 2% H2O) and secondly with 60% trifluoroacetic acid (in dichlormethane, containing 3% triisobutylsilane and 2% H2O). To remove trifluoroacetic acid salts the membrane was washed several times with H2O, ethanol, Tris-buffered saline, and ethanol again, and finally dried. Finally, the membrane was blocked in blocking buffer from Sigma (catalog no. B6429) dilated in Tris-buffered saline (pH 8.0) and supplemented with 5% saccharose (Merck, catalog no. K32055087-422) for 2 h before the predefined peptide library is ready for ligand binding analysis.
Binding Studies of Cellulose-bound Peptides
The membrane-bound libraries were incubated with the combined S-peptide and polyhistidine-tagged pro-domains (10 μg/ml) in blocking buffer overnight at 4 °C, followed by a second incubation with 1 μg/ml of HRP-conjugated S-protein from Novagen (catalog no. 69047-3) also in blocking buffer but for 3 h at room temperature. Subsequently, the membrane was washed three times for 10 min with Tris-buffered saline before quantitative characterization of bound ligand was carried out using the UptiLight chemiluminescence substrate from Uptima (catalog no. UP99619A) and the LumiImager instrument from Roche Diagnostics, providing the spot signal intensities in Boehringer light units. Alternatively, detection of bound ligand was performed by an immunochemical assay with an antibody against the histidine tag from Sigma (catalog no. H-1029) and a secondary HRP-conjugated anti-mouse antibody from Calbiochem (catalog no. 401207). Incubations followed standard Western blotting procedures and spot detection as above.
The method of substitution analysis and length analysis to identify unique single amino acid residues and to determine the minimal peptide sequence, respectively, for binding HisS pro-domains to the sortilin peptide, followed similar protocols as for the initial testing of ligand binding to the spotted membrane.
Pulldown Assay
The expressed extracellular domains of sortilin-WT or sortilin-4A was incubated with either GST-tagged NGFpro or the propeptide of sortilin and precipitated using 100 μl of glutathione (GSH)-Sepharose beads (Amersham Biosciences, catalog no. 17-0756-01). The amount of applied receptor domains was determined by precipitation using Talon beads as control. Bound protein was separated by SDS-PAGE analysis and visualized using anti-histidine antibody by standard Western blotting.
Ligand Internalization by HEK 293 Cells
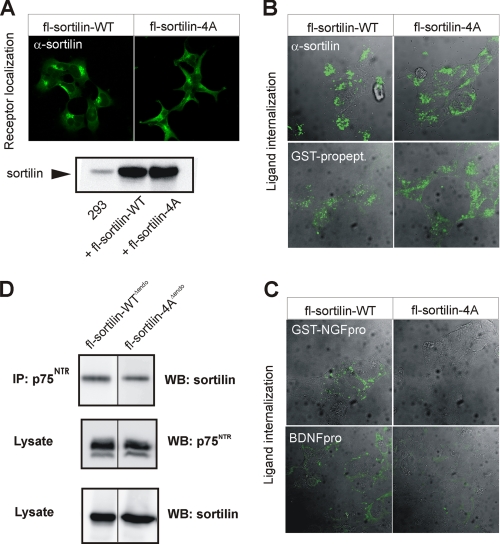
To study the effect of disrupting the identified binding site in full-length sortilin, HEK 293 cells were stably transfected with plasmids encoding either fl-sortilin-wt or fl-sortilin-4A. Receptor expression was confirmed by Western blotting using a sortilin-specific monoclonal antibody and by immunofluorescence. For immunofluorescence, cells were fixed in 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and subsequently incubated with rabbit anti-sortilin polyclonal antibodies followed by donkey anti-rabbit Alexa-488-conjugated secondary antibodies (1:300). Internalization of bound ligands was studied by the incubation of cells for 30 min at 37 °C with anti-sortilin polyclonal antibodies (10 μg/ml), GST-NGFpro (1 μg/ml), or 100 nm BDNF pro-domain (Alomone) diluted in culture medium. Internalized GST-NGFpro was visualized by the incubation with goat anti-GST (Amersham Biosciences, 1:300) followed by donkey anti-goat Alexa-488-conjugated secondary antibodies. Internalized BDNF pro-domain was visualized with rabbit polyclonal antibodies raised against the BDNF pro-domain (Alomone, 1:200).
p75NTR:Sortilin Co-immunoprecipitation
HEK 293 cells stably transfected with constructs encoding fl-sortilin-WTΔendo or fl-sortilin-4AΔendo were incubated in phosphate-buffered saline (with 1 mm CaCl2 and MgCl2) for 90 min at room temperature and then treated with 5 nm reducible protein cross-linker dithiobis(succinimidylpropionate) (Pierce) according to the manufacturer's instructions. After wash, cells were lysed on ice for 10 min in TNE buffer (20 mm Tris-HCl, pH 8.0, 1% Nonidet P-40, 10 mm EDTA) supplemented with complete protease inhibitor mixture. Samples were immunoprecipitated overnight at 4 °C by use of Gammabind G-Sepharose beads (Amersham Biosciences) coupled with anti-p75NTR (10494) from Abcam. Unspecific binding was removed by washing five times in Tris-buffered saline containing 0.05% Tween 20, and proteins were eluted by boiling samples in reducing sample buffer (20 mm dithioerythritol, 2.5% SDS). Protein samples were subjected to SDS-PAGE and Western blotted using anti-sortilin from BD Transduction Laboratories (catalog no. 612100) and anti-p75NTR.
Apoptosis Assay
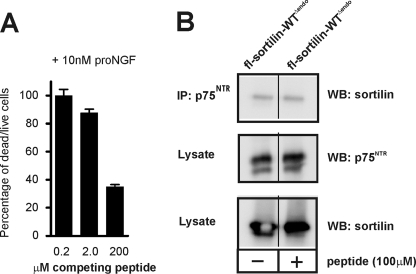
The effect of sortilin-derived peptides on pro-NGF-induced cell death was studied in RN22 schwannoma cells. Cells were cultured in Dulbecco's modified Eagle's medium added 10% fetal bovine serum, and seeded in 96-well plates at a density of 10,000 cells/well. After 24 h, the cells were washed twice in serum-free medium and incubated for 72 h in the presence of increasing concentrations of the sortilin-derived sort166–181 peptide diluted in Dulbecco's modified Eagle's medium without serum and phenol red, containing 30 nm selenium, 5 μg/ml insulin, 5 μg/ml transferrin, 30 nm triiodothyronine, and 10 nm pro-NGF (Chemicon). Dead and live cells were subsequently quantified using the MultiTox-Fluor Multiplex cytotoxicity assay (Promega, Madison, WI).
RESULTS
Identification of a Linear Pro-NT Binding Sequence in Sortilin
We first produced the pro-domains of NGF and BDNF as recombinant fusion proteins containing two N-terminal tags, a polyhistidine and an S-peptide tag, HisS-NGFpro and HisS-BDNFpro, respectively, to facilitate protein purification and detection of ligand binding to immobilized sortilin peptides (supplemental Fig. S1A). The S-tag system is based on the interaction of the 15-amino acid S-tag peptide with a 104-amino acid-long S-protein derived from pancreatic ribonuclease A (37). To confirm the receptor-binding properties of the recombinant pro-domains, we tested their binding to immobilized sortilin using surface plasmon resonance (SPR) analysis. The calculated affinities of HisS-NGFpro (KD ∼ 9 nm) and HisS-BDNFpro (KD ∼ 1.6 nm) were in accordance with past findings (supplemental Fig. S1B) (25, 27).
