Abstract
The α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid (AMPA) subtype of ionotropic glutamate receptors mediates much of the fast excitatory neurotransmission in the central nervous system. The ability of these receptors to shape such responses appears to be due in part to dynamic processes induced by agonists in the ligand-binding domain. Previous studies employing fluorescence spectroscopy and whole cell recording suggest that agonist binding is followed by sequential transitions to one or more distinct conformational states. Here, we used hydrogen-deuterium exchange to determine the mechanisms of binding of glutamate and kainate (full and partial agonists, respectively) to a soluble ligand-binding domain of GluR2. Our results provide a structural basis for sequential state models of agonist binding and the free energy changes of the associated state-to-state transitions. For glutamate, a multi-equilibrium binding reaction was discerned involving distinct ligand docking, domain isomerization, and lobe-locking steps. In contrast, kainate binding involves a simpler dock-isomerization process in which the isomerization equilibrium is shifted dramatically toward open domain conformations. In light of increasing evidence that the stability, in addition to the extent, of domain closure is a critical component of the channel activation mechanism, the differences in domain opening and closing equilibria detected for glutamate and kainate should be useful structural measures for interpreting the markedly different current responses evoked by these agonists.
Keywords: Glutamate Receptors Ionotropic (AMPA, NMDA); Neuroscience; NMR; Receptor Desensitization; Receptor Structure-Function
Introduction
As a consequence of glutamate binding, AMPA2 receptors produce a rapid flux of cations into post-synaptic neurons that is vital for information transfer in the central nervous system. In addition, these receptors undergo fast deactivation and desensitization along with slower recovery kinetics, which together shape the time course of current decay and restoration (1–6). Structurally, AMPA receptors are tetrameric membrane-bound proteins composed of modular subunits having two large extracellular domains, a membrane-spanning ion channel, and a C-terminal intracellular region (4, 7–12). One of the extracellular domains, S1S2, is a bilobed structure that binds agonists in a cleft between its lobes (13). Domain closure of S1S2 around the agonist leads to channel opening, and maximum currents have been attributed to complete lobe closure upon complex formation with full agonists (13–17). However, among the full agonists examined, those like glutamate, which are less potent and which bind to S1S2 with lower affinity, produce higher rates of channel deactivation and resensitization (18), despite inducing the same degree of cleft closure in S1S2. This suggests that AMPA receptor function is dependent not only on the extent of lobe closure caused by the agonist but also on the lobe closing and opening dynamics associated with agonist binding and dissociation (19).
A prevailing model for the binding reaction is
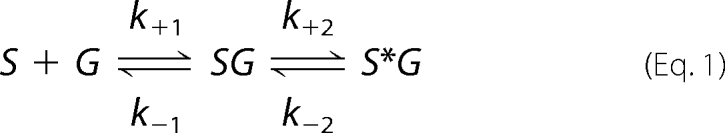 |
where SG represents a state in which glutamate (G) has rapidly docked to sites in lobe 1 of S1S2 (S) (20). The species S*G establishes key contacts between glutamate and lobe 2, which brings about domain closure and triggers the opening of a gate in the ion channel. This has been referred to as the dock-isomerization model and is consistent with data from stopped flow (20) and time-resolved Fourier transform infrared spectroscopy experiments (21). However, the following model, which was recently proposed based on whole cell recordings of GluR2 channels, implies a related but more complicated binding reaction (18),
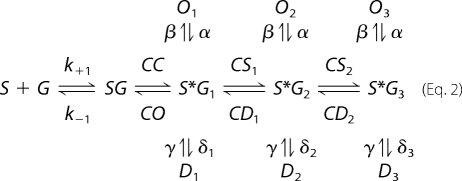 |
where Oi and Di (where i = 1, 2, or 3) denote open and desensitized channel states, respectively. Although this model accounts for structural changes occurring throughout a subunit of the tetrameric receptor, SG, S*G1, S*G2, and S*G3 were suggested to represent unique conformational states in the binding cleft of S1S2, with S*G3 possessing the highest degree of cleft closure stability (18, 19). The sequential nature of this mechanism was found to be critical to reproduce current responses for full agonists of varying potency and binding affinity for S1S2. For glutamate, receptors primarily occupy S*G1, whereas higher potency full agonists show increased occupation of S*G2 and S*G3. Importantly, though, longer application of glutamate followed by its sudden removal under nondesensitizing conditions leads to slower current decays corresponding to increased sampling of S*G2 and S*G3. According to Equation 2, these deactivation rates are dependent on agonist binding affinity and application time because increases in either of these variables increase the probability that the binding cleft of S1S2 is closed and populating S*G2 or S*G3 (18). However, solution studies of S1S2 have yet to answer the more fundamental question of whether lobe opening and closure occur by way of a single domain isomerization event, according to Equation 1, or whether there are indeed additional conformational equilibria in the binding cleft distinct from lobe isomerization that have measurable free energies, as suggested by Equation 2.
In the present study, this question was addressed by performing hydrogen-deuterium (HD) exchange experiments on complexes of glutamate and kainate with S1S2J, a 30-kDa agonist-binding domain excised from a full-length GluR2 subunit (14) (Fig. 1A). To evaluate binding mechanisms, we determined free energy changes, ΔGE = −RT ln KE, associated with the solvent exposure of backbone NH protons (HNs) in S1S2J that are protected, in most cases by H-bonds, as a consequence of agonist binding and cleft closure. Assuming EX2-type Linderstrøm-Lang kinetics (22, 23), equilibrium constants, KE, were estimated from KE = kHDX/k̃HDX, where kHDX is the measured HD exchange rate, and k̃HDX is the exchange rate in solvent-exposed states (see “Experimental Procedures”). Importantly, KE, in addition to providing a thermodynamic measure of structural stability, supplies mechanistic detail, because the OD−-catalyzed exchange reaction requires complete disruption of pre-existing H-bonds, including ones made to water molecules (24, 25). Previous studies have argued that small thermal fluctuations are not expected to lead to successful reactions but rather that separations of at least a few angstroms are required (25, 26). Based on this view, domain opening and agonist dissociation events were inferred from the detection of hydrogen exchange at key sites protected through ligand binding and lobe closure.
