Abstract
Different nicotinic acetylcholine receptor (nAChR) subtypes are implicated in learning, pain sensation, and disease states, including Parkinson disease and nicotine addiction. α-Conotoxins are among the most selective nAChR ligands. Mechanistic insights into the structure, function, and receptor interaction of α-conotoxins may serve as a platform for development of new therapies. Previously characterized α-conotoxins have a highly conserved Ser-Xaa-Pro motif that is crucial for potent nAChR interaction. This study characterized the novel α-conotoxin LtIA, which lacks this highly conserved motif but potently blocked α3β2 nAChRs with a 9.8 nm IC50 value. The off-rate of LtIA was rapid relative to Ser-Xaa-Pro-containing α-conotoxin MII. Nevertheless, pre-block of α3β2 nAChRs with LtIA prevented the slowly reversible block associated with MII, suggesting overlap in their binding sites. nAChR β subunit ligand-binding interface mutations were used to examine the >1000-fold selectivity difference of LtIA for α3β2 versus α3β4 nAChRs. Unlike MII, LtIA had a >900-fold increased IC50 value on α3β2(F119Q) versus wild type nAChRs, whereas T59K and V111I β2 mutants had little effect. Molecular docking simulations suggested that LtIA had a surprisingly shallow binding site on the α3β2 nAChR that includes β2 Lys-79. The K79A mutant disrupted LtIA binding but was without effect on an LtIA analog where the Ser-Xaa-Pro motif is present, consistent with distinct binding modes.
Keywords: Peptides, Peptides/Chemical Synthesis, Peptides/Interactions, Peptides/Neuropeptide, Receptors, Toxins, Toxin/Channels
Introduction
Neuronal nAChRs2 are found throughout the central and peripheral nervous system. These nAChRs, located both pre- and post-synaptically, modulate the release of neurotransmitters and/or mediate fast synaptic transmission (1, 2). In addition, nAChRs are present in a variety of non-neuronal tissue, including epithelium and immune cells. In mammals, there are nine α and four β nAChR subunits that can combine to form multiple subtypes of hetero-pentamers (3). The array of physiological functions mediated by nAChRs is correspondingly broad. Compounds acting on nicotinic receptors are currently utilized as medications for treating nicotine addiction and are being explored as therapeutic options for a variety of conditions, including cognitive disorders and pain (4). Understanding the factors contributing to ligand-nAChR subtype specificity will play a crucial role in developing compounds that have the desired therapeutic effects but that lack side effects (5).
Many phyla utilize acetylcholine for neurotransmission. Predatory organisms have exploited this by developing toxins that act at nAChRs to disrupt the flight and fight of their prey (6). Predatory marine snails of the genus Conus synthesize complex venoms that contain an arsenal of peptides. Over the past several years, these “conotoxins” have been shown to have a remarkable diversity of pharmacological function and utility (7, 8). Based on molecular structure, the conotoxins have been grouped into A, O, M, P, R, S, and T superfamilies (9, 10). A subgroup of compounds found within the A superfamily is the α-conotoxins. These are small, 13–21-residue, cysteine-rich peptides that functionally block nAChRs (11, 12). Structural studies utilizing NMR spectroscopy and x-ray crystallography have provided insight into the role and spatial location of residues important for function (13).