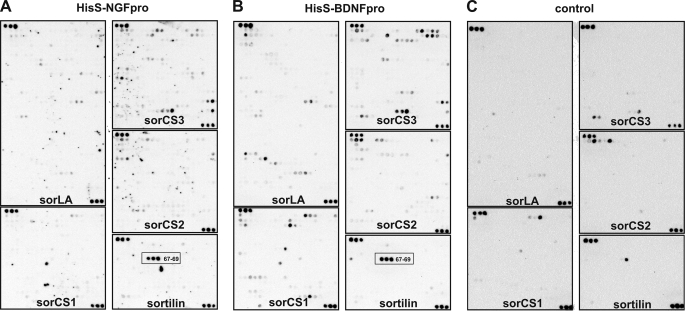
To screen for potential linear ligand binding sites in the VPS10p-domain receptors, we synthesized a peptide library in which each of the receptors were dissected into consecutive 16-mer peptides overlapping by 13 amino acids and subsequently spotted onto filters. Binding of HisS-NGFpro and HisS-BDNFpro was then tested for their abilities to interact with the receptor peptide libraries by a so-called SPOT binding analysis (38–40). A specific sequential triplet of signals was observed for peptides derived from sortilin, suggesting that this particular VPS10p-domain contains a linear binding epitope for the neurotrophin pro-domains. Interestingly, the HisS-NGFpro and HisS-BDNFpro gave virtually identically results, suggesting that they may share a common (linear) binding site (Fig. 1).
FIGURE 1.
Analysis of the NGF and the BDNF pro-domain binding to the mammalian VPS10p receptors by SPOT analysis. A peptide library containing a total of 2181 peptides represented by 734 from sorLA, 403 from sorCS3, 389 from sorCS1, 382 from sorCS2, and 273 from sortilin as overlapping 16-mer peptides of the five human VPS10p-domain-containing receptors sortilin (CAA66904, 831 aa), sorLA (AAC50891, 2214 aa), sorCS1(AAM43811, 1179 aa), sorCS2 (Q96PQ0, 1159 aa), and sorCS3(Q9UPU3, 1222 aa). The membrane was incubated either in the presence of 10 μg/ml HisS-NGFpro (A), 10 μg/ml HisS-BDNFpro (B), or in the absence of ligand (C). Detection was carried out by incubation of the membrane with an HRP-conjugate of S-protein that recognizes the S-peptide tag of bound NT pro-domains. Each receptor library is cornered at the upper left and lower right corner by three control peptides equivalent to the S-peptide tag (amino acid sequence KETAAAKFERQHMS). The specific binding site for both HisS-NGFpro and HisS-BDNFpro (A and B), which is not seen for the control experiment in absence of ligand (C), is indicated for the three sortilin peptides (SPOT numbers 67–69; red box).
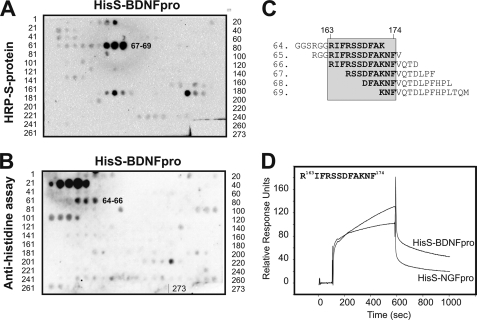
We focused the subsequent studies on sortilin and prepared a second peptide array comprising 273 immobilized sortilin-derived peptides. In this set of experiments we took advantage of both tags present in the HisS-BDNFpro ligand as we used either a peroxidase-conjugate of the S-protein or an immunoassay with an antibody against the polyhistidine tag for detection. The analysis of the two assays gave slightly different, but overlapping, results. Whereas detection with S-protein resulted in strong positive signals for peptides 67–69, spanning sortilin residues 166–187 (sort166–187) (Fig. 2, A and C), the immunoassay identified peptides 64–66, comprising residues 157–178, as the ligand binding site (Fig. 2, B and C). The shift in signal between the two detection methods likely represents different sterical hindrance from the S-protein and anti-polyhistidine antibody. We considered the signal corresponding to peptides 22–26 in the anti-histidine blot unspecific, because a similar spot pattern was observed on membranes incubated with the secondary antibody alone (data not shown). Collectively, these experiments suggest that amino acids 163–174 in sortilin, corresponding to the overlapping residues in the peptide sequences, constitute a binding epitope for pro-NT (Fig. 2C).
FIGURE 2.
Identification of a linear pro-NT binding site within sortilin. A and B, sortilin represented as 273 overlapping peptides generated by SPOT synthesis on a cellulose membrane with peptides 64–69 binding to HisS-BDNFpro, as detected by either HRP-S-protein (A, SPOT numbers 67–69) or antibody immunoassay against the polyhistidine tag (B, SPOT numbers 64–66). C, peptide sequences of the six peptides 64–69 found to interact with the pro-domains of BDNF and NGF. Residues corresponding to the sortilin fragment 163–174 are boxed. D, SPR analysis of the 12-mer sortilin-derived peptide (residues 163RIFRSSDFAKNF174) binding to flow cells with immobilized HisS-BDNFpro and HisS-NGFpro.
To confirm binding of HisS-NGFpro and HisS-BDNFpro to sort163–174, we synthesized a peptide comprising these 12 amino acid residues. The peptide was subsequently subjected to SPR analysis (BIAcore) using a Sensorchip coupled with HisS-NGFpro and HisS-BDNFpro, thereby reversing the system from immobilized peptides to peptides in solution. We found that the higher affinity of the BDNF pro-domain as compared with the NGF pro-domain for sortilin (supplemental Fig. S1B and Refs. 25, 27) could be replicated when using the sortilin peptide. This suggests that the linear ligand recognition sequence sort163–174 may account for the difference in binding properties between pro-BDNF and pro-NGF (Fig. 2D).
Identification of Key Residues in Sortilin for Pro-domain Binding
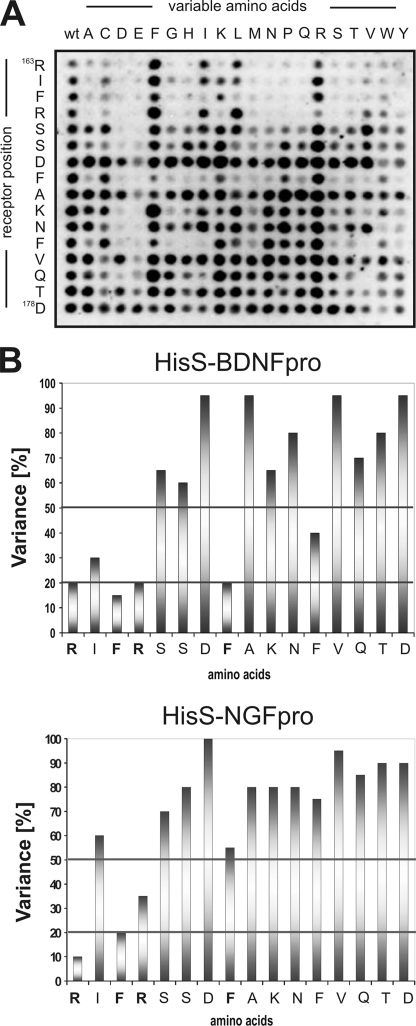
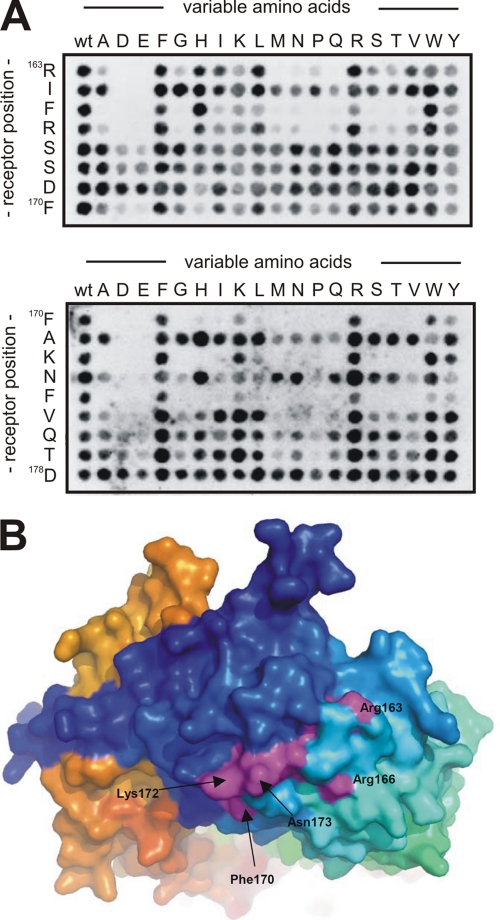
Using substitutional analysis we next set out to map key residues in the linear sortilin sequence that contribute to pro-neurotrophin binding. A variant of the SPOT synthesis method was applied to generate a new library in which every residue in the sequence 163RIFRSSDFAKNFVQTD178 had been replaced by all 20 naturally occurring l-amino acid residues. The SPOT filters were then incubated with ligand, as exemplified by HisS-BDNFpro (Fig. 3A), and the intensity of the binding signals were quantified and used to calculate the replacement variability for each amino acid. Incubation with HisS-BDNFpro and HisS-NGFpro showed that both ligands were critically dependent on amino acids Arg163, Phe165, and Arg166 inasmuch as the variance was low, i.e. within 10–35% (Fig. 3, B and C). Interestingly, while the variance of residues Phe170 and Phe174 amounted to 60–75% for HisS-NGFpro, indicating little impact on binding, the same amino acids were noticeable more important for the interaction between HisS-BDNFpro and the sortilin peptide as reflected by a variance between 30 and 35%.
