Abstract
hPEBP4 (human phosphatidylethanolamine-binding protein 4) has been identified to be able to potentiate the resistance of breast, prostate, and ovarian cancers, with the preferential expression of hPEBP4, to tumor necrosis factor-α (TNF-α) or tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis, suggesting that inhibitors targeting the anti-apoptotic protein hPEBP4 may be useful to increase the sensitivity of hPEBP4-expressing cancer cells to TNF-α or TRAIL-induced apoptosis. By structure-based virtual screening and following surface plasmon resonance-based binding assay, seven small compounds were found to potently bind with hPEBP4. The hit compounds were further functionally screened for their ability to inhibit cancer cell growth, and one small compound, IOI-42, was identified to be able to promote TNF-α-mediated growth inhibition of MCF-7 breast cancer cells. IOI-42 could potentiate TNF-α-induced apoptosis of MCF-7 cells by inhibiting hPEBP4 and could suppress anchorage-independent cell growth of MCF-7 cells. We further demonstrated that IOI-42 could reduce the endogenous association of hPEBP4 with Raf-1/MEK1 and enhance the activation of ERK1/2 and JNK while inhibiting Akt activation. Furthermore, IOI-42 also promoted TRAIL-induced cell apoptosis of prostate cancer cells. Taken together, our data suggest that IOI-42, as the first chemical inhibitor of anti-apoptotic protein hPEBP4, may serve as a potential anti-tumor drug by sensitizing tumor cells to apoptotic inducers.
Keywords: Anticancer Drug, Apoptosis, Breast Cancer, Phosphatidylethanolamine, Tumor Necrosis Factor (TNF)
Introduction
Evasion of apoptosis is an important hallmark of tumor cells (1–4), for which many tumor cells become insensitive to radiation and chemotherapy. Therefore, alternative protocols aiming to induce apoptosis or block anti-apoptotic pathway need to be explored for the treatment of cancer. Enhancing chemosensitivity of tumor cells by targeting anti-apoptotic proteins represents one of the most important strategies in current cancer therapy. Now, several studies attempting to induce tumor cell apoptosis by targeting some well known anti-apoptotic proteins have shown promising results (5–9). However, the general expression of the most anti-apoptotic molecules with essential physical functions in a wide spectrum of cells or their nonspecific expression in cancer cells leads to serious side effects because of killing of normal cells when these targets were inhibited in vivo. Therefore, much effort now has been devoted to search for target agents that can specifically induce apoptosis in tumor cells, ideally with no or less toxicity to normal cells.
Human phosphatidylethanolamine (PE)4-binding protein 4 (hPEBP4) was identified as a novel member of the human phosphatidylethanolamine-binding protein (PEBP) family by our laboratory in 2004 (10). hPEBP4 is preferentially expressed in cancer cells including breast cancer cells, ovary cancer cells, and prostate cancer cells. Our further experiments show that hPEBP4 normally co-localizes with the lysosome, and TNF-α stimulation triggers its transfer to the cell membrane, where it binds to Raf-1 and MEK1. hPEBP4 can promote cellular resistance to TNF-α-induced apoptosis by inhibiting activation of JNK and the Raf-1/MEK/ERK pathway. hPEBP4, with preferential expression in cancer, functions as an antiapoptotic protein, thus being a potential target for cancer treatment (10).
TNF-α induces a broad range of cellular effects, including inflammatory responses and apoptosis, by activating signal transduction pathways, including NF-κB (5, 11, 12). Apoptosis induced by TNF is initiated at the membrane where engagement of the TNF receptor results in the recruitment of TNF receptor-associated death domain and then Fas-associated death domain. The death effector domain in FADD serves as a docking site for procaspase-8, which initiates apoptosis upon activation (13–15). We have demonstrated that silencing hPEBP4 significantly enhances both TNF-α-induced apoptosis of breast cancer cells and TRAIL-mediated apoptosis of ovarian and prostate tumor cells (16–18). Together, these studies indicate that hPEBP4 could be a candidate target to increase the sensitivity of cancer cells to cytotoxic effect of chemotherapy.
Considering that hPEBP4 is particularly abundant in tumor tissues (16, 17) and potentiates resistance to TNF-α- and TRAIL-induced apoptosis in certain kinds of cancer cells. In this study, we aimed to find a lead compound that specifically targets hPEBP4 to inhibit its anti-apoptotic function and subsequently increase the sensitivity of cancer cells to the killing of chemical drugs. By applying virtual screening based on a three-dimensional homology model of hPEBP4 together with functional confirmations, we identified for the first time a novel small molecule inhibitor, IOI-42, for hPEBP4 and demonstrated that IOI-42 could promote apoptosis of cancer cells induced by TNF-α and TRAIL by targeting hPEBP4. Our results suggest that IOI-42 may be a potential lead compound for the development of an antitumor chemical drug.
EXPERIMENTAL PROCEDURES
Reagents and Cell Lines
MCF-7, LNCaP, and L929 cells, obtained from ATCC, were grown in Dulbecco's modified Eagle's medium and RPMI 1640 medium, respectively, supplemented with 10% (v/v) fetal calf serum, 4.5 g/liter d-glucose, nonessential amino acids (100 μm), 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mm glutamine at 37 °C in a 5% CO2 atmosphere. Recombinant human TNF-α and TRAIL were from R&D Systems and Peprotech, respectively.
