Abstract
LXXLL/leucine zipper-containing alternative reading frame (ARF)-binding protein (LZAP) was recently shown to function as a tumor suppressor through inhibition of the NF-κB signaling pathway. LZAP is also known as a negative regulator of cell invasion, and its expression was demonstrated to be reduced in several tumor tissues. However, the molecular mechanism of the negative effect of LZAP on cell invasion is unclear. In this study, we identify NLBP as a novel LZAP-binding protein using tandem affinity purification. We demonstrate the negative effects of NLBP on cell invasion and the NF-κB signaling pathway. NLBP expression was not detected in hepatocellular carcinoma cells with strong invasive activity, whereas its expression was detected in a hepatocellular carcinoma cell line with no invasive activity. We also demonstrate that these two proteins mutually affect the stability of each other by inhibiting ubiquitination of the other protein. Based on these results, we suggest that NLBP may act as a novel tumor suppressor by inhibiting cell invasion, blocking NF-κB signaling, and increasing stability of the LZAP protein.
Keywords: Cancer, Cell/Migration, Gene, Protein/Degradation, Tumor, Tumor/Suppressor
Introduction
Multistep progressions are involved in tumor formation. Many proteins, including tumor suppressor genes and oncogenes, affect the tumorigenesis of normal cells (1). One of the most important factors for tumor formation is mutations in tumor suppressors or oncogenes, which affect expression levels and/or protein activity (2). Therefore, maintaining oncogenes and tumor suppressor genes in a wild type state is crucial to blocking tumorigenesis.
LZAP3 (also known as Cdk5rap3 or C53 protein) was originally identified as an ARF-binding protein and was found to have numerous functions as a tumor suppressor. These include activation of p53, induction of apoptosis mediated by genotoxic agents leading to inhibition of tumor cell growth, and negative regulation of the checkpoint response by antagonizing checkpoint kinases to promote cyclin-dependent kinase 1 activation (3–6). Recently, it was shown that upon depletion of LZAP expression, NF-κB-dependent MMP-9 expression, and cellular invasion were increased (7). In addition, LZAP protein was reduced in ∼30% of human head and neck squamous cell carcinomas (7).
Although LZAP has been functionally shown to be a tumor suppressor, the molecular mechanism of how LZAP blocks tumorigenesis is not yet clear. To gain further insight into the molecular mechanism of LZAP functions on tumorigenesis, we performed biochemical tandem repeat affinity purification and identified KIAA0776 as a novel LZAP-binding protein (NLBP). NLBP encodes an uncharacterized protein of 794 amino acids and does not contain any conserved domains. In this study, we show that NLBP is a novel LZAP-binding protein that may function as a tumor suppressor by inhibiting cell invasion, blocking NF-κB signaling, and increasing the stability of the LZAP protein.
EXPERIMENTAL PROCEDURES
Plasmids
Human NLBP (KIAA0776), CT116, and LZAP cDNA were purchased from the American Type Culture Collection (Manassas, VA). Full-length and deletion mutants of human NLBP were generated by PCR and subcloned into a modified pIRES-EGFP mammalian expression vector, as described previously (8), to create constructs that are S-FLAG-SBP (streptavidin-binding peptide)-tagged (SFB-tagged). A GST-LZAP fusion construct was generated by PCR of LZAP and subcloned into pGEX-5T-1 (GE Healthcare). Full-length and deletion mutants of human LZAP were generated by PCR and subcloned into a modified mammalian expression vector, as described previously (8), to create constructs encoding Myc-tagged full-length or deletion mutants of LZAP. NF-κB-Luc promoter plasmid (9) and Myc-RelA expression plasmid (10) were previously described.
Cell Culture
HeLa, U2OS, 293T, HepG2, Hep3B, HLE, PLC, and Huh7 cell lines were purchased from American Type Culture Collection (Manassas, VA) and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum at 37 °C in 5% CO2 (v/v).
siRNA
All siRNA duplexes used in this study were purchased from Dharmacon Research (Lafayette, CO). The sequences of LZAP siRNA1 and -2 are 5′-AAGGATTGGCAGGAGATTATA-3′ and 5′-CAAGGTATGTGGACCGAGT-3′, respectively. The sequences of NLBP siRNA1 and -2 are 5′-AGAAATGAGAGATGAGCTA-3′ and 5′-AGAAGAGGTCAATGATAAA-3′, respectively. The sequence of control siRNA is 5′-UUCAAUAAAUUCUUGAGGUUU-3′. siRNAs were transfected into cells using Oligofectamine (Invitrogen) according to the manufacturer's instructions.