FIGURE 3.
Substitution analysis of the linear sortilin sequence. A, a representative substitutional binding analysis of HisS-BDNFpro to peptides with the wild-type sortilin sequence 163RIFRSSDFAKNFVQTD178 listed to the left on the membrane and detection using the anti-histidine immunoassay. B, binding assays as demonstrated in panel A were carried out for both HisS-BDNFpro and HisS-NGFpro, and signal intensities, measured in Boehringer Light Units (BLU), were quantified for each peptide. The percentage of replacement variability (V) of each sequence position have been calculated (V = BLU/20 × 100%) and plotted against each amino acid within the peptide (residues 163–178).
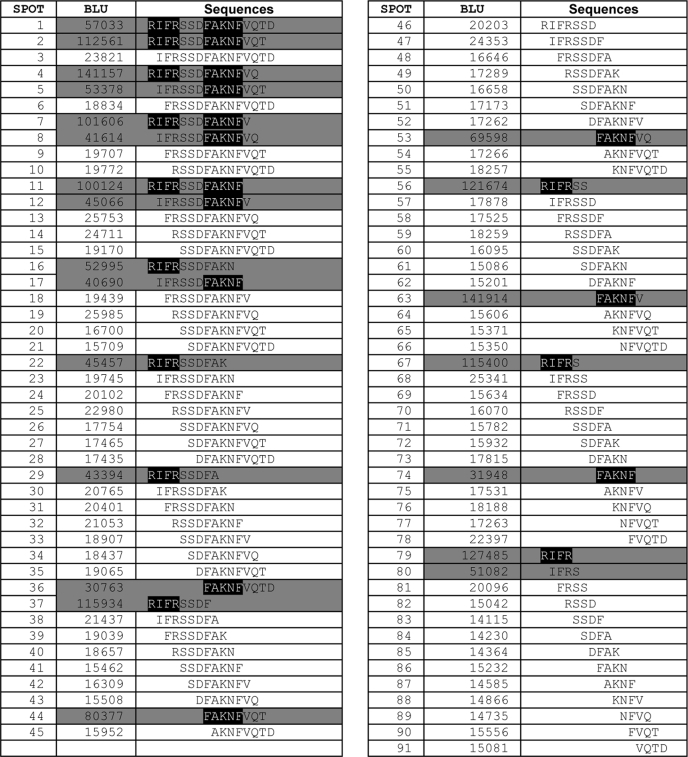
To determine the minimal linear sortilin sequence capable of binding the pro-NT pro-domains we carried out a so-called length analysis. In practice, we synthesized the peptide sort163–178 and removed consecutive single residues from the N- and/or C-terminal ends of the peptide (Table 1). Using this assay we narrowed down the length of the binding epitope to a central region of the sequence corresponding to the 12-mer fragment 163RIFRSSDFAKNF174. This result is in accordance with the substitution analysis, which also showed that residues 175VQTD178 do not significantly influence ligand binding (Fig. 3B). Interestingly, by further trimming the peptide we found that both halves of this fragment corresponding to the tetramer 163RIFR166 and the pentamer 170FAKNF174, respectively, were able to associate independently with HisS-BDNFpro (Table 1) and HisS-NGFpro (data not shown).
TABLE 1.
Length analysis for trimming the 163RIFRSSDFAKNFVQTD178 peptide
The quantification of a representative experiment testing the influence of the length of the sort163–178 peptide binding to HisS-BDNFpro in a modified SPOT analysis. Peptides with a high ligand affinity are shown on a grey background. For this presentation, an intensity threshold for the signal intensity of 30,000 BLU have been chosen as indication for high affinity. The presence of the two identified minor binding sites 163RIFR166 and 170FAKNF174 is presented on a black background.
This observation prompted us to repeat the substitution analysis, but this time with peptide libraries of 163RIFRSSDF170 and 170FAKNFVQTD178, because each of these fragments contains one of the two putative binding epitopes. Using this approach we confirmed that an exchange of Arg163, Phe165, or Arg166 within the N-terminal half and Phe170, Lys172, or Phe174 within the C-terminal half significantly decreased binding of the HisS-BDNFpro ligand (Fig. 4A). Similar results were obtained for HisS-NGFpro (data not shown), further corroborating the presence of a shared binding site in sortilin for both pro-NTs. In further support of this notion, binding of HisS-BDNFpro to immobilized sortilin, as determined by SPR analysis, was completely prevented by preincubation with saturating concentrations of HisS-NGFpro (supplemental Fig. S2). Collectively, the above data suggest the existence of a common binding epitope comprising two interaction sites in which three residues in the second epitope may account for the stronger binding of HisS-BDNFpro as compared with HisS-NGFpro (Fig. 4B).
FIGURE 4.
Mutational binding analysis of HisS-pro-BDNF to short sortilin fragments. A, HisS-BDNFpro binding analysis to peptides with the wild-type sortilin sequences 163RIFRSSDF170 and 170FAKNFVQTD178 listed to the left on each membrane. Binding to mutant peptides where each amino acid has been substituted with the 20 naturally occurring amino acids is used for identification of specific residues important for interaction with the immature part of pro-BDNF. The substitution analysis clearly identifies the two sequences 163RIFR166 (upper part) and 170FAKNF174 (lower part) as specific binding sites for HisS-BDNFpro. B, crystal structure presentation of sortilin showing the surface exposure of side chains from several identified key residues in purple (e.g. Arg163, Arg166, Phe170, and Lys172). Structure was drawn from coordinates present in the Protein Data Bank with accession code 3F6K.
Ligand Binding to Sortilin-4A
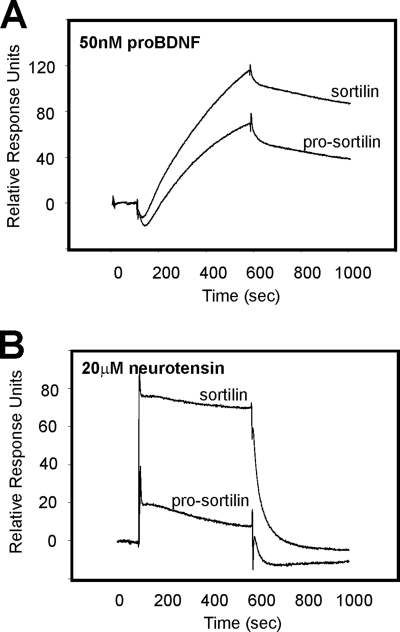
Sortilin is produced with a propeptide that needs to be cleaved off to allow entrance of ligands into the tunnel (41). We therefore expressed a non-cleavable soluble sortilin variant as described (41), immobilized it on a biosensor chip, and compared binding of unprocessed pro-BDNF and neurotensin to the mutant with that of the fully processed wild-type receptor. We found that both receptors bound pro-BDNF (Fig. 5A), in agreement with the existence of a binding site located away from the blocked central cavity. For comparison, binding of neurotensin was completely abolished in the non-processed sortilin variant (Fig. 5B), demonstrating that binding of this ligand entirely depends on access to the tunnel epitope.
