Abstract
A fundamental biologic principle is that diverse biologic signals are channeled through shared signaling cascades to regulate development. Large scaffold proteins that bind multiple proteins are capable of coordinating shared signaling pathways to provide specificity to activation of key developmental genes. Although much is known about transcription factors and target genes that regulate cardiomyocyte differentiation, less is known about scaffold proteins that couple signals at the cell surface to differentiation factors in developing heart cells. Here we show that AKAP13 (also known as Brx-1, AKAP-Lbc, and proto-Lbc), a unique protein kinase A-anchoring protein (AKAP) guanine nucleotide exchange region belonging to the Dbl family of oncogenes, is essential for cardiac development. Cardiomyocytes of Akap13-null mice had deficient sarcomere formation, and developing hearts were thin-walled and mice died at embryonic day 10.5–11.0. Disruption of Akap13 was accompanied by reduced expression of Mef2C. Consistent with a role of AKAP13 upstream of MEF2C, Akap13 siRNA led to a reduction in Mef2C mRNA, and overexpression of AKAP13 augmented MEF2C-dependent reporter activity. The results suggest that AKAP13 coordinates Gα12 and Rho signaling to an essential transcription program in developing cardiomyocytes.
Keywords: Actin, Heart, Myosin, Protein Kinase A (PKA), Rho, AKAP13, Brx, RhoA, SRF
Introduction
We previously reported cloning and characterization of a human 5.3-kb brx-1 (breast cancer nuclear hormone receptor auxiliary factor 1) transcript that encoded a 170-kDa Dbl family member (1) and later localized the gene to chromosome 15q24-25 (2). Larger transcripts of the gene were subsequently isolated (3), and based on its AKAP region, the gene we initially called BRX is now known as AKAP13.
The central Dbl homology (DH)3 or guanine nucleotide exchange region (GEF) is present in all native transcripts of AKAP13 (Fig. 1). The lbc oncogene (4), derived from two chromosomes, 15 and 7 (5), contains the GEF region of AKAP13 but lacks the N terminus and carboxyl regions. The GEF domain of AKAP13 was shown to bind RhoA and activate Rho family GTPases (3, 5, 6). Rho GTPases have been reported to influence the cell cytoskeleton and sarcomere development in cardiomyocytes (7–9). RhoA, a target of AKAP13, influences at least 11 downstream effectors (10), including two factors essential for cardiomyocyte differentiation, serum response factor (SRF) and GATA-4 (8, 11, 12). AKAP13 was also shown to play a role in cardiac hypertrophy (13) via MEF2 (myocyte enhancer factor-2). Despite the established role for RhoA in cardiac development, the importance of AKAP13 in the developing heart has not been described.
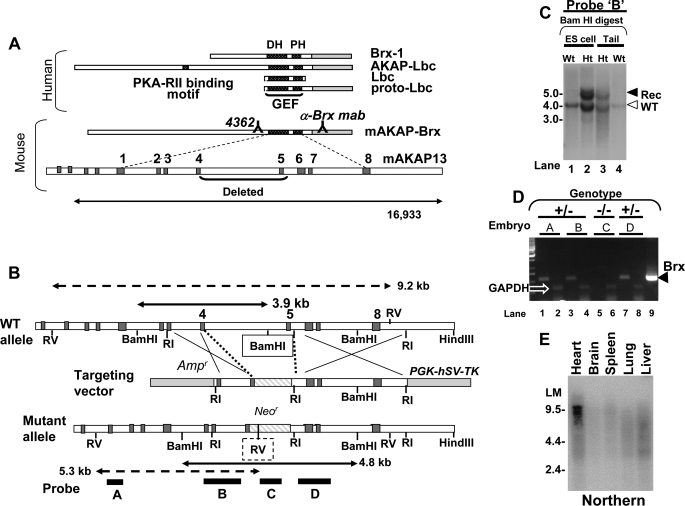
FIGURE 1.
Diagram of murine Akap13 gene and deletion strategy. A, the human AKAP13 gene encodes a 5.3-kb brx-1 transcript and an 8.5- and 10-kb transcript. Shown is the murine AKAP-Brx transcript and genomic intron-exon structure with DH and pleckstrin homology (PH) regions as indicated. Binding sites of antiserum 4362 and anti-BRX monoclonal antibody mab are shown. Exons are numbered consecutively beginning at the GEF region. B, partial restriction map of the wild-type murine Akap13 GEF region, the targeting vector, and the mutant Akap13 allele. Shown are the neomycin cassette (striped) insertion with deletion of part of exon 4, the entire exon 5, and 5398 bp of flanking genomic sequence. Dark gray, exons; white, introns. Probes A–D are shown as black bars. Double-headed solid arrows, restriction fragments generated by BamHI digestion of wild-type (3.9 kb) and mutant alleles (4.8 kb) with 5′-probe B (black bar). The BamHI site deleted in the mutant allele is boxed. Double-headed dashed arrows depict restriction fragments for EcoRV digestion probed with 5′-probe A. The inserted EcoRV site is shown as a dashed box. C, Southern analysis of targeted BamHI-digested embryonic stem cell DNA (lanes 1 and 2) or tail DNA (lanes 3 and 4) with 5′-probe B revealed a recombinant 4.8 kb band and 3.9 kb wild-type bands (open arrowhead) in heterozygous (Ht) embryonic stem cells and mice. D, RT-PCR amplification of mRNA from E9.0–9.5 embryos. 90–99 ng of cDNA product from embryos was amplified with 33 pmol of primers for either murine Akap13 (odd lanes) or glyceraldehyde-3-phosphate dehydrogenase (even lanes). Murine cDNA was used as a positive control (lane 9). The expected 550 bp band was present in lanes 1, 3, and 7, consistent with the heterozygous genotype (black triangle). An Akap13 transcript was not detectable in mRNA harvested from an embryo homozygous for the Akap13 null allele (lane 5). Results were confirmed in four independent experiments. E, Northern analysis of a multi-tissue mRNA blot using an EcoRI-EcoRI fragment from the 3′-region of the murine Akap13 cDNA. A 9.5–10.0 kb band was detected in RNA harvested from heart tissues.
The function of the carboxyl region of proteins encoded by AKAP13 has remained enigmatic. The carboxyl region of the brx-1 transcript encoded an LXXLL nuclear receptor-interacting domain, and BRX-1 protein was found to specifically interact with nuclear hormone receptors (1, 14, 15). Homodimerization of the carboxyl region has been reported to regulate Rho-GEF activity of the protein (16). These observations suggest that the carboxyl region of AKAP13 functions to regulate GEF activity and couple protein kinase A and Rho-GEF activity to downstream effectors.
AKAP13 is differentially spliced and encodes several modular proteins of varying length in the N-terminal region that comprises the protein kinase A anchoring motif (Fig. 1). The brx-1 transcript (1) encodes the smallest naturally occurring protein of AKAP13, due to an abbreviated AKAP domain (3). AKAP13 proteins have been shown to coordinate signals originating from the cell membrane, including thrombin (17), lysophosphatidic acid (LPA) receptors (3, 15), and the osmoreceptor (18). Specifically, AKAP13 proteins have been shown to interact with Gα12 (3, 17), Gα13 (19), Gα14 (20), Gq (19–21), and possibly Gα15 (15). Despite considerable evidence suggesting the importance of AKAP13 in vitro, the in vivo role of the protein has remained unclear. To assess the in vivo physiologic role of BRX-1 (AKAP13), we pursued a targeted loss of function approach. Here we report that loss of function of AKAP13 in the mouse resulted in a thin myocardium and an embryonic lethal phenotype. We show that Akap13 is required for cardiac development and induction of normal levels of MEF2C during cardiomyocyte differentiation.