Antibodies, Transfection, and Immunoprecipitation
Rabbit anti-NLBP antibody was raised by immunizing rabbits with mixed peptides (649CDIMVKRGDKKRER669 and 755ELDKEQEDVASTTRK770). The resulting rabbit polyclonal antibodies were affinity-purified using the SulfoLink or AminoLink Plus immobilization and purification kit (Pierce). Anti-FLAG, -hemagglutinin, -Myc, and -β-actin antibodies were purchased from Sigma, and polyclonal human LZAP and p65 antibodies were purchased from Oncogene Science. Transient transfection was performed using FuGENE 6 reagent (Roche Applied Science). For immunoprecipitation, cells were washed with ice-cold PBS and then lysed in NETN buffer (0.5% Nonidet P-40, 20 mm Tris (pH 8.0), 50 mm NaCl, 50 mm NaF, 100 μm Na3VO4, 1 mm dithiothreitol, and 50 μg/ml phenylmethylsulfonyl fluoride) at 4 °C for 10 min. Crude lysates were cleared by centrifugation at 14,000 rpm at 4 °C for 5 min, and supernatants were incubated with protein A-agarose-conjugated primary antibodies. The immunocomplexes were washed three times with NETN buffer and subjected to SDS-PAGE. Western blotting was performed using the antibodies indicated in the figure legends.
Establishment of Stable Cell Lines and Affinity Purification of SFB-tagged LZAP-containing Complexes
The establishment of stable cell lines was previously described (11). To establish cell lines stably expressing epitope-tagged proteins, 293T cells were transfected with plasmids encoding SFB-LZAP and puromycin-resistant protein. Forty-eight hours after transfection, the cells were split at a 10:1 ratio and cultured in medium containing puromycin (10 μg/ml) for 3 weeks. Individual puromycin-resistant colonies were isolated and screened by Western blotting for expression of the LZAP protein. 293T cells stably expressing SFB-LZAP protein were lysed with 4 ml of NETN buffer on ice for 10 min. Crude lysates were cleared by centrifugation at 14,000 rpm at 4 °C for 10 min, and supernatants were incubated with 300 μl of streptavidin-conjugated beads (Amersham Biosciences). The immunocomplexes were washed three times with NETN buffer, and bead-bound proteins were eluted with 500 μl of NETN buffer containing 2 mg/ml biotin (Sigma). The eluted supernatant was incubated with 60 μl of S-protein beads (Novagen). The immunocomplexes were washed three times with NETN buffer and subjected to SDS-PAGE. Silver staining was performed to visualize the protein bands. Specific bands were excised and digested, and the peptides were analyzed by a mass spectrometer.
GST Pulldown Assay
The GST fusion protein was expressed in Escherichia coli and purified as described previously (12). Two μg of GST fusion protein or GST was immobilized on glutathione-Sepharose 4B beads and incubated with lysates prepared from cells that were transiently transfected with plasmids encoding the indicated proteins in NETN buffer for 2 h at 4 °C. After washing with NETN buffer, the samples were analyzed by Western blotting analysis.
Immunofluorescence Staining
Immunofluorescence staining was previously described (11). Cells grown on coverslips were fixed with 3% paraformaldehyde at room temperature for 15 min. Next, the cells were permeabilized with PBS containing 0.5% Triton X-100 at room temperature for 5 min and blocked with PBS containing 5% goat serum at room temperature for 30 min. The coverslips were incubated with primary antibodies at room temperature for 20 min. After washing with PBS, cells were incubated with one of the following secondary antibodies: fluorescein isothiocyanate-conjugated goat anti-mouse IgG, rhodamine-conjugated goat anti-rabbit IgG, or rhodamine-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Laboratories) at room temperature for 20 min. 4′,6-Diamidino-2-phenylindole (DAPI) was used to counterstain the nuclei. After a final wash with PBS, coverslips were mounted with glycerin containing p-phenylenediamine. All images were obtained with a Nikon ECLIPSE E800 fluorescence microscope.
In Vitro Invasion Assays
The cell invasion assay was performed using 24-well Transwell permeable supports (8-μm pore size, Corning Life Sciences) coated with 60 μl (1 mg/ml) of Matrigel (BD Biosciences). Cells were starved in serum-free medium overnight, trypsinized, and washed three times in Dulbecco's modified Eagle's medium containing 1% fetal bovine serum. Twenty thousand cells in 1% fetal bovine serum-Dulbecco's modified Eagle's medium were seeded into the upper chamber, and 600 μl of Dulbecco's modified Eagle's medium containing 10 or 1% fetal bovine serum was placed in the lower chamber. After a 16-h incubation, the Matrigel and cells remaining in the upper chamber were removed. Cells on the lower surface of the membrane were fixed in 4% paraformaldehyde and stained with 0.5% crystal violet. For the quantitative assay, the cells that invaded through the Matrigel were stained with 4 μg/ml calcein-AM (Molecular Probes) in PBS for 30 min at 37 °C and scanned for fluorescence with the Victor 3 multiplate reader (PerkinElmer Life Sciences); fluorescent cells were counted. All experiments were run in duplicate and were repeated three times.
In Vitro Migration Assays
U2OS cells were transfected with indicated siRNAs. Cells were plated 48 h after treatment onto 6-well plates and allowed to grow into confluent monolayers. Scrape wounds were generated using a micro tip, and cell medium was replaced. After 6 h, cells that had migrated were counted. All experiments were repeated three times.