FIGURE 5.
Differential binding of pro-BDNF and neurotension to sortilin and pro-sortilin. A, SPR analysis showing that unprocessed pro-BDNF (50 nm) binds nearly as efficiently to the receptor in the presence (pro-sortilin) as in the absence (sortilin) of the receptor propeptide. Pro-sortilin is a receptor variant unable to cleave off the propeptide due to mutation of the furin recognition site. B, binding of neurotensin (20 μm) is strongly ablated for pro-sortilin as compared with binding to sortilin without the presence of its propeptide.
To determine the impact of the linear pro-NT binding epitope in the full-length VPS10p-domain, we expressed in EBNA 293 cells a set of single residue alanine substitution mutants (R163A, F165A, R166A, F170A, K172A, and F174A) of the sortilin ectodomain. As a control, we included the mutation R160A, a residue outside the ligand-binding peptide sequence. All mutants were effectively secreted from the cells in similar quantities and showed a migration pattern in non-reducing gel analysis identical to that of the non-mutated sortilin domain, suggesting correct disulfide bridging of the mutants (supplemental Fig. S3). To further demonstrate the intact structure of the VPS10p β-propeller, we determined the CD spectra for each variant and found that none of the mutations resulted in aberrant folding of sortilin (supplemental Fig. S3). Binding of GST-tagged NGFpro was subsequently tested using SPR analysis with each receptor mutant immobilized on individual flow cells at similar protein densities, thus allowing for direct comparison of bound ligand. However, none of the single residue mutants displayed any substantial reduction in ligand affinity (maximally 2- to 3-fold reduced binding) as deduced by fitting a series of ligand concentration sensorgrams (supplemental Fig. S3B and Table 2).
TABLE 2.
Binding kinetics of GST-NGFpro binding to sortilin mutants
Concentration series of 10, 20, 30, 40, and 50 nm of GST-NGFpro was applied to biosensor chips with immobilized sortilin single residue mutants. Affinities and rate constants were estimated using the BIAevaluation software, using the 1:1 Langmuir binding isotherm.
| Mutation | Amino acids 160–174 | 10 nm to 50 nm series of sensorgrams |
||
|---|---|---|---|---|
| ka | kd | KD | ||
| s−1 | m s−1 | |||
| WT | RGGRIFRSSDFAKNF | 1.72 × 105 | 3.45 × 10−4 | 2.12 nM |
| R160A | AGGRIFRSSDFAKNF | 1.47 × 105 | 2.27 × 10−4 | 1.89 nM |
| R163A | RGGAIFRSSDFAKNF | 1.85 × 105 | 4.21 × 10−4 | 2.70 nM |
| F165A | RGGRIARSSDFAKNF | 1.10 × 105 | 4.22 × 10−4 | 3.86 nM |
| R166A | RGGRIFASSDFAKNF | 6.48 × 104 | 2.85 × 10−4 | 4.38 nM |
| F170A | RGGRIFRSSDAAKNF | 1.30 × 105 | 5.91 × 10−4 | 5.59 nM |
| K172A | RGGRIFRSSDFAANF | 1.01 × 105 | 3.46 × 10−4 | 3.91 nM |
| F174A | RGGRIFRSSDFAKNA | 2.35 × 105 | 1.03 × 10−3 | 4.27 nM |
Based on the existence of two independent motives within the linear binding site (cf. Table 1) we speculated that failure of the single residue mutants to show reduced GST-NGFpro affinity was accounted for by binding to the non-mutated residues in the second site. We therefore hypothesized that disrupting an entire motif in combination with substitution of a single critical residue from the adjacent motif might be required to efficiently abolish ligand binding. To address this premise, we mutated the four most important residues within the 163RIFRSSDFAKNF174 motif (cf. Fig. 4) thereby generating a construct with a R163A,F165A,R166A,F170A quadruple mutation, denoted sortilin-4A. This receptor mutant was also efficiently expressed by EBNA 293 cells, able to be purified using Ni2+-affinity chromatography, and folded properly as demonstrated by CD analysis (supplemental Fig. S3C).
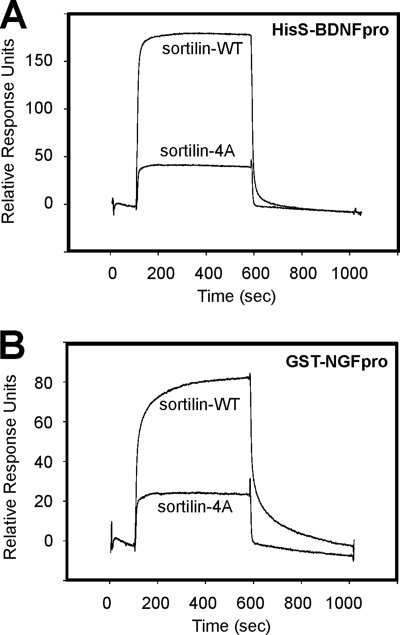
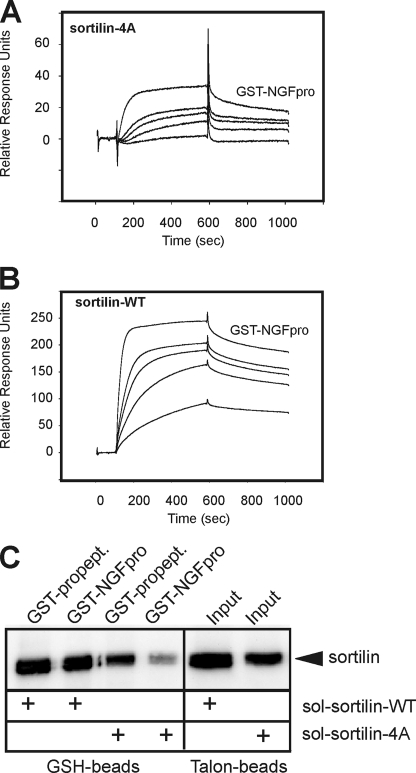
The interaction of sortilin-4A with NT pro-domains was studied by SPR analysis and using a ligand precipitation assay. HisS-BDNFpro and GST-NGFpro were immobilized on the sensor chip, and equal amounts (100 nm) of wild-type sortilin (sortilin-WT) and sortilin-4A were applied. We found that HisS-BDNFpro and GST-NGFpro exhibited considerable stronger binding to sortilin-WT as compared with sortilin-4A (Fig. 6, A and B). In another set of SPR experiments, where sortilin-WT and sortilin-4A were the immobilized parts, we found that GST-NGFpro bound to the mutant receptor with a >10-fold lower affinity, i.e. KD ∼26 nm versus ∼2 nm, corroborating an important role of the linear binding site also in the VPS10p holo-domain (Fig. 7, A and B). A similar decrease in GST-NGFpro binding to sortilin-4A was revealed when comparing the ability of GST-NGFpro to precipitate wild-type and mutant sortilin in solution. In practice, the two VPS10p-domains were incubated with GST-NGFpro, allowed to form complexes, and then precipitated by glutathione beads. The precipitates were resolved by SDS-PAGE analysis, and bound sortilin was subsequently detected by Western blotting (Fig. 7C). In line with the SPR analysis the mutated sortilin domain exhibited considerably less interaction with GST-NGFpro. To assure that binding of ligands that engage the tunnel of the β-propeller far away from the site of the quadruple mutation was unperturbed, we repeated the experiment using the GST-tagged sortilin propeptide as a ligand (33). Importantly, this ligand efficiently precipitated sortilin-4A, confirming that the mutated domain is capable of interacting with other ligands independent of the linear binding site.
FIGURE 6.
Mutation of the linear binding site specifically impairs binding of both the NGF and the BDNF pro-domains. SPR analysis showing reduced binding of equal amounts (analyte concentration: 200 nm) of the soluble extracellular domains of sortilin-4A compared with sortilin-WT when binding is tested to both the immobilized BDNF pro-domain (HisS-BDNFpro, A) and the immobilized NGF pro-domain (GST-NGFpro, B).
