Abstract
The cytokine interleukin (IL)-21 exerts pleiotropic effects acting through innate as well as adaptive immune responses. The activities of IL-21 are mediated through binding to its cognate receptor complex composed of the IL-21 receptor private chain (IL-21Rα) and the common γ-chain (γC), the latter being shared by IL-2, IL-4, IL-7, IL-9, and IL-15. The binding energy of the IL-21 ternary complex is predominantly provided by the high affinity interaction between IL-21 and IL-21Rα, whereas the interaction between IL-21 and γC, albeit essential for signaling, is rather weak. The design of IL-21 analogues, which have lost most or all affinity toward the signaling γC chain, while simultaneously maintaining a tight interaction with the private chain, would in theory represent candidates for IL-21 antagonists. We predicted the IL-21 residues, which compose the γC binding epitope using homology modeling and alignment with the related cytokines, IL-2 and IL-4. Next we systematically analyzed the predicted binding epitope by a mutagenesis study. Indeed two mutants, which have significantly impaired γC affinity with undiminished IL-21Rα affinity, were successfully identified. Functional studies confirmed that these two novel hIL-21 double mutants do act as hIL-21 antagonists.
Keywords: Cytokine, Cytokine Action, Mutagenesis Mechanisms, Receptor Structure-Function, Receptors, Cytokine Antagonist
Introduction
Interleukin (IL),2-21 which is produced by activated CD4+ T cells, T-follicular helper cells, and natural killer T (NKT) cells, (1) has been demonstrated to exert pleiotropic effects on the proliferation, differentiation, and cytotoxicity of various classes of lymphoid cells. More recently, IL-21 has furthermore been shown to play a crucial role in the differentiation of CD4+ T cells into T-helper 17 (TH17) cells, a subset of T cells associated with development of inflammatory conditions and autoimmune diseases (2, 3).
IL-21 signals through its receptor complex composed of the private chain IL-21Rα and the common chain, γC, the latter of which is shared by five other cytokines: IL-2, IL-4, IL-7, IL-9, and IL-15 (1). Together these cytokines constitute the so-called γC family of cytokines. Despite the rather limited sequence homology (on average 15% sequence identity), these γC cytokines share a highly conserved overall four-helix bundle topology.
Previously, we have reported the three-dimensional structure of human IL-21 (hIL-21) resolved by heteronuclear NMR spectroscopy (4). The structure of hIL-21 was shown to comprise a typical up-up-down-down four-α-helical-bundle topology similar to that observed for several other type I cytokines, including the γC cytokines IL-2, IL-4, and IL-15. Noteworthy, a segment of hIL-21, including the helix C that by modeling is presumed to be important for IL-21Rα binding, was demonstrated to represent a structurally unstable segment, not observed in the structures of other γC cytokines. A chimeric variant in which this flexible segment of hIL-21 had been exchanged with the corresponding IL-4 sequence was constructed and demonstrated to have a 10-fold increase in potency in a cell-based assay (4).
Functional receptor complexes for this family of cytokines consist of heterodimeric structures, which besides the γC receptor chain include a private chain. Within this complex, the γC chain provides the crucial capacity for signaling, while the specificity and high affinity of the cytokine-receptor interaction is conferred predominantly by the private chain. The active IL-21 receptor complex thus constitutes the ternary complex IL-21/IL-21Rα/γC (5, 6).
The corresponding signaling complex of IL-4: IL-4/IL-4Rα/γC has been shown to be assembled in a sequential fashion, initiated by high affinity binding of IL-4 to the private chain, IL-4Rα, followed by recruitment of γC thus leading to the formation of the ternary complex (7). Recently the structure of this complex has been resolved by x-ray crystallography revealing specific amino acid residues of IL-4 that are involved in the binding of each of the receptor chains (7). Interestingly, preceding this structural knowledge of the IL-4 cytokine/cytokine receptor complex, IL-4 antagonists had been identified through systematic mutagenesis, seeking to abolish binding to the signaling receptor chain γC while preserving high affinity binding to the IL-4Rα chain (8).
A crystal structure has furthermore been reported for the γC cytokine IL-2 (9). In contrast to IL-4 and IL-21, the high affinity signaling complex of this cytokine is composed of a quaternary complex: IL-2/IL-2Rα/IL-2Rβ/γC of which the IL-2Rβ receptor chain is homologous to the private chains of the IL-4 and IL-21 signaling complexes, while the IL-2Rα receptor chain is a non-signaling receptor chain with no (known) homologous equivalent for the IL-21 receptor complex.
The IL-2 and IL-4 interfaces toward γC in both of these complexes are strikingly similar, and characterized by a rather conserved apolar “canyon” on the cytokines, which accommodate the protruding γC binding loops through near-ideal shape complementarity (7). Furthermore the structurally equivalent side chains of the IL-2 residues GluIL-215, LeuIL-219, GlnIL-2126, and SerIL-2130 and IL-4 residues GlnIL-48, LysIL-412, ArgIL-4121, and SerIL-4125, respectively, make analogous interactions with γC. The cytokine contact residues donated by γC furthermore maintain similar conformations in both of these IL-2 and IL-4 complexes indicating that cross-reactivity is not occurring through structural plasticity of γC. Residues of the cytokines, which were identified by structure resolution as contacts toward γC are in close agreement with results previously obtained through mutagenesis studies (10). Overall, mutagenesis and BIAcore studies have shown that the γC epitope for the cytokines, IL-4 and IL-21, are not identical although partially overlapping (11). Together these data strongly indicated that a similar structural basis for γC binding exists within the γC-cytokine family.
Similar to IL-4, IL-21 has been shown to bind with high affinity to its cognate private chain IL-21Rα (KD 70 pm) and with a considerably lower affinity to the γC chain (KD 160 μm), implicating a sequential binding mechanism similar to that of IL-4 (7, 11). By analogy, an IL-21 variant in which binding to γC had been eliminated, while binding to the Rα chain was retained, would thus be considered a likely candidate for an IL-21 antagonist.