Homology Modeling
The sequence of hPEBP-4 was retrieved from GenBankTM (NM_144962). BLAST was used to search the Protein Data Bank for homologous proteins of hPEBP-4. The crystal structures of bovine PEBP-1 (Protein Data Bank code 1A44), human PEBP-1 (Protein Data Bank code 1BEH), and murine PEBP-2 (Protein Data Bank code 1KN3) were selected as reference structures. Multiple structure alignment algorithm encoded in InsightII was used to align reference sequences based on the similarity of their corresponding three-dimensional structures. The sequence of hPEBP-4 was aligned to Protein Data Bank code 1A44 by pairwise alignment, and then the alignment was manually adjusted to obtain a fine alignment for three-dimensional structure construction. The three-dimensional model of hPEBP-4 was generated by using the MODELLER program (19) encoded in InsightII. Finally, the whole structure was optimized by 200 steps of steepest descent, followed by Powell minimization to a root-mean-square energy gradient of 0.05 kcal/(mol·Å). The AMBER force field with the Kollman all-atom charges was employed during the optimization. The quality of the model was checked by Procheck (20).
Virtual Screening
The SPECS database containing the structural information of >270,000 compounds was used for the virtual screening. DOCK 4.0 program (21) was used for primary screening. Based on a crystal structure of the PE-binding domain of human PEBP1/RKIP (SWISS-MODEL code 1BD9) (22), the PE-binding pocket formed by residues within a radius of 6 Å around the binding site of hPEBP4 was used to construct the grids for the docking procedure. During the docking calculation, Kollman all-atom charges were assigned to the protein, and Gasteiger-Marsili charges were assigned to the small molecules of SPECS database. The candidate compounds were rescored using the program FlexX (23), AutoDock 3.05 (24), and our in-house drug-like filter (25), consecutively. At the last step, visual inspection was applied to pick the final candidates.
Surface Plasmon Resonance (SPR)-based Binding Assay
The measurement was performed using the dual flow cell Biacore 3000 instrument (Biacore AB) as described previously (26). The hPEBP4 protein was expressed by vector pGEX4T-1 in Escherichia coli strain BL21 with an N-terminal glutathione S-transferase tag. Subsequently, hPEBP4 protein was purified and diluted in 10 mm sodium acetate buffer (pH 4.5) to a final concentration of ∼50 μg/ml, and then covalently immobilized to the hydrophilic carboxymethylated dextran matrix of the sensor chip by the standard primary amine coupling method (26). Biacore data were collected at 25 °C with HBS-EP (10 mm HEPES, 150 mm NaCl, 3.4 mm EDTA, and 0.005% [v/v] surfactant P20, pH 7.4) as the running buffer at a constant flow of 30 μl/min.
Cell Growth Analysis
To determine the cell growth, MCF-7 cells were seeded into 96-well plates at 3 × 103 cells per well and treated with 20 ng/ml TNF-α in the presence or absence of small molecules for various incubation periods and concentrations. On the day of harvest, 200 μl medium were replaced with an equal volume of fresh medium containing 10% 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, 5 mg/ml stock. Plates were incubated at 37 °C for 4 h, and then 100 μl of dimethyl sulfoxide (Sigma) was added to each well, and plates were shaken at room temperature for 10 min. The cell growth was determined by measuring the absorbance of the converted dye at a wavelength of 570 nm (27).
Cell Apoptosis Assay
For apoptosis assay, cells were washed, resuspended in the staining buffer, and stained with propidium iodide (Sigma) and Vybrant apoptosis assay (Invitrogen) or R123 (R-302, Molecular Probes) according to the instruction of the manufacturer (28, 29). Stained cells were analyzed by fluorescence-activated cell sorting (FACSCalibur, Becton Dickinson).
Western Blot Analysis
A BCA protein assay reagent kit (Pierce) was used to measure protein concentration. Samples containing equal amounts of protein were separated by 12% SDS-PAGE and transferred to Protran nitrocellulose membranes (Schleicher & Schuell, Keene, NH). Blots were probed with antibodies specific for phospho-ERK1/2, phospho-JNK, and phospho-p38 (Cell Signaling Technology, Beverly, MA) and MEK1, Raf-1, (Santa Cruz Biotechnology, Santa Cruz, CA) with appropriate horseradish peroxidase-conjugated antibodies as secondary antibodies (Cell Signaling Technology). SuperSignal West Femto maximum sensitivity substrate (Pierce) was used for the chemiluminescent visualization of proteins (27, 30).
Co-immunoprecipitation
Cell lysates were precleared with protein A-Sepharose beads (Sigma), and immunoprecipitation was done using anti-hPEBP4 polyclonal antibody (10) cross-linked to protein A-Sepharose beads. Samples were subjected to Western blot analysis directly.
hPEBP4 RNA Interference Assay
21-nucleotide sequences of hPEBP4-specific siRNA and hPEBP4 mutation control siRNA were synthesized by Proligo as described previously (16). For annealing, 20 μm single-stranded 21-nucleotide RNAs were incubated in annealing buffer (100 mm potassium acetate, 30 mm HEPES-KOH at pH 7.4, 2 mm magnesium acetate) for 1 min at 90 °C and then for 1 h at 37 °C. siRNA duplexes were transfected into MCF-7 breast cancer cells using Oligofectamine reagent (Invitrogen), as described previously (16). All of the data shown in this article are representative of at least three independent experiments.
Anchorage-independent Growth Assay
Cells (1 × 104/well) were seeded into 0.3% Bacto-agar (Difco) over a 0.6% agar bottom layer in triplicate in six-well plates. Both layers contained 1× Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Plates were incubated at 37 °C in 5% CO2 for 3 weeks. IOI-42 and/or TNF-α were added every 3 days. Clones with >50 cells were counted.