FIGURE 7.
GST-NGFpro binding to sortilin-WT and sortilin-4A. SPR analysis showing concentration series of GST-NGFpro (10, 20, 30, 40, and 50 nm) tested for binding to immobilized extracellular domains of sortilin-4A (A) and sortilin-WT (B), demonstrating a strong decrease in the binding capacity for the NGF pro-domain upon the quadruple mutation in sortilin-4A. A higher than 10-fold decrease in affinity (WT: KD ∼2 nm versus 4A: KD ∼26 nm) was estimated using the BIAevaluation software. C, medium from EBNA 293 cells producing either the soluble ectodomain of sortilin-WT or sortilin-4A were incubated with a GST-tagged variant of the receptor propeptide (GST-propept) or GST-NGFpro and precipitated by glutathione (GSH) beads. The amount of total secreted sortilin-WT and sortilin-4A (i.e. input) was determined by precipitation using Talon beads binding to the histidine tag within the sortilin domains. The precipitated proteins were subjected to SDS-PAGE analysis and visualized by Western blot analysis for sortilin.
To provide evidence for a functional role for the linear pro-NT binding site in the full-length and membrane-associated receptor, we expressed the entire wild-type and mutated sortilin receptors, designated fl-sortilin-WT and fl-sortilin-4A, respectively, in HEK 293 cells. The mutated receptor was mainly localized in perinuclear vesicles in a pattern indistinguishable from that of wild-type sortilin (Fig. 8A). Identical expression levels of fl-sortilin-WT and -4A were documented by Western blot analysis of cell extracts (Fig. 8A). Normal internalization of an antibody against the sortilin extracellular domain confirmed that the mutant was also capable of endocytic uptake. Notably, the GST-propeptide of sortilin was equally well taken up by cells expressing wild-type and mutant receptor (Fig. 8B). However, whereas fl-sortilin-WT efficiently internalized the pro-domains of both NGF and BDNF, cells expressing fl-sortilin-4A did not (Fig. 8C). This demonstrates that the linear and surface-exposed pro-NT binding site is also functional in the full-length and membrane-anchored receptor.
FIGURE 8.
Decreased binding of pro-NT to sortilin-4A within cells. A, immunostaining of HEK 293 cells transfected with constructs for full-length (fl)-sortilin-WT or sortilin-4A with an antibody against sortilin. Protein expression levels tested by Western blot analysis of cell lysates. B and C, cells were labeled with ligands at 4 °C followed by endocytosis for 30 min at 37 °C before fixation and internalized ligand visualization by staining. Ligands applied were either a sortilin antibody (against the receptor extracellular domain; α-sortilin), the receptor propeptide (GST-propept) (B), GST-NGFpro, or BDNFpro (C). D, cells co-transfected with constructs for sortilin variants devoid of endocytosis (fl-sortilin-WTΔendo or fl-sortilin-4AΔendo) together with p75NTR were immunoprecipitation using an antibody against p75NTR. Lysates and precipitated proteins were subjected to SDS-PAGE and analyzed by Western blotting for either p75NTR or sortilin as indicated.
Because sortilin and p75NTR can physically associate in the plasma membrane (11, 25), likely by a mechanism that is independent of the β-propeller tunnel, we next examined if fl-sortilin-4A were impaired in p75NTR binding. To increase cell surface expression and the co-immunoprecipitation signal, we deleted a previously well described endocytosis signal (34) in the sortilin cytoplasmic tail of both receptors generating fl-sortilin-WTΔendo and fl-sortilin-4AΔendo, respectively. Interestingly, the two receptor variants precipitated p75NTR equally well, suggesting that hetero-oligomerization between sortilin and p75NTR is independent of the surface-exposed pro-NT binding epitope (Fig. 8D).
The Linear Sortilin Epitope Prevents Pro-NT Binding and Death Induction
The importance of the linear epitope for pro-NT binding was further substantiated in a competition assay where the pro-domains of BDNF and NGF were immobilized on a biosensor chip, and the interaction with saturating concentrations of 200 nm soluble sortilin-WT was measured in the absence or presence of a competing peptide spanning the ligand-binding sequence. A clear decrease in receptor-ligand complex formation was observed in the presence of peptide. Thus, the response for sortilin-WT binding to HisS-BDNFpro was lowered from 300 RU in the absence of competitor to 100 RU in the presence of 200 μm sort166–181 (Fig. 9A). Quantification of the equilibrium response at increasing amounts of peptide showed a concentration-dependent competition profile with the inhibition being stronger for the interaction of sortilin with HisS-BDNFpro than for HisS-NGFpro, in line with a higher affinity of the former pro-domain (Fig. 9B). Moreover, the peptide also blocked the interaction of pro-BDNF and pro-NGF with sortilin immobilized on the chip surface (Fig. 10, A and B). As a control for the specificity of the inhibitor, binding of sortilin to an unrelated ligand, the RAP (42, 43), was not affected by the sortilin peptide (Fig. 10C).
FIGURE 9.
Sortilin-derived peptide specifically competes binding of pro-NT to sortilin. A, recombinant sortilin was purified from 293 cells (indicated to the right by silver-stained SDS-PAGE analysis), and used for SPR studies to immobilized HisS-BDNFpro. The signal of 300 RU observed for binding in the absence of the peptide sort166–181 (− peptide) was significantly lowered to 100 RU in the presence of 200 μm linear sortilin antagonist (+ peptide). B, sortilin binding to either HisS-NGFpro or HisS-BDNFpro was determined by SRP analysis (as exemplified in A) and the inhibition by increasing amounts of sort166–181 (at 2, 20, and 200 μm) was plotted relative to the observed interaction in the absence of competitor (in percentage). Values represent the mean ± S.E. from three experiments. *, p = 0.0007; two-tailed Student's t test.
FIGURE 10.
Selective competition of ligands by sortilin-derived peptide antagonist. SPR binding analysis of 50 nm unprocessed pro-BDNF (A), 50 nm unprocessed pro-NGF (B), and 90 nm RAP (C) to immobilized sortilin in the absence and presence of the sort166–181 peptide (100 μm). Specific inhibition of pro-NGF and pro-BDNF is observed, whereas the interaction between sortilin and RAP is largely intact.
We finally asked if the sortilin-derived peptide was capable of blocking pro-NGF-induced apoptosis by the sortilin:p75NTR complex. To this end we incubated the schwannoma cell line RN22 with 10 nm pro-NGF in the absence or presence of the peptide sort166–181 and scored apoptosis after 72 h. Indeed, the peptide inhibited pro-NGF-induced cell death in a concentration-dependent manner with >60% reduction in the dead/live cell ratio at the highest peptide concentration used (Fig. 11A). Because reduced killing could also arise from a deficient physical interaction between sortilin and p75NTR, we also examined if peptide sort166–181 would interfere with receptor hetero-oligomerization. We observed no effect on the ability of p75NTR to precipitate sortilin in the presence of 100 μm of the peptide inhibitor (Fig. 11B). These data show that blocking the interaction with the linear epitope in sortilin is sufficient to specifically prevent binding of pro-NTs to sortilin, leaving ligand binding inside the tunnel as well as receptor hetero-oligomerization intact.
FIGURE 11.
A sortilin-derived peptide blocks pro-NGF-induced cell death. A, RN22 schwannoma cells were incubated in the presence of 10 nm pro-NGF and increasing concentrations (0.2, 2.0, and 200 μm) of the sort166–181 peptide. The amount of pro-NGF-induced cell death after 72 h were quantified using a fluorescence-based assay, and plotted against competitor concentrations as the ratio between dead and live cells in percentage. B, cells co-transfected with constructs for a sortilin variant devoid of endocytosis (fl-sortilin-WTΔendo) together with p75NTR were incubated in the absence or presence of 100 μm sort166–181 peptide, and immunoprecipitated using an antibody against p75NTR. Lysates and precipitated proteins were subjected to SDS-PAGE and analyzed by Western blotting for either p75NTR or sortilin as indicated.