Previously characterized α-conotoxins have a highly conserved Ser-Xaa-Pro motif in the first intercysteine loop. The Pro residue is essential for high affinity binding to nAChRs (14). This study characterizes the novel α4/7 conotoxin LtIA, which lacks the highly conserved Ser-Xaa-Pro sequence but maintains high potency. (The 4/7 terminology refers to the number of non-Cys residues in the first and second intercysteine loops present in all α-conotoxins.) Structural and functional studies provide insight into the mechanism of action and the nature of the nAChR-binding site of LtIA. nAChRs are pentamers made up of α and β subunits. Previous reports indicated that the ACh-binding site is composed of a hydrophobic group of conserved aromatic residues from both the α and β subunits in proximity to the two disulfide-linked vicinal cysteines of the α subunit C loop. High conservation of residues that form the ligand-binding interface among subtypes of nAChRs hinders the ability of ligands to discriminate among these subtypes. However, nonconserved residues appear to line a binding cleft, and functional understanding of these residues could be utilized to develop subtype-specific ligands (15, 16). Mutation of one of these residues, nAChR β2 subunit Phe-119, substantially reduces the block by LtIA. This decrease in activity is the largest effect described thus far for a β subunit mutant affecting α-conotoxin block and was supported by molecular docking simulations. α-Conotoxin LtIA, the first α-conotoxin to be identified from Conus litteratus of the South China Sea, provides a novel probe of nAChR structure and function. This study suggests there is a novel, shallow microsite for α-conotoxins lacking the conserved Ser-Xaa-Pro motif.
EXPERIMENTAL PROCEDURES
Materials
Acetylcholine chloride, atropine, and bovine serum albumin were obtained from Sigma. Reversed-phase HPLC analytical Vydac C18 column (5 μm, 4.6 × 250 mm) and preparative C18 Vydac column (10 μm, 22 × 250 mm) were obtained from Shenyue (Shanghai City, China). Reagents for peptide synthesis were from Applied Biosystems (Guangzhou City, China). Acetonitrile was purchased from Fisher. Trifluoroacetic acid was from Tedia Co. (Fairfield, OH). All other chemicals used were of analytical grade. The sequence of α-conotoxin LtIA (GCCARAACAGIHQELC) was based on a cDNA clone (LeD2P) from C. litteratus from Hainan, China (Table 1) (17). A highly similar cDNA sequence was also reported from Conus leopardus (18). Clones of rat α2–α7 and β2–β4 as well as mouse muscle α1β1δϵ cDNAs were kindly provided by S. Heinemann (Salk Institute, San Diego). Clones of β2 and β3 subunits in the high expressing pGEMHE vector were kindly provided by C. W. Luetje, University of Miami, Miami, FL).
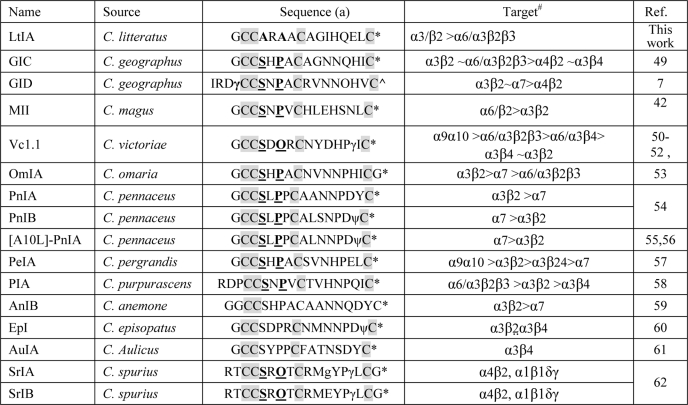
TABLE 1.
Selected neuronally active α-4/7 conotoxin sequences showing a highly conserved serine and proline residue in the first intercysteine loop (loop 1)
(a) O = 4-trans-hydroxyproline; γ = γ-carboxyglutamate; ∧ = C-terminal carboxylate; ψ = sulfated tyrosine; * = C-terminal carboxamide. The highly conserved Ser and Pro in loop 1 are highlighted in boldface and underlined. The conserved Cys residues are shaded. #, note that all nAChR subtypes were not tested in each instance. Refer to cited literature for full details.