In the present report, we have applied a rational approach toward the generation of hIL-21 antagonists. Firstly, residues predicted to constitute part of the γC binding interface, and thus possibly being implicated in the binding of this receptor chain, were identified by homology modeling based on the structures of IL-2 and IL-4 in complex with γC, and through knowledge of the NMR structure of IL-21. Subsequently, through mutagenesis, these residues constituting the predicted γC binding epitope were explored with respect to their effect on the binding of γC and IL-21Rα. Finally, through the combination of mutants demonstrated to have impaired γC affinity, while undiminished IL-21Rα affinity, we have identified two hIL-21 double mutants as novel hIL-21 antagonists.
EXPERIMENTAL PROCEDURES
Homology Modeling
The NMR structure of human IL-21 (PDB code 2oqp) and the crystal structures of human IL-2/IL-2Rβ/γC (PDB code 2b5i) and IL-4/IL-4Rα/γC (PDB code 3bpl) were superimposed using the program Discovery Studio. According to this structural superimposition, the sequences of IL-21, IL-2, and IL-4 were aligned, and the alignment adjusted by hand. IL-21Rα was aligned to IL-2Rβ and IL-4Rα based on the primary sequence. Structural information of γC was taken from the IL-2 and IL-4 complexes. Based on the alignment, a homology model was built for the hIL-21/IL-21Rα/γC complex using the Modeler program integrated in Discovery Studio. The model quality was examined through Profiles-3D. Finally, using a 5-Å cut-off, the potential γC binding residues on hIL-21 were identified.
Expression of hIL-21 Variants
A full-length cDNA of human IL-21 including a C-terminal HA epitope tag (YPYDVPDYA) was inserted into the pcDNA3.1(+) vector to construct a eukaryotic expression plasmid.
Site-directed mutagenesis was performed on the pcDNA3.1(+)/hIL-21HA plasmid using a QuikChange® mutagenesis kit (Stratagene) according to the manufacturer's instructions to create the hIL-21 variants. DNA sequencing was subsequently used to confirm the integrity of the mutants.
Plasmid DNA encoding the respective recombinant protein was transfected with 293fectinTM reagent (Invitrogen) into FreeStyle HEK293 cells. For protein production, cells were grown in serum-free FreeStyle 293 medium containing 4 mm glutamine, 1% PLURONIC® F68 and penicillin-streptomycin antibiotics at 1 × 106 cells per ml and incubated with shaking for 3 days at 37 °C, 8% CO2. Supernatants were collected and concentrated by ultrafiltration.
Relative concentrations of IL-21 fusion proteins were determined by an AlphaScreen® HA Detection Kit (PerkinElmer Life Sciences, Cat. No. 6760612C) and performed in triplicate in 96-well white opaque half-area plates (PerkinElmer) as follows: First, 15 μl of biotinylated-HA (30 nm final concentration) was incubated with decreasing concentrations of hIL-21HA variants, prepared by serial dilutions in binding buffer. After 10 min, 10 μl of anti-HA acceptor beads (1:100 dilution) were added to each well and incubated for 60 min at room temperature. Then, 10 μl of streptavidin-coated donor beads (1:100 dilution) were added to each well and incubated for 60 min at room temperature. All additions and incubations were made in subdued lighting conditions due to photosensitivity of the beads. The assay was measured on an EnVisionTM microplate analyzer.
Receptor Extracellular Domain Expression and Purification
Expression vectors encoding hIL-21Rα (residues 1–232) and γC (residues 1–254), both including at the C-terminal end a His6 tag, were transiently expressed in FreeStyle HEK293 cells. Supernatants were collected for analysis on day 4 post-transfection. Similarly, IL-21Rα (residues 1–232) including a C-terminal Avi-tag (GLNDIFEAQKIEWHE), IL-21Rα-Avi (12) was expressed as described above.
Purification steps: (a) supernatant of His6-tagged IL-21Rα or γC was concentrated and buffer exchanged into PBS (10 mm sodium phosphate, 0.15 m NaCl, pH 7.4) by ultrafiltration; (b) target proteins were captured by Ni2+-chelating Sepharose Fast Flow (Amersham Biosciences) equilibrated with PBS buffer, and eluted with PBS plus 500 mm imidazole; (c) the eluted proteins were concentrated to 5–10 mg/ml by ultrafiltration (10 kDa cutoff, Amicon) and applied to a 16/60 column of Superdex75 (GE Healthcare) equilibrated with the same PBS buffer. The major protein peak was collected and concentrated to 5–10 mg/ml by ultrafiltration (10 kDa cutoff; Amicon). The protein concentration was estimated by the absorption at 280 nm using the molar extinction coefficient (IL-21Rα-His6: 49860 cm−1 m−1 and γC-His6: 61810 cm−1 m−1) calculated by the GPMAW7.0 program.
Biotinylation of hIL-21HA and Receptor EC Domains
hIL-21HA was biotinylated using the Sulfo-NHS-Biotin reagent (Pierce). Briefly, protein was incubated with Sulfo-NHS-Biotin at a 1:1 molar ratio on ice for 2 h. Free biotin was removed via a PD-10 column (Amersham Biosciences).
Supernatant of IL-21Rα-Avi was concentrated and buffer exchanged into PBS and then biotinylated by RTS AviTag Biotinylation Kit (Roche, Cat. No. 03514935), followed by removal of the free biotin via a PD-10 column (Amersham Biosciences).
NK92 Proliferation Test
NK92 cell stock was obtained from the American Type Tissue Collection and propagated in MyeloCultTM (MyeloCultTM 5100, StemCell Inc, Cat. No. 05150) supplemented with 150 units/ml of IL-2 (Chemicon Cat. No. IL002), and penicillin-streptomycin; maintained at 37 °C and 5% CO2; and passaged every 48 h. For IL-2 starvation, NK92 cells were plated in the absence of IL-2 for 12–16 h prior to hIL-21 variant stimulation. A total of 105 cells/80 μl/well were seeded in 96-well plates, followed by addition of 20 μl of hIL-21 variant at variable concentrations. After 3 days in culture, 20 μl Alamar-BlueTM (Serotec, UK) was added to each well. Six hours later, fluorescence was measured at an excitation wavelength of 530 nm and a fluorescence wavelength of 590 nm using FLUOstar OPTIMA plate reader (BMG LABTECH). Analysis was performed in triplicate, and data were analyzed using GraphPad Prism.