RESULTS
Identification of NLBP as an LZAP-binding Protein
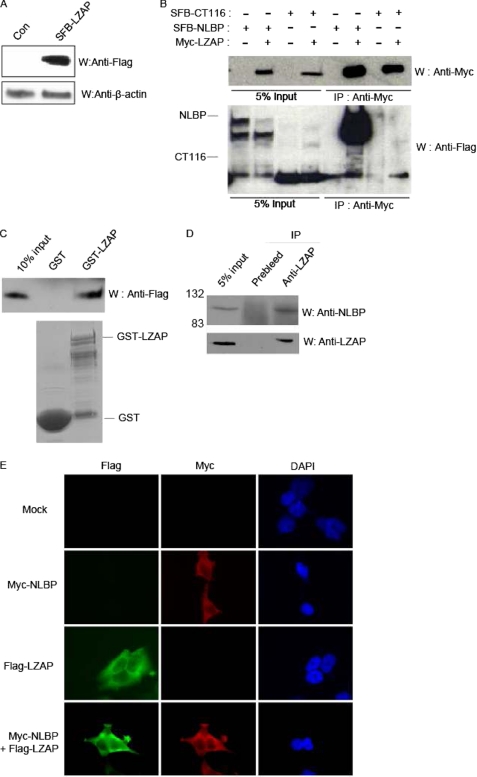
To identify new LZAP-binding proteins, we performed tandem repeat affinity purification using 293T cells stably expressing SFB-LZAP, the LZAP protein triply tagged at the N terminus with SBP (SFB tag) (Fig. 1A). After sequential affinity chromatography with streptavidin agarose and S-agarose beads and mass spectrometry analysis using the cell lysates prepared from cells expressing SFB-LZAP, we identified several putative LZAP-binding proteins (Table 1). Among these proteins, we focused on KIAA0776 (subsequently named NLBP) and CT116 (uncharacterized protein c20orf116 precursor) because peptides from these two proteins were the most abundantly recovered from the mass spectrometry analysis. First, we checked for binding between LZAP and NLBP or CT116 by overexpression in 293T cells. As shown in Fig. 1B, LZAP associated strongly with NLBP and showed weak binding to CT116. To further confirm the interaction between LZAP and NLBP, GST pulldown assays were performed using the GST protein alone or the GST-LZAP fusion protein with FLAG-NLBP-transfected 293T cell lysates. This experiment showed that the NLBP protein specifically bound to GST-LZAP but not GST alone (Fig. 1C). Physical interaction between endogenous LZAP and NLBP was also confirmed (Fig. 1D). Finally, NLBP was shown to colocalize with LZAP in the cytoplasm (Fig. 1E). These data indicate that NLBP is a bona fide LZAP-binding partner.
FIGURE 1.
Identification of NLBP as a novel LZAP-binding protein. A, establishment of human embryonic kidney 293T cell lines stably expressing SFB-LZAP. Cell extracts prepared from 293T cells stably expressing a control (Con) plasmid or SFB-LZAP fusion protein were subjected to Western blotting analysis (W) using anti-FLAG antibody. B, interaction between exogenous Myc-tagged LZAP and SFB triple-tagged NLBP or CT116. Immunoprecipitation (IP) reactions were performed using anti-Myc antibody and subjected to Western blot analysis using anti-Myc and anti-FLAG antibodies. C, GST pulldown assay. GST only or GST-LZAP protein was incubated with cell lysates containing exogenously expressed SFB triple-tagged wild type NLBP (SFB-NLBP). After extensive washing, bound NLBP proteins were analyzed by Western blotting analysis with anti-FLAG antibody. The amounts of GST and GST-LZAP are shown in the lower panel. D, binding between endogenous LZAP and NLBP. Immunoprecipitation reactions were performed using preimmune serum or anti-LZAP antibodies and subjected to Western blot analysis using anti-NLBP (upper panel) or anti-LZAP (lower panel) antibody. E, colocalization of NLBP with LZAP. 293T cells were transfected with Myc-tagged NLBP and SFB triple-tagged LZAP expression plasmids. Next, immunofluorescence assays were performed using anti-FLAG and -Myc antibodies. 4′,6-Diamidino-2-phenylindole (DAPI) was used as an indicator for the nucleus. Mock, mock-transfected.
TABLE 1.