Statistical Analysis
The Student's t test was used to determine the statistical significance of the data obtained and to compare the means between groups. A p value of < 0.05 represented a statistically significant difference.
RESULTS
Identification of IOI-42 as an Inhibitor of Anti-apoptotic Protein hPEBP4
The DOCK program was employed as the first step in the preliminarily screening of the potential inhibitors of hPEBP4 based on the three-dimensional structure model. The top 8,700 molecules with the highest score as obtained by DOCK search were subsequently rescored using the FlexX program, and then the top 600 molecules were subjected to AutoDock 3.05 and our in-house drug-like filter for rescoring. Finally, 100 molecules were manually selected from the top molecules of the last step as inhibitor candidates. Of those 100 candidates, 83 compounds could be purchased from the SPECS Company for further experimental assay.
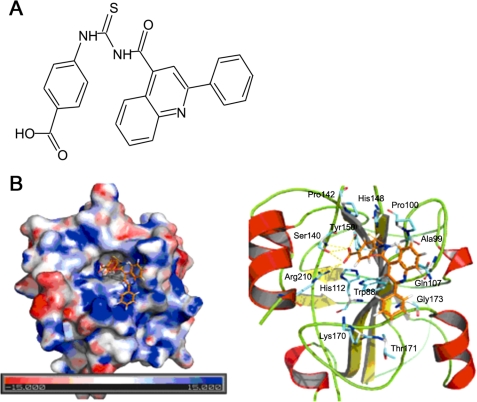
Next, the SPR biosensor technique was adopted as a method to screen compounds for receptor binding in vitro (31) to determine the binding capacity of those hit compounds with hPEBP4. In sum, seven compounds were found to actively interact with hPEBP4 in vitro with efficient estimated KD values (data not shown). Considering that silencing of hPEBP4 significantly enhances TNF-α-induced cell death of MCF-7 human breast cancer cells (16), we then used 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay to screen functionally active compounds that could potentiate TNF-α-induced cell growth inhibition at various concentrations. We found that two of these seven compounds could significantly enhance TNF-α-induced growth inhibition at concentrations of 5–10 μm; however, one compound showed serious cytotoxic effect even when used alone (data not shown). Thus, we only carried out further experiments with IOI-42, which alone showed no significant cytotoxic effect on the growth of MCF-7 cells, as silencing of hPEBP4 alone did not influence spontaneous growth of MCF-7 cells (10). Structure of IOI-42 was shown in Fig. 1A. Examining the interaction of hPEBP4 and IOI-42 on the three-dimensional structure model (Fig. 1B), we found that hPEBP4 consisted of one central β-sheet flanked by a smaller β-sheet on one side, an α-helix on other side, and another α-helix at the end of the C-terminal. The surface of hPEBP4 shows a significant positive charged area around the binding pocket. The negatively charged carboxyl head group of IOI-42 stretches down into bottom of the positively charged pocket and makes strong interactions with hPEBP4 by forming six hydrogen bonds with Ser140, Tyr150, and Arg210, whereas the benzene ring adjacent to the carboxyl group makes interactions with residues in the pocket by hydrophobic interactions. Moreover, the left part of IOI-42, mainly comprised of aromatic rings, also forms hydrophobic interactions with residues on the surface area of hPEBP4 near the entrance of the pocket. Thus, IOI-42 may be the inhibitor for hPEBP4 by targeting at the conserved PE-binding domain.
FIGURE 1.
Identification of small compound IOI-42 as the inhibitor of anti-apoptotic protein hPEBP4. A, chemical structure of IOI-42. B, simulated docking model of IOI-42 (colored in brown and shown in sticks mode) and hPEBP4. The docking model was generated with the AutoDock program and was visualized with PyMOL (DeLano Scientific LLC). The left panel is an overview of the binding pocket. The electrostatic potential surface map of hPEBP4 was generated by the Adaptive Poisson-Boltzmann Solver software package. On the right panel are the predicted structural contacts of IOI-42 in the binding pocket of hPEBP4. The hydrogen bonds were represented by yellow dashed lines. Hydrophobic interactions are not shown.