FIGURE 1.
Key backbone HNs and selected H-bonds involved in glutamate binding and lobe closure of GluR2-S1S2J; adapted from Protein Data Bank entry 1FTJ (14). A, S1S2J (ribbons) shown bound to glutamate in a closed cleft conformation. B, section of the glutamate binding site of protomer C of 1FTJ, in which the backbone of Asp651–Gly653 adopts a conformation that makes interlobe H-bonds with HY450N and HG451N. Additional crystal structures (27) and solution NMR data (28) suggest that this is the dominant conformation. The yellow spheres are oxygen atoms of crystallographic water molecules. C, alternate backbone conformations of Asp651–Gly653 in 1FTJ.
The key sites considered were HY450N, HG451N, HT480N, HS654N, and HT655N (Fig. 1B). HT480N in lobe 1 and HS654N and HT655N in lobe 2 are solvent-protected as a result of H-bonding to glutamate. In contrast, HY450N and HG451N are protected via interlobe H-bonds with the backbone COs of Asp651 and Ser652, respectively, where the former interaction is mediated by a water molecule. However, these interlobe H-bonds are not made in all of the glutamate-bound S1S2J crystal structures because of the ability of Asp651–Gly653 to adopt distinct backbone conformations (14) (Fig. 1C). Notably, for full agonists that have higher binding affinity than glutamate, the Asp651–Gly653 peptide tends to favor more the conformer that enables interlobe H-bond formation (14–16), and this apparent shift in equilibrium may be related to the increase in occupation probabilities of S*G2 and S*G3 for such agonists over glutamate (18).
Understanding the relative conformational free energies associated with the interlobe H-bonds and the interactions between glutamate and sites on lobe 2 is central to determining the binding mechanism. Assuming that glutamate docking to lobe 1 is rapid, similar free energy changes for these equilibria would provide strong evidence for a single-step mechanism of domain opening and closure, such as that implied in Equation 1. On the other hand, distinct free energy changes for these equilibria would be more consistent with Equation 2. Below, we provide evidence for the latter view, namely, that the mechanism of cleft closure for glutamate binding comprises multiple equilibria. However, our results for kainate binding, in which the interlobe H-bonds are absent (13, 14, 28), are consistent with Equation 1 and hence with previous results reported for kainate binding to GluR4-S1S2 (20).
EXPERIMENTAL PROCEDURES
All experiments were performed on the 263-residue protein S1S2J (14) derived from the rat GluR2-flop subunit (4). S1S2J was expressed in BL-21(DE3) Escherichia coli grown in fully deuterated minimal medium, and perdeuterated samples of the native protein were prepared via refolding from inclusion bodies and extensive purification (29, 30). NMR spectroscopic measurements were made using a Varian 500 MHz spectrometer fitted with a cryoprobe. The spectra were processed using Lorentzian-to-Gaussian window functions and modeled using Gaussian line shapes for the estimation of peak volumes (V) (31). 1H-15N peaks were identified based on assignments made previously (30, 32).
HD Exchange Measurements
The HD exchange rates, kHDX, for ∼70 binding cleft HNs were determined by fitting V(t) = V0 exp(−kHDXt) to measured V(t) results. The measured peak intensity profiles were obtained from series of 1H-15N heteronuclear single quantum coherence-transverse relaxation optimized (33) spectra acquired after purified samples were transferred into deuterated buffers. Uncertainties in V were estimated based on the spectral noise, and errors in the fitted parameters were calculated from Monte Carlo simulations (31). For each spectrum, 192 × 2048 total (real plus imaginary) points were recorded with 32 scans performed per increment, which corresponded to ∼2 h. HD exchange measurements were carried out with solutions of 0.3 mm S1S2J, 25 mm sodium acetate, 25 mm NaCl, 3 mm sodium azide, and either glutamate (0.45, 1, 4, or 10 mm) or kainate (4 or 10 mm).
In most experiments, single protein samples were used to collect multiple series of spectra at progressively increasing temperatures (T). This procedure helped identify conditions yielding appreciable intensity decays for accurately computing kHDX. Furthermore, for sites experiencing modest peak intensity loss at lower temperatures, it enabled kHDX determination for additional temperatures. Estimates for higher temperatures benefited from increased line sharpening. Specifically, the data in Figs. 2 (A–C), 2 (D and E), 3 (A–C), 3 (D–F), 3G, 3H, 5 (C and D), and 5E were taken from eight experimental runs carried out at {10, 14, 18, 21, 25 °C}10 mm,5.1,F, {10, 18 °C}1 mm,5.1,S, {14, 20, 25 °C}10 mm,6.65,F, {14, 20, 25 °C}1 mm,6.4,F, {25 °C}4 mm,6.68,S, {25 °C}0.45 mm,6.68,S, {10, 14 °C}10 mm,5.1,F, and {25 °C}4 mm,6.63,S, respectively. The subscripts specify the agonist concentration, pD (34), and whether freeze-drying (F) or solution exchange (S) was used to bring the sample into a deuterated buffer immediately prior to recording NMR spectra. Freeze-dried samples of agonist-bound S1S2J were resuspended in pure D2O buffered by deuterated acetic acid, whereas solution exchanges were carried out to >99% via two or three 15-min dilution-concentration cycles using Millipore Amicon Ultra-4 concentrators at 2 °C.
FIGURE 2.
A–E, 1H-15N peak intensity versus time curves from two HD exchange experiments conducted at pD 5. 1 with 0.3 mm GluR2-S1S2J and either 10 mm (A–C) or 1 mm (D and E) glutamate. In each experiment, runs at different temperatures were performed consecutively without removing the sample from the magnet (see “Experimental Procedures”). Fitted τHDX are shown in parentheses. The increasing peak intensity profiles presumably arise from reduced magnetic relaxation due to more rapid HD substitution at nearby sites, such as in the Arg485 side chain for HT480N (see Fig. 1B). F, 1H-15N magnetization build-up curves resulting from CLEANEX-PM exchange experiments (37). Intensity ratios (V/Vref) are plotted as a function of the mixing time τm. τHHX estimated from initial slope analysis are given in parentheses.
FIGURE 3.