EXPERIMENTAL PROCEDURES
Nomenclature
AKAP13 is the gene that encodes the brx-1 transcript of 5.3 kilobases that encodes BRX (1) and a larger transcript of 10.0 kilobases that encodes the protein AKAP13 (also known AKAP-BRX or AKAP-LBC (3–5)). The relationship of the different transcripts is shown in Fig. 1A.
Gene Targeting
Murine cDNA clones were isolated from a spleen cDNA library (Stratagene, La Jolla, CA) and sequenced, and cDNA regions from the carboxyl and GEF domain of murine Brx were used to identify a genomic fragment containing the GEF region of murine Akap13 from a BAC library (GenomeSystems Inc., St. Louis, MO). Overlapping genomic clones were verified by Southern analysis and DNA sequencing. The targeting vector RMV1-pPNT was constructed by insertion of a 1.6-kb NotI-XhoI fragment into the 5′-arm and a 5.0-kb EcoRI-EcoRI fragment as the 3′-arm of the murine Akap13 gene, replacing a critical exon in the DH domain and 5.39 kb of flanking genomic sequence with the Neor cassette. The targeting vector was sequenced, linearized with HindIII, and electroporated into 129 embryonic stem cells. The targeted event on the 5′-end of the gene was confirmed by Southern blot hybridization of BamHI-digested DNA and hybridization with two 5′-probes (Fig. 1). Targeting of the 3′-region was confirmed using Southern analysis of PCR products (supplemental Fig. S1, A and B). Additional Southern and PCR analyses substantiated correct recombination events and excluded non-homologous events (not shown). Northern analysis of the multi-tissue mRNA blot using the EcoRI-EcoRI fragment from the 3′-region of the murine Akap13 cDNA was as described (1). A 9.5–10.0 kb band was detected in RNA harvested from heart tissues.
Genotyping and Embryo Preparation
Genotypes were analyzed using primers 5′-AAC CCA GTG TGA GCC CAA TAG TC-3′ and 5′-TGC AAA GTA GAG CGG GAA AGA GA-3′ for the wild-type allele and 5′-GGG CGC CCGGTTCTT TTT-3′ and 5′-CTG CCG CGC TTG TTC TCC TCTTCC-3′ for the mutant allele.
Reverse Transcription (RT)-PCR
RT-PCR for Akap13 mRNA in mutant embryos was performed using the Absolutely RNA kit (Stratagene), followed by the Ambion two-step method as per the manufacturer's instructions. The 5′-primer for RT-PCR was 5′-CGG GAG TGT GTT TCT GAA-3′; the 3′-primer was 5′-GGT CCT CCT TTG AGA GCC TC-3′.
Real-time RT-PCR
Embryonic hearts were immediately snap-frozen. Total RNA was isolated using the RLT buffer (RNeasy) isolation method according to the manufacturer's instructions (Qiagen, Gaithersburg, MD). Quantitative PCR was performed using an ABI PRISM 7500 real-time PCR system (Applied Biosystems, Foster City, CA) with duplicates of the 25-μl reaction mixture in Micro Amp optical 96-well reaction plates and sealed with Optical Adhesive Film (Applied Biosystems). Real-time RT-PCR was conducted with oligonucleotide primers for mouse Akap13, Mef2C, and 18 S as follows: Akap13 forward, 5′-TGA GAT TTA CAC AAA GAC AG-3′; Akap13 reverse, 5′-TCA GTG AGT AGA ACT GCT TG-3′; Mef2C forward, 5′-TGG ATG AGC GTA ACA GAC AG-3′; Mef2C reverse, 5′-TGA GCA ACA CCT TAT CCA TG-3′; 18 S forward, 5′-CTT AGA GGG ACA AGT GGC G-3′; 18 S reverse, 5′-ACG CTG AGC CAG TCA GTG TA-3′.
TUNEL Assay
Embryos were embedded in paraffin and sectioned in 5-μm slices (Histoserv Inc., Gaithersburg, MD), and TUNEL assays were performed as per the manufacturer's instructions (In Situ Cell Death Detection Kit, Roche Applied Science). Samples treated with DNase I (Worthington Biochemical) were used as a positive control.
Antibodies and Antisera
A peptide corresponding to the murine AKAP-BRX protein (REKEKDKIKEKEKDS KEKEKDK KTLNGHTF) was used to generate polyclonal antiserum 4362 using standard techniques, and binding to BRX was confirmed using an enzyme-linked immunosorbant assay (Covance Laboratories, Sterling, VA). Mouse anti-BRX monoclonal antibody (05-759; Millipore, Billerica, MA) was used as indicated. Antibodies to SRF (G-20) and GATA-4 were obtained commercially (Santa Cruz Biotechnology, Inc., Santa Cruz, CA).
Whole Mount Immunohistochemistry
After fixation and dehydration, embryos were bleached in 3% peroxide with 80% methanol and 20% DMSO for 3 h. Embryos were rehydrated, incubated in phosphate-buffered saline plus 1% Tween 20 (PBST) overnight, and blocked in buffer (5% milk, PBST, 0.5% Triton X-100, 5% DMSO, 0.1% NaN3) twice for 1 h each. Embryos were incubated with primary antibody at a 1:15 dilution in blocking buffer for 2 days with gentle shaking, washed, and incubated in anti-rabbit horseradish peroxidase-conjugated IgG (Amersham Biosciences) at a 1:200 dilution in blocking buffer overnight at 4 °C. After washing, embryos were developed with one tablet of DAB Fast (Sigma-Aldrich), washed, and photographed. Genotypes were confirmed by analysis of yolk sacs and tail fragments before staining and/or analysis of the entire embryo after staining and photography.
Immunohistochemistry
Studies were performed as described (2) except as noted. Slides were incubated overnight at 4 °C in primary Ki67 antibodies (Abcam Inc., Cambridge, MA) or 4362 at a dilution of 1:1000–1:3000 in Tris-buffered saline with 1% bovine serum albumin as indicated. Slides were developed using DAB Fast for 10 min and rinsed under a cold tap for 5 min, mounted, and photographed.
In Situ Hybridization
Whole mount in situ hybridization was performed using digoxygenin-labeled (Roche Applied Science) probes. Antisense probes were prepared from partial mouse cDNAs (Hand1 (nucleotides 769–1792), Nkx2.5 (nucleotides 1023–1593), and murine Akap (nucleotides 1778–1792)) inserted in pBluescript (Stratagene) or pcDNA1 (Invitrogen), followed by linearizing the phagemid and transcription with RNA polymerase. Antisense probes for Gata-4, Mef2C, Tbx2, Anf (atrial naturetic factor), Cx40, and Mlc2a were generously provided by Drs. Christoffels and Moorman (Amsterdam, The Netherlands). Embryos were treated with Proteinase K, fixed in 4% paraformaldehyde, and then hybridized with probe overnight at 70 °C. After hybridization, embryos were treated with RNase A at 37 °C for 30 min. After blocking, anti-digoxygenin AP was added overnight at 4 °C. Embryos were then incubated in purple AP substrate and photographed.