Promotion of TNF-α- or TRAIL-induced Tumor Cell Apoptosis by IOI-42
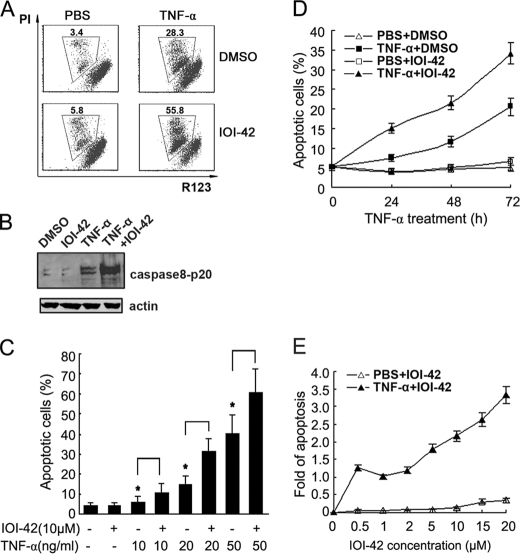
hPEBP4 is regarded as an anti-apoptotic protein for its role in apoptosis resistance of tumor cells to TNF-α and TRAIL (10, 16, 17). Thus, we wondered whether IOI-42 could potentiate TNF-α-induced tumor cell apoptosis. We first used rhodamine 123 (R123) and phosphatidylinositol (PI) labeling for the detection of apoptotic cells. Consistent with the result observed after hPEBP4 was silenced by siRNA in MCF-7 cells (16), IOI-42 pretreatment could significantly enhance the TNF-α-induced apoptosis of MCF-7 cells, as shown by increased percentage of apoptotic cells (Fig. 2, A and C) and downstream caspase-8 activation (Fig. 2B), whereas IOI-42 alone did not cause any apoptosis of MCF-7 cells compared with dimethyl sulfoxide control. To further confirm the phenomenon we observed above, fluorescein isothiocyanate-conjugated annexin V/PI staining was also performed to detect apoptotic cancer cells induced with different concentrations of TNF-α. IOI-42 could significantly potentiate the cellular apoptosis of MCF-7 cells induced by 10, 20, and 50 ng/ml TNF-α, respectively (Fig. 2C). IOI-42 pretreatment and 20 ng/ml TNF-α treatment for 24–48 h could increase the apoptosis of MCF-7 cells by ∼1-fold when compared with treatment with TNF-α alone (Fig. 2D). In addition, when cells were pretreated with different concentrations of IOI-42, IOI-42 could exert its proapoptotic effect in a dose-dependent manner (Fig. 2E). These data suggest that IOI-42 could promote TNF-α-induced tumor cell apoptosis.
FIGURE 2.
IOI-42 potentiates TNF-α-induced apoptosis of human breast cancer cells. A and B, MCF-7 cells were pretreated with IOI-42 (10 μm) for 4 h and then treated with TNF-α (20 ng/ml) for 30 h. Cells were stained with rhodamine 123 (R123) and PI and then followed by FACS analysis. The percentages of the gated cells represented the dead cells (A). Cell lysates were immunoblotted by anti-caspase-8 p20 antibody (B). C, MCF-7 cells were pretreated with IOI-42 (10 μm) for 4 h and then treated with increasing concentration of TNF-α (as indicated) for 30 h, and then the cells were stained with AnnexinV and PI for analysis of cell apoptosis by FACS analysis. Data presented the percentage of AnnexinV positive cells. D, MCF-7 cells were treated with 10 μm IOI-42 for 4 h followed by TNF-α (20 ng/ml) treatment for various times. E, MCF-7 cells were treated with various concentrations of IOI-42 for 4 h followed by TNF-α (20 ng/ml) stimulation for 30 h. Cellular apoptosis was then determined by Annexin V/PI staining as in C. Data represent the fold of increase over apoptosis induced by TNF-α alone. Data show means ± S.D. of triplicates from one experiment representative of three experiments. *, p < 0.05. PBS, phosphate-buffered saline; DMSO, dimethyl sulfoxide.
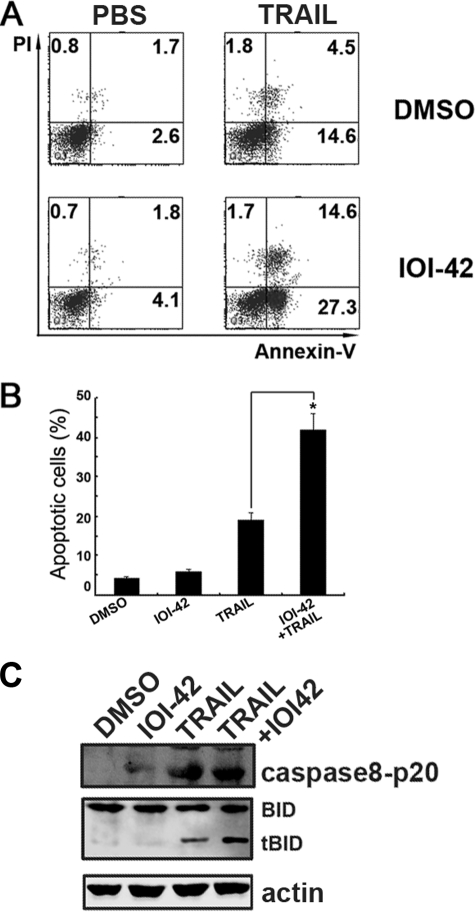
Because hPEBP4 has been proved to play a role in the resistance of tumor cells to the apoptosis induction by TRAIL as well (17, 18), we then tested whether IOI-42 could sensitize LNCaP cells to TRAIL-induced apoptosis. LNCaP is a human prostate cancer cell line with high expression of hPEBP4 and resistance to TRAIL-induced apoptosis, while IOI-42 pretreatment significantly potentiated TRAIL-induced apoptosis of LNCaP cells than when TRAIL used alone (Fig. 3, A and B; p < 0.05), accompanied with increased caspase-8 and BH3-interacting domain death agonist cleavage (Fig. 3C). The data indicate that IOI-42 also sensitizes tumor cells to TRAIL-induced apoptosis.
FIGURE 3.
IOI-42 promotes TRAIL-induced apoptosis of human prostate cancer cells. LNCaP cells were pretreated with IOI-42 (10 μm) for 4 h and then treated with TRAIL (200 ng/ml) for 48 h. Cells were stained with Annexin V/PI staining and followed by FACS analysis. A, the representative dot plot of apoptosis assay. Data presented the percentage of Annexin V-positive cells. B, data show means ± S.D. of triplicates from one experiment representative of three experiments. *, p < 0.05. C, cell lysates from A were immunoblotted by anti-caspase-8 p20 antibody. DMSO, dimethyl sulfoxide; PBS, phosphate-buffered saline. tBID, truncated BID.