In conclusion, we find that sortilin harbors a linear binding epitope that selectively engages pro-NTs. We propose that this sequence may be used to derive a peptide antagonist or small organic molecule that can prevent apoptosis.
DISCUSSION
Pro-neurotrophins were initially considered inert precursors that merely serve as a reservoir for their mature counterparts, but it is now well established that they also exhibit activities on their own (16, 23). Although mature NTs stimulate neuronal survival and differentiation by engaging their specific tyrosine-receptor kinases (Trks) in conjunction with p75NTR, pro-NTs can elicit apoptosis in cells expressing sortilin and p75NTR (1, 24, 44, 45). Although the signaling pathway has not been fully elucidated, it has been shown that the simultaneous binding of the pro-NT pro-domain to sortilin and the mature part to p75NTR induces formation of a tripartite death-inducing complex capable of activating the Jun-kinase signaling pathway (46–49). We here identified a linear and surface-exposed amino acid sequence in sortilin that partakes in pro-NT binding and plays a pivotal role in death induction by pro-NTs.
The molecular structure of the sortilin ectodomain in complex with the neuropeptide neurotensin was recently solved at atomic resolution. The structure revealed an unprecedented 10-bladed β-propeller encircling a large tunnel that encompasses the binding site for neurotensin, the sortilin propeptide, and RAP (33, 41, 50). Intriguingly, we found that the surface of the receptor may engage in binding of pro-NTs. First, the soluble sortilin peptide sort163–174 bound to immobilized NT pro-domains and vice versa. Second, this interaction critically depended on the surface-exposed amino acids Arg163, Phe165, and Arg166 for both NGFpro and BDNFpro, whereas Phe170 and Phe174 also contributed to the binding of the BDNF pro-domain, an observation that may explain the higher affinity of sortilin for pro-BDNF than pro-NGF (Fig. 2D and supplemental Fig. S1) (25, 27). Third, mutated sortilin harboring alanine substitutions in the surface-exposed residues Arg163, Phe165, Arg166, and Phe170 showed only little interaction with pro-NTs as determined by SPR analysis, co-immunoprecipitation, and cellular uptake studies, whereas binding of the sortilin propeptide was unperturbed. Fourth, the peptide sort166–181 competitively inhibited sortilin binding of pro-NTs, but not of p75NTR, which likely engages an alternative surface epitope on the β-propeller, nor RAP or the sortilin propeptide that selectively target the tunnel. In agreement, the sortilin peptide was a potent inhibitor of pro-NT induced apoptosis in RN22 cells.
In the SPOT analysis, application of the two detection methods gave similar, though not identical, results confirming the specificity of the binding between the receptor peptides and the NT pro-domains. Although HisS-NGFpro and HisS-BDNFpro binding to the sortilin peptides gave rise to the same spots when both were assayed with the S-protein (peptides 67–69) (Fig. 1), we saw a consistent shift toward earlier sequences (peptides 64–66) when developed with the anti-histidine antibody (Fig. 2). We speculate this difference could be accounted for by sterical hindrance of the detection system from the solid support. Thus, the S-protein is only 104 residues, whereas the anti-histidine immunoassay requires binding of two large antibodies, each roughly of 1500 amino acids.
Interestingly, the identification of two positively (Arg163 and Arg166) but no negatively charged residues in the sortilin epitope suggests that electrostatic interactions may contribute to the binding between the receptor and pro-NTs. In agreement with this model, the BDNF pro-domain contains 17 acidic residues many of which are located in a region suggested to be vital for the interaction with sortilin (51).
Sequence analysis of sortilin predicted the presence of several Asp-box motives throughout the VPS10p-domain (52). Indeed, x-ray analysis of sortilin revealed that Asp-boxes constitute a well defined structure that folds into a surface-exposed hairpin-loop bridging strands 3 and 4 in each of the 10 β-blades of the propeller (33). The Asp-box consensus sequence can be expressed as X-X-Ser-X-Asp-X-Gly-X-Thr-Trp/Phe-X, where X represents any amino acid (53). Interestingly, this motif, 163RIFRSSDFAKNF174, is part of the identified pro-NT binding in blade 2 of the receptor. Asp-boxes normally serve a structural function in β-propellers, where the conserved residues participate in folding and ensure the stability of the domain (54). The variable residues are free to carry out other functions, e.g. ligand binding as observed for nucleotide binding to microbial ribonucleases (53), in line with our finding that these amino acids are involved in binding of the NT pro-domain. It is noteworthy that, although most β-propellers bind their ligands at the top face, as is also the case for neurotensin and the sortilin propeptide (33), binding to the bottom face or side domain has also been reported (55–57).
Our data do not exclude that NGFpro and BDNFpro may also target the epitope in the tunnel of the β-propeller. Although the residues identified in the current study are exposed on the rim and outer surface of the barrel, it is possible that the >100-amino acid-long NT pro-domains simultaneously can associate with additional binding epitopes, including the tunnel. This would be in accordance with the suggested highly flexible and extended conformation of the pro-NGF pro-domain (58, 59) and in line with the recently reported crystal structure of the pro-NGF:p75NTR complex that shows the pro-domain to accommodate an exposed and disordered conformation free to interact with sortilin (49).
By SPOT peptide mapping we only identified one linear binding site in sortilin comprising amino acids 163–174. We believe this fact is due to the tunnel surface binding site for neurotensin being comprised of residues located far away from each other in the primary sequence, e.g. Lys227, Ser283, Arg292, and Tyr318 (33). These residues are separated by up to 90 amino acids within the primary structure thereby representing a discontinuous binding epitope not suitable for detection by a peptide mapping approach.
There are experimental data to support as well to refute an additional pro-NT binding epitope in the tunnel. We found that, following disruption of the four most critical amino acids (Arg163, Phe165, Arg166, and Phe170), for the interaction with pro-NT there was an ∼25% residual binding still present, arguing for one or more additional interaction sites (Fig. 6). On the contrary, mutations in residue Ser283, which is indispensable for binding of the sortilin propeptide inside the tunnel, completely abrogated binding of this peptide while pro-NGF binding was unperturbed (33). Thus the experimental evidence in support of a composite pro-NT binding site, that overlaps with that of the sortilin propeptide and neurotensin in the tunnel, is contradictory. However, it is possible that pro-NT might engage the tunnel via an epitope in the vicinity of but independent of amino acid Ser283. This is supported by the observation that binding of the pro-NGF pro-domain is significantly better inhibited by the 13-amino acid-long full-length neurotensin than by residues 11–13, which constitute the sortilin binding site in the neuropeptide (33). These data may suggest that neurotensin competes with pro-NGF for binding due to steric hindrance in the tunnel. Collectively, the above data underscore the requirement of elucidating the atomic structure of the sortilin:pro-NT complex to precisely and unanimously identify the number and location of all binding epitopes.
Recent evidence has suggested that pro-NT-induced cell deaths play an important role in conditions characterized by apoptosis such as aging, and in pathological conditions, including seizures, spinal cord injury, retinal dystrophy, and Alzheimer disease (15, 18, 24, 28, 29, 60–65). In accordance, recent reports have described changes in the levels of pro-NTs in many of these conditions, including early stages of Alzheimer disease (60, 66–68). Of particular interest, increased glycation and lipoxidation of pro-NGF in the hippocampus and entorhinal cortex of Alzheimer disease patients makes it less susceptible to processing into mature NGF. Furthermore, in these patients TrkA expression is decreased, whereas the expression of p75NTR and sortilin are unchanged (69, 70).4 Possibly, an imbalance between the relationship of receptor expression and pro- versus mature NGF changes the equilibrium between survival and apoptosis signaling in favor of the latter (71, 72). Prompted by the biological significance of pro-NTs in cell death, we tested if the ligand-binding peptide of sortilin might by capable of preventing apoptosis. Indeed, we found that the peptide offered efficient protection against pro-NGF-induced apoptosis in cultured RN22 schwannoma cells, a finding that holds promise for the idea that an antagonist that can prevent pro-NGF binding to sortilin might be an efficient target to prevent pro-apoptotic conditions.