List of proteins associated with SFB-LZAP identified by mass spectrometry analysis
| Protein name | No. of peptides obtained per experiment |
|---|---|
| LZAP | 48 |
| KIAA0776 | 31 |
| Uncharacterized protein c20orf116 precursor (CT116) | 9 |
| β-1,4-Galactosyltransferase 1 (B4GT1) | 6 |
| Heat shock 70-kDa protein 1-like | 6 |
| Heat shock 70-kDa protein 1 | 5 |
| Highly similar to Rattus norvegicus ubiquitin c (UBC) | 4 |
| 60 S acidic ribosomal protein p0 | 3 |
| Ubiquitin-fold modifier-conjugating enzyme 1 | 3 |
| Heat shock cognate 71-kDa protein | 2 |
| Ubiquitin fold modifier 1 precursor | 1 |
| Hect, uba, and wwe domain-containing protein 1 | 1 |
| Adenine nucleotide translocator 1 | 1 |
| 60 S ribosomal protein I3 | 1 |
| Rpl10a protein | 1 |
| Transmembrane protein 55a | 1 |
| Nucleolar phosphoprotein b23 | 1 |
| β-1,4-Galactosyltransferase 5 (B4GT5) | 1 |
| Rps3 protein | 1 |
Identification of the Reciprocal Binding Regions of LZAP and NLBP
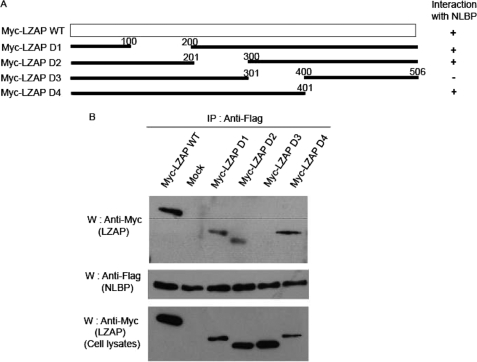
To identify the region of NLBP that interacts with LZAP, several deletion mutants of NLBP were constructed (Fig. 2A). NLBP wild type protein and deletion mutants were tested for the ability to interact with full-length LZAP by coexpression in 293T cells. As shown in Fig. 2, B and C, NLBP amino acid residues 121–250 were found to be important for association with full-length LZAP protein. Similarly, we generated a series of LZAP deletion mutants and examined which regions of LZAP might be required for interaction with NLBP. We found that LZAP amino acid residues 301–400 are capable of binding NLBP (Fig. 3, A and B).
FIGURE 2.
Identification of the LZAP-binding regions of NLBP. A, diagram of wild type (WT) NLBP and serial deletion mutants (D1–D6). B and C, 293T cells were transfected with plasmids encoding Myc-LZAP and wild type SFB-NLBP or serial deletion mutants. Cell lysates were subjected to immunoprecipitation (IP) with anti-Myc (B) or anti-FLAG (C) antibodies and immunoblotted (W) with the antibodies on the left. The amounts of SFB triple-tagged NLBP and Myc-tagged LZAP in the lysates were analyzed by immunoblotting and shown in the bottom panels.
FIGURE 3.
Identification of the NLBP-binding region of LZAP. A, diagram of wild type (WT) LZAP and serial deletion mutants (D1–D4). B, 293T cells were transfected with plasmids encoding SFB-NLBP and wild type Myc-LZAP or serial deletion mutants. Cell lysates were subjected to immunoprecipitation (IP) with anti-FLAG antibody and immunoblotted (W) with the antibodies indicated on the left. Myc-tagged LZAP in the lysates were analyzed by immunoblotting and shown in the bottom panel. Mock, mock-transfected.
NLBP Is Important for Inhibition of Cell Invasion
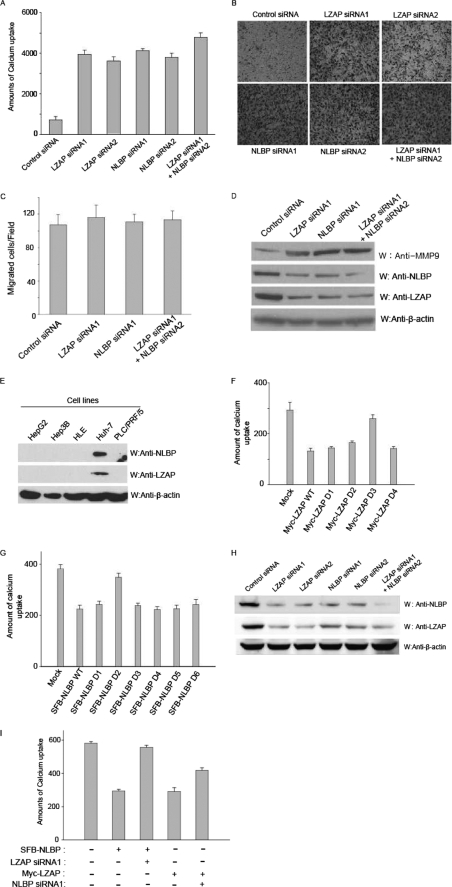
Decreased LZAP expression was previously shown to enhance cell invasion and increase MMP-9 expression levels (7). Therefore, we tested whether NLBP can also affect cell invasion and MMP-9 expression using NLBP or LZAP siRNA-transfected U2OS cells. We measured in vitro cell-invasive activity, either by measuring calcium uptake (Fig. 4A) or by measuring invasion through a Matrigel barrier (Fig. 4B), after LZAP or NLBP expression was decreased by siRNA transfection. Following siRNA-mediated inhibition of LZAP or NLBP expression, invasion was increased by ∼5-fold (Fig. 4A), and invasive activity was slightly enhanced when cotransfected with both LZAP and NLBP siRNAs (Fig. 4, A and B). Down-regulation of LZAP or NLBP expression did not affect cellular migration through the Transwell permeable supports in the absence of Matrigel (Fig. 4C). Next, we performed Western blotting analysis to check MMP-9 expression levels in control, LZAP, or NLBP siRNA-transfected U2OS cell lines. MMP-9 expression levels were increased following knockdown of LZAP or NLBP (Fig. 4D), consistent with the increase in cell-invasive activity. Our in vitro data showed that NLBP likely inhibits