Effect of IOI-42 on Anchorage-independent Growth of Tumor Cells
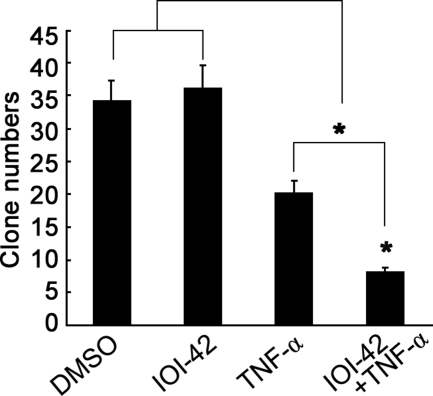
Due to the lack of a generally accepted in vivo model to study TNF-α-based tumor therapy, we instead observed the effect of IOI-42 on the anchorage-independent survival of MCF-7 cells under a long term treatment of TNF-α in vitro. TNF-α alone could inhibit colony formation of tumor cells dramatically, and if combined with IOI-42 pretreatment, TNF-α-mediated inhibition of colony formation was much more significant than TNF-α used alone (Fig. 4). These data further suggest that IOI-42 may be effective in inhibiting tumor growth if used together with TNF-α.
FIGURE 4.
IOI-42 enhances the inhibitory effect of TNF-α on anchorage-independent growth of human breast cancer cells. MCF-7 cells were seeded into a six-well plate as described under “Experimental Procedures” and subjected to treatment with IOI-42 (10 μm) and TNF-α (20 ng/ml) once every 2 days for a total of 3 weeks before clones were counted. Data show means ± S.D. of triplicates from one experiment representative of three experiments. *, p < 0.05. DMSO, dimethyl sulfoxide.
IOI-42 Potentiates TNF-α-induced Apoptosis by Targeting hPEBP4
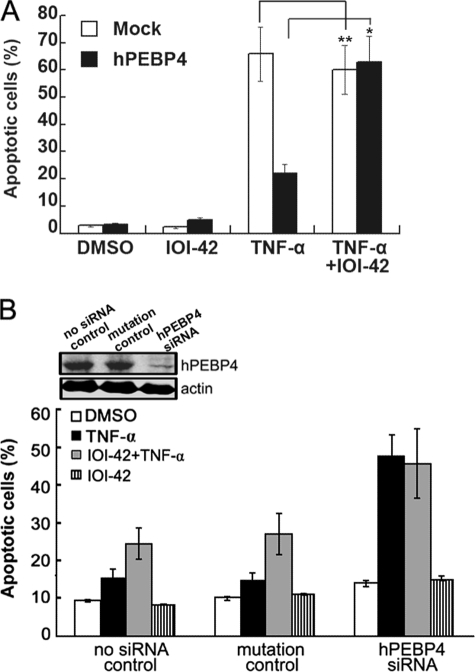
Although IOI-42, initially identified as a hPEBP4-targeted small compound, has been proved to be able to potentiate TNF-α-induced apoptosis as silencing of hPEBP4 does, we still wondered whether hPEBP4 was the real intracellular target of IOI-42. For this purpose, we first observed the effect of IOI-42 on TNF-α-induced apoptosis of L929 cells, a general model cell used to study TNF-α-mediated apoptosis but without endogenous hPEBP4 expression. TNF-α caused dramatic apoptosis of L929 cells, but IOI-42 pretreatment failed to further increase the apoptosis percentage, whereas overexpression of hPEBP4 significantly decreased TNF-α-induced apoptosis, which can be reversed by IOI-42 pretreatment (Fig. 5A). To rule out false effect caused by the genetic difference between MCF-7 cells and L929 cells, we silenced the expression of hPEBP4 by RNA interference in MCF-7 cells and then observed the effect of IOI-42 on TNF-α-induced apoptosis. IOI-42 pretreatment yielded no promotion of TNF-α-induced apoptosis in MCF-7 cells when hPEBP4 expression was efficiently depleted by siRNA, which is consistent with that observed in L929 cells (Fig. 5B). Furthermore, if IOI-42 was added after TNF-α, the cellular apoptosis was significantly reduced to about the level of cells treated with TNF-α alone (data not shown). This is consistent with our previous reports (10, 15, 16,) that hPEBP4 exerts its effect at the early stage of TNF-α-induced signal pathway. Taken together, these data suggest that IOI-42, as purposely designed, targeted hPEBP4 protein to exert its anti-tumor effect by sensitizing tumor cells to the TNF-α-induced apoptosis.
FIGURE 5.
IOI-42 potentiates TNF-α-induced cell apoptosis by targeting hPEBP4. A, L929 cells were transfected as indicated, and 48 h later, cells were pretreated with or without 10 μm IOI-42 for 4 h followed by TNF-α (20 ng/ml) stimulation for 24 h. Cellular apoptosis was determined by Annexin V/PI staining followed by FACS analysis. B, MCF-7 cells were transfected with hPEBP4-specific siRNA. 48 h later, cells were treated with 10 μm IOI-42 in presence or absence of 20 ng/ml TNF-α for 30 h. Cellular apoptosis was determined by Annexin V/PI staining followed by FACS analysis. Cells were also subjected to Western blot analysis to monitor the expression of hPEBP4. Data show means ± S.D. of triplicates from one experiment representative of three experiments. *, p < 0.05 and **, p > 0.05. DMSO, dimethyl sulfoxide.