ALPHAScreen Competition Test
The affinities of IL-21HA variants toward the hIL-21Rα EC domain were determined using a modified ALPHAScreen assay (PerkinElmer) and performed in triplicate in 96-well white opaque half-area plates (PerkinElmer) as follows: First, 15 μl of biotinylated hIL-21HA (30 nm final concentration) was incubated with decreasing concentrations of hIL-21HA variants, prepared by serial dilution in binding buffer. After 10 min, 15 μl of hIL-21Rα-His6 (30 nm final concentration) were added to each well and incubated for 30 min at room temperature. Then 10 μl of Ni2+-chelating acceptor beads (1:100 dilution) were added to each well and incubated for 60 min at room temperature. Finally, 10 μl of streptavidin-coated donor beads (1:100 dilution) were added to each well and incubated for 60 min at room temperature. All additions and incubations were made under subdued lighting conditions because of photosensitivity of the beads. The assay was measured on an EnVisionTM microplate analyzer.
Biosensor Experiments
All experiments were carried out on a BIAcore T100 system (Pharmacia Biosensor) at 25 °C at a flow rate of 30 μl min−1 in HBS running buffer (10 mm Hepes, pH 7.4, 150 mm NaCl, 3.4 mm EDTA, 0.005% surfactant P20) with a data collection rate of 2.5 s−1.
The IL-21Rα EC was immobilized onto Sensor chip SA through the C-term biotinylated Avi-tag at a density of 75–150 RU as detailed in the BIAapplications handbook (Pharmacia Biosensor AB, 1994). To measure the affinity between the γC ectodomain and hIL-21mut/IL-21Rα, IL-21Rα was first saturated with the IL-21 mutant and the resultant complex was then perfused with variable concentration of γC in the presence of a fixed concentration of IL-21 mutant in the running buffer.
RESULTS
Identification of hIL-21 γC Binding Interface by Homology Modeling
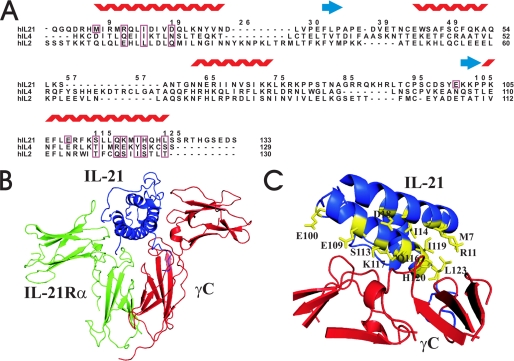
Although the average sequence homology between the γC cytokines hIL-21, hIL-2, and hIL-4 is only about 17%, the topology of the four-helical bundle, which forms the core structure of these γC cytokines, is highly conserved. Still, some variation is seen with respect to the exact lengths of the rather flexible loop regions connecting the four helices. We have applied two structure-based approaches toward the identification of IL-21 residues likely to constitute part of the IL-21/γC binding interface. First, a structure-based alignment of IL-2, IL-4, and IL-21 was conducted (Fig. 1A). From this alignment, one set of putative γC-interacting residues could be identified directly on the basis of alignment to positions in IL-2 and IL-4, which had been demonstrated to form part of the γC binding interface in crystal structures of IL-2 and IL-4 receptor complexes, respectively (Fig. 1A). Secondly, using the structure of IL-21 resolved by NMR analysis together with crystal structures of the IL-2/IL-2Rβ/γC (PDB code 2b5i) and IL-4/IL-4Rα/γC complexes (PDB code 3bpl), homology modeling was employed to build a model of the IL-21/IL-21Rα/γC complex. IL-21 residues present as part of the IL-21/γC binding interface in this model were similarly included as putative γC-interacting residues. From this analysis the following residues of IL-21 were identified as positions potentially critical for γC binding: Met7, Arg11, Ile14, Asp18, Glu100, Glu109, Ser113, Gln116, Lys117, Ile119, His120, and Leu123.
FIGURE 1.
A, sequence alignment of hIL-21, hIL-2, and hIL-4 based on the structural alignment and adjusted by hand. γC binding residues on IL-21 predicted from alignment with IL-2 and IL-4 receptor complexes are boxed in the alignment together with the corresponding residues in IL-2 and IL-4. Three additional γC binding residues identified from the model (M7, E100, E109) are only boxed in the sequence of hIL-21. The numbering follows hIL-21. B, homology model of the IL-21/IL-21Rα/γC complex generated by the Modeler program. The structures of IL-21, IL-21Rα, and γC are depicted as blue, green, and red ribbons, respectively. C, potential γC binding residues of hIL-21 are depicted as yellow sticks.
In the model of the ternary IL-21 receptor complex, the two receptor chains, IL-21Rα and γC, converge to form a Y shape with IL-21 positioned in close contact with both chains within the fork (Fig. 1) similarly to the known structures of other type 1 cytokines. The IL-21 residues facing the γC chain are mainly located on the helices A and D. In contrast, modeling revealed that the binding surface toward the IL-21Rα predominantly is formed by residues present within the helices A and C of IL-21 (not shown in Fig. 1).
Expression of hIL-21 Mutants
To study the contribution of each of the IL-21 residues proposed to participate the binding of the γC chain, these residues were first studied by alanine substitution. Wt hIL-21 and the hIL-21 variants carrying single substitutions were transiently expressed by HEK293 cells in serum-free medium. All constructs included a C-terminal HA tag, which has been shown to affect neither binding nor biological activity of the hIL-21 protein (data no shown). The HA tag provided the ability to estimate concentration of the hIL-21 variants, by ALPHA screen, independent of the hIL-21 protein sequence. All of the hIL-21 variants were expressed in the monomer form, according to analysis on non-reducing SDS gels, and at rather low expression levels (∼200 ng/ml, data not shown). Following expression, supernatants of the variants were collected and concentrated prior to further analysis.