Cell Culture and Transfection
Embryo hearts were dissected, and dispersed cells were cultured in DMEM plus 20% fetal bovine serum at 37 °C in 5% CO2. All primary cultures were verified to be spontaneously contracting or were stained positively for cardiac myosin (Abcam). After 2–4 days, cells were replated on collagen or fibronectin (FNC coating mix (AthenaES, Baltimore, MD))-coated slides (Labtek II chamber slides, Nalge Nunc International (Naperville, IL)) for confocal studies. Transfection studies were performed as described using expression constructs for Brx and Gα12QL and the c-fos SRE-luciferase reporter (14). Following overnight incubation, cells were lysed, lysates were normalized for protein content, and luciferase assays were performed. H9C2 cells derived from embryonic heart tissue were obtained from the American Tissue Type Collection (Manassas, VA). A Mef2c cDNA was amplified from a skeletal muscle cDNA library (Clontech, Mountain View, CA) and subcloned into plasmid pM (Clontech) to generate pM-MEF2C. Cells for transfection were grown on collagen-coated plates.
Transmission Electron Microscopy
Dissected embryo hearts were placed directly into Teflon baskets, fixed with glutaraldehyde, and prepared for scanning electron microscopy using standard techniques. Grids were examined with a Philips CM10 electron microscope.
Confocal Microscopy
Primary cardiomyocyte cultures were fixed, permeabilized with 0.2% Triton-X 100, blocked using 1% bovine serum albumin and 10% normal goat serum, and incubated either with 4362 murine anti-BRX polyclonal antibody or monoclonal anti-BRX (Millipore) at a 1:50 dilution overnight at 4 °C. For secondary antibody, either FITC (anti-rabbit; 1:100) or Alexa-594 (anti-mouse, 1 μg/ml; Invitrogen) was used. DAPI (Sigma) was added at 0.1 μg/ml in phosphate-buffered saline before the cells were examined with an Axiovert 405M epifluorescence inverted light microscope (Carl Zeiss, Oberkochen, Germany). Images were acquired with a CCD camera (Hamamatsu Orca, Shizuoka, Japan). To study filamentous actin, primary cultures of cardiac cells were fixed with 4% formaldehyde. Phalloidin-fluorescein isothiocyanate (Sigma) antibody or Alexa 546-phalloidin (Invitrogen) was used per the manufacturer's instructions. Mouse monoclonal antibody (BA-G5) specific for heavy chain cardiac myosin (Abcam) was used at 1:200 to detect cardiac myosin. MEF2C was detected using E-17 (Santa Cruz Biotechnology, Inc.) at 1:50 overnight at 4 °C. Images were examined as described, and the intensity of staining was quantified using ImageJ analysis (National Institutes of Health, Bethesda, MD) of ×64 magnification images.
Knockdown
After overnight incubation of H9C2 cells, 0.45 μg of the siRNA for Akap13 or AllStars negative control siRNA (Qiagen) was transfected with x-tremeGENE siRNA (Roche Applied Science). Cells were harvested at 24 h, and RNA was extracted using an RNeasy minikit (Qiagen) as per the manufacturer's instructions. The reverse transcription reaction was performed with random hexamers with the TaqMan reverse transcriptase kit (Applied Biosystems, Foster City, CA). The following primer pairs were used: Akap13 forward, 5′-AGG ATC ACC AAG TAC CCA GT-3′; Akap13 reverse, 5′-CAT CTG CCC ACT CTT CAT TC-3′; SRF forward, 5′-GGA GTT CAT CGA CAA CAA GC-3′; SRF reverse, 5′-GTA TAC ACA TGG CCT GTC TC-3′; GATA-4 forward, 5′-TCT CAC TAT GGG CAC AGC AG-3′; GATA-4 reverse, 5′-CGA GCA GGA ATT TGA AGA GG-3′; glyceraldehyde-3-phosphate dehydrogenase forward, 5′-ATG GTG AAG GTC GGT GTG AA-3′; glyceraldehyde-3-phosphate dehydrogenase reverse, 5′-GGG TAG AGT CAT ACT GGA AC-3′. Reactions were performed in triplicate using SYBR Green PCR Master Mix (Applied Biosystems) in an ABI PRISM 7700 SDS light cycler (Applied Biosystems).
Glutathione S-Transferase (GST) Binding Studies
The 35S-labeled in vitro transcribed and translated proteins were tested for binding to GST-fused BRX proteins bound to GST-Sepharose beads essentially as described (1). pEXP-RXRβ and constructs expressing GST-fused full-length BRX or its C-terminal fragment (2.10) have been described (1).
Statistics
Results are reported as means ± S.E. Values were compared using an unpaired, two-tailed Student's t test with α = 0.05 or 0.01 and were reported as p values. Tests were performed with SAS (Cary, NC).
RESULTS
Loss of Akap13 Is Associated with an Arrest of Heart Development
Following cloning and characterization of the human BRX-1 (1), we cloned the murine cDNA and murine BRX gene (hereafter called Akap13). The mouse Akap13 gene was targeted in embryonic stem cells using homologous recombination, resulting in a deletion of critical residues in the GEF region of Akap13 (Fig. 1, A–D and supplemental Fig. S1, A–C). Mice heterozygous for the null Akap13 allele appeared grossly normal but exhibited impaired glucocorticoid action (15) and an altered lymphocyte response to osmotic stress (18). Examination of wild-type and heterozygous embryos using whole mount immunohistochemistry revealed staining for AKAP13 protein in the forebrain, limb bud, first brachial arch, and developing heart (Fig. 2E). Mice heterozygous for the null Akap13 allele, while subfertile, were intercrossed with C57/BL6 mice to yield embryos homozygous for the null Akap13 allele. Amplification of Akap13 transcripts using RT-PCR from embryos with two null Akap13 alleles (−/−) failed to reveal any Akap13 transcripts (Fig. 1D; see also real-time RT-PCR results in Fig. 6A), indicating that either the mutant gene was not transcribed or the mutant mRNA transcript was unstable.
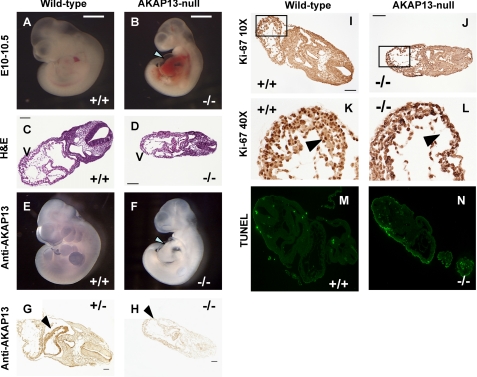
FIGURE 2.