IOI-42 Inhibits the Association of hPEBP4 with Raf-1/MEK1 in Tumor Cells
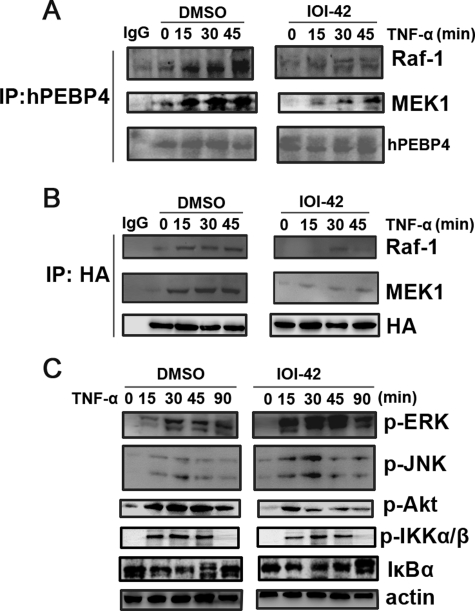
Previously, we have found that hPEBP4 depletion by siRNA significantly sensitized MCF-7 cells to TNF-α-induced apoptosis accompanied by abortion of endogenous association of hPEBP4 with Raf-1-MEK1 as well as greater activation of ERK1/2 and JNK (10, 16). It is assumed that an effective inhibitor of hPEBP4 should also have a similar effect. The results showed that upon TNF-α stimulation, IOI-42 treatment significantly decreased the interaction of Raf-1/MEK1 with both endogenous hPEBP4 in MCF-7 cells (Fig. 6B) and exogenous expressed hemagglutinin-tagged hPEBP4 in L929 cells (Fig. 6C). And IOI-42 significantly attenuated Akt activation, promoted TNF-α-induced activation of ERK and JNK, while it only slightly prolonged activation duration of JNK and slightly decreased activation of IκB kinase α/β and IκBα (Fig. 6A). The data suggest that IOI-42 could impair the association of hPEBP4 with Raf-1 and MEK1 specifically and potentially and therefore block the suppressive effect of hPEBP4 on the TNF-α-induced activation of Raf-1/MEK1/ERK cascade.
FIGURE 6.
Effect of IOI-42 pretreatment on the association of hPEBP4 with Raf-1/MEK1 and ERK activation. A, MCF-7 cells were pretreated with 10 μm IOI-42 for 4 h, followed by TNF-α (20 ng/ml) stimulation for the indicated time. Cell lysates were immunoprecipitated (IP) with anti-hPEBP4 antibody and then immunoblotted using anti-Raf-1, MEK1, or hPEBP4 antibody. B, L929 cells were transfected with hemagglutinin (HA)-tagged hPEBP4 or mock plasmid. 48 h later, the cells were with 10 μm IOI-42 for 4 h, followed by TNF-α (20 ng/ml) stimulation as indicated. Cell lysates were immunoprecipitated with anti-hemagglutinin antibody and then immunoblotted using anti-Raf-1, MEK1, or hemagglutinin antibodies. C, cell lysates in A were immunoblotted by the indicated antibodies. DMSO, dimethyl sulfoxide.
DISCUSSION
As a novel member of human PEBP family that we identified, hPEBP4 is selectively expressed in several types of cancer cells and could inhibit TNF-α/TRAIL-induced apoptosis by inhibiting mitogen-activated protein kinase activation (MAPK) (10, 17). We also have reported that sensitization of breast cancer cells to TNF-α-induced apoptosis by hPEBP4-targeted siRNA, and the increase of TNF-α-induced cell death by hPEBP4 silencing was significantly inhibited by PD98059 or SP600125, inhibitors of ERK and JNK activation, respectively (16). All of these results strongly imply that the anti-apoptotic protein hPEBP4 could be a potential target for drug design for cancer therapy. In the present study, we have adopted a three-dimensional structure-based virtual screening approach coupled with functional experiments to successfully identify IOI-42 as the first chemical inhibitor of hPEBP4. We show that this compound was able to interact with hPEBP4 in vitro, weaken endogenous association of hPEBP4 with Raf-1 and MEK1, increase ERK1/2 and JNK activation, and potentiate TNF-α/TRAIL-induced tumor cell apoptosis. We also have excluded the possibility that apoptosis induction was possibly due to the promotion by off-target effect because the proapoptotic effect of the compound disappeared either in hPEBP4-silenced MCF-7 cells or hPEBP4-negative cells. Data from this study suggest that IOI-42, as a first generation inhibitor for hPEBP4, is effective in sensitizing cancer cells to cytotoxic agents by targeting specially to hPEBP4. Given the restricted and high expression in tumor tissues of hPEBP4, IOI-42 may be able to provide a wide therapeutic window for the potential application of TNF-α in cancer therapy.
To understand the mechanisms by which IOI-42 sensitizes cancer cells to TNF-α and TRAIL-induced apoptosis, we evaluated the simulated model of IOI-42 docking on hPEBP4. In this model, most of the interacting residues of hPEBP4 are in or near the two highly conserved motifs across the PEBP family, DPDxPxnH (residues 96–112, where n is 11 in all mammalian proteins), and GxHR (residues 146–149). Both motifs are within the PE-binding domain (residues 84–191), which has been proved to bind Raf-1 and MEK (10). The major interaction involved in the binding is hydrogen bonding. The carboxyl acid head group of IOI-42 forms strong hydrogen bonds with Ser140, Tyr150, and Arg210 of hPEBP4. But hydrophobic contacts also make significant contributions to the interaction. We suspect that the interactions between hPEBP4 and the benzoic acid group of IOI-42 govern the binding specificity of IOI-42. Because the volume of the binding pocket is rather small, there is not much room to accommodate more groups other than the benzoic acid group. To further improve the inhibitory activity of IOI-42 on hPEBP4, it may be applicable to modify other parts of IOI-42 to achieve better fitting to the surface area near the entrance of the binding pocket to make more interactions while retaining the key interactions essential to the binding specificity.