Peptide Synthesis by One-step Oxidation
α-Conotoxin LtIA was assembled by solid-phase methodology on an ABI 433A peptide synthesizer using FastMoc chemistry and standard side chain protection. All cysteines were protected with S-trityl groups. The peptides were removed from a solid support by treatment with reagent K (trifluoroacetic acid/water/ethanedithiol/phenol/thioanisole, 82.5:5:2.5:5:5, v/v). The released peptide was precipitated and washed several times with cold ether. The reduced peptides were purified by reverse-phase HPLC using a preparative C18 Vydac column with a linear gradient of acetonitrile in 0.1% (v/v) trifluoroacetic acid. The flow rate was 10 ml/min, and elution was monitored by UV detection at 215 nm. Identity of each peptide was confirmed by ESI-MS analysis. After purification, the linear peptide was lyophilized. Folding was carried out in a buffered solution of 0.1 m NH4HCO3, pH 7.5, containing 1 mm EDTA. The final peptide concentration was 20 μm. The reaction was allowed to proceed at room temperature (25 °C) for 72 h. The reaction mixture was then quenched by acidification with formic acid (8% final concentration), and the samples were separated by analytical reverse-phase C18 HPLC, using the following gradient of acetonitrile at a flow rate of 1 ml/min: 0–50 min 0–50% solvent B, 50–55 min 50–100% solvent B. Solvent B is 0.05% trifluoroacetic acid in 90% acetonitrile.
The analogs LtIA(A4S/A6P) and LtIA(A4S) were manually synthesized by t-butoxycarbonyl chemistry coupled with in situ neutralization protocol on 4-methylbenzhydrylamine amide resin (Peptide Institute). 0.5 m 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate in N,N-dimethylformamide was the activation agent; diisopropylethylamine was the neutralization agent, and the terminal amide protection groups were removed using trifluoroacetic acid. The peptides were cleaved off the resin using HF with p-cresol and p-thiocresol as scavengers (HF/p-cresol/p-thiocresol, 10:1:1, v/v) at 0 °C for 1 h. The peptides were then precipitated with cold diethyl ether, dissolved in 50% acetonitrile containing 0.05% trifluoroacetic acid, and lyophilized. The mass of the crude peptides was verified by ESI-MS on Applied Biosystems API 2000 liquid chromatography/mass spectrometry. The crude peptides were purified by preparative reverse-phase HPLC using a Vydac C18 column (22 × 250 mm, 10 μm) on a Waters 600E solvent delivery system using a linear gradient of 1% buffer B per min at a flow rate of 10 ml/min (buffer A, H2O, 0.05% trifluoroacetic acid; buffer B, 90% acetonitrile, 10% H2O, 0.043% trifluoroacetic acid), and the eluant was monitored at 230 nm.
Disulfide bonds were formed by air oxidation of free thiol groups on cysteine residues in 0.1 m NH4HCO3 buffer with 30% isopropyl alcohol. The reaction was carried out overnight at room temperature, and the peptide concentration was 0.1 mg/ml. The molecular weight of the folded peptide was verified by ESI-MS, which was then purified and lyophilized. The purity of the product obtained after each purification step was determined by analytical reverse-phase HPLC using Shimadzu LC-2010 system using a Vydac C18 column (4.6 × 250 mm, 5 μm) at the flow rate of 1 ml/min.
Peptide Synthesis by Two-step Oxidation
α-Conotoxin LtIA was synthesized on an amide resin using Fmoc chemistry and standard side protection, except for cysteine residues. Cys residues were protected in pairs with either S-trityl on Cys-2 and Cys-8 or S-acetamidomethyl on Cys-3 and Cys-16. The crude peptide was cleaved from the resin and precipitated. A two-step oxidation protocol was used to fold the peptides selectively as described previously (19). Briefly, the disulfide bridge between Cys-2 and Cys-8 was closed by dripping the peptide into an equal volume of 20 mm potassium ferricyanide, 0.1 m Tris, pH 7.5. The solution was allowed to react for 30 min, and the monocyclic peptide was purified by reverse-phase HPLC. Simultaneous removal of the S-acetamidomethyl groups and closure of the disulfide bridge between Cys-3 and Cys-16 were carried out by iodine oxidation as follow; the monocyclic peptide in HPLC eluent was dripped into an equal volume of iodine (10 mm) in H2O/trifluoroacetic acid/acetonitrile (78:2:20 by volume) and allowed to react for 10 min. The reaction was terminated by the addition of ascorbic acid and diluted 20-fold with 0.1% trifluoroacetic acid, and the bicyclic peptide was purified by HPLC on a reverse-phase C18 Vydac column using a linear gradient of 0.1% and 0.092% trifluoroacetic acid, 60% acetonitrile, and the remainder H2O. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry was utilized to confirm the identity of the products.