cell invasion and suggested the possibility that NLBP expression would be reduced in highly invasive cancer cell lines when compared with non-invasive cancer cell lines. Therefore, we checked the expression levels of NLBP and LZAP in several hepatocellular carcinoma cell lines by Western blotting with anti-NLBP or anti-LZAP antibodies. As shown in Fig. 4E, expression levels of LZAP and NLBP were reduced in several invasive hepatocellular carcinomas, including HepG2, Hep3B, HLE, and PLC, but not in the non-invasive Huh7 cell line (13). To identify the regions of NLBP or LZAP that are important for cell invasion, NLBP or LZAP wild type protein or serial deletion mutants were transfected into Hep3B cells and tested for their ability to affect cell invasion. As shown in Fig. 4, F and G, NLBP amino acid residues 121–250 and LZAP amino acid residues 301–400 were found to be important for inhibition of cell invasion. Because NLBP and LZAP are binding partners, we tested to see whether reducing the levels of one binding partner by siRNA treatment would affect the levels of the other protein. Using antibodies against NLBP or LZAP, we found that treatment with either or both siRNAs can efficiently down-regulate both LZAP and NLBP expression (Fig. 4H). To further verify the functional importance of the presence of both proteins in cell invasion, we overexpressed one protein in the invasive Hep3B cell line and looked at the effect of siRNA treatment of the other protein. LZAP siRNA treatment was found to restore the levels of Hep3B cell-invasive activity that had been inhibited by overexpression of NLBP (Fig. 4I). Similar results were observed for NLBP siRNA-treated LZAP overexpression cells (Fig. 4I).
FIGURE 4.
NLBP is important for inhibition of cell invasion. A, B, C, D, and H, control, LZAP, NLBP, or LZAP + NLBP siRNAs were transfected into U2OS cells. Invasion assays using the transfected U2OS cells were performed using the calcium uptake method (A) or the Matrigel assay (B). These experiments were performed in duplicate, and the results shown are the average of three independent experiments. S.D. is shown on each bar. C, down-regulation of LZAP or NLBP expression did not affect cellular migration through the Transwell permeable supports in the absence of Matrigel. The migration assay was performed in the absence of Matrigel using U2OS cells transfected with control, LZAP, NLBP, or LZAP + NLBP siRNAs. D, MMP-9 expression levels were increased following knockdown of LZAP or NLBP, as shown by Western blot analysis (W) with control, LZAP, NLBP, or LZAP + NLBP siRNA-transfected U2OS cells. E, NLBP and LZAP protein expression levels in hepatocellular carcinomas. The lysates of various hepatocellular carcinoma cell lines were subjected to immunoblotting with the indicated antibodies. F and G, identification of the LZAP and NLBP regions important for cell-invasive activity. Hep3B cells were transfected with plasmids encoding Myc-LZAP or serial deletion mutants (F) (D1–D4) or wild type SFB-NLBP (WT) or serial deletion mutants (G) (D1–D6). Calcium uptake was measured to assay for invasive activity. These experiments were performed in duplicate, and the results shown are the average of three independent experiments. S.D. is shown on each bar. Mock, mock-transfected. H, treatment with LZAP or NLBP siRNA can efficiently down-regulate both LZAP and NLBP expression. Expression levels of endogenous LZAP and NLBP proteins in LZAP or NLBP siRNA-transfected U2OS cells were confirmed using the indicated antibodies. I, both NLBP and LZAP proteins are needed to inhibit cell invasion. As indicated, control or LZAP or NLBP expression plasmids and LZAP or NLBP siRNAs were transfected into Hep3B cells, and invasion assays were performed using the calcium uptake method. These experiments were performed in duplicate, and the results shown are the average of three independent experiments. S.D. is shown on each bar.