Phenotype of Akap13-null mutant embryos. Sibling embryos are shown in A and B, C and D, E and F, and I and J. A, dissecting microscope view of wild-type embryo at E10–10.5 (bar for comparison). B, mutant sibling embryo with an enlarged heart and pericardial effusion (white arrow). Somite development of mutant embryos was within two somite pairs of wild-type embryos at E9.0–9.5. C, transverse section (hematoxylin and eosin (H&E)) through wild-type embryo at E9.5–10.0. V, ventricle. Bar, 100 μm. Magnification was ×10. D, transverse section through mutant sibling embryo. V, ventricle. Note the thin myocardium. E, whole mount immunohistochemical staining of wild-type embryo using 4362 antiserum. AKAP13 protein was present in heart, limb bud, first brachial arch, and forebrain. F, whole mount immunohistochemical staining of mutant embryo. No staining was observed. The developing heart shows a pericardial effusion and a constriction corresponding to the primary ring (arrowhead). G, transverse section of heterozygous embryo E10.0–10.5 reacted with 1:1000 antiserum 4362. Strong brown staining was present in the developing heart identical to wild-type embryos. Bar, 50 μm. H, transverse section of mutant embryo stained with 1:1000 dilution of antiserum 4362. Intensity of staining of Akap13-null embryos resembled the intensity of staining observed in wild-type embryos reacted with preimmune serum (not shown). Shown is transverse section of wild-type embryo (I) or mutant sibling embryo (J) stained with antiserum against Ki67. K and L, ×40 magnification of the section boxed in I and J, respectively. A reduction in cardiomyocyte number in the developing ventricle was noted at ×40 magnification (compare black arrowheads in K and L). M and N, TUNEL assay of cardiac tissues at E9.5 from either a wild-type (M), or Akap13-null (N) embryos.
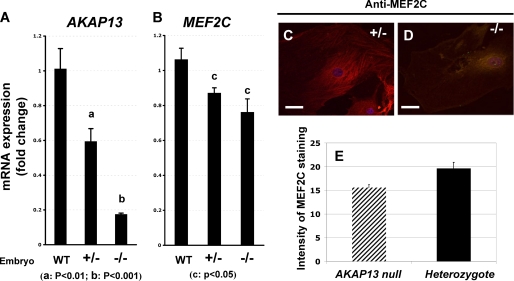
FIGURE 6.
AKAP13 is required for expression of normal levels of MEF2C. A, quantification of Akap13 mRNA levels in developing hearts at E9.0 using real-time RT-PCR. Levels of Akap13 transcripts in the hearts were compared in embryos that were wild type (WT), haploinsufficient, or homozygous null for the Akap13 gene. Transcript levels in the −/− embryos resembled background. y axis, relative expression. B, quantification of Mef2C transcript levels in embryonic hearts at E9.0. Genotypes on the x axis refer to Akap13 genotypes. Mef2C transcripts in hearts of −/− embryos were reduced 25–30% from the levels present in wild-type mice. C, representative cardiomyocyte from heterozygote embryo plated on collagen and stained for MEF2C (E-17) and Alexa-594. D, representative cardiomyocyte from Akap13-null mouse stained for MEF2C and Alexa-594. E, quantification of confocal staining intensity for MEF2C from heterozygote or Akap13-null cardiomyocytes. y axis, pixels/area; error bars, ±S.E.; p < 0.05.
Genotypes of 438 offspring from 74 heterozygous crosses revealed all newborn mice to be either wild-type or heterozygous, indicating that homozygous mutation of the Akap13 allele resulted in embryonic lethality (Table l). Heterozygous adult mice haploinsufficient for the allele exhibited reduced mRNA and protein (18), as expected. Genotypes of embryos from timed pregnancies confirmed that embryos homozygous for the null Akap13 allele were present at the predicted Mendelian frequency before E9.0–10.0; however, after E10.5–11.0, resorbing nidation sites were observed (Table 2). Dissection of Akap13−/− embryos revealed normal development until E8.0–8.5. Mutant −/− embryos at E9.0–9.5 were typically within 2 somite pairs of wild-type embryos (Fig. 2), but by E10, homozygous null embryos appeared developmentally delayed compared with wild-type littermates with reduced size of the forebrain, constriction of the nuchal region, and a hypoplastic first brachial arch (Fig. 2F). For this reason, experiments were performed at E9.0–9.5.
TABLE 2.
Gestational age at time of embryonic loss in 26 heterozygous-heterozygous matings
Mice were heterozygous for the null Akap13 allele. Pregnant dams were sacrificed at the days noted (column 1), and nidation sites were scored as normal (column 2) or being resorbed (column 3). Genotyping by PCR confirmed that resorbing embryos were homozygous for the Akap13-null allele, where it was possible to distinguish embryonic tissue from resorbing decidua. Gestations were scored according to the most advanced embryo development.
| Days postcoitum | Normal embryos | Resorbed embryos | Percent resorbed |
|---|---|---|---|
| % | |||
| E7–7.5 | 54 | 0 | 0 |
| E8–8.5 | 77 | 1 | 1.3 |
| E9.5–10.5 | 30 | 10 | 25 |
| E12.5+ | 13 | 6 | 31.6 |
TABLE 1.
Genotypes of 438 newborn mice from 74 heterozygous (+/−) × heterozygous (+/−) matings
Mice were heterozygous for wild-type and null Akap13 allelles.
| Genotype of newborn mice | Expected Mendelian ratio | Observed ratio (number of pups) |
|---|---|---|
| % | % | |
| +/+ | 25 | 36 (n = 157) |
| +/− | 50 | 64 (n = 281) |
| −/− | 25 | 0 (n = 0) |
Importantly, cardiac development of homozygous embryos was abnormal because the hearts became enlarged and developed a pericardial effusion. Hematoxylin and eosin (H&E) sections of Akap13-null embryos revealed the developing myocardium to be thinned with reduced trabeculation (Fig. 2, D, K, and L). The arrest of heart development after E9.0–9.5 suggested that Akap13 was required for cardiomyocyte differentiation. Immunohistochemical analysis revealed strong staining for AKAP13 in newborn and adult hearts (supplemental Fig. 2). Northern and Western blots indicated that the primary cardiac transcript size was 9.5–10.0 kb (Fig. 1E) and that it encoded a 210-220-kDa protein (supplemental Fig. S2D). Staining of developing hearts from wild-type and heterozygous mice at E9.0–9.5 with an antiserum directed against AKAP13 revealed strong staining in the myocardium (Fig. 2, E and G). The primary ring was visible in hearts of Akap13-null embryos (Fig. 2F, arrowhead); however, chamber development slowed and arrested by E9.5–10.0.
As an initial approach to examine the mechanism responsible for arrest of cardiac development, we assessed markers of proliferation and apoptosis. Intensity of staining for the Ki67 proliferation antigen did not differ between wild-type and homozygous null embryos (Fig. 2, I–L). Analysis of embryos at E9.0–9.5 using a TUNEL assay showed no increase in apoptosis in Akap13−/− embryos, consistent with an arrest in development rather than an increase in programmed cell death (Fig. 2, M and N, and supplemental Fig. S3).
AKAP13 Is Co-localized to Actin and Myosin Filaments in Developing Cardiomyocytes
To further define the mechanism responsible for the failure of heart development, we examined the subcellular localization of AKAP13 in primary cultures of cardiomyocytes from wild-type or Akap13−/− embryos. AKAP13 protein was localized to filaments in cardiac cells of wild-type mice. An identical pattern of staining was observed for both a monoclonal antibody (Fig. 3C) and polyclonal antisera directed against different regions of AKAP13 (Fig. 3A), confirming specificity of detection.
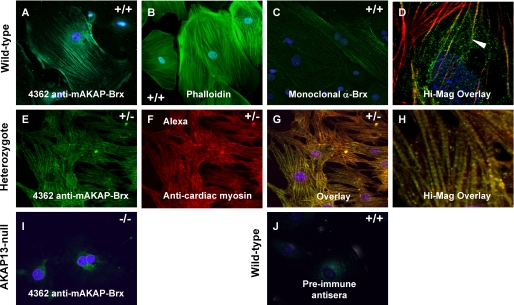
FIGURE 3.