1H-15N peak intensity-time profiles from four HD exchange experiments involving 0. 3 mm GluR2-S1S2J. A–C, 10 mm glutamate, pD 6.65, and 14, 20, or 25 °C. D–F, 1 mm glutamate, pD 6.4, and 14, 20, or 25 °C. G, 4 mm glutamate, pD 6.68, and 25 °C. H, 0.45 mm glutamate, pD 6.68, and 25 °C. The dashed lines in H derive from fits that excluded the first six data points or approximately 12 h, over which time small intensity losses occurred for strongly protected sites. For sites that exchange substantially at these conditions (e.g. HY450N, HT480N, and HD651N), such losses do not alter τHDX significantly.
FIGURE 5.
A and B, ligand-binding cleft of GluR2-S1S2J bound to either glutamate (A) or kainate (B). Glutamate induces full cleft closure with interlobe H-bonds formed by HY450N and HG451N, and several side chain-side chain interactions (Protein Data Bank entry 1FTJ, protomer C) (14). In contrast, kainate prevents full cleft closure and formation of most of these interlobe interactions (Protein Data Bank entry 1FW0) (13, 14). C–E, 1H-15N peak intensity-time profiles from two HD exchange experiments. C and D, 10 mm kainate, pD 5.1, and 10 or 14 °C. E, 4 mm kainate, pD 6.63, and 25 °C. The dashed lines in E derive from fits that excluded the first six data points (see caption to Fig. 3H).
Sequential Binding Reaction Mechanism with HD Exchange
Expressions were considered for relating measured kHDX to equilibrium constants in sequential reactions. Our aim was to formulate a general scheme that reduces to Equations 1 or 2 for two or four agonist-bound states of S1S2J, respectively. However, we note that whereas Equations 1 and 2 describe forward reactions for agonist binding followed by domain closure, we considered the reverse reaction in our HD exchange analysis. This view was taken because it is conventional and convenient to consider transitions from solvent-protected to solvent-exposed states.
In particular, the general reaction
 |
was employed, which progresses from a native complex (P0) through a series of conformational transition states (Pi) ultimately to a state where the ligand (L) dissociates (PM). M is equal to both the number of transition states and the number of agonist-bound states. The Xi states denote that the HN being monitored has undergone HD exchange, which was assumed to occur irreversibly in solutions containing nearly 100% D2O. Structurally, P0 contains the maximum number of protective H-bonds (or other interactions), and movement in the direction of P0 to PM corresponds to the successive disruption of H-bonds.
HD exchange rates are typically used (26, 35) to determine equilibrium constants for simpler structural opening events as described by Equation 4 (22, 23),
 |
whose general solution is as follows.
In this case, K1 = kE1/kP1, where kE1 and kP1 are transition rates into solvent-exposed and protected states, respectively. k̃HDX is the HD exchange rate in the solvent accessible state. In the EX2 limit (kP1 ≫ kE1, k̃HDX) (23), Equation 5 reduces to K1 = kHDX/k̃HDX. This ratio of HD exchange rates can be computed directly from experimental data and is defined here as KE = kHDX (T, pD)/k̃HDX (T, pD). KE was calculated for several HNs in S1S2J and related to the conformational equilibrium constants (Ki) of Equation 3.
By extension, for any M in Equation 3,
 |
and the overall binding dissociation constant is
 |
where [L] is the free ligand concentration. Equations 6 and 7 assume that k̃HDX ≡ k̃HDX,1 = k̃HDX,2 = … = k̃HDX,M, exchange is slow relative to the conformational transition rates, and ΔGE ≫ RT (or KE ≪ 1), which implies that the HN is well protected from solvent exposure (see supplemental materials).
For the determination of KE, k̃HDX was estimated using the random coil approximation, i.e. k̃HDX = k̃D[D+] + k̃OD[OD−] + k̃D2O[D2O], where k̃D, k̃OD, and k̃D2O are tabulated, sequence-specific HD exchange rates for HNs in model peptides (36). For particular HNs in S1S2J, this approximation was found to be inaccurate, and corrected values were estimated as described under “Results.”
HH Exchange Measurements
Although this study focused primarily on HD exchange, we also conducted several HH exchange experiments to obtain an independent assessment of H-bond stability. For example, a CLEANEX-PM pulse sequence (37) was used to test whether 1H-15N magnetization could be built up over millisecond time scale windows by HH transfer from water (for which coherent 1H magnetization was maintained) to S1S2J HN sites. Such experiments were applied to solutions of 0.3 mm S1S2J, 25 mm NaCl, 3 mm sodium azide, 10 mm sodium glutamate, and 25 mm of either sodium acetate (pH 5.1, 6.0, 6.9, 7.5, and 8.7), EPPS (pH 8.0), CHES (pH 9.5), or CAPS (pH 10.3). In general, as the buffer pH is increased typically to above 10, the EX1 limit (k̃HHX ≫ kP1, kE1) (23) is approached for protected HNs, and Equation 5 becomes kHHX = kE1. However, at such pHs, S1S2J was observed to be unstable. Thus, kHHX measurements using CLEANEX at pHs nearer to 9 were made to obtain lower bounds for structural opening rates. Although kHHX could be measured for HG451N and HG653N using this approach, these pHs were too low to detect HH transfer for the more stable key binding site HNs. The particular 1H-15N magnetization build-up curves shown in Fig. 2F were acquired from experiments involving 256 × 1024 total points and 64 scans/increment. Peak intensities, V(τm), were obtained for seven mixing times, τm, involving 30, 40, 60, 80, 100, 120, or 150 mixing cycles of ∼350 μs. These V(τm), along with intensities from reference heteronuclear single quantum coherence spectra, Vref, were used to estimate kHHX according to established procedures (37). Uncertainties in V(τm) and Vref were estimated from the spectral noise and were propagated to obtain the uncertainties in V/Vref.