The peptide used in the present work is unlikely to be suitable for therapeutic use due to its modest affinity and accordingly high concentrations required to inhibit pro-NT binding. Yet, the peptide may serve as a lead for rational drug design aimed at increasing the binding affinity for NT pro-domains as well as to enhance the pharmacological properties in terms of solubility, stability, and efficacy. Ideally, such a compound should specifically prevent pro-NT-induced apoptosis, leaving the trophic actions of mature neurotrophins unchanged.
Supplementary Material
Acknowledgments
We thank Peder Madsen for the neurotrophin cDNAs, Lars Sottrup-Jensen for help doing the CD analysis, and Claus M. Petersen for valuable discussions throughout the preparation of the manuscript. Søren Thirup is greatly acknowledged for preparing the picture in Fig. 4B. Anja Aagaard, Anne Marie Bundsgaard, and Marit Nyholm Nielsen are thanked for excellent technical support.
This work was supported in part by grants from the Danish Medical Research Council and the Lundbeck Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
Mufson, E. J., Wuu, J., Counts, S. E., and Nykjaer, A. (2010) Neurosci. Lett. 473, 129–133.
- NT
- neurotrophin
- pro-NT
- pro-neurotrophin
- NGF
- nerve growth factor
- BDNF
- brain-derived neurotrophic factor
- RAP
- receptor-associated protein
- SPR
- surface plasmon resonance
- VPS10p
- vacuolar protein sorting 10-protein
- RU
- response unit(s)
- HRP
- horseradish peroxidase
- GST
- glutathione S-transferase
- SPR
- surface plasmon resonance
- wt
- wild type
- Trk
- tyrosine-receptor kinase
- aa
- amino acid(s).
REFERENCES
- 1.Willnow T. E., Petersen C. M., Nykjaer A. (2008) Nat. Rev. Neurosci. 9, 899–909 [DOI] [PubMed] [Google Scholar]
- 2.Andersen O. M., Reiche J., Schmidt V., Gotthardt M., Spoelgen R., Behlke J., von Arnim C. A., Breiderhoff T., Jansen P., Wu X., Bales K. R., Cappai R., Masters C. L., Gliemann J., Mufson E. J., Hyman B. T., Paul S. M., Nykjaer A., Willnow T. E. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13461–13466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rogaeva E., Meng Y., Lee J. H., Gu Y., Kawarai T., Zou F., Katayama T., Baldwin C. T., Cheng R., Hasegawa H., Chen F., Shibata N., Lunetta K. L., Pardossi-Piquard R., Bohm C., Wakutani Y., Cupples L. A., Cuenco K. T., Green R. C., Pinessi L., Rainero I., Sorbi S., Bruni A., Duara R., Friedland R. P., Inzelberg R., Hampe W., Bujo H., Song Y. Q., Andersen O. M., Willnow T. E., Graff-Radford N., Petersen R. C., Dickson D., Der S. D., Fraser P. E., Schmitt-Ulms G., Younkin S., Mayeux R., Farrer L. A., St George-Hyslop P. (2007) Nat. Genet. 39, 168–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scherzer C. R., Offe K., Gearing M., Rees H. D., Fang G., Heilman C. J., Schaller C., Levey A. I., Lah J. J. (2004) Arch. Neurol. 61, 1200–1205 [DOI] [PubMed] [Google Scholar]
- 5.Clee S. M., Yandell B. S., Schueler K. M., Rabaglia M. E., Richards O. C., Raines S. M., Kabara E. A., Klass D. M., Mui E. T., Stapleton D. S., Gray-Keller M. P., Young M. B., Stoehr J. P., Lan H., Boronenkov I., Raess P. W., Flowers M. T., Attie A. D. (2006) Nat. Genet. 38, 688–693 [DOI] [PubMed] [Google Scholar]
- 6.Baum A. E., Akula N., Cabanero M., Cardona I., Corona W., Klemens B., Schulze T. G., Cichon S., Rietschel M., Nöthen M. M., Georgi A., Schumacher J., Schwarz M., Abou Jamra R., Höfels S., Propping P., Satagopan J., Detera-Wadleigh S. D., Hardy J., McMahon F. J. (2008) Mol. Psychiatry 13, 197–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewin G. R., Barde Y. A. (1996) Annu. Rev. Neurosci. 19, 289–317 [DOI] [PubMed] [Google Scholar]
- 8.Snider W. D. (1994) Cell 77, 627–638 [DOI] [PubMed] [Google Scholar]
- 9.Huang E. J., Reichardt L. F. (2001) Annu. Rev. Neurosci. 24, 677–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies A. M. (2003) EMBO J. 22, 2537–2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teng K. K., Hempstead B. L. (2004) Cell Mol. Life Sci. 61, 35–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee R., Kermani P., Teng K. K., Hempstead B. L. (2001) Science 294, 1945–1948 [DOI] [PubMed] [Google Scholar]
- 13.Chao M. V., Bothwell M. (2002) Neuron 33, 9–12 [DOI] [PubMed] [Google Scholar]
- 14.Yang J., Siao C. J., Nagappan G., Marinic T., Jing D., McGrath K., Chen Z. Y., Mark W., Tessarollo L., Lee F. S., Lu B., Hempstead B. L. (2009) Nat. Neurosci. 12, 113–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beattie M. S., Harrington A. W., Lee R., Kim J. Y., Boyce S. L., Longo F. M., Bresnahan J. C., Hempstead B. L., Yoon S. O. (2002) Neuron 36, 375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chao M. V. (2003) Nat. Rev. Neurosci. 4, 299–309 [DOI] [PubMed] [Google Scholar]
- 17.Lu B. (2003) Neuron 39, 735–738 [DOI] [PubMed] [Google Scholar]
- 18.Volosin M., Song W., Almeida R. D., Kaplan D. R., Hempstead B. L., Friedman W. J. (2006) J. Neurosci. 26, 7756–7766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pagadala P. C., Dvorak L. A., Neet K. E. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17939–17943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Domeniconi M., Hempstead B. L., Chao M. V. (2007) Mol. Cell Neurosci. 34, 271–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pang P. T., Teng H. K., Zaitsev E., Woo N. T., Sakata K., Zhen S., Teng K. K., Yung W. H., Hempstead B. L., Lu B. (2004) Science 306, 487–491 [DOI] [PubMed] [Google Scholar]
- 22.Woo N. H., Teng H. K., Siao C. J., Chiaruttini C., Pang P. T., Milner T. A., Hempstead B. L., Lu B. (2005) Nat. Neurosci. 8, 1069–1077 [DOI] [PubMed] [Google Scholar]
- 23.Lu B., Pang P. T., Woo N. H. (2005) Nat. Rev. Neurosci. 6, 603–614 [DOI] [PubMed] [Google Scholar]
- 24.Nykjaer A., Willnow T. E., Petersen C. M. (2005) Curr. Opin. Neurobiol. 15, 49–57 [DOI] [PubMed] [Google Scholar]
- 25.Nykjaer A., Lee R., Teng K. K., Jansen P., Madsen P., Nielsen M. S., Jacobsen C., Kliemannel M., Schwarz E., Willnow T. E., Hempstead B. L., Petersen C. M. (2004) Nature 427, 843–848 [DOI] [PubMed] [Google Scholar]
- 26.Harrington A. W., Leiner B., Blechschmitt C., Arevalo J. C., Lee R., Mörl K., Meyer M., Hempstead B. L., Yoon S. O., Giehl K. M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6226–6230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teng H. K., Teng K. K., Lee R., Wright S., Tevar S., Almeida R. D., Kermani P., Torkin R., Chen Z. Y., Lee F. S., Kraemer R. T., Nykjaer A., Hempstead B. L. (2005) J. Neurosci. 25, 5455–5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jansen P., Giehl K., Nyengaard J. R., Teng K., Lioubinski O., Sjoegaard S. S., Breiderhoff T., Gotthardt M., Lin F., Eilers A., Petersen C. M., Lewin G. R., Hempstead B. L., Willnow T. E., Nykjaer A. (2007) Nat. Neurosci. 10, 1449–1457 [DOI] [PubMed] [Google Scholar]