Localization of AKAP13 to the cytoskeleton. A, confocal image of primary culture of cardiomyocytes from a wild-type (+/+) embryos at E9.0–9.5 plated on collagen and stained with antiserum 4362 directed against AKAP13 and DAPI. B, confocal image of cardiomyocytes of wild-type embryo plated on collagen and stained with FITC-phalloidin and DAPI. C, wild-type cardiomyocytes stained with a monoclonal antibody directed against AKAP13 and DAPI (×40, oil). D, high magnification; overlay (+/+) cardiomyocytes stained with 4362 antiserum (green) and actin (red). The yellow signal (arrowhead) shows co-localization of AKAP13 and actin. E, heterozygote cardiomyocytes plated on fibronectin stained with antiserum 4362 and DAPI. Intensity of staining from cardiomyocytes from heterozygous mice resembled that in cells from wild-type mice. Cells plated on fibronectin showed more robust actin-myosin filaments. F, heterozygote cardiomyocytes plated on fibronectin stained with monoclonal antibody specific for heavy chain cardiac myosin (ab50967) (×40, oil). G, overlay of heterozygote cell in E and F. The yellow signal shows co-localization of AKAP13 and cardiac myosin (×40, oil). H, high magnification overlay showing myosin bands. I, cardiomyocytes from an Akap13 null embryo at E9.0–9.5 stained with 4362 antiserum showed only background levels of signal. J, wild-type cardiomyocytes stained with preimmune serum showed no staining of cytoskeleton (×40, oil).
Of particular interest, confocal microscopic studies of doubly stained cells co-localized AKAP13 to filaments that stained positively for actin (Fig. 3D). Co-localization of AKAP13 with the cytoskeleton had not been previously reported. We noted that most, but not all, of the AKAP13 protein was closely associated with cytoskeletal filaments. Staining intensity for AKAP13 protein in heart cells derived from Akap13−/− embryos (Fig. 3I) resembled background staining with preimmune sera (Fig. 3J), consistent with the absence of protein, as expected based on RT-PCR results (Fig. 1D). To further explore the co-localization of AKAP13 with cytoskeletal filaments, cardiomyocytes derived from E9.5 mice were stained with a monoclonal antibody specific for cardiac myosin and antisera directed against AKAP13 (Fig. 3, E–H). AKAP13 protein also co-localized to filaments that stained positively for cardiac myosin.
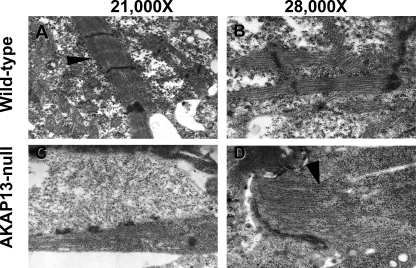
The co-localization of AKAP13 with actin and myosin led us to examine sarcomere development in the Akap13-null mice because 1) Rho GTPases have been shown to be required for normal sarcomere development in cardiomyocytes (7–9), and 2) SRF promotes actin polymerization following serum deprivation, and targeted disruption of SRF was accompanied by altered sarcomere structure (22–25). Electron micrographs of wild-type embryonic hearts at E9.0–9.5 showed normal sarcomere structure (Fig. 4, A and B). In contrast, sarcomeres in the hearts of Akap13-null embryos were abnormally formed with incompletely produced myofilaments that ended blindly and failed to form Z-discs (Fig. 4, C and D). Although mitosis of developing cardiomyocytes is associated with a transient dedifferentiation of contractile apparatus (26), results of Ki-67 staining did not support that explanation for the altered sarcomere structure in the Akap13-null mice.
FIGURE 4.
Altered sarcomere development in Akap13-null mice. A, transmission electron micrograph from developing heart at E9.0–9.5 of wild-type mice exhibited normal sarcomere structure. Arrowhead, Z-disc. B, ×28,000 magnification of sarcomere from wild-type mouse. C, transmission electron micrograph of mutant heart at E9.0–9.5 showed few sarcomere structures and structures lacking distinct Z-discs. D, ×28,000 magnification of mutant sarcomere. Note that the sarcomere ended blindly (black arrowhead).
Loss of Akap13 Is Associated with Altered Expression of Genes Involved in Heart Development
A failure of cardiac formation was consistent with inactivation of AKAP13 because downstream targets of Rho influence cardiomyocyte differentiation (reviewed in Ref. 27). For this reason, we examined markers of cardiac development using whole mount in situ hybridization (Fig. 5). The results suggested that expression of Mef2C was reduced in Akap13−/− embryos. Levels of Nkx2.5 and Gata-4 transcripts (and GATA-4 protein; supplemental Fig. 4) were not reduced in Akap13-null embryos. Tbx2 transcripts were reduced in mutant embryos compared with wild-type littermates.
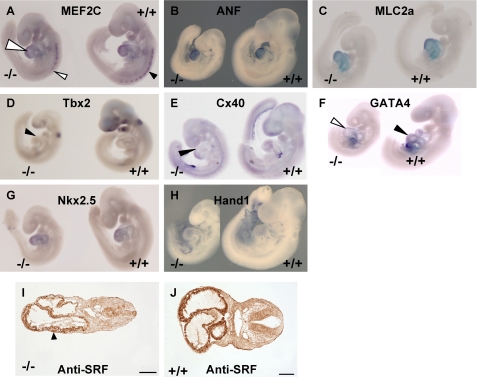
FIGURE 5.
Altered expression of heart markers in Akap13-null embryos. A–H, whole mount in situ hybridization of sibling embryos E9–9.5 with either a mutant genotype (−/−) or a wild-type (+/+) genotype. Each panel is a representative of three independent experiments with sibling mutant embryos. A, staining for Mef2C mRNA was reduced in AKAP13-null embryos (left) compared with wild-type littermates. Reduced staining was noted in somites (arrowheads) and heart. B, Anf transcripts did not differ between wild-type littermates and mutant embryos. C, Mlc2a transcripts were similarly expressed. D, Tbx2 staining was reduced in Akap13-null embryos (black triangle) compared with wild-type littermates. E, connexin 40 (Cx40) transcripts were localized to the extreme anterior ventricle in the Akap13-null embryos (black triangle) compared with wild-type littermates. F, Gata-4 transcripts were slightly reduced, in developing hearts of Akap13−/− embryos (open triangle) compared with wild-type embryos (black triangle). G, staining for Nkx2.5 transcripts was similar in −/− embryos compared with wild-type littermates. H, Hand1 transcripts in Akap13−/− embryos were comparable with wild-type littermates. I, transverse section of mutant embryo reacted with 1:500 antiserum directed against SRF. SRF protein was reduced in the developing hearts of mutant embryos. Bar, 100 μm. J, transverse section of wild-type embryo reacted with 1:500 antiserum directed against SRF. Strong staining is observed in myocardium. Bar, 100 μm.
Markers were chosen to assess the status of anterior-posterior positioning in mutant embryos. Mutant embryos displayed similar expression of Mlc2a in the ventricle compared with wild-type littermates at E9.0. Likewise, Hand1 transcripts were not reduced in Akap13-null embryos. Although expression patterns for Cx40 were detected at the proper temporal stage, spatial localization occurred at the extreme anterior ventricle and extreme posterior of the atria in Akap13−/− embryos, suggesting that Cx40 expression at E9.0 was inhibited in Akap13-null embryos. Anf transcripts were observed in these embryos at E9.0 with heavy expression in the periphery of the ventricle in Akap13-null mutants. Collectively, the results (Fig. 5) reveal that the absence of Akap13 resulted in alterations in expression of the genes essential for normal cardiac development, in particular Mef2C.