Effect of hIL-21 Mutations on γC Binding
A BIAcore based binding assay was developed in order to examine whether the alanine exchange of the putative γC binding interface of hIL-21 had affected the binding affinity of the IL-21:γC interaction. Because formation of the binary IL-21/γC complex is characterized by a very low binding affinity (KD 160 μm), BIAcore analysis of the direct binding between the hIL-21 variants and the γC receptor chain was not feasible. Also, the γC ectodomain did not measurably bind to immobilized hIL-21Rα in the absence of hIL-21, even at a 10 μm concentration of γC present in the perfusate (11). Thus, to analyze the interaction of γC with the hIL-21 variants, the immobilized ectodomain of the private chain, IL-21Rα, was first saturated with IL-21 wt or mutant protein, and the resultant complex was subsequently perfused with γC as well as the IL-21 protein, both present in the running buffer. A fast association rate (kon = 2.4 × 107 m−1 s−1) was observed for the formation of the complex between hIL-21 and the private chain EC domain (data not shown), similarly to what has been observed for IL-4 and viewed as a reflection of a binding event involving predominantly electrostatic interactions (11, 13). The calculated KD values for the interaction between the hIL-21 variants, immobilized via hIL-21Rα, and the ectodomain of γC are compiled in Table 1. These mutants may be viewed as falling into three categories: One mutant (E109A) showed at least a 2-fold decrease in γC binding affinity. Eight mutants (M7A, R11A, I14A, E100A, Q116A, I119A, H120A, and L123A) showed an at least 2-fold decrease in γC binding affinity, while one particular mutant, namely Q116A, showed an immeasurable low γC binding affinity. Surprisingly, three mutants (D18A, S113A, and K117A) showed a slightly increased γC binding affinity, mainly resulting from a slower dissociation rate.
TABLE 1.
Affinities of γC toward immobilized IL-21Rα saturated by either wt IL-21 or variants as determined by BIAcore analysis
Four variants (Q116A, I14D, Q116D, and I119D) resulted in very low affinities, which could not be measured in this experiment, and were thus marked as unmeasurable (u.m). All of the variants were measured three times. The relative affinities of IL-21 variants to γC is presented by the KDmut/wt value. mut, IL-21 variants.
| Alanine variant | KD ± S.D. | KDmut/wt | Charge variant | KD ± S.D. | KDmut/wt |
|---|---|---|---|---|---|
| nm | nm | ||||
| M7A | 166 ± 15 | 2.4 | M7D | 382 ± 40 | 5.6 |
| R11A | 264 ± 20 | 3.9 | R11D | 1063 ± 152 | 15.6 |
| I14A | 203 ± 12 | 3.0 | I14D | u.m | u.m |
| D18A | 37 ± 5 | 0.5 | D18K | 57 ± 3 | 0.8 |
| E100A | 137 ± 12 | 2.0 | E100R | 230 ± 13 | 3.4 |
| E109A | 95 ± 15 | 1.4 | E109R | 107 ± 16 | 1.6 |
| S113A | 59 ± 3 | 0.9 | S113D | 396 ± 54 | 5.8 |
| Q116A | u.m | u.m | Q116D | u.m | u.m |
| K117A | 30 ± 4 | 0.4 | K117D | 1091 ± 147 | 16.0 |
| I119A | 187 ± 32 | 2.8 | I119D | u.m | u.m |
| H120A | 142 ± 23 | 2.1 | H120D | 2510 ± 480 | 36.9 |
| L123A | 561 ± 52 | 8.3 | L123D | 3100 ± 236 | 45.6 |
| WT | 68 ± 2 | 1.0 |
According to our premise, variants with severely impaired γC binding are favorable in this approach toward the rational design of hIL-21 antagonists. However, in this alanine-scan analysis, most of the mutants generated showed at best only a slightly reduced γC binding affinity. Some of these hIL-21 residues may indeed be part of the hIL-21/γC interface however with just a minor contribution for each single residue to the total binding energy. Thus changing the side chain of such γC interface residues into alanine does not necessarily alter the γC binding very much. More pronounced effects compared with alanine mutations could potentially be obtained by the introducing of either steric hindrance, repulsion, or mismatch through the substitution of a given amino acid side chain with a larger one or alternatively into a side chain representing a distinctly different chemical property. All of the predicted residues were therefore next mutated to either aspartic acid, or if the wt amino acid was charged, into an amino acid of the opposite charge. As expected, these more dramatically exchanged mutants showed a more severe effect on γC binding affinity compared with the alanine-substituted mutants. Three mutants (M7D, E100R, and S113D) showed ∼3–6-fold decreases in γC binding affinity and four mutants (R11D, K117D, H120D, L123D) showed ∼10–50-fold decreases in γC binding affinity mainly due to a faster dissociation rate. Interestingly, three mutants (I14D, Q116D, and I119D) did not show kinetic rate constants within the measurable range because of very low affinities.
Effect of hIL-21 Mutations on hIL-21Rα Binding
According to the premise, a hIL-21 antagonist should have a markedly reduced affinity toward the signaling receptor chain, γC, while at the same time have a similar or even improved affinity to the overall receptor complex, as compared with the wt IL-21. In this setting, the affinity of the hIL-21 variants toward the hIL-21Rα private chain should be undiminished, thus allowing the antagonist variants to compete efficiently with the wt hIL-21 agonist. An ALPHAscreen binding assay was employed to test the affinity of hIL-21 variants toward hIL-21Rα. In this assay, the hIL-21 variants were analyzed by competition with wt hIL-21 for binding to a bead-immobilized EC of hIL-21Rα. Relative affinities of IL-21 variants toward IL-21Rα are presented in Table 2.
TABLE 2.
Relative affinities of IL-21 variants to IL-21Rα determined in ALPHAscreen competition test
Increasing concentrations of IL-21 variants were added to compete the binding of biotinylated wt IL-21 to IL-21Rα-His6. The relative affinities of IL-21 variants to IL-21Rα is expressed by the IC50mut/wt value. Values of IC50mut/wt greater than one indicate decreased binding affinity by the variants toward IL-21Rα relative to wt IL-21. IC50 values were measured in three independent experiments. mut, IL-21 variants.