cRNA Preparation and Injection
Capped cRNA for the various subunits were made using the mMessage mMachine in vitro transcription kit (Ambion, Austin, TX) following linearization of the plasmid. The cRNA was purified using the Qiagen RNeasy kit (Qiagen, Valencia, CA). The concentration of cRNA was determined by absorbance at 260 nm. cRNA of chimera α6/α3 was combined with cRNA of high expressing β2 and β3 subunits or β4 (in the pGEMHE vector) to give 167–500 ng/μl of each subunit cRNA. Fifty nl of this mixture was injected into each Xenopus oocyte with a Drummond microdispenser (Drummond Scientific, Broomall, PA), as described previously, and incubated at 17 °C. Oocytes were injected within 1 day of harvesting, and recordings were made 2–4 days post-injection.
Voltage Clamp Recording
Oocytes were voltage-clamped and exposed to ACh and peptide as described previously (20). Briefly, the oocyte chamber consisting of a cylindrical well (∼30 μl in volume) was gravity-perfused at a rate of ∼2 ml/min with ND-96 buffer (96.0 mm NaCl, 2.0 mm KCl, 1.8 mm CaCl2, 1.0 mm MgCl2, 5 mm HEPES, pH 7.1–7.5) containing 1 μm atropine and 0.1 mg/ml bovine serum albumin. In the case of the α9α10 and mouse muscle α1β1δϵ subtypes, the ND-96 contained no atropine when recording. The oocyte was subjected once a minute to a 1-s pulse of 100 μm ACh. In the case of the α9α10 and mouse muscle α1β1δϵ subtypes, there is a 1-s pulse of 10 μm ACh. 200 μm ACh pulse is for the α7 subtype. For screening of receptor subtypes, for toxin concentrations of 10 μm and lower, once a stable base line was achieved, either ND-96 alone or ND-96 containing varying concentrations of the α-conotoxins was manually pre-applied for 5 min prior to the addition of the agonist. All recordings were done at room temperature (∼22 °C).
Data Analysis
The average of five control responses just preceding a test response was used to normalize the test response to obtain “% response.” Each data point of a dose-response curve represents the average ± S.E. of at least four oocytes. The dose-response data were fit to the equation, % response = 100/(1 + ([toxin]/IC50)⋀nH), where nH is the Hill coefficient, by nonlinear regression analysis using GraphPad Prism (GraphPad Software, San Diego).
Circular Dichroism Analysis
CD spectra were acquired on a Jasco J-810 spectropolarimeter, which was routinely calibrated using 0.6% (w/v) ammonium-d-camphor-10-sulfonate. All experiments were conducted at room temperature under a nitrogen atmosphere (15 ml/min). The scanning speed was set to 50 nm/min with response time of 1 s, sensitivity range of 100 millidegrees, and a step resolution of 1 nm. Absorbance was measured in the far-UV region (190–260 nm) using a cell with a 1-mm path length, and each recording was an accumulation of four scans. CD spectra were obtained for peptides dissolved in 20 mm ammonium bicarbonate buffer at pH 7. Spectra were also obtained after addition of 10 and 30% (v/v) trifluoroethanol in phosphate buffer. CD spectra of the pure solvents were subtracted from those of the peptide samples to eliminate interference due to solvent, cell, or spectropolarimeter optics. The molar ellipticity [θ] was calculated using Equation 1,
where θ is ellipticity in millidegrees; C is molar concentration of the peptide; Np is number of peptide units (Np = 15 for LtIA), and l is the path length of the cell.