RESULTS
Key Binding Site HNs in Glutamate-bound S1S2J Undergo HD Exchange over a Wide Range of Time Scales
HD exchange lifetimes, τHDX = 1/kHDX, for S1S2J backbone HNs were measured over 10–25 °C, pD 5.1–6.68, and several glutamate concentrations. Under these conditions, exchange is base catalyzed (k̃HDX = k̃OD [OD−]), and in the EX2 limit where kHDX is proportional to k̃HDX, kHDX changes by a factor of 10ΔpD for a change ΔpD at a given T. At the slowest exchange conditions considered (pD 5.1, 10 °C), all of the key HNs involved directly in binding and lobe closure were found to be sufficiently protected for HD exchange analysis.
Figs. 2 and 3 demonstrate that these sites exchange over distinct time scales. In particular, HG653N and HG451N undergo rapid exchange with τHDX of 3.4 and 9.3 h, respectively, at pD 5.1 and 10 °C (Fig. 2A). As shown in Fig. 2F, exchange at these sites is fast enough to be detected from HH transfer experiments (37) performed under alkaline conditions at which 1H-15N magnetization was built up over 50 ms via proton exchange from water. The kHHX reported for pH 9.5 may be considered as lower bounds for the structural transition rates that expose HG653N and HG451N to solvent. In contrast, HY450N and HT480N do not experience significant HD exchange for any T explored at pD 5.1. For these HNs, higher pDs of ∼6.5, where the driving force for exchange is 25 (101.4) times greater, were required to obtain quantifiable τHDX (Fig. 3). Finally, HS654N and HT655N exchange on intermediate time scales with τHDX comparatively long at pD 5.1 (Fig. 2, A–E) and short at pDs near 6.5 (Fig. 3, A, D, and G).
τHDX for HNs That Make H-bonds with Glutamate Depends Strongly on Ligand Concentration, whereas τHDX for HNs That Form Interlobe H-bonds Shows an Apparent Independence
When glutamate is bound to both lobes of S1S2J through H-bonding as shown in Fig. 1B, HT480N, HS654N, and HT655N are protected from undergoing HD exchange. Hence, the τHDX of these HNs should decrease significantly for decreasing concentrations of glutamate because, after dissociation, τHDX is determined largely by a competition between ligand rebinding and HD exchange. We tested for such a dependence first through HD exchange measurements at pD 6.4 and 1 mm glutamate. Compared with pD 6.65 and 10 mm glutamate, τHDX for sites insensitive to the ligand on-rate should increase by a factor of ∼1.8 (100.25), whereas τHDX for sensitive sites should increase less or show appreciable decreases. The results in Fig. 3 (A–F) demonstrate that such a distinction is observed for several HNs. Specifically, the τHDX of HT480N, HS654N, and HT655N are reduced by a factor of 2 (or 4 when the difference caused by changes in pD is corrected) at 1 mm glutamate, indicating a sensitivity to glutamate concentration. This dependence is also seen in Fig. 3 (G and H), which plot intensity decays for pD 6.68, 25 °C, and either 4 or 0.45 mm glutamate, respectively. At 0.45 mm glutamate, τHDX for HT480N is reduced by a factor of 7, and HS654N and HT655N exchange too rapidly to yield detectable peaks. Finally, Fig. 2 (A, B, D, and E) present results for pD 5.1 and either 1 or 10 mm glutamate, which show 3–7-fold smaller τHDX for HS654N and HT655N at 1 mm glutamate.
In striking contrast to what is observed for HT480N, HS654N, and HT655N, the τHDX of HY450N and HG451N, as well as HD651N and HG653N located directly across the cleft, exhibit essentially no dependence on glutamate concentration (Figs. 2 and 3).
Estimated KE Indicate That the Binding Reaction Involves Multiple Equilibria
Fig. 4A presents KE and ΔGE results for the key binding site HNs based on the τHDX given in Figs. 2 and 3 and k̃HDX set to random coil values (36). Notably, particular pairs of HNs that occupy similar regions of the binding site have similar KE, which suggests that their solvent exposure arises from related conformational opening events. For example, the KE of HG451N (1.9 × 10−3) and HG653N (2.9 × 10−3) are relatively large and comparable in magnitude. Structurally, HG653N resides directly across the cleft from HG451N and shares a peptide bond with the CO of Ser652; this CO in turn forms an interlobe H-bond with HG451N (Fig. 1, B and C). The KE results imply that this H-bond is relatively unstable and that its breakage is associated with backbone flexibility in lobe 2 near Gly653, a view consistent with previous interpretations (14).
FIGURE 4.
KE and ΔGE for key binding site HNs in S1S2J. The arrows point in the direction of increasing glutamate concentration. A, results for glutamate binding, where k̃HDX was assigned to the HD exchange rates of model random coil peptides (36). B, corrected results for glutamate and kainate binding. For HY450N in glutamate-bound S1S2J, k̃HDX was determined from experiments with kainate (see text). For HT480N, KE was back-predicted from KDs. For HG451N, HG653N, HS654N, and HT655N, k̃HDX was based on random coil rates as in A.
In addition, at several conditions tested, HS654N exchanges approximately two times faster than HT655N (Figs. 2 and 3), and such rates lead to similar KE because random coil predictions for HS654N are two times greater than those for HT655N (36). As these HNs form the only direct interactions between glutamate and the backbone of lobe 2 (Fig. 1B), the results suggest that the HD transfer events emanate from a structural equilibrium involving detachment of glutamate from lobe 2. That the KE of HS654N and HT655N are larger than those of HT480N is suggestive of unique ligand docking and lobe isomerization equilibria (Fig. 4A). However, as discussed in the next subsection, the high degree of protection of HT480N appears to be due in part to departure of solvent-exposed states from random coil behavior.
Although the random coil approximation is commonly applied to solvent-exposed states of HNs in proteins, it must be used carefully because departure from random coil behavior can clearly lead to erroneous k̃HDX and hence ΔGE. Furthermore, it may be difficult to identify and correct for such discrepancies when ΔGE corresponds to local backbone unfolding events. However, because the key HNs considered in the present study presumably become solvent-exposed upon ligand detachment or domain opening, it is possible in principle to estimate their k̃HDX from experiments employing either the Apo protein or agonists that stabilize the HNs in solvent-exposed states. As discussed below, the latter approach was found to be useful for evaluating the exchange behavior of HY450N.