Given the qualitative reduction of Mef2C in whole mount in situ experiments, we tested whether absence of AKAP13 might be associated with a reduction in Mef2C transcripts in developing hearts at E9.0. As expected, levels of Akap13 transcripts were reduced in heterozygous and present only at background levels in Akap13-null hearts (Fig. 6A). Levels of Mef2C transcripts were significantly reduced (Fig. 6B). MEF2C protein levels were also significantly reduced in cardiomyocytes derived from Akap13-null mice compared with cardiomyocytes derived from heterozygous mice or wild-type mice (not shown).
The reduction in Mef2C led us to test whether knockdown of AkaP13 might cause a reduction in endogenous levels of transcripts encoding MEF2 or SRF (Fig. 7). The addition of siRNA specific for Akap13 resulted in an 80–90% reduction in Akap13 transcripts (Fig. 7A, left). Gata-4 transcripts were not reduced (supplemental Fig. S3), but transcripts of Mef2C were significantly reduced to 50% of control (Fig. 7A, middle). In contrast, Srf transcripts were reduced to 30% of control following LPA stimulation (Fig. 7). These findings indicate that AKAP13 is required for expression of normal levels of endogenous Mef2C mRNA in heart cells, a conclusion consistent with the results in Figs. 5 and 6.
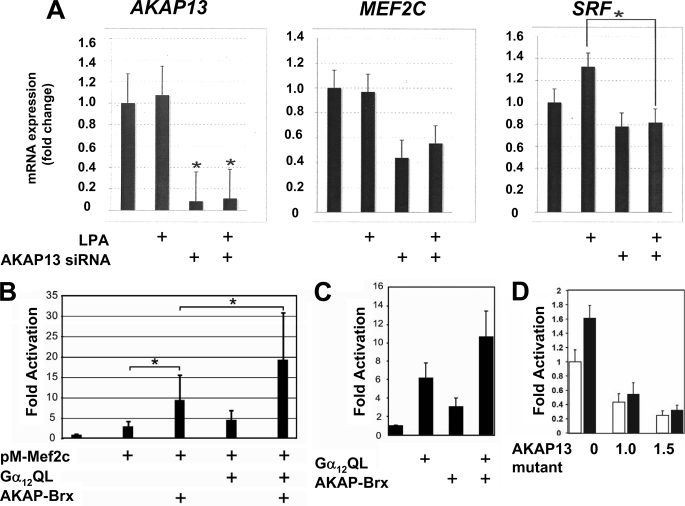
FIGURE 7.
AKAP13 augments MEF2C-dependent gene activation via Gα12. A, endogenous AKAP13 is required for optimal MEF2C expression. H9C2 cells were transfected with control or Akap13 siRNA and treated with LPA as indicated. Total RNA was purified, and amounts of Akap13, Mef2C, or Srf were determined by real-time RT-PCR. Relative expression levels of mRNA are shown as -fold induction over base line. Results were confirmed in two separate experiments. Error bars, S.E. Mef2C mRNA levels were significantly reduced. *, p < 0.01. B, H9C2 cells were grown to 50% confluence on 12-well plates and transfected with 1.0 μg of Gal4E1b-Luc, 600 ng of pM-MEF2C, 50 ng of Gα12QL, and 1.2 μg of AKAP13 expression vectors as indicated. Following overnight incubation, cells were lysed, and luciferase assays were performed. Results are from three independent experiments. *, p < 0.05. Basal levels of reporter activity were not increased by AKAP13 alone (supplemental Fig. S5). C, transfection of serum-starved COS-7 cells (COS-7 cells were chosen because of low levels of endogenous AKAP13) with an SRE-luciferase reporter, 500 ng of an AKAP-BRX expression vector, and 400 ng of a Gα12QL expression vector, as indicated. y axis, -fold activation. Error bars, ±S.E. D, an AKAP13 mutant lacking the GEF region functions in a dominant-negative manner to inhibit activation of an SRE reporter. Shown is transient transfection of OVCAR-3 cells (shown here because of high levels of endogenous AKAP13) with 1.0 μg of SRE-luciferase reporter and the indicated amounts of an AKAP13 expression construct lacking the GEF region (AKAP13 mutant) in the presence (solid bars) or absence (open bars) of 1 μm LPA. Results of two experiments repeated in quadruplicate are shown. Error bars, ±S.E. The addition of the AKAP13 mutant lacking the GEF region inhibited SRF activation by endogenous levels of AKAP13.
Next we tested whether AKAP13 might contribute to MEF2-dependent or SRF-dependent activation through Gα12 because the contribution of Gα12 to either SRF (28) or MEF2-dependent activation (13) had not been examined. The possible contribution of Gα12 was of particular interest because AKAP13 has been shown to act downstream of Gα12 in mature cardiomyocytes (29). Transfection experiments revealed that AKAP13 caused a specific and dose-dependent increase in MEF2-dependent activation that was significantly augmented by the addition of a constitutively active Gα12 mutant (Fig. 7B). Furthermore, activation of an SRF-dependent reporter was also enhanced, albeit to a lesser extent, in the presence of Gα12 and was dependent upon the GEF region of AKAP13 (Fig. 7, C and D). In addition, GST-binding experiments revealed that the carboxyl region of AKAP13 bound to MEF2C, suggesting that a direct physical interaction may be involved (Fig. 8) that required the carboxyl region of AKAP13 (BRX). Together, the findings indicate that AKAP13 significantly enhanced MEF2-dependent gene activation through a mechanism involving Gα12 and suggest that MEF2C activity may be dependent, at least in part, upon AKAP13 in developing cardiomyocytes.
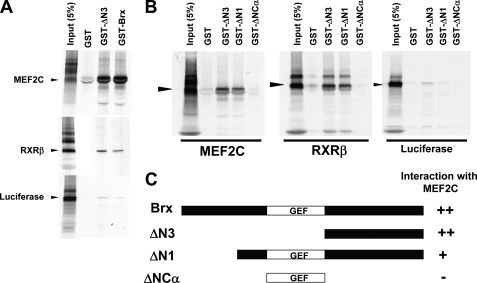
FIGURE 8.
The carboxyl region of AKAP13 (BRX) binds to MEF2C. A, 35S-labeled MEF2C and RXR bound avidly to GST fusion proteins of BRX or a GST fusion protein containing only the carboxyl region (ΔN3). B, 35S-labeled MEF2C and RXR bound avidly to BRX mutants (black arrowhead). The carboxyl-terminal region of BRX was sufficient for MEF2C binding. The 35S-labeled RXRβ or luciferase was used and positive and negative controls, respectively. C, schematic diagram of BRX mutants (see Ref. 1). GST-BRX includes amino acids 47–1428. ΔN1 contains amino acids 236–1428 of BRX. ΔN3 includes amino acids 960–1428. ΔNCα contains amino acids 527–950.