NMR Spectroscopy
1H NMR measurements were recorded on a 1 mm solution of the globular isomer of LtIA in 10% D2O, 90% H2O at ∼pH 3. Spectra were obtained on a Bruker Avance 600-MHz spectrometer at 280, 290, 300, and 310 K. The spectra for LtIA(A4S) and LtIA(A4S/A6P) were recorded at 308 and 290 K, respectively. The two-dimensional NMR experiments consisted of total correlation spectroscopy using an MLEV17 spin lock sequence with an isotropic mixing time of 80 ms and nuclear Overhauser effect spectroscopy (NOESY) experiments with mixing times of 200, 300, and 350 ms. Spectra were recorded in a phase-sensitive mode using time proportional phase incriminations. A modified WATERGATE sequence was used for suppression of the water signal. All spectra obtained were processed using Topspin (Bruker). Resonance assignment was carried out using the program SPARKY.
Molecular Modeling and Docking
Molecular models of rat α3 and β2 subunits were built individually using α1-nicotinic acetylcholine receptor (α1-nAChR; PDB code 2QC1) (21) as the template, and the interface was created using Lymnaea stagnalis acetylcholine-binding protein (Ls-AChBP; PDB code 1IBG) (22) as the template, with the program Modeler 9, version 2 (23). These templates were chosen as they share a high degree of conservation in protein sequence identities with rat α3 and β2 subunits as determined by BLAST (24).
The native LtIA and its analogs LtIA(A4S) and LtIA(A4S/A6P) vary in their α-helical tendencies in solution as suggested by both NMR and CD analyses. Although the bound state of the peptide is unknown, as an initial approach, we built the LtIA, LtIA(A4S) and LtIA(A4S/A6P) models using the TxIA (A10L) template extracted from the Aplysia californica AChBP-TxIA(A10L) co-crystal structure (PDB code 2UZ6) (25) All sequence alignments were generated using ClustalW (26) and were further corrected by hand based on the secondary structure alignment to maximize the accuracy of sequence alignment and protein sequence identity. The structural models were validated using the on-line server Verify3D (27) and Ramachandran plot available from ProFunc (28) data base. LtIA, LtIA(A4S), and LtIA(A4S/A6P) and MII (PDB code 1MII) (29, 30) were docked to the α3β2 nAChR homology model and Ls-AChBP crystal structure using the program HEX 5.0 (31), and the solutions that disagreed with the known interactions were excluded. The optimal hydrogen bonds between ligands and α3β2 nAChR that are likely to contribute to the complex stability were predicted using the on-line server WHAT IF (32).
RESULTS
Chemical Synthesis and Oxidative Folding
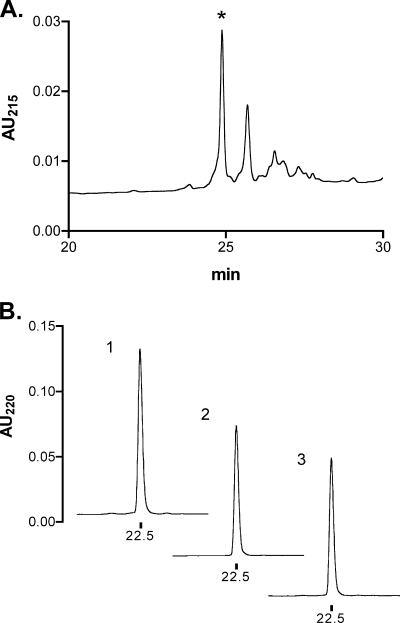
α-CTX LtIA is a 16-amino acid C-terminally amidated peptide with four Cys residues (Table 1) (17). Fmoc chemistry was used to synthesize the reduced peptide. In the initial synthesis, Cys side chains were protected with the acid-labile S-trityl group. Cleavage of the peptide from the resin simultaneously removed the S-trityl groups. Subsequently, Cys residues were air-oxidized in 1 m NH4HCO3, pH 7.5 buffer, containing 1 mm EDTA and purified by HPLC (Fig. 1A).