NLBP Inhibits the NF-κB-mediated Signaling Pathway
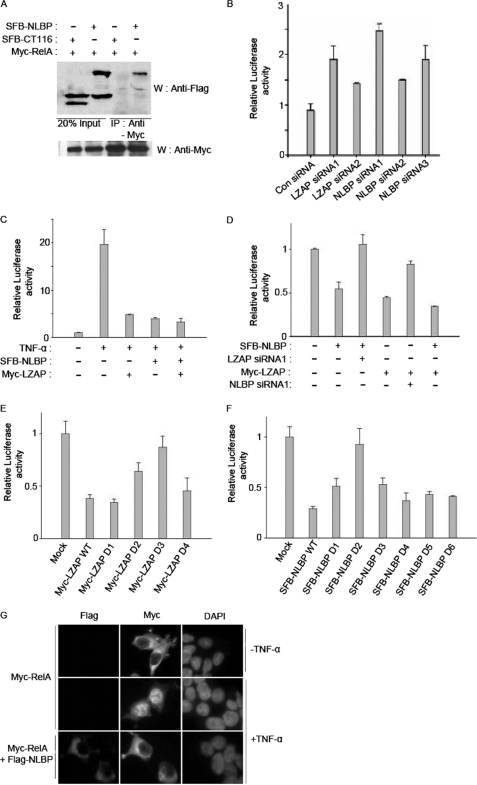
Decreased LZAP expression was previously shown to activate NF-κB-mediated transcription, and LZAP was found to bind to the NF-κB p65/RelA protein (7). Therefore, we tested whether NLBP also binds to the NF-κB p65/RelA protein. In 293T cells overexpressing the proteins, NLBP specifically bound to the NF-κB p65/RelA protein, whereas the CT116 protein did not (Fig. 5A). NF-κB-mediated transcription was also observed to increase after the reduction of LZAP or NLBP protein expression levels by LZAP or NLBP siRNA transfection (Fig. 5B). TNF-α is known to activate NF-κB-mediated transcription. To test whether NLBP can also affect TNF-α-induced NF-κB activation, NF-κB transcriptional activity was measured in cells transfected with the NLBP or LZAP expression plasmids after treatment with TNF-α. NLBP was found to inhibit TNF-α-induced NF-κB activation, similar to what had been previously described for LZAP (Fig. 5C) (7). We further determined the importance of NLBP in LZAP-mediated inhibition of NF-κB activation and vice versa. Hep3B cells were transfected with the LZAP expression plasmid in the absence or presence of NLBP siRNA. Overexpressed LZAP inhibits NF-κB-mediated transcription in Hep3B cells, but NF-κB transcriptional activity is restored in the presence of NLBP siRNA (Fig. 5D). To identify the regions of NLBP or LZAP that affect NF-κB-mediated transcription, NLBP or LZAP wild type protein or serial deletion mutants were transfected into Hep3B cells and tested for their ability to affect NF-κB-mediated transcription. As shown in Fig. 5, E and F, we found that NLBP amino acid residues 121–250 and LZAP amino acid residues 301–400 are important for inhibition of NF-κB-mediated transcription. Overexpressed NLBP inhibits TNF-α-induced p65/RelA nuclear translocation (Fig. 5G).
FIGURE 5.
NLBP inhibits the NF-κB signaling pathway. A, exogenously expressed NLBP binds to the NF-κB p65 protein. 293T cells were transfected with plasmids encoding Myc-p65 with or without the SFB-LZAP or SFB-CT116 plasmid. The transfected cell lysates were subjected to immunoprecipitation (IP) with anti-Myc antibody and immunoblotted (W) with the indicated antibodies. Myc-tagged p65 in these lysates was analyzed by immunoblotting and shown in the bottom panels. B, knockdown of NLBP increases endogenous NF-κB signaling. Control (Con), LZAP, or NLBP siRNAs were transfected with the NF-κB response element-containing plasmid into 293T cells. After 48 h, the transfected 293T cells were assayed using a Dual-Luciferase reporter assay system kit (Promega). These experiments were performed in duplicate, and the results shown are the average of three independent experiments. S.D. is shown on each bar. C, NLBP inhibits TNF-α-mediated NF-κB activation. NLBP or LZAP expression plasmids were transfected with the NF-κB response element-containing plasmid into 293T cells. After 24 h, the transfected 293T cells were treated with 50 ng/ml TNF-α and assayed using a Dual-Luciferase reporter assay system kit. D, both NLBP and LZAP proteins are needed to inhibit the activity of the NF-κB signaling pathway. The NF-κB response element-containing plasmid and LZAP or NLBP expression plasmids with control, LZAP, or NLBP siRNAs were transfected into Hep3B cells as indicated. After 48 h, the transfected Hep3B cells were assayed using a Dual-Luciferase reporter assay system kit. E and F, identification of the regions of LZAP and NLBP affecting the inhibition of NF-κB activation. The NF-κB response element-containing plasmid was transfected with plasmids encoding Myc-LZAP or serial deletion mutants (E) (D1–D4) and wild type SFB-NLBP or serial deletion mutants (F) (D1–D6) into Hep3B cells. After 48 h, the transfected Hep3B cells were assayed using a Dual-Luciferase reporter assay system kit. G, expression of NLBP inhibits RelA nuclear translocation. 293T cells were transfected with Myc-tagged RelA with/without SFB triple-tagged NLBP expression plasmids and then treated with TNF-α for 1 h, and then immunofluorescence assays were performed using anti-FLAG and -Myc antibodies. 4′,6-Diamidino-2-phenylindole (DAPI) was used as an indicator for the nucleus.