Surprisingly, the KE of HY450N (based on random coil values for k̃HDX) suggest a high degree of protection (Fig. 4A). The increased stability relative to HS654N and HT655N is particularly interesting given that the latter two HNs make H-bonds to glutamate that are buried beneath where HY450N forms an interlobe H-bond (Figs. 1B and 5A). In addition, the KE of HY450N are independent of glutamate concentration, unlike those of HS654N and HT655N. There are seemingly only two explanations for these results: either (i) the lobes of Apo S1S2J adopt closed conformations that protect HY450N in a way that leads to the observed glutamate concentration independence or (ii) HY450N can become solvent accessible in closed cleft conformations with glutamate bound but residual structure slows k̃HDX well below random coil estimates. Although solution x-ray scattering studies suggest that Apo S1S2J adopts a mixture of open and closed domain states (38), the rapid docking of agonist to the protein (20, 21) argues against explanation (i). Similar arguments against extended closed states in the Apo protein can be made from free energy calculations as a function of the degree of lobe closure (39, 40).
Further support for explanation (ii) was obtained from additional HD exchange experiments with kainate as the agonist. Relative to glutamate, the lobes of S1S2J are substantially more open in the presence of kainate, and formation of the interlobe H-bonds involving HY450N and HG451N is prevented (Fig. 5, A and B) (13, 14, 28). As a first approximation, we assigned kHDX for HY450N measured with kainate to k̃HDX for HY450N in glutamate-bound S1S2J. Fig. 5 (C and D) shows that HY450N exchanges with τHDX,kai of 33 and 17 h for pD 5.1, 10 mm kainate, and 10 or 14 °C, respectively. The τHDX,kai for 14 °C, in combination with the τHDX,glu in Fig. 3 (A and D), provides an estimate of KE,glu ≈ τHDX,kai (T, pD)/τHDX,glu(T, pD) for HY450N of 0.0033 ± 0.0004 (the estimates in Fig. 4B for 20 and 25 °C assume τHDX,kai increases with T by similar factors as those observed for τHDX,glu, namely 3.2 and 6.6, respectively; see Fig. 3). Remarkably, this KE is comparable in magnitude with the KE obtained for HG451N and HG653N (Fig. 4B). Thus, these results imply that, like the interlobe H-bond formed by HG451N, the interactions protecting HY450N have similar stability.
Although the random coil approximation appears to be poor for HY450N, several points suggest that it is realistic for HG451N, HG653N, HS654N, and HT655N. First, as discussed earlier in this section, the similarity in the KE of HG451N and HG653N on the one hand and HS654N and HT655N on the other agrees with our expectation that conformational openings expose both HNs in these pairs approximately equally. Second, the KE for HS654N and HT655N predict realistic dissociation constants (KD) (see below). Finally, the values of τHDX for these four HNs measured using kainate seem to be consistent with random coil approximations, which predict τ̃HDX of ∼1 min for pD 5.1 and 10 °C (36). For example, at these conditions (Fig. 5C), we found no detectable peak intensity for HG451N and HG653N in the first spectrum in the series. We also did not detect peaks for these HNs in similar experiments with shorter NMR acquisition times of 30 min. These observations suggest an upper bound for τHDX on the order of minutes consistent with the random coil values. HS654N and HT655N also exchange very rapidly at pD 5.1 and 10 °C despite being protected by kainate.
Based on the corrected KE results in Fig. 4B, we next sought to determine the mechanisms of binding of glutamate and kainate to S1S2J. This was achieved by using Equations 6 and 7 to relate KE to conformational equilibria and agonist binding constants for different numbers of agonist-bound states (M).
M = 2; Distinct Dock-isomerization Equilibria Detected for Glutamate- and Kainate-bound S1S2J
We first applied the KE results to the dock-isomerization model given by Equation 1. In this case, M = 2 in Equation 3, and Equations 6 and 7 simplify to Equations 8 and 9.
K1 and K2 are the equilibrium constants for domain opening and ligand dissociation from lobe 1, respectively. In the next section, it is argued utilizing the KE of HY450N and HG451N that M > 2 for glutamate binding because of the presence of significant interlobe interactions. Here, these interactions are ignored, and the relative lobe closure stabilities of glutamate- and kainate-bound S1S2J are analyzed based solely on the KE of HT480N, HS654N, and HT655N.
Application of KE for HS654N and HT655N to Equation 8, which assumes exchange from both P1 and P2, results in K1 = 8.1 × 10−5 ± 4.1 × 10−5 and K2 = 4.0 ± 0.6 mm for glutamate binding. This calculation was based on five pairs of KE in Fig. 4B corresponding to two glutamate concentrations, identical temperatures, and similar pDs. By Equation 9, KD = 320 ± 120 nm, which is consistent with competition binding results for this protein (KD 450 nm) (14) and for two related GluR2-S1S2 proteins that differ only in the regions where the domains are excised from the full subunit (120 and 250 nm) (41, 42).
It was also possible to estimate KD from the KE of HT480N. However, calculations assuming either exchange from both P1 and P2, or from P2 only, lead to values of ∼4 nm, which are too small. Thus, as with HY450N, random coil HD exchange rates (36) seemingly overestimate k̃HDX for HT480N. This is not surprising though because, unlike HS654N and HT655N, HT480N resides more deeply in the binding cleft and in a more stable section of the protein (29, 43, 44).
Despite the apparent error in the random coil values of HT480N, its measured τHDX was used to correctly estimate the KD for kainate (based on the KD for glutamate determined above) via Equation 10.
 |
In accordance with a dock-isomerization mechanism, this expression assumes that HT480N exchanges only from P2 in Equation 3 with M = 2, i.e. KE = KD/[L] (τ̃HDX does not appear in Equation 10 by τ̃HDX,glu = τ̃HDX,kai). Application of the τHDX in Figs. 3 (C and F–H) and 5E to Equation 10 leads to KD,kai = 4.1 ± 2.3 μm, which agrees well with the competition binding results of 8.0, 1.3, and 2.3 μm (41, 42, 14).