FIGURE 1.
HPLC analysis of the oxidative folding of α-CTX LtIA and comparison of the one-step and two-step oxidation methods. A, linear peptide was air-oxidized in one step and purified by analytical HPLC. The asterisk denotes the major product that was subjected to co-elution with the two-step oxidation product. B, 1, HPLCs of 1 nmol of material from A; 2, ∼1 nmol of peptide obtained from the two-step directed folding of the peptide; and 3, co-elution of 0.5 nmol each of 1 and 2. Peptides in B were analyzed on a reverse-phase analytical Vydac C18 HPLC using a linear gradient of 90% buffer A to 50% buffer B over 40 min, where A = 0.1% trifluoroacetic acid and B = 0.92%, 60% acetonitrile, remainder water. Absorbance was monitored at 220 nm. AU, absorbance units.
With four Cys residues, there are three possible disulfide bond connectivities. Previously characterized α-conotoxins purified from venom typically have a disulfide bond connectivity linking the 1st Cys to the 3rd Cys and the 2nd Cys to the 4th Cys, which is referred to as the “globular” form to distinguish it from the alternative “ribbon” and “beads” connectivities (33). We therefore also synthesized α-CTX LtIA using a directed two-step folding. Cys pairs as found in native purified peptides were orthogonally protected using acid-labile S-trityl and acid-stable S-acetamidomethyl groups. Cleavage from the resin selectively removed the S-trityl groups, and the deprotected Cys residues were oxidized with potassium ferricyanide. After HPLC purification of the monocyclic peptide, acetamidomethyl groups were removed from the second Cys pair by treatment with iodine that also oxidized the peptide to form the second disulfide bond. The identity of the bicyclic peptide was confirmed by mass spectrometry, average calculated as follows: 1600.66 Da; observed 1600.4 Da. A co-elution experiment was performed with an ∼1:1 ratio of the major product of the one-step air oxidation and the major product of the two-step directed oxidation. As shown in Fig. 1B, the two products have identical elution profiles. Thus, the major product of the equilibrium air oxidation folding of the peptide has the disulfide bond configuration of native α-conotoxins. Synthetic peptide with this disulfide bond arrangement was used for all further analysis.
LtIA lacks the conserved Ser-4 and Pro-6 of other α-conotoxins (see Table 1). Accordingly, as part of our study of native LtIA, we also synthesized and characterized one analog in which LtIA Ala-4 was replaced by Ser and another analog in which Ala-4 and Ala-6 of LtIA were replaced by Ser and Pro, respectively. t-Butoxycarbonyl chemistry/in situ neutralization protocol (34) was used to synthesize the LtIA analogs. Syntheses were carried out on a 0.5 mmol scale with an overall coupling efficiency of >99.7%, as determined by quantitative ninhydrin tests. During assembly, the Cys side chains were protected with HF-labile methylbenzyl groups, which were removed upon cleavage. The reduced peptides were folded in 0.1 m NH4HCO3 buffer with 30% isopropyl alcohol, overnight at room temperature. For each analog, ESI-MS analysis showed a decrease in molecular mass by 4 Da, confirming the formation of two disulfide bonds.