DISCUSSION
These results reveal an essential role for Akap13 in cardiomyocyte differentiation and heart morphogenesis. Two GEFs, Trio and Obscurin, have been implicated in control of skeletal muscle development (6), but no GEFs have formerly been reported to be essential for cardiac development. A requirement for Akap13 in the developing myocardium is supported by recent evidence that AKAP13 influences cardiac hypertrophy through MEF2 by coupling activation of protein kinase D with phosphorylation-dependent class II histone deacetylase HDAC5 (13). Our findings extend those observations to reveal a critical requirement for Akap13 in the developing heart.
MEF2C is the first MEF2 gene product to appear in mesodermal cells that develop into the heart (30). MEF2 proteins belong to the MADS-box family of transcriptional regulators that includes SRF. Co-transfection experiments showed that AKAP13 increased both SRF- and MEF2C-dependent reporter activity. Activation of MEF2 transcription factors leads to cardiac, smooth, and skeletal muscle gene regulation (for a review, see Ref. 31). Of note, MEF2C-null embryos developed normally until E9.0, when a pericardial effusion developed, and there was a failure of looping morphogenesis resulting in an embryonic lethal mutation (32). Thus, the cardiac phenotype of the Akap13-null mutant resembled the cardiac phenotype of the Mef2C-null mice.
Based on activation of AKAP13 by Gα12, our current working model is that splice variants of AKAP13 function in a hierarchical fashion as platforms to integrate signals emanating from cell surface receptors to transcription factors through Rho (Fig. 9). This interpretation is in keeping with the reported role of AKAP13 downstream of the α1 adrenergic receptor (29), the role of AKAP13 in cardiac hypertrophy (13), and prior studies of cardiac differentiation (see below). Two caveats are that the heart phenotype of the Akap13 knock-out could possibly be due to selective loss of MEF2-expressing cells in the heart or a consequence of a defect in other organ systems (e.g. blood or vessels). However, evidence against an indirect etiology for the observed lethal phenotype is that AKAP13 is most heavily expressed in the heart, both during development (Fig. 2) and in adult tissues (Fig. 1E and supplemental Fig. S2). Definitive experiments to exclude the possibility of contribution from another organ system will require a tissue-specific deletion of AKAP13 solely in the heart.
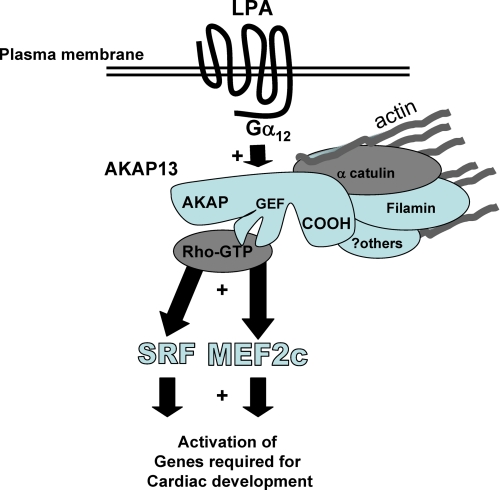
FIGURE 9.
Model of AKAP13 action in developing heart cells. AKAP13 is activated by extracellular molecules, such as LPA, through Gα12, leading to augmentation of MEF2C and SRF activation. AKAP13 is co-localized with actin in cardiomyocytes, possibly through previously reported interactions with α-catulin (18, 28) and filamins A and B (18, 43). MEF2C and SRF are known to play critical roles in cardiomyocyte differentiation (32, 37).
It is worth noting that different sized forms of AKAP13 appear to have distinct roles. In the heart, the larger 10-kb transcript is expressed. In lymphocytes of mice, the smaller 6.7-kb transcript is present and regulates the response to osmotic stress (18) through activation of NFAT5 (nuclear factor of activated T cells 5). Interestingly, several studies have shown a role for NFAT5 in endocardial cushion development in the heart (33–35).
Because AKAP13 is stimulated by G-proteins and in turn activates Rho, our transfection experiments are also supported by studies indicating an essential role of G-proteins (36) and Rho in cardiac development (37–39). Furthermore, the Akap13-null phenotype is supportive of previous reports indicating an important role of AKAPs in heart development (40, 41), but our findings extend the understanding to reveal a specific developmental requirement for Akap13.
In addition, we show by confocal microscopy and immunogold studies that AKAP13 was closely associated with actin and myosin filaments in embryonic cardiac cells. This suggests that AKAP13 functions as a scaffold protein in these cells and supports prior reports that an actin cytoskeleton was essential for RhoA-dependent activation of SRF in cardiac myocytes (37) and that SRF was required for actin assembly (42). Thus, it is very likely that reduced activation of SRF may contribute substantially to the observed phenotype. Our results are also supported by prior reports that AKAP13 associated with the actin-binding protein filamin (43). It is notable that targeted deletion of FLNA (filamin A) also resulted in a severe and lethal cardiac defect (44). In support of the localization studies in Fig. 3, a yeast two-hybrid screen using the C terminus of Brx as bait (18) indicated that this region was capable of interaction with actin-associated proteins, including filamin A, filamin B, α-catulin, and vimentin.
In consideration of the marked phenotype observed in Akap13-null mice, we note that AKAP13 is the only AKAP-Rho-GEF described to date, and no redundancy of the gene was apparent on our Southern analyses (1). Moreover, AKAP13 has been reported to have a 660-fold greater affinity for dephosphorylated RII (regulatory unit of protein kinase A) than AKAP 15/18 (40, 45), suggesting that substrate affinity may explain, at least in part, the severity of the phenotype observed. More specific proof of the regions in Akap13 required for cardiomyocyte differentiation will require a heart-specific conditional inactivation of this protein.
Supplementary Material
Acknowledgments
We recognize the unwavering support and advice of Drs. George Chrousos and Alan H. DeCherney, which helped to make the work possible. The authors also thank Dr. Matthew Phillips and Joy Britten-Webb for essential technical assistance and suggestions. We express gratitude to Drs. Vincent Christoffels and Antoon Moorman for antisense probes for GATA-4, MEF2C, Tbx2, ANF, Cx40, and MLC2a. G±12QL was a kind gift of Dr. Silvio Gutkind.
This work was supported, in whole or in part, by the Intramural Research Program of NICHD, National Institutes of Health, Grant ZO1-HD-008737-07-URE.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- DH
- Dbl homology
- SRF
- serum response factor
- AKAP
- protein kinase A-anchoring protein
- GEF
- guanine nucleotide exchange factor
- LPA
- lysophosphatidic acid
- RXR
- retinoid X receptor
- RT
- reverse transcription
- TUNEL
- terminal deoxynucleotidyltransferase dUTP nick end labeling
- DAPI
- 4′,6-diamidino-2-phenylindole
- siRNA
- small interfering RNA
- GST
- glutathione S-transferase
- En
- embryonic day n.