Both TNF-α and TRAIL are promising drugs for cancer treatment (32, 33). However, serious systematic toxicities of TNF-α and the broad spectrum resistance of TRAIL dampened the hope for their wide application (34, 35). TNF-α-induced activation of NF-κB protects cells from apoptosis, whereas prolonged activation of JNK and increased production of ROS potentiate cell death. Here, we found that IOI-42 promoted apoptosis with a slightly inhibited NF-κB activation and slightly prolonged JNK activation. The data are in accordance with our previous reports (10, 15, 16) that hPEBP4 functions mainly in early signal transduction of TNF-α by inhibiting ERK and JNK activation, while promoting Akt activation (10, 16, 17). Furthermore, we found that preincubation of IOI-42 dramatically weakened the association of hPEBP4 with Raf-1/MEK1 and promoted TNF-α-induced apoptosis in MCF-7 cells, but not in another more malignant breast cancer cell line, MDA-MB-231 (data not shown). This difference is probably caused by differential activation of NF-κB in the two cell lines (8, 36). Overactivated NF-κB can both inhibit the complex II-mediated programmed cell death and suppress JNK activation by up-regulating anti-apoptotic proteins (14, 37). Thus, the effect of IOI-42 may be abrogated in MDA-MB-231 cells. This fact suggests that curing tumors with a single-target-based drug is too difficult due to the heterogeneous nature of the tumor cell population. It was reported that overexpression of mPEBP4, the murine counterpart of hPEBP4, can promote breast cancer metastasis to lung by up-regulating COX-2 (38). Therefore, we speculate that IOI-42 might have a potential application in other aspects of tumor behaviors if further investigation could clarify the effect of hPEBP4 on tumor biology.
In summary, we have identified IOI-42 as the first chemical inhibitor of hPEBP4 and have shown that IOI-42 can sensitize tumor cells to TNF-α- and TRAIL-mediated apoptosis. The data suggest that IOI-42 may be one potential candidate to be used for tumor therapy in combination with cytotoxic drugs. Further modification is necessary to achieve a better fitting into the groove and to increase the docking affinity in the future.
Acknowledgments
We sincerely thank Drs. X. Wang and H. Li for helpful discussion and Y. Zhang, M. Jin, and Y. Li for technical assistance.
This work was supported by grants from the National High Biotechnology Development Program of China (2009ZX09503-003, 2009AA02Z101, and 2006AA02A305), the National Natural Science Foundation of China (30721091 and 30772504), the National Key Basic Research Program of China (2010CB911903), and the Shanghai Committee of Science and Technology (09QH1402800 and 09SG35).
- PE
- phosphatidylethanolamine
- PEBP
- phosphatidylethanolamine-binding protein
- TRAIL
- tumor necrosis factor-related apoptosis-inducing ligand
- PI
- phosphatidylinositol
- siRNA
- small interfering RNA
- ERK
- extracellular signal-regulated kinase
- JNK
- c-Jun N-terminal kinase
- MEK
- mitogen-activated protein kinase/extracellular signal-regulated kinase kinase
- FACS
- fluorescence-activated cell sorter
- TNF
- tumor necrosis factor.
REFERENCES
- 1.Jana S., Paliwal J. (2007) Curr. Med. Chem. 14, 2369–2379 [DOI] [PubMed] [Google Scholar]
- 2.Rodier F., Campisi J., Bhaumik D. (2007) Nucleic Acids Res. 35, 7475–7484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanahan D., Weinberg R. A. (2000) Cell 100, 57–70 [DOI] [PubMed] [Google Scholar]
- 4.Viatour P., Merville M. P., Bours V., Chariot A. (2005) Trends Biochem. Sci. 30, 43–52 [DOI] [PubMed] [Google Scholar]
- 5.Li L., Thomas R. M., Suzuki H., De Brabander J. K., Wang X., Harran P. G. (2004) Science 305, 1471–1474 [DOI] [PubMed] [Google Scholar]
- 6.Van Molle W., Wielockx B., Mahieu T., Takada M., Taniguchi T., Sekikawa K., Libert C. (2002) Immunity 16, 685–695 [DOI] [PubMed] [Google Scholar]
- 7.Vassilev L. T., Vu B. T., Graves B., Carvajal D., Podlaski F., Filipovic Z., Kong N., Kammlott U., Lukacs C., Klein C., Fotouhi N., Liu E. A. (2004) Science 303, 844–848 [DOI] [PubMed] [Google Scholar]
- 8.Eddy S. F., Guo S., Demicco E. G., Romieu-Mourez R., Landesman-Bollag E., Seldin D. C., Sonenshein G. E. (2005) Cancer Res. 65, 11375–11383 [DOI] [PubMed] [Google Scholar]
- 9.Shoemaker A. R., Oleksijew A., Bauch J., Belli B. A., Borre T., Bruncko M., Deckwirth T., Frost D. J., Jarvis K., Joseph M. K., Marsh K., McClellan W., Nellans H., Ng S., Nimmer P., O'Connor J. M., Oltersdorf T., Qing W., Shen W., Stavropoulos J., Tahir S. K., Wang B., Warner R., Zhang H., Fesik S. W., Rosenberg S. H., Elmore S. W. (2006) Cancer Res. 66, 8731–8739 [DOI] [PubMed] [Google Scholar]
- 10.Wang X., Li N., Liu B., Sun H., Chen T., Li H., Qiu J., Zhang L., Wan T., Cao X. (2004) J. Biol. Chem. 279, 45855–45864 [DOI] [PubMed] [Google Scholar]
- 11.Gaur U., Aggarwal B. B. (2003) Biochem. Pharmacol. 66, 1403–1408 [DOI] [PubMed] [Google Scholar]
- 12.Aggarwal B. B., Shishodia S., Ashikawa K., Bharti A. C. (2002) Curr. Drug Targets. Inflamm. Allergy 1, 327–341 [DOI] [PubMed] [Google Scholar]
- 13.Wajant H., Pfizenmaier K., Scheurich P. (2003) Cell Death. Differ. 10, 45–65 [DOI] [PubMed] [Google Scholar]
- 14.Aggarwal B. B. (2003) Nat. Rev. Immunol. 3, 745–756 [DOI] [PubMed] [Google Scholar]
- 15.Muppidi J. R., Tschopp J., Siegel R. M. (2004) Immunity 21, 461–465 [DOI] [PubMed] [Google Scholar]
- 16.Wang X., Li N., Li H., Liu B., Qiu J., Chen T., Cao X. (2005) Clin. Cancer Res. 11, 7545–7553 [DOI] [PubMed] [Google Scholar]
- 17.Li H., Wang X., Li N., Qiu J., Zhang Y., Cao X. (2007) J. Biol. Chem. 282, 4943–4950 [DOI] [PubMed] [Google Scholar]
- 18.Li P., Wang X., Li N., Kong H., Guo Z., Liu S., Cao X. (2006) Int. J. Mol. Med. 18, 505–510 [PubMed] [Google Scholar]
- 19.Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 20.Morris A. L., MacArthur M. W., Hutchinson E. G., Thornton J. M. (1992) Proteins 12, 345–364 [DOI] [PubMed] [Google Scholar]
- 21.Ewing T. J., Makino S., Skillman A. G., Kuntz I. D. (2001) J. Comput. Aided Mol. Des. 15, 411–428 [DOI] [PubMed] [Google Scholar]
- 22.Banfield M. J., Barker J. J., Perry A. C., Brady R. L. (1998) Structure. 6, 1245–1254 [DOI] [PubMed] [Google Scholar]
- 23.Hoffmann D., Kramer B., Washio T., Steinmetzer T., Rarey M., Lengauer T. (1999) J. Med. Chem. 42, 4422–4433 [DOI] [PubMed] [Google Scholar]
- 24.Park H., Lee J., Lee S. (2006) Proteins 65, 549–554 [DOI] [PubMed] [Google Scholar]
- 25.Zheng S., Luo X., Chen G., Zhu W., Shen J., Chen K., Jiang H. (2005) J. Chem. Inf. Model. 45, 856–862 [DOI] [PubMed] [Google Scholar]
- 26.Chen L., Gui C., Luo X., Yang Q., Günther S., Scandella E., Drosten C., Bai D., He X., Ludewig B., Chen J., Luo H., Yang Y., Yang Y., Zou J., Thiel V., Chen K., Shen J., Shen X., Jiang H. (2005) J. Virol. 79, 7095–7103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang C., Li N., Liu X., Zheng Y., Cao X. (2008) J. Biol. Chem. 283, 11565–11574 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Li N., Wang C., Wu Y., Liu X., Cao X. (2009) J. Biol. Chem. 284, 3021–3027 [DOI] [PubMed] [Google Scholar]
- 29.Li N., Zheng Y., Chen W., Wang C., Liu X., He W., Xu H., Cao X. (2007) Cancer Res. 67, 11176–11185 [DOI] [PubMed] [Google Scholar]
- 30.Wang C., Chen T., Zhang J., Yang M., Li N., Xu X., Cao X. (2009) Nat. Immunol. 10, 744–752 [DOI] [PubMed] [Google Scholar]
- 31.Cooper M. A. (2002) Nat. Rev. Drug Discov. 1, 515–528 [DOI] [PubMed] [Google Scholar]
- 32.Johnstone R. W., Frew A. J., Smyth M. J. (2008) Nat. Rev. Cancer 8, 782–798 [DOI] [PubMed] [Google Scholar]
- 33.Shi R. X., Ong C. N., Shen H. M. (2004) Oncogene 23, 7712–7721 [DOI] [PubMed] [Google Scholar]
- 34.Lejeune F. J., Liénard D., Matter M., Rüegg C. (2006) Cancer Immun. 6, 6. [PubMed] [Google Scholar]
- 35.Ashkenazi A. (2002) Nat. Rev. Cancer 2, 420–430 [DOI] [PubMed] [Google Scholar]
- 36.Cao Z., Xiong J., Takeuchi M., Kurama T., Goeddel D. V. (1996) Nature 383, 443–446 [DOI] [PubMed] [Google Scholar]
- 37.Hayden M. S., Ghosh S. (2008) Cell 132, 344–362 [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y., Wang X., Xiang Z., Li H., Qiu J., Sun Q., Wan T., Li N., Cao X., Wang J. (2007) Int. J. Mol. Med. 19, 55–63 [PubMed] [Google Scholar]