Effect of α-CTX LtIA on ACh-evoked Currents through nAChRs
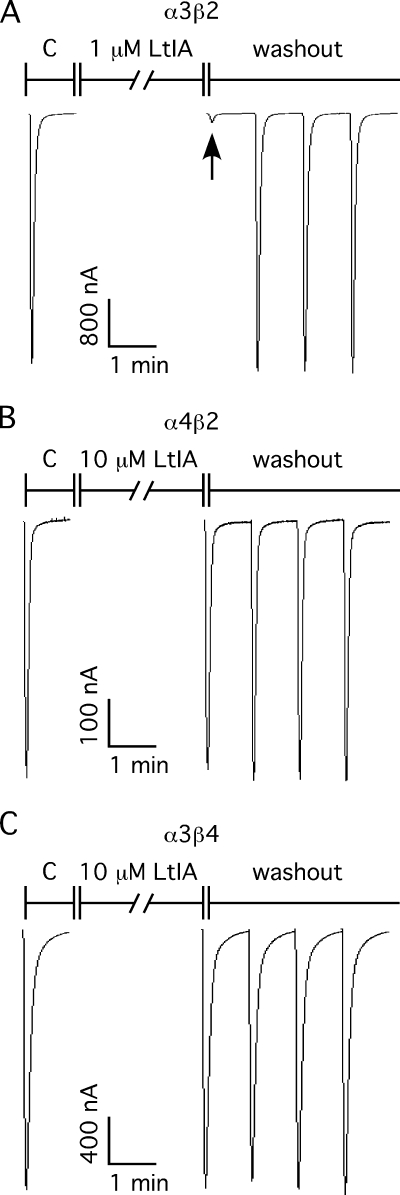
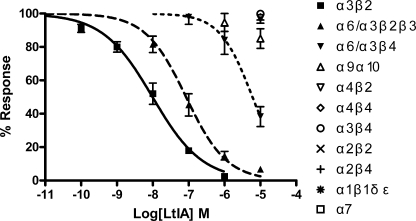
α-Conotoxins have been widely used as pharmacological tools to characterize nAChRs. The effects of α-CTX LtIA on different nAChRs heterologously expressed in Xenopus oocytes were analyzed. Fig. 2 shows representative responses to ACh of α3β2, α4β2, and α3β4 nAChRs in the presence and absence of α-CTX LtIA. Complete block of ACh-evoked currents was obtained with 1 μm α-CTX LtIA on α3β2 nAChRs (Fig. 2A) compared with little or no block of α4β2 (Fig. 2B) and α3β4 (Fig. 2C) nAChRs by 10 μm LtIA. The block of α3β2 nAChRs by LtIA is rapidly reversible (Fig. 2A). Concentration-response curves for α-CTX LtIA are shown in Fig. 3. α3β2 nAChRs were most potently blocked by α-CTX LtIA with an IC50 of 9.8 nm (Table 2). When a β4 rather than β2 nAChR subunit was co-expressed with the α3 subunit, the IC50 for α-CTX LtIA was >1000-fold higher, thus indicating that amino acid residue differences between the homologous β subunits significantly influence toxin potency. Likewise, LtIA was substantially less potent on α6/α3β4 versus α6/α3β2β3 nAChRs. In contrast, there was little or no block by LtIA at 10 μm LtIA on other nAChR subtypes, including α1β1δϵ, α2β2, α2β4, α4β2, α4β4, α7, and α9α10, (Table 2).
FIGURE 2.
α-CTX LtIA differentially blocks α3β2 (A), α4β2 (B), and α3β4 (C) nAChRs. Oocytes expressing α3β2 nAChRs (A), α4β2 nAChRs (B), and α3β4 nAChRs (C) were voltage-clamped at −70 mV and subjected to a 1-s pulse of 100 μm ACh every minute as described under “Experimental Procedures.” In each panel, the C response is control, following which the oocyte was exposed to the peptide for 5 min as indicated under “Experimental Procedures.” The 1st trace by the ACh pulse is the peptide response on corresponding nAChRs subtype. A, arrow denotes the current response trace of 1 μm LtIA on α3β2 nAChRs. B and C, 1st trace after the control is the current response of 10 μm LtIA on 4β2 and α3β4 nAChRs, respectively. The ND-96 perfusion and ACh pulses were then resumed to monitor the recovery from block during washout of peptide. The peptide was potent in blocking α3β2 nAChR and no block on α4β2 and α3β4 nAChRs. Likewise, the block of α3β2 nAChRs was quickly reversible.