REFERENCES
- 1.Rubino D., Driggers P., Arbit D., Kemp L., Miller B., Coso O., Pagliai K., Gray K., Gutkind S., Segars J. (1998) Oncogene 16, 2513–2526 [DOI] [PubMed] [Google Scholar]
- 2.Miller B. T., Rubino D. M., Driggers P. H., Haddad B., Cisar M., Gray K., Segars J. H. (2000) Am. J. Obstet. Gynecol. 182, 286–295 [DOI] [PubMed] [Google Scholar]
- 3.Diviani D., Soderling J., Scott J. D. (2001) J. Biol. Chem. 276, 44247–44257 [DOI] [PubMed] [Google Scholar]
- 4.Toksoz D., Williams D. A. (1994) Oncogene 9, 621–628 [PubMed] [Google Scholar]
- 5.Sterpetti P., Hack A. A., Bashar M. P., Park B., Cheng S. D., Knoll J. H., Urano T., Feig L. A., Toksoz D. (1999) Mol. Cell. Biol. 19, 1334–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmidt A., Hall A. (2002) Genes Dev. 16, 1587–1609 [DOI] [PubMed] [Google Scholar]
- 7.Hall A. (1998) Science 279, 509–514 [DOI] [PubMed] [Google Scholar]
- 8.Charron F., Tsimiklis G., Arcand M., Robitaille L., Liang Q., Molkentin J. D., Meloche S., Nemer M. (2001) Genes Dev. 15, 2702–2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wettschureck N., Offermanns S. (2002) J. Mol. Med. 80, 629–638 [DOI] [PubMed] [Google Scholar]
- 10.Teramoto H., Malek R. L., Behbahani B., Castellone M. D., Lee N. H., Gutkind J. S. (2003) Oncogene 22, 2689–2697 [DOI] [PubMed] [Google Scholar]
- 11.Sahai E., Alberts A. S., Treisman R. (1998) EMBO J. 17, 1350–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei L., Zhou W., Croissant J. D., Johansen F. E., Prywes R., Balasubramanyam A., Schwartz R. J. (1998) J. Biol. Chem. 273, 30287–30294 [DOI] [PubMed] [Google Scholar]
- 13.Carnegie G. K., Soughayer J., Smith F. D., Pedroja B. S., Zhang F., Diviani D., Bristow M. R., Kunkel M. T., Newton A. C., Langeberg L. K., Scott J. D. (2008) Mol. Cell 32, 169–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Driggers P. H., Segars J. H., Rubino D. M. (2001) J. Biol. Chem. 276, 46792–476797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kino T., Souvatzoglou E., Charmandari E., Ichijo T., Driggers P., Mayers C., Alatsatianos A., Manoli I., Westphal H., Chrousos G. P., Segars J. H. (2006) J. Biol. Chem. 281, 9118–9126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baisamy L., Jurisch N., Diviani D. (2005) J. Biol. Chem. 280, 15405–15412 [DOI] [PubMed] [Google Scholar]
- 17.Majumdar M., Seasholtz T. M., Buckmaster C., Toksoz D., Brown J. H. (1999) J. Biol. Chem. 274, 26815–26821 [DOI] [PubMed] [Google Scholar]
- 18.Kino T., Takatori H., Manoli I., Wang Y., Tiulpakov A., Blackman M. R., Su Y. A., Chrousos G. P., DeCherney A. H., Segars J. H. (2009) Sci. Signal. 2, ra5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan J., Slice L. W., Rozengurt E. (2001) J. Biol. Chem. 276, 38619–38627 [DOI] [PubMed] [Google Scholar]
- 20.Dutt P., Kjoller L., Giel M., Hall A., Toksoz D. (2002) FEBS Lett. 531, 565–569 [DOI] [PubMed] [Google Scholar]
- 21.Sagi S. A., Seasholtz T. M., Kobiashvili M., Wilson B. A., Toksoz D., Brown J. H. (2001) J. Biol. Chem. 276, 15445–15452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arsenian S., Weinhold B., Oelgeschläger M., Rüther U., Nordheim A. (1998) EMBO J. 17, 6289–6299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parlakian A., Tuil D., Hamard G., Tavernier G., Hentzen D., Concordet J. P., Paulin D., Li Z., Daegelen D. (2004) Mol. Cell. Biol. 24, 5281–5289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miano J. M., Ramanan N., Georger M. A., de Mesy Bentley K. L., Emerson R. L., Balza R. O., Jr., Xiao Q., Weiler H., Ginty D. D., Misra R. P. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 17132–17137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balza R. O., Jr., Misra R. P. (2006) J. Biol. Chem. 281, 6498–6510 [DOI] [PubMed] [Google Scholar]
- 26.Engel F. B., Schebesta M., Duong M. T., Lu G., Ren S., Madwed J. B., Jiang H., Wang Y., Keating M. T. (2005) Genes Dev. 19, 1175–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pikkarainen S., Tokola H., Kerkelä R., Ruskoaho H. (2004) Cardiovasc. Res. 63, 196–207 [DOI] [PubMed] [Google Scholar]
- 28.Park B., Nguyen N. T., Dutt P., Merdek K. D., Bashar M., Sterpetti P., Tosolini A., Testa J. R., Toksoz D. (2002) J. Biol. Chem. 277, 45361–45370 [DOI] [PubMed] [Google Scholar]
- 29.Appert-Collin A., Cotecchia S., Nenniger-Tosato M., Pedrazzini T., Diviani D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10140–10145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edmondson D. G., Lyons G. E., Martin J. F., Olson E. N. (1994) Development 120, 1251–1263 [DOI] [PubMed] [Google Scholar]
- 31.Black B. L., Olson E. N. (1998) Annu. Rev. Cell Dev. Biol. 14, 167–196 [DOI] [PubMed] [Google Scholar]
- 32.Lin Q., Schwarz J., Bucana C., Olson E. N. (1997) Science 276, 1404–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ichida M., Finkel T. (2001) J. Biol. Chem. 276, 3524–3530 [DOI] [PubMed] [Google Scholar]
- 34.Chang C. P., Neilson J. R., Bayle J. H., Gestwicki J. E., Kuo A., Stankunas K., Graef I. A., Crabtree G. R. (2004) Cell 118, 649–663 [DOI] [PubMed] [Google Scholar]
- 35.Uhlén P., Burch P. M., Zito C. I., Estrada M., Ehrlich B. E., Bennett A. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2160–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams J. W., Brown J. H. (2001) Oncogene 20, 1626–1634 [DOI] [PubMed] [Google Scholar]
- 37.Wei L., Wang L., Carson J. A., Agan J. E., Imanaka-Yoshida K., Schwartz R. J. (2001) FASEB J. 15, 785–796 [DOI] [PubMed] [Google Scholar]
- 38.Sah V. P., Hoshijima M., Chien K. R., Brown J. H. (1996) J. Biol. Chem. 271, 31185–31190 [DOI] [PubMed] [Google Scholar]
- 39.Hoshijima M., Sah V. P., Wang Y., Chien K. R., Brown J. H. (1998) J. Biol. Chem. 273, 7725–7730 [DOI] [PubMed] [Google Scholar]
- 40.Ruehr M. L., Russell M. A., Bond M. (2004) J. Mol. Cell. Cardiol. 37, 653–665 [DOI] [PubMed] [Google Scholar]
- 41.Zakhary D. R., Moravec C. S., Bond M. (2000) Circulation 101, 1459–1464 [DOI] [PubMed] [Google Scholar]
- 42.Schratt G., Philippar U., Berger J., Schwarz H., Heidenreich O., Nordheim A. (2002) J. Cell Biol. 156, 737–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pi M., Spurney R. F., Tu Q., Hinson T., Quarles L. D. (2002) Endocrinology 143, 3830–3838 [DOI] [PubMed] [Google Scholar]
- 44.Feng Y., Chen M. H., Moskowitz I. P., Mendonza A. M., Vidali L., Nakamura F., Kwiatkowski D. J., Walsh C. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 19836–19841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zakhary D. R., Fink M. A., Ruehr M. L., Bond M. (2000) J. Biol. Chem. 275, 41389–41395 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.