NLBP and LZAP Proteins Mutually Affect the Stability of Each Other
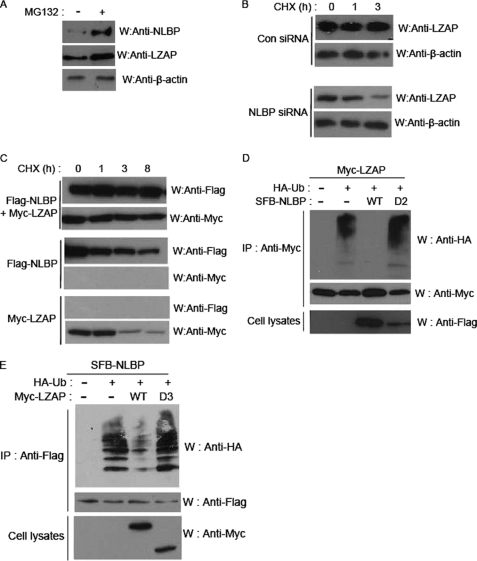
The data in Fig. 4, E and H, provided us with the possibility that the NLBP and LZAP proteins mutually regulate the stability of each other. We therefore checked the effect of the proteasome inhibitor MG132 on the expression levels of NLBP and LZAP proteins in U2OS cell lines. Expression levels of NLBP and LZAP proteins increased after treatment with MG132 (Fig. 6A), and similar results were obtained using 293T cells (data not shown). We also checked the effect of NLBP expression on the turnover of LZAP protein by measuring LZAP levels after blocking protein synthesis with cycloheximide (CHX). Degradation of the LZAP protein in control siRNA-transfected cells was delayed when compared with NLBP siRNA-transfected cells (Fig. 6B). Moreover, overexpression of both proteins together appeared to increase the half-life of both proteins (Fig. 6C). These data are consistent with the possibility that NLBP and LZAP proteins are degraded in a proteasome-dependent manner. Therefore, LZAP and NLBP were both examined for ubiquitination. The ubiquitination levels of the LZAP and NLBP proteins increased when cotransfected with an ubiquitin expression plasmid, but when the expression plasmids for both proteins were cotransfected, ubiquitination levels decreased (Fig. 6, D and F). This increase in protein stability appears to be mediated by binding between LZAP and NLBP because coexpression of LZAP with a NLBP deletion mutant lacking binding activity to LZAP did not influence ubiquitination levels of LZAP (Fig. 6D), and similar results were obtained when NLBP was coexpressed with a LZAP deletion mutant lacking NLBP binding activity (Fig. 6F).
FIGURE 6.
NLBP and LZAP proteins mutually affect the stability of each other. A–C, NLBP and LZAP proteins are degraded in a proteasome-dependent manner. Western blot analysis (W) of endogenous NLBP and LZAP expression levels in U2OS cells treated with 10 μm MG132 for 4 h was performed using the indicated antibodies (A). For cycloheximide (CHX) analysis, control (Con) or NLBP siRNA1 were transfected into U2OS cells (B), and NLBP or LZAP expression plasmids were transfected singly or together into U2OS cells (C). Cell lysates were prepared after the indicated chase incubation times, and Western blot analysis was performed using the indicated antibodies. D and E, coexpression of NLBP and LZAP reduces ubiquitination of both proteins. 293T cells were transfected with indicated expression plasmids. Transfected cell lysates were immunoprecipitated (IP), and Western blotting analysis was performed using the indicated antibodies. HA-Ub, hemagglutinin-ubiquitin. D2, deletion mutant.
DISCUSSION
In this study, we identified NLBP as a novel LZAP-binding protein using the tandem repeat affinity purification method. We confirmed the physical association between NLBP and LZAP by overexpression in 293T cells and also at endogenous levels. Similar to LZAP, knockdown of NLBP expression increased MMP-9 expression and cell invasion but did not affect cell migration. NLBP was found to function as a negative regulator of NF-κB-mediated transcription, also similar to LZAP. In addition, NLBP and LZAP expression was reduced in certain hepatocellular carcinomas that have highly invasive activity. These data strongly suggest that NLBP may function as a tumor suppressor together with LZAP through inhibition of cell invasion and NF-κB-mediated transcription.
Several studies have shown that LZAP localized to different subcellular compartments, including the nucleus, nucleolus, or cytoplasm (3, 4, 6, 7). However, our data show that NLBP protein colocalizes with LZAP in the cytoplasm. These data suggest that the cytoplasm may be the compartment where the NLBP and LZAP proteins function together.
Here, we show that NLBP or LZAP expression inhibits cell invasion and NF-κB activation in Hep3B cells, which have high invasive activity and low expression levels of both the NLBP and the LZAP proteins. The inhibition of cell invasion and NF-κB activation seen upon overexpression of NLBP or LZAP was restored upon LZAP or NLBP siRNA treatment, respectively. Furthermore, deletion mutants of LZAP or NLBP, which lack the region binding to the other protein, could not inhibit cell-invasive activity or NF-κB activation in Hep3B cells. Taken together, these findings indicate that the interaction between LZAP and NLBP is required for tumor suppression through inhibition of cell invasion and NF-κB activation. We speculate that the effects of NLBP and LZAP on cell invasion and MMP-9 expression may be mediated by effects on the NF-κB signaling pathway. The Matrigel chamber model that we used to quantify invasive activity reflects an early step of metastasis where tumor cells invade basement membranes. Data from our Matrigel assays and the reduced expression levels of NLBP and LZAP observed in non-invasive tumor cell lines lead us to conclude that the NLBP-LZAP complex is a potent regulator that inhibits local invasion of cells in the early steps of metastasis.