FIGURE 3.
α-CTX LtIA concentration-response on native nAChR subtypes. Values are means ± S.E. from 5 to 7 separate oocytes. Oocytes expressing 1 of 11 different nAChRs were voltage-clamped and subjected to ACh pulses as described in Fig. 2 and under “Experimental Procedures.”
TABLE 2.
IC50 and Hill slope values for block of various rat nAChR subtypes by α-CTX LtIA
| Subtypes | IC50a | Ratiob | Hill slopea | Subtypes | IC50c |
|---|---|---|---|---|---|
| nm | nm | ||||
| α3β2 | 9.79 (7.3–13.2) | 1 | 0.63 (0.52–0.73) | α4β4 | >10,000 |
| α6/α3β2β3 | 84.4 (58–123) | 8.6 | 0.76 (0.57–0.96) | α3β4 | >10,000 |
| α6/α3β4 | 5990 (3400–10,500) | 612 | 0.95 (0.43–1.48) | α2β2 | >10,000 |
| α9α10 | >10,000 | α2β4 | >10,000 | ||
| α4β2 | >10,000 | α1β1δϵ | >10,000 | ||
| α7 | >10,000 |
a Numbers in parentheses are 95% confidence intervals.
b nAChR subtype IC50/α3β2 IC50 is shown.
c No block at 10−5 m was seen.
Block of α3β2 nAChRs by α-CTX MII Is Prevented by α-CTX LtIA
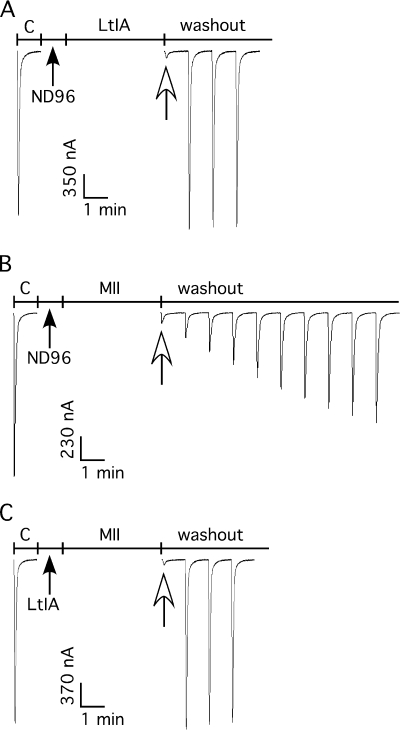
Where tested, the binding of previously isolated α4/7 conotoxins competitively prevents the binding of other nAChR agonists and antagonists. In contrast, the α4/3 conotoxin ImII is a noncompetitive antagonist. Blocking α3β2 nAChRs by α-LtIA is rapidly reversible, in contrast to the relatively slow reversibility of α-CTX MII (Fig. 4, A and B) (35). We utilized this difference in koff values to assess whether LtIA could prevent the binding of the competitive antagonist α-CTX MII. Pre-application of 10 μm LtIA for 1 min prior to application of MII led to a block of α3β2 nAChRs that was rapidly reversed upon toxin washout, comparable with the reversible block by LtIA alone. Thus, LtIA is able to prevent the slowly reversible block by MII indicating that LtIA and MII may have overlapping binding sites.
FIGURE 4.
Pre-block of α3β2 nAChRs with α-CTX LtIA prevented the slowly reversible block associated with α-CTX MII. In each instance, toxin or control solution of ND-96 was applied to α3β2 nAChRs for 1 min followed by a 4-min exposure to toxin or ND-96. A, 10 μm α-CTX LtIA was followed