This value of KD,kai was used to estimate K1,kai and K2,kai for comparison with K1 and K2 determined for glutamate. Figs. 2A and 5C show that HS654N and HT655N exchange 60–80 times faster for kainate relative to glutamate at pD 5.1, 10 °C, and 1 mm agonist. Equation 10 assumes that exchange occurs from P2 only and requires that τHDX,glu/τHDX,kai = KD,kai/KD,glu for equal concentrations of glutamate and kainate. However, for HS654N and HT655N, τHDX,glu/τHDX,kai ≫ KD,kai/KD,glu, suggesting that exchange occurs from P1, i.e. from domain opening, more readily for kainate than glutamate. By applying the KE for HS654N and HT655N to Equation 8, we estimated that K1,kai = 0.0050 ± 0.0005 and K2,kai = 0.8 ± 0.08 mm. Thus, relative to kainate, these results suggest that glutamate binds to the open domain state of S1S2J with roughly five times lower affinity but shifts the domain isomerization equilibrium 60-fold toward cleft closure.
M > 2; Ligand Concentration Independence of KE for HY450N and HG451N Suggests Lobe Locking Equilibria Distinct from Domain Isomerization in Glutamate-bound S1S2J
We showed above that the KE for HY450N and HG451N are relatively large and essentially independent of glutamate concentration. Clearly, it is impossible to fit these KE to the dock-isomerization scheme (M = 2) with K1 = 8.1 × 10−5 and K2 = 4 mm obtained in the preceding section. This implies that the binding reaction for glutamate consists of M > 2 states and that the domain-isomerization step must be decomposed into subequilibria. Thus, we considered the next simplest model, M = 3, shown in Equations 11 and 12.
Here, K1, K2, and K3 characterize lobe unlocking, domain opening, and agonist dissociation equilibria, respectively.
In accordance with our calculations for M = 2, we assumed that HT480N exchanges only from P3 (KE = KD/[L]), HS654N and HT655N exchange from P2 and P3 (KE = K1K2 + KD/[L]), and HY450N and HG451N exchange from P1, P2, and P3 (Equation 11). In this way, K1, K2, and K3 could be determined using the KE given in Fig. 4B. In particular, application of HG451N KE leads to 0.0019 ± 0.0002, 0.043 ± 0.004, and 4.0 ± 0.6 mm, respectively; HY450N KE yield 0.0031 ± 0.0002, 0.026 ± 0.002, and 4.0 ± 0.6 mm. These results imply that the KE of HY450N and HG451N are essentially ligand concentration-independent because K1 is much greater than K1K2 and K1K2K3/[L]. This is a direct consequence of the sequential mechanism and the distinctness of the lobe-locking and domain isomerization steps.
The K1 correspond to ΔG of roughly 6 RT, which suggests that lobe locking is a significant step in the binding reaction for glutamate. In this analysis, we calculated K1 separately for HY450N and HG451N based on a parsimonious choice of M = 3. Of course, differences in the KE for these two HNs could plausibly support M = 4, but we cannot discriminate such differences because of the experimental errors involved. Other stabilizing interlobe interactions, such as those operating through interactions of the side chains of Tyr450 and Leu650, Lys449 and Ser652, and Thr686 and Glu402 (Fig. 5A), could also conceivably constitute additional equilibria because they are present in crystal structure protomers lacking the H-bonds mediated by HY450N and HG451N (14). Moreover, a number of mutagenic studies have shown that alteration of these side chain interactions impacts domain closure stability and receptor function (19, 20, 45–49).
DISCUSSION
The lobe dynamics occurring in S1S2 of AMPA receptors appear to be tailored for the efficacious binding and fast release of glutamate to produce large activating currents in combination with efficient channel deactivation and resensitization (14, 19, 18). Models based on channel recordings suggest a sequential multi-state binding mechanism comprising rapidly and slowly equilibrating components (18). Here, using hydrogen-deuterium exchange, we have provided evidence for such a mechanism for glutamate binding and have estimated the thermodynamic stabilities of the detected states. Furthermore, for kainate binding, which lacks the interlobe interactions constituting distinct states in the complex with glutamate, we have confirmed the two-state dock-isomerization mechanism first proposed from stopped flow experiments (20) and have shown that this reaction has a higher proclivity for sampling agonist-bound open domain conformations.
The HD exchange rates presented in this study have uncertainties typically much less than 10%. Yet the associated KE were evaluated carefully due to the use of random coil exchange rates for estimating k̃HDX. Although random coil rates have been validated for HNs in denatured proteins (36, 50, 51) and appear to be realistic measures for many solvent-exposed HNs in native proteins (52), examples of poor predictions have been reported (53, 54). Thus, for the key binding site HNs in S1S2J, we either justified the use of the random coil approximation or provided corrections through experiments. It is important to note, however, that our main conclusions can also be justified without invoking this approximation. First, the significance of the lobe-locking equilibrium in the binding reaction for glutamate is evident by the much higher degree of protection of HY450N and HG451N in complexes with glutamate compared with kainate. Second, the uniqueness of the lobe-unlocking and domain opening equilibria is established by the insensitivity of the HD exchange rates of HY450N and HG451N to changes in glutamate concentration. For if these equilibria were not unique, then these HNs would become solvent-exposed only upon domain opening and a strong glutamate concentration dependence similar to that exhibited by HS654N and HT655N would have been observed. Finally, the higher tendency for glutamate binding to induce closed cleft states in S1S2J relative to kainate is apparent in the 60–80-fold smaller exchange rates of HS654N and HT655N measured for glutamate.
We have demonstrated that the binding reaction for glutamate is composed of multiple states, but the question arises as to how this relates to the functional model in Equation 2 (18). One might speculate that, for the HD exchange model of Equation 3 with M = 3, P3 is the state S, and P2 is the state SG, i.e. glutamate docked to lobe 1 without H-bonding to HS654N and HT655N in lobe 2. P1 would then correspond to the state S*G1, for which glutamate interacts with lobe 2 in the absence of the interlobe H-bonds mediated by HY450N and HG451N. Whether the side chain interactions shown in Fig. 5A are made in this state or in S*G2 or S*G3 is unclear (all of the crystal structure protomers of glutamate-bound S1S2J contain these side chain interactions, whereas only some of the protomers show the interlobe main chain H-bonds) (14, 27). Finally, P0 would be associated with S*G2 and/or S*G3 in some way and include the H-bonds formed by HY450N and HG451N. In the present study, we could only detect the breakage of H-bonds made to backbone HNs, but clearly the side chain interactions are an essential component of the binding mechanism and may be energetically and kinetically separable from the main chain interactions.