- 29.Volosin M., Trotter C., Cragnolini A., Kenchappa R. S., Light M., Hempstead B. L., Carter B. D., Friedman W. J. (2008) J. Neurosci. 28, 9870–9879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan Y. J., Wu L. L., Li H. Y., Wang Y. J., Zhou X. F. (2008) Eur. J. Neurosci. 27, 2380–2390 [DOI] [PubMed] [Google Scholar]
- 31.Al-Shawi R., Hafner A., Olsen J., Chun S., Raza S., Thrasivoulou C., Lovestone S., Killick R., Simons P., Cowen T. (2008) Eur. J. Neurosci. 27, 2103–2114 [DOI] [PubMed] [Google Scholar]
- 32.Yano H., Torkin R., Martin L. A., Chao M. V., Teng K. K. (2009) J. Neurosci. 29, 14790–14802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quistgaard E. M., Madsen P., Grøftehauge M. K., Nissen P., Petersen C. M., Thirup S. S. (2009) Nat. Struct. Mol. Biol. 16, 96–98 [DOI] [PubMed] [Google Scholar]
- 34.Nielsen M. S., Madsen P., Christensen E. I., Nykjaer A., Gliemann J., Kasper D., Pohlmann R., Petersen C. M. (2001) EMBO J. 20, 2180–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frank R. (1992) Tetrahedron 48, 9217–9232 [Google Scholar]
- 36.Scharn D., Wenschuh H., Reineke U., Schneider-Mergener J., Germeroth L. (2000) J. Comb. Chem. 2, 361–369 [DOI] [PubMed] [Google Scholar]
- 37.Kim J. S., Raines R. T. (1993) Protein Sci. 2, 348–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frank R., Overwin H. (1996) Methods. Mol. Biol. 66, 149–169 [DOI] [PubMed] [Google Scholar]
- 39.Reineke U., Volkmer-Engert R., Schneider-Mergener J. (2001) Curr. Opin. Biotechnol. 12, 59–64 [DOI] [PubMed] [Google Scholar]
- 40.Frank R. (2002) J. Immunol. Methods 267, 13–26 [DOI] [PubMed] [Google Scholar]
- 41.Munck Petersen C., Nielsen M. S., Jacobsen C., Tauris J., Jacobsen L., Gliemann J., Moestrup S. K., Madsen P. (1999) EMBO J. 18, 595–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petersen C. M., Nielsen M. S., Nykjaer A., Jacobsen L., Tommerup N., Rasmussen H. H., Roigaard H., Gliemann J., Madsen P., Moestrup S. K. (1997) J. Biol. Chem. 272, 3599–3605 [DOI] [PubMed] [Google Scholar]
- 43.Tauris J., Ellgaard L., Jacobsen C., Nielsen M. S., Madsen P., Thøgersen H. C., Gliemann J., Petersen C. M., Moestrup S. K. (1998) FEBS Lett. 429, 27–30 [DOI] [PubMed] [Google Scholar]
- 44.Schweigreiter R. (2006) BioEssays 28, 583–594 [DOI] [PubMed] [Google Scholar]
- 45.Chao M. V., Rajagopal R., Lee F. S. (2006) Clin. Sci. 110, 167–173 [DOI] [PubMed] [Google Scholar]
- 46.Kaplan D. R., Miller F. D. (2004) Nature 427, 798–799 [DOI] [PubMed] [Google Scholar]
- 47.Bandtlow C., Dechant G. (2004) Sci. STKE 2004, pe24. [DOI] [PubMed] [Google Scholar]
- 48.Bronfman F. C., Fainzilber M. (2004) EMBO Rep. 5, 867–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feng D., Kim T., Ozkan E., Light M., Torkin R., Teng K. K., Hempstead B. L., Garcia K. C. (2010) J. Mol. Biol. 396, 967–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Westergaard U. B., Sørensen E. S., Hermey G., Nielsen M. S., Nykjaer A., Kirkegaard K., Jacobsen C., Gliemann J., Madsen P., Petersen C. M. (2004) J. Biol. Chem. 279, 50221–50229 [DOI] [PubMed] [Google Scholar]
- 51.Chen Z. Y., Ieraci A., Teng H., Dall H., Meng C. X., Herrera D. G., Nykjaer A., Hempstead B. L., Lee F. S. (2005) J. Neurosci. 25, 6156–6166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paiardini A., Caputo V. (2008) Neuropeptides 42, 205–214 [DOI] [PubMed] [Google Scholar]
- 53.Copley R. R., Russell R. B., Ponting C. P. (2001) Protein Sci. 10, 285–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quistgaard E. M., Thirup S. S. (2009) BMC Struct. Biol. 9, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stamos J., Lazarus R. A., Yao X., Kirchhofer D., Wiesmann C. (2004) EMBO J. 23, 2325–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paoli M., Anderson B. F., Baker H. M., Morgan W. T., Smith A., Baker E. N. (1999) Nat. Struct. Biol. 6, 926–931 [DOI] [PubMed] [Google Scholar]
- 57.Beisel H. G., Kawabata S., Iwanaga S., Huber R., Bode W. (1999) EMBO J. 18, 2313–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paoletti F., Konarev P. V., Covaceuszach S., Schwarz E., Cattaneo A., Lamba D., Svergun D. I. (2006) Biochem. Soc. Trans. 34, 605–606 [DOI] [PubMed] [Google Scholar]
- 59.Kliemannel M., Golbik R., Rudolph R., Schwarz E., Lilie H. (2007) Protein Sci. 16, 411–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fahnestock M., Michalski B., Xu B., Coughlin M. D. (2001) Mol. Cell Neurosci. 18, 210–220 [DOI] [PubMed] [Google Scholar]
- 61.Nakamura K., Namekata K., Harada C., Harada T. (2007) Cell Death. Differ. 14, 1552–1554 [DOI] [PubMed] [Google Scholar]
- 62.Yune T. Y., Lee J. Y., Jung G. Y., Kim S. J., Jiang M. H., Kim Y. C., Oh Y. J., Markelonis G. J., Oh T. H. (2007) J. Neurosci. 27, 7751–7761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arnett M. G., Ryals J. M., Wright D. E. (2007) Brain Res. 1183, 32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wei Y., Wang N., Lu Q., Zhang N., Zheng D., Li J. (2007) Neurosci. Lett. 429, 169–174 [DOI] [PubMed] [Google Scholar]
- 65.Provenzano M. J., Xu N., Ver Meer M. R., Clark J. J., Hansen M. R. (2008) Laryngoscope 118, 87–93 [DOI] [PubMed] [Google Scholar]
- 66.Michalski B., Fahnestock M. (2003) Brain Res. Mol. Brain Res. 111, 148–154 [DOI] [PubMed] [Google Scholar]
- 67.Pedraza C. E., Podlesniy P., Vidal N., Arévalo J. C., Lee R., Hempstead B., Ferrer I., Iglesias M., Espinet C. (2005) Am. J. Pathol. 166, 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peng S., Wuu J., Mufson E. J., Fahnestock M. (2005) J. Neurochem. 93, 1412–1421 [DOI] [PubMed] [Google Scholar]
- 69.Counts S. E., Nadeem M., Wuu J., Ginsberg S. D., Saragovi H. U., Mufson E. J. (2004) Ann. Neurol. 56, 520–531 [DOI] [PubMed] [Google Scholar]
- 70.Counts S. E., Mufson E. J. (2005) J. Neuropathol. Exp. Neurol. 64, 263–272 [DOI] [PubMed] [Google Scholar]
- 71.Podlesniy P., Kichev A., Pedraza C., Saurat J., Encinas M., Perez B., Ferrer I., Espinet C. (2006) Am. J. Pathol. 169, 119–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kichev A., Ilieva E. V., Piñol-Ripoll G., Podlesniy P., Ferrer I., Portero-Otín M., Pamplona R., Espinet C. (2009) Am. J. Pathol. 175, 2574–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.