The expression levels of the NLBP and LZAP proteins increased after treatment with the proteasome inhibitor MG132 (Fig. 6A), and the half-life of each protein increased in the presence of the other protein (Fig. 6, B and C). In addition, the levels of the ubiquitinated forms of these proteins were reduced when both protein expression plasmids were cotransfected, whereas binding region deletion mutants did not reduce the levels of the ubiquitinated form of the other protein (Fig. 6, D and F). These results suggest that binding between these two proteins is required for mutual stabilization. Because these two proteins are ubiquitinated in vivo (Fig. 6, D and F), we reason that one or more ubiquitin E3 ligase proteins may ubiquitinate these proteins and may have a major role in the interplay between LZAP and NLBP. The identification of the E3 ligase(s) and the molecular mechanisms underlying the interplay between LZAP and NLBP remain to be determined.
LZAP was previously shown to have reduced expression in head and neck squamous cell carcinomas (7), and many reports show that the 6q16.1 locus, where the NLBP gene is located, was deleted in several tumor tissues (14–17). These data provide additional conviction that NLBP may be a bona fide new tumor suppressor protein and may also imply that the loss of the NLBP gene is initiating events leading to the loss of LZAP.
Because overexpression of NLBP can efficiently block cell invasion, the activation or supply of NLBP may be a useful mode of therapy to inhibit the cell invasion of tumors with non-functional NLBP. In conclusion, the identification of NLBP, a new LZAP-binding protein, provides new implications for the interplay between signaling pathways and protein networks in tumor development.
Acknowledgment
We thank members of Dr. Kim's laboratory for helpful discussions and technical support.
This work was supported by the Korea Research Foundation Grant KRF-2008-313-C00257 funded by the Korean Government (MOEHRD, Basic Research Promotion Fund) (to H. K.) and Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (Grant 2009-0074228).
- LZAP
- LXXLL/leucine zipper-containing ARF-binding protein
- NLBP
- novel LZAP-binding protein
- siRNA
- small interfering RNA
- GST
- glutathione S-transferase
- SBP
- streptavidin-binding peptide
- SFB
- S-Flag-SBP
- PBS
- phosphate-buffered saline
- TNF
- tumor necrosis factor
- ARF
- alternative reading frame.
REFERENCES
- 1.Hahn W. C., Weinberg R. A. (2002) N. Engl. J. Med. 347, 1593–1603 [DOI] [PubMed] [Google Scholar]
- 2.Wiedemann L. M., Morgan G. J. (1992) Eur. J. Cancer 28, 248–251 [DOI] [PubMed] [Google Scholar]
- 3.Jiang H., Luo S., Li H. (2005) J. Biol. Chem. 280, 20651–20659 [DOI] [PubMed] [Google Scholar]
- 4.Jiang H., Wu J., He C., Yang W., Li H. (2009) Cell Res. 19, 458–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X., Ching Y. P., Lam W. H., Qi Z., Zhang M., Wang J. H. (2000) J. Biol. Chem. 275, 31763–31769 [DOI] [PubMed] [Google Scholar]
- 6.Wang J., He X., Luo Y., Yarbrough W. G. (2006) Biochem. J. 393, 489–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J., An H., Mayo M. W., Baldwin A. S., Yarbrough W. G. (2007) Cancer Cell 12, 239–251 [DOI] [PubMed] [Google Scholar]
- 8.Kim H., Chen J., Yu X. (2007) Science 316, 1202–1205 [DOI] [PubMed] [Google Scholar]
- 9.Kim H., Lee Y. H., Won J., Yun Y. (2001) Biochem. Biophys. Res. Commun. 286, 886–894 [DOI] [PubMed] [Google Scholar]
- 10.Park S. G., Chung C., Kang H., Kim J. Y., Jung G. (2006) J. Biol. Chem. 281, 31770–31777 [DOI] [PubMed] [Google Scholar]
- 11.Kim H., Huang J., Chen J. (2007) Nat. Struct. Mol. Biol. 14, 710–715 [DOI] [PubMed] [Google Scholar]
- 12.Hofer B., Backhaus S., Timmis K. N. (1994) Gene 144, 9–16 [DOI] [PubMed] [Google Scholar]
- 13.Masumoto A., Arao S., Otsuki M. (1999) Hepatology 29, 68–74 [DOI] [PubMed] [Google Scholar]
- 14.Kasahara K., Taguchi T., Yamasaki I., Kamada M., Yuri K., Shuin T. (2002) Cancer Genet. Cytogenet. 137, 59–63 [DOI] [PubMed] [Google Scholar]
- 15.Saito S., Ghosh M., Morita K., Hirano T., Miwa M., Todoroki T. (2006) Oncol. Rep. 16, 949–956 [PubMed] [Google Scholar]
- 16.Sun M., Srikantan V., Ma L., Li J., Zhang W., Petrovics G., Makarem M., Strovel J. W., Horrigan S. G., Augustus M., Sesterhenn I. A., Moul J. W., Chandrasekharappa S., Zou Z., Srivastava S. (2006) DNA Cell Biol. 25, 597–607 [DOI] [PubMed] [Google Scholar]
- 17.van Gils W., Kilic E., Brüggenwirth H. T., Vaarwater J., Verbiest M. M., Beverloo B., van Til-Berg M. E., Paridaens D., Luyten G. P., de Klein A. (2008) Melanoma Res. 18, 10–15 [DOI] [PubMed] [Google Scholar]