In the whole cell experiments for measuring deactivation upon which Equation 2 is partially based, glutamate was applied to GluR2 restrained in the nondesensitizing state by cyclothiazide (18). Seemingly, for this reason S*G1 was primarily populated with lower percentages observed for S*G2 and S*G3. In contrast, our HD exchange measurements were performed on S1S2J, which lacks structural restraints present in the nondesensitizing state. Because the ΔGE of HY450N and HG451N are significant, free S1S2J appears to favor transitions to states with stronger interactions across the cleft, which may be active in the desensitized state. This is consistent with the finding that the EC50 for glutamate activation of GluR2 (in cyclothiazide) is 300 μm (18) but that the binding affinity for the desensitized state is much higher (1 μm) (55). Likewise, glutamate binds to GluR2 S1S2J with a KD of roughly 300 nm (41, 42, 14), suggesting that this complex represents the desensitized state, as proposed previously (14).
Thus, the process of successive H-bond disruption measured via HD exchange may account for structural changes in S1S2 that occur during recovery from desensitization. On the other hand, the responses of nondesensitizing (or resting) GluR2 to brief glutamate applications would seemingly be dominated by agonist docking and domain isomerization events (consistent with occupation of S*G1) (18) with less lobe locking than that observed in S1S2J. This would account for the ability of AMPA receptors to activate and deactivate rapidly. The lobe-locking equilibria presumably become more functionally active during extended glutamate applications (consistent with increased occupation of S*G2 and S*G3) (18) and aid in the stabilization of glutamate binding in desensitized states geared to abolish macroscopic currents.
Because full and partial agonists sample similar unitary conductance levels in single channels (17) and the extent of channel activation can be correlated with the degree of cleft closure in S1S2J (17, 28, 56), a more significantly closed cleft apparently acts to increase the probability that a channel gate opens rather than promote a gradually more open gate. Such a mechanism would also seemingly depend on the probability that the cleft is closed when agonist is bound, and this is consistent with recent studies showing that domain closure stability also influences activation (18, 19, 48). However, this form of structural stability is difficult to characterize energetically from crystal structures and ps–μs dynamics because domain opening in S1S2J occurs on slower time scales. Thus, the methods established in the present study, which probe ms and longer time scale motions and yield equilibrium constants for lobe opening in agonist-bound S1S2J, should be useful for such characterizations.
As a case in point, kainate binding to nondesensitizing GluR2 generates currents that are 50 times smaller than those evoked by glutamate (45), and this has been attributed to the smaller degree of cleft closure in S1S2 induced by kainate (13, 14). However, a broader view that includes domain closure stability measures is obtained from the K1 of our two-state binding model (Equation 3; M = 2), which indicates that S1S2J adopts agonist-bound, open cleft states (identified by the HD exchange of HS654N and HT655N) much more often for kainate than glutamate. Although it is difficult to assess the relative importance of cleft closure extent and stability on channel activation for kainate, these results support a dynamic view of agonism consistent with an activation mechanism that depends not only on the extent of cleft closure but also on the probability that the cleft is closed when agonist is bound.
Supplementary Material
Acknowledgments
We thank Eric Gouaux (Vollum Institute) for the S1S2J construct, Lewis Kay (Univ. Toronto) for the CLEANEX NMR pulse sequence, and Dean Madden (Dartmouth), Greg Weiland, Alexander Maltsev, Christopher Ptak, Kinning Poon, Adrienne Loh, Madeline Martinez, Linda Nicholson, Ahmed Ahmed, and Linda Nowak (Cornell) for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants T32CA09682 (M. K. F.) and GM068935 (to R. E. O.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, equations, and Tables S1 and S2.
- AMPA
- α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid
- GluR2
- ionotropic glutamate receptor 2
- HD
- hydrogen-deuterium
- HH
- hydrogen-hydrogen
- HN
- protein backbone NH proton
- EPPS
- 4-(2-hydroxyethyl)-1-piperazinepropanesulfonic acid
- CHES
- 2-(cyclohexylamino)ethanesulfonic acid
- CAPS
- 3-(cyclohexylamino)propanesulfonic acid.