| Alanine variant | IC50mut/wt ± S.D. | Charge variant | IC50mut/wt ± S.D. |
|---|---|---|---|
| M7A | 1.2 ± 0.2 | M7D | 2.1 ± 0.2 |
| R11A | 0.5 ± 0.1 | R11D | 13.0 ± 1.3 |
| I14A | 2.5 ± 0.2 | I14D | 97.0 ± 4.1 |
| D18A | 13.2 ± 0.5 | D18K | 15.9 ± 1.5 |
| E100A | 0.9 ± 0.1 | E100R | 23.2 ± 2.7 |
| E109A | 2.3 ± 0.3 | E109R | 20.1 ± 2.7 |
| S113A | 1.2 ± 0.2 | S113D | 2.3 ± 0.3 |
| Q116A | 0.9 ± 0.1 | Q116D | 0.9 ± 0.1 |
| K117A | 3.9 ± 0.4 | K117D | 7.1 ± 0.8 |
| I119A | 5.7 ± 0.4 | I119D | 27.1 ± 1.9 |
| H120A | 1.0 ± 0.1 | H120D | 0.3 ± 0.0 |
| L123A | 0.8 ± 0.2 | L123D | 0.4 ± 0.1 |
| WT | 1.0 ± 0.1 |
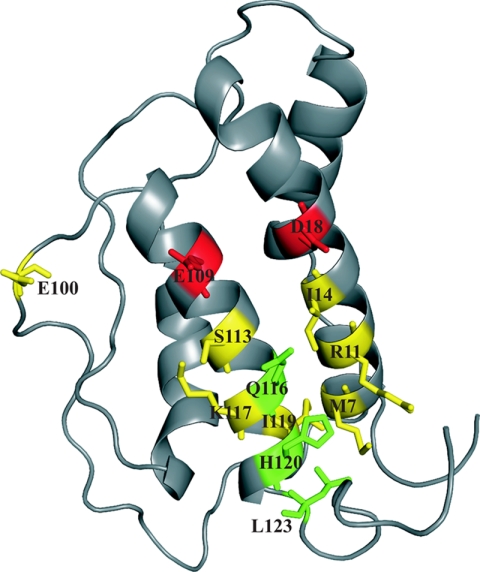
At only one position, Asp18, did the exchange with alanine markedly reduce the binding to hIL-21Rα (∼13-fold). Similarly to the observation with binding to the γC, more significant effects were observed upon exchange into larger and charged side chains. Here several positions exhibited highly reduced binding toward IL-21Rα (>10-fold): Arg11, Ile14, Asp18, Glu100, Glu109, and Ile119. All together, three classes of positions were identified using 2-fold impaired affinity as a cut-off; one representing a class of positions which when mutated into larger and charged side chains impaired the binding to both receptor chains (Met7, Arg11, Ile14, Glu100, Ser113, Lys117, and Ile119); and two classes of positions which when mutated into larger and charged side chains selectively impaired binding to either the IL-21Rα (Asp18 and Glu109) or the γC (Gln116, His120, and Leu123) (Fig. 2). The latter class of variants mutated at positions Gln116, His120, and Leu123 displays the binding profile presumed to characterize (novel) putative antagonists.
FIGURE 2.
Binding interface of IL-21Rα depicted on IL-21. IL-21 residues mutated to aspartic acid or residues containing opposite charge are labeled on the structure and color-coded according to effect on IL-21Rα and γC binding affinity. Residues shown in red and green have ≥2-fold impaired binding affinity toward either IL-21Rα or γC, respectively, whereas residues having ≥2-fold impaired IL-21Rα and γC binding affinities are shown in yellow (See Tables 1 and 2).
Effect of hIL-21 Mutations on Biological Activity
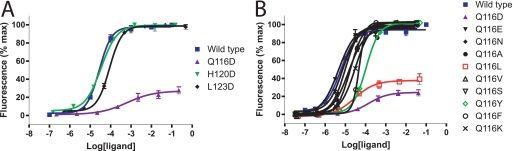
Human NK92 cells proliferate efficiently in response to hIL-21 (14). With a focus on the identification of novel antagonists, hIL-21 variants, selectively impaired in binding to the γC chain (Q116D, H120D, and L123D), were evaluated in an NK92 proliferation assay. Of these variants, H120D and L123D showed a slightly increased EC50 value, whereas Q116D showed a more than 40-fold increased EC50 value, and furthermore a ∼4-fold decreased Rmax value (Fig. 3A).
FIGURE 3.
Biological activity of IL-21 variants in a NK92 cell proliferation assay. A, variants with aspartate mutations in positions 116, 120, and 123 compared with wt IL-21. B, screen of position 116 variants compared with wt IL-21. In each case, NK92 cells were incubated with serial 10-fold dilutions of wt IL-21 or variants for 72 h, and thereafter proliferation was quantified by Alamar-Blue reduction. The curves represent one experiment performed in triplicate, and the error bars represent the S.D. Experiments were repeated at least three times with similar results. Activity is expressed as a percentage of maximal response. IL-21 variants were expressed by HEK293 cells and used as crude supernatants. Protein concentrations were measured relative to wt IL-21 using an ALPHAscreen-based assay.
Through this analysis, residue Gln116 was identified as crucially important for γC binding and signal generation, but not for the binding of IL-21 to IL-21Rα, neither upon substitution by alanine nor aspartic acid. A Q116D mutation showed a more severely reduced biological activity than seen for Q116A. To determine whether any single substitution at position 116 of hIL-21 can totally abolish biological activity, further substitutions by a series of other amino acid residues representing different sizes and/or chemically distinct side chains (Glu, Asn, Leu, Val, Ser, Tyr, Phe, Lys) were introduced. All of these mutants showed a less pronounced alteration of Rmax value compared with Q116D (Fig. 3B). Substitution of position Gln116 by leucine resulted in a 30-fold increased EC50 value, which is similar to that observed for Q116D, and a ∼3-fold decreased Rmax value. Remarkably, mutants Q116N and Q116E showed very similar activity profile to wt hIL-21. Thus, while the activity of the two Gln116 mutants, Q116D and Q116L, was significantly reduced, no single point mutants within our proposed γC epitope set were totally inactive.
hIL-21[Q116D/H120D] and hIL-21[Q116D/L123D] Behave as Antagonists in Vitro
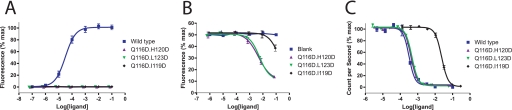
Seeking the generation of hIL-21 antagonist(s) with fully abolished biological activity, we next generated two double mutants through the combination of Q116D with H120D and L123D, respectively. The binding of both of these double mutants, hIL-21[Q116D/H120D] and hIL-21[Q116D/L123D] to hIL-21Rα equaled that of wt hIL-21, yet both double mutants failed to induce the proliferation of NK92 cells (Fig. 4, C and A). Moreover, these hIL-21 variants inhibited hIL-21-induced proliferation of NK92 cells in a dose-dependent manner (Fig. 4B). Thus, in this bioassay, both mutants acted as full antagonists of the hIL-21 wt cytokine.
FIGURE 4.
Characterization of antagonistic IL-21 variants. A, biological activity of IL-21 variants compared with wt IL-21 in a NK92 cell proliferation assay. B, inhibition of wt IL-21 induced NK92 cell proliferation by antagonistic IL-21 variants; NK92 cells were incubated with wt IL-21 at the IL-21 concentration inducing 50% of maximal response, followed by adding serial dilutions of antagonistic IL-21 variants or PBS as blank control. After 72 h of incubation, proliferation was quantified by Alamar-Blue reduction. C, relative affinities of antagonistic IL-21 variants toward IL-21Rα were measured using an ALPHAscreen competition test. Increasing concentrations of IL-21 variants were added to compete the binding of biotinylated IL-21 to IL-21Rα-His6. The curves represent one experiment performed in triplicate, and the error bars represent the S.D. All of the experiments were repeated at least three times with similar results. IL-21 variants were expressed by HEK293 cells and used as crude supernatants. Protein concentrations were measured relative to wt IL-21 using an ALPHAscreen-based assay.
hIL-21[Q116D/H120D] and hIL-21[Q116D/L123D] Show a Better Inhibitory Effect than hIL-21[Q116D/I119D]
A putative hIL-21 antagonist including two point mutations, hIL-21[Q116D/I119D], has previously been reported (15). To further evaluate the novel hIL-21 antagonists reported in this communication, we performed a direct comparison of all three variants, which interestingly share the mutation Q116D. In the NK92 proliferation assay, neither antagonist demonstrated any measurable activity (Fig. 4A). However, in the inhibitory competition assay, the effect of hIL-21[Q116D/I119D] was in the order of 100 times lower than that of our newly identified antagonists, hIL-21[Q116D/H120D] and hIL-21[Q116D/L123D] (Fig. 4B). According to our analysis of receptor binding, the I119D mutant has a dramatically reduced binding affinity to the γC receptor chain. In contrast to the H120D and L123D mutations included in our new antagonists, the I119D mutation also results in a significantly reduced affinity toward the hIL-21Rα chain, which in turn decreases the affinity of hIL-21[Q116D/I119D] toward hIL-21Rα (Fig. 4C). The selective elimination of γC binding is thus a unique characteristic of the two novel antagonists, hIL-21[Q116D/H120D] and hIL-21[Q116D/L123D], reported in this work.
DISCUSSION
Rationale for the Identification of Antagonists
A common feature of the γC cytokines, which have been subjected to structural analysis, is that the binding energy of the cytokine:receptor complex is provided predominantly by the interaction between the cytokine and the private receptor chain (e.g. IL-2Rβ, IL-4Rα, IL-21Rα), whereas the interaction between the cytokine and γC, albeit essential for signaling, is rather weak. Also whereas formation of the cytokine:private chain complex is driven mostly by electrostatic interactions, the binding interactions between the cytokine and the γC chain are largely apolar in nature. Mutational analysis and crystal structures of IL-2 and IL-4 suggest and support these conclusions (16).
In the present report, we have performed a mutational scan of hIL-21 to elucidate residues involved in binding of the γC chain and especially such positions which upon mutagenesis strongly diminish the γC interaction, while at the same time leave the high affinity binding to the hIL-21Rα chain unaffected. In theory, such mutants do not have the capacity to induce signaling but can still compete for IL-21 receptor binding with wt IL-21, thereby representing putative antagonists. Importantly, we have taken advantage of structural information available for the IL-2 and IL-4 receptor complexes, thus allowing a rational structure-based strategy in the analysis of hIL-21. Further, all of the residues, which according to our structural alignment correspond to positions homologous to residues which within IL-2 and/or IL-4 have been reported as γC binding residues, were investigated.
γC Epitope in Different Cytokines
Resolution of the crystal structures of receptor-bound IL-2 and IL-4 as well as mutational analysis, including IL-21, suggest that binding of the cytokines by the shared receptor subunit, γC, employs overlapping, yet also distinct sets of epitopes (7, 9, 11, 17).
X-ray analysis of the IL-2 and IL-4 receptor complexes has revealed structurally highly similar interfaces between each of the cytokines and γC, in both complexes burying an area of about 1000 Å2 and with a similar ratio of polar/apolar surfaces (7). Conserved apolar canyons on both IL-2 and IL-4 accept the protruding γC residue TyrγC103 and the disulfide Cys160-CysγC209 through a good shape complementarity. These residues have also been shown as important for binding to IL-7, IL-15, and IL-21 (16). According to the crystal structure, the γC binding interface on IL-4 comprises residues IleIL-411, AsnIL-415 on helix A and residues GluIL-4114, LysIL-4117, ThrIL-4118, ArgIL-4121, GluIL-4122, TyrIL-4124, SerIL-4125 on helix D. Of these only a subset, namely IleIL-411, AsnIL-415, and TyrIL-4124 showed >5-fold decrease in binding affinity upon alanine exchange (10). This observation agrees with the general notion that the functional epitope of a protein-protein interface, as deduced from a mutational scan, may only include a subset of the structural epitope revealed by resolution of the crystal structure of the protein complex (18). More residues may, however, be included in this functional epitope when charged or steric hindrance substitutions are investigated. Residue ArgIL-4121, which according to our structural alignment corresponds to GlnIL-21116, thus had no large effect when exchanged into alanine, whereas the R121DIL-4 mutant did show a markedly reduced activity (10, 19). GlnIL-21116 was identified as a particularly sensitive epitope in this report, being the only single position which upon alanine exchange by itself had a marked effect on γC binding, while leaving IL-21Rα binding unchanged. This would suggest that for the IL-21 cytokine, this position provides a greater contribution to the cytokine:γC interaction than seen for the comparable site in the IL-4 complex. Similar to our result on IL-21, the IL-2 residue GlnIL-2126, which is structural equivalent to GlnIL-21116 and ArgIL-4121, has been shown by mutagenesis to constitute a critical energetic hotspot (10).
Mutational analysis of the cytokine:γC interaction has been directed at the cytokine as well as the γC receptor subunit. Within the cytokine critically important residues for γC binding have been located to the A and D helices. Alanine exchange has previously identified IleIL-411, and AsnIL-415 of helix A in IL-4 as hotspots for γC binding (10). Exchange by alanine of the corresponding IL-21 positions, IleIL-2114 and AspIL-2118, had in contrast only a minor effect on binding. Although, the introduction of a charged residue at position IleIL-2114 did produce a markedly reduced binding of γC, altering the charge at position AspIL-2118 (D18K) still did not significantly change the γC binding properties of IL-21. Within helix D, residue LeuIL-21123 was, in addition to the GlnIL-21116 position mentioned above, identified as IL-21 hot spot for γC binding showing an ∼8-fold decrease in γC binding affinity upon alanine substitution.
Within the γC chain, positions that when mutated, affect the binding of all γC cytokines (e.g. TyrγC103) have been observed (11, 20), as have positions that selectively affect the binding of the individual cytokines, e.g. AsnγC44, which has been demonstrated to be essential for the binding of IL-21, while not for the binding of IL-4 (11).
Thus, although the hotspot for γC binding differ between IL-21 (GlnIL-21116 and LeuIL-21123) and IL-4 (IleIL-411, AsnIL-415, and TyrIL-4124) as revealed by mutagenesis, the γC binding epitopes on IL-21 and IL-4 do largely overlap as observed by superposition of the three-dimensional structures of IL-21 and IL-4. Mutagenesis studies on the γC receptor chain similarly showed that binding sites for IL-21 and IL-4 do overlap, yet are not identical (11). Our data further support the notion that a common structural basis for γC binding does indeed exist within the γC cytokine family, however distinct differences within the functional γC epitope are observed between the different γC cytokines.
Separation of the γC and the IL-21Rα Binding Epitopes of IL-21
For all of the γC cytokines that have been analyzed, namely IL-2 and IL-4, residues critical for binding of the private chain are predominantly found in the helix A and C segments, partially separate from the γC epitopes residing in the helix A and D segments. Binding epitopes for the two receptor chains are thus intertwined on helix A. Furthermore the basic assumption in the interpretation of mutational mapping, that the substitutions change only the chemical property of the side chain without disturbing the local structure, is not always fulfilled. Therefore, the γC binding residues, especially those located in helix A, are likely to affect the binding of both receptor chains, γC and IL-21Rα, upon mutagenesis. We have in the present report analyzed not only the effect on γC binding following the exchange of residues predicted to form the binding epitope for this receptor subunit, but also the effect on IL-21Rα binding. This is in contrast to the studies of IL-4 and IL-2 where only binding to the γC was analyzed. Three classes of mutants were obtained in our study based on the observed effect of alanine substitution on γC and IL-21Rα affinity, using 2-fold impaired affinity as a cut-off. The first class of mutants (M7A, R11A, E100A, Q116A, H120A, and L123A) showed a decrease of γC binding affinity without impairing IL-21Rα binding. The second class of mutants (I14A and I119A) were characterized by impairing both γC and IL-21Rα binding affinity. The last class of mutants (D18A, E109A, and K117A) only impaired the IL-21Rα binding affinity. When charged/or chemically distinct side chains were introduced at potential γC binding sites, three additional mutants were identified (Q116D, H120D, and L123D), which showed more severely impaired γC binding affinity than their corresponding alanine mutants, and yet unimpaired IL-21Rα binding affinity.
Mutational analysis of the γC cytokine IL-4 parallels observations reported here for IL-21. In IL-4, mutational exchange has thus demonstrated the feasibility of functionally separating the high affinity binding mediated predominantly via the private chain, IL-4Rα, and receptor signaling via the γC receptor chain. Two distinct sites, site I and site II, are critical for each of these functional characteristics. Site I is composed of residues present in helices A and C. Site I variants of IL-4 are affected in receptor binding as reflected in higher EC50 values, while efficacies similar to those observed for the wt IL-4 are achieved at sufficiently high concentrations. In contrast, site II mutants, are characterized by EC50 values similar to that of the wt protein but highly reduced Rmax values, or even mutants completely devoid of detectable activity (19, 21). These mutants show unaffected high affinity binding to the private IL-4Rα chain, yet impaired in γC binding, as subsequently directly demonstrated by cross-linking analysis (22). Interestingly, site II was noticed to be dominated by residues which together form a hydrophobic patch. This observation has later been fully corroborated by the resolution of the x-ray structure of the IL-4/IL-4Rα/γC complex (7). Clearly, the introduction of an aspartic acid residue within this hydrophobic patch represents a dramatic change, spatially as well as physio-chemically, a common observation at this site in IL-2, IL-4, and IL-21.
IL-4 antagonists have been generated by abolishment of γC binding through the simultaneous mutagenesis of two critical γC binding residues, ArgIL-4121 and TyrIL-4124, into charged, aspartic acid residues (8). Here, synergistic antagonism was achieved through the combination of two single point mutations (8). By analogy the structurally equivalent IL-21 variant, [Q116D/I119D]IL-21, has been reported as an IL-21 antagonist (15). In our experiments, the hIL-21 mutant I119DIL-21 showed a significantly decreased (27-fold) IL-21Rα binding affinity, thus compromising its potential as an IL-21 antagonist.
Solution structures of the IL-4 mutants, Y124GIL-4 and Y124DIL-4, which are both partial antagonists, the latter most efficaciously, have been determined by NMR (23). The overall structures of these variants were observed to be only minutely perturbed relative to the wt protein, including specifically the helical protein structures immediately surrounding the mutated residues. It thus seems plausible that the changed activity of these mutants can be ascribed to changes introduced within the hydrophobic interaction patch. This interpretation gained further support from the observation that mutational exchange of a Tyr residue with either of the hydrophobic substituents, Phe or His, had but a minor effect, while removal of the hydrophobic residue through the introduction of a Gly residue resulted in a markedly reduced activity (21). It is at present not known whether the mutations in helix D that confers antagonism to hIL-21 disturb the structure of the protein.
Phenotype of Antagonistic Mutants
The helix D mutants Q116D, H120D, and L123D, which were identified here as representing selective γC mutations, were tested for biological activity in the NK92 proliferation assay. Two of the mutants, H120D and L123D, showed slightly increased EC50 value, ∼2-fold and ∼4-fold respectively, and unchanged Rmax values. This rather minor decrease in potency, surprising in view of a significantly reduced γC binding (37-fold and 46-fold respectively), may be compensated by the increased IL-21Rα affinity seen for these mutants, (∼3.3-fold for H120D and ∼2.5-fold for L123D), thus reflecting the importance of IL-21Rα affinity for signal generation. Alternatively, the γC binding affinity of wt hIL-21 may surpass the requirements for cellular signaling, as seen in the growth hormone system where native hGH receptor affinity significantly exceeds the basic requirement for cellular activity (24). The third selective γC mutant identified in helix D, Q116D, did not show any measurable affinity to γC, yet could still stimulate the proliferation of NK92 cells although with a ∼40-fold increased EC50 value and ∼4-fold decreased Rmax value.
Position 116 was identified as a γC epitope particularly sensitive to mutagenesis, as even alanine substitution at this position had a marked effect. Thus, both of the exchanges, Q116A and Q116D, resulted in significantly decreased γC affinity in our analysis. To further elucidate this position, a thorough mutagenesis was performed, including the exchange into each of the following residues: Asn, Glu, Lys, Leu, Val, Phe, Tyr, and Ser. Although several of these mutants were affected in the affinity toward γC, none of them had completely lost the competence for induction of NK92 proliferation. Among the mutants, Q116D, Q116L, and Q116Y showed significantly decreased efficacy without changing the affinity to IL-21Rα. The mutants Q116E and Q116N exhibited very similar activity profiles to the wt hIL-21. Mutation Q116D exhibited larger effect than any of the other mutants, an observation that could not simply be explained by the introduction of a negative charge or the variation in size of the different side chains. Additional conformational effects may contribute to the large effect in this case. As Q116D showed the most significantly impaired potency and Rmax value within the series of Gln116 mutants, it was chosen as the critical constituent for the generation of IL-21 antagonists, which we obtained through the construction of double substitution mutants. Such antagonists could serve as beneficial tools in the elucidation of mechanisms and treatment of autoimmune diseases by inhibition of IL-21 activity.
Acknowledgments
We thank Dorthe Pedersen, Jørgen Steen Jensen, and Xianwei Zhang for expert technical assistance.
Footnotes
- IL
- interleukin
- γC
- common γ-chain
- hIL-XRy
- hIL-X receptor chain y
- wt
- wild type
- NMR
- nuclear magnetic resonance
- PBS
- phosphate-buffered saline
- PDB
- Protein Data Bank
- HA
- hemagglutinin.
REFERENCES
- 1.Spolski R., Leonard W. J. (2008) Annu. Rev. Immunol. 26, 57–79 [DOI] [PubMed] [Google Scholar]
- 2.Korn T., Bettelli E., Gao W., Awasthi A., Jäger A., Strom T. B., Oukka M., Kuchroo V. K. (2007) Nature 448, 484–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nurieva R., Yang X. O., Martinez G., Zhang Y., Panopoulos A. D., Ma L., Schluns K., Tian Q., Watowich S. S., Jetten A. M., Dong C. (2007) Nature 448, 480–483 [DOI] [PubMed] [Google Scholar]
- 4.Bondensgaard K., Breinholt J., Madsen D., Omkvist D. H., Kang L., Worsaae A., Becker P., Schiødt C. B., Hjorth S. A. (2007) J. Biol. Chem. 282, 23326–23336 [DOI] [PubMed] [Google Scholar]
- 5.Asao H., Okuyama C., Kumaki S., Ishii N., Tsuchiya S., Foster D., Sugamura K. (2001) J. Immunol. 167, 1–5 [DOI] [PubMed] [Google Scholar]
- 6.Habib T., Senadheera S., Weinberg K., Kaushansky K. (2002) Biochemistry 41, 8725–8731 [DOI] [PubMed] [Google Scholar]
- 7.LaPorte S. L., Juo Z. S., Vaclavikova J., Colf L. A., Qi X., Heller N. M., Keegan A. D., Garcia K. C. (2008) Cell 132, 259–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tony H. P., Shen B. J., Reusch P., Sebald W. (1994) Eur. J. Biochem. 225, 659–665 [DOI] [PubMed] [Google Scholar]
- 9.Wang X., Rickert M., Garcia K. C. (2005) Science 310, 1159–1163 [DOI] [PubMed] [Google Scholar]
- 10.Letzelter F., Wang Y., Sebald W. (1998) Eur. J. Biochem. 257, 11–20 [DOI] [PubMed] [Google Scholar]
- 11.Zhang J. L., Foster D., Sebald W. (2003) Biochem. Biophys. Res. Commun. 300, 291–296 [DOI] [PubMed] [Google Scholar]
- 12.Beckett D., Kovaleva E., Schatz P. J. (1999) Protein Sci. 8, 921–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y., Shen B. J., Sebald W. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 1657–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toomey J. A., Gays F., Foster D., Brooks C. G. (2003) J. Leukoc. Biol. 74, 233–242 [DOI] [PubMed] [Google Scholar]
- 15.Presnell S. R., West J. W., Novak J. E. (2007) United States Patent 7, 186, 805B2 [Google Scholar]
- 16.Wang X., Lupardus P., LaPorte S. L., Garcia K. C. (2009) Annu. Rev. Immunol. 27, 29–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J. L., Buehner M., Sebald W. (2002) Eur. J. Biochem. 269, 1490–1499 [DOI] [PubMed] [Google Scholar]
- 18.Clackson T., Wells J. A. (1995) Science 267, 383–386 [DOI] [PubMed] [Google Scholar]
- 19.Kruse N., Shen B. J., Arnold S., Tony H. P., Müller T., Sebald W. (1993) EMBO J. 12, 5121–5129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olosz F., Malek T. R. (2002) J. Biol. Chem. 277, 12047–12052 [DOI] [PubMed] [Google Scholar]
- 21.Kruse N., Tony H. P., Sebald W. (1992) EMBO J. 11, 3237–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duschl A. (1995) Eur. J. Biochem. 228, 305–310 [PubMed] [Google Scholar]
- 23.Müller T., Dieckmann T., Sebald W., Oschkinat H. (1994) J. Mol. Biol. 237, 423–436 [DOI] [PubMed] [Google Scholar]
- 24.Pearce K. H., Jr., Cunningham B. C., Fuh G., Teeri T., Wells J. A. (1999) Biochemistry 38, 81–89 [DOI] [PubMed] [Google Scholar]