REFERENCES
- 1.Trussell L. O., Fischbach G. D. (1989) Neuron 3, 209–218 [DOI] [PubMed] [Google Scholar]
- 2.Colquhoun D., Jonas P., Sakmann B. (1992) J. Physiol. 458, 261–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hestrin S. (1992) Neuron 9, 991–999 [DOI] [PubMed] [Google Scholar]
- 4.Hollmann M., Heinemann S. (1994) Annu. Rev. Neurosci. 17, 31–108 [DOI] [PubMed] [Google Scholar]
- 5.Jonas P., Spruston N. (1994) Curr. Opin. Neurobiol. 4, 366–372 [DOI] [PubMed] [Google Scholar]
- 6.Jones M. V., Westbrook G. L. (1996) Trends Neurosci. 19, 96–101 [DOI] [PubMed] [Google Scholar]
- 7.Wo Z. G., Oswald R. E. (1995) Trends Neurosci. 18, 161–168 [DOI] [PubMed] [Google Scholar]
- 8.Rosenmund C., Stern-Bach Y., Stevens C. F. (1998) Science 280, 1596–1599 [DOI] [PubMed] [Google Scholar]
- 9.Dingledine R., Borges K., Bowie D., Traynelis S. F. (1999) Pharm. Rev. 51, 7–61 [PubMed] [Google Scholar]
- 10.Madden D. R. (2002) Nat. Rev. Neurosci. 3, 91–101 [DOI] [PubMed] [Google Scholar]
- 11.Oswald R. E. (2004) Adv. Protein Chem. 68, 313–349 [DOI] [PubMed] [Google Scholar]
- 12.Mayer M. L. (2006) Nature 440, 456–462 [DOI] [PubMed] [Google Scholar]
- 13.Armstrong N., Sun Y., Chen G. Q., Gouaux E. (1998) Nature 395, 913–917 [DOI] [PubMed] [Google Scholar]
- 14.Armstrong N., Gouaux E. (2000) Neuron 28, 165–181 [DOI] [PubMed] [Google Scholar]
- 15.Hogner A., Kastrup J. S., Jin R., Liljefors T., Mayer M. L., Egebjerg J., Larsen I. K., Gouaux E. (2002) J. Mol. Biol. 322, 93–109 [DOI] [PubMed] [Google Scholar]
- 16.Jin R., Horning M., Mayer M. L., Gouaux E. (2002) Biochemistry 41, 15635–15643 [DOI] [PubMed] [Google Scholar]
- 17.Jin R., Banke T. G., Mayer M. L., Traynelis S. F., Gouaux E. (2003) Nat. Neurosci. 6, 803–810 [DOI] [PubMed] [Google Scholar]
- 18.Zhang W., Robert A., Vogensen S. B., Howe J. R. (2006) Biophys. J. 91, 1336–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robert A., Armstrong N., Gouaux J. E., Howe J. R. (2005) J. Neurosci. 25, 3752–3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abele R., Keinanen K., Madden D. R. (2000) J. Biol. Chem. 275, 21355–21363 [DOI] [PubMed] [Google Scholar]
- 21.Cheng Q., Du M., Ramanoudjame G., Jayaraman V. (2005) Nat. Chem. Biol. 1, 329–332 [DOI] [PubMed] [Google Scholar]
- 22.Berger A., Linderstrom-Lang K. (1957) Arch. Biochem. Biophys. 69, 106–118 [DOI] [PubMed] [Google Scholar]
- 23.Hvidt A., Nielsen S. O. (1966) Adv. Protein Chem. 21, 287–386 [DOI] [PubMed] [Google Scholar]
- 24.Eigen M. (1964) Angew. Chem. Int. Ed. 3, 1–19 [Google Scholar]
- 25.Milne J. S., Mayne L., Roder H., Wand A. J., Englander S. W. (1998) Protein Sci. 7, 739–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Englander S. W., Kallenbach N. R. (1983) Q. Rev. Biophys. 16, 521–655 [DOI] [PubMed] [Google Scholar]
- 27.Ahmed A. H., Wang Q., Sondermann H., Oswald R. E. (2009) Proteins 75, 628–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maltsev A. S., Ahmed A. H., Fenwick M. K., Jane D. E., Oswald R. E. (2008) Biochemistry 47, 10600–10610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McFeeters R. L., Oswald R. E. (2002) Biochemistry 41, 10472–10481 [DOI] [PubMed] [Google Scholar]
- 30.Fenwick M. K., Oswald R. E. (2007) Biomol. NMR Assign. 1, 241–243 [DOI] [PubMed] [Google Scholar]
- 31.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 32.McFeeters R. L., Swapna G. V., Montelione G. T., Oswald R. E. (2002) J. Biomol. NMR 22, 297–298 [DOI] [PubMed] [Google Scholar]
- 33.Pervushin K., Riek R., Wider G., Wüthrich K. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 12366–12371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glasoe P. K., Long F. A. (1960) J. Phys. Chem. 64, 188–190 [Google Scholar]
- 35.Dempsey C. E. (2001) Prog. NMR Spectrosc. 39, 135–170 [Google Scholar]
- 36.Bai Y., Milne J. S., Mayne L., Englander S. W. (1993) Proteins 17, 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hwang T. L., van Zijl P. C., Mori S. (1998) J. Biomol. NMR 11, 221–226 [DOI] [PubMed] [Google Scholar]
- 38.Madden D. R., Armstrong N., Svergun D., Pérez J., Vachette P. (2005) J. Biol. Chem. 280, 23637–23642 [DOI] [PubMed] [Google Scholar]
- 39.Lau A. Y., Roux B. (2007) Structure 15, 1203–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mamonova T., Yonkunas M. J., Kurnikova M. G. (2008) Biochemistry 47, 11077–11085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arvola M., Keinänen K. (1996) J. Biol. Chem. 271, 15527–15532 [DOI] [PubMed] [Google Scholar]
- 42.Chen G. Q., Gouaux E. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 13431–13436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Valentine E. R., Palmer A. G., 3rd (2005) Biochemistry 44, 3410–3417 [DOI] [PubMed] [Google Scholar]
- 44.Fenwick M. K., Oswald R. E. (2008) J. Mol. Biol. 378, 673–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Armstrong N., Mayer M., Gouaux E. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 5736–5741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holm M. M., Lunn M. L., Traynelis S. F., Kastrup J. S., Egebjerg J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 12053–12058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holm M. M., Naur P., Vestergaard B., Geballe M. T., Gajhede M., Kastrup J. S., Traynelis S. F., Egebjerg J. (2005) J. Biol. Chem. 280, 35469–35476 [DOI] [PubMed] [Google Scholar]
- 48.Weston M. C., Gertler C., Mayer M. L., Rosenmund C. (2006) J. Neurosci. 26, 7650–7658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mankiewicz K. A., Rambhadran A., Du M., Ramanoudjame G., Jayaraman V. (2007) Biochemistry 46, 1343–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roder H., Wagner G., Wüthrich K. (1985) Biochemistry 24, 7407–7411 [DOI] [PubMed] [Google Scholar]
- 51.Robertson A. D., Baldwin R. L. (1991) Biochemistry 30, 9907–9914 [DOI] [PubMed] [Google Scholar]
- 52.Molday R. S., Englander S. W., Kallen R. G. (1972) Biochemistry 11, 150–158 [DOI] [PubMed] [Google Scholar]
- 53.Maity H., Lim W. K., Rumbley J. N., Englander S. W. (2003) Protein Sci. 12, 153–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anderson J. S., Hernández G., Lemaster D. M. (2008) Biochemistry 47, 6178–6188 [DOI] [PubMed] [Google Scholar]
- 55.Kessler M., Suzuki E., Montgomery K., Arai A. C. (2008) Neurochem. Int. 52, 1383–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramanoudjame G., Du M., Mankiewicz K. A., Jayaraman V. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10473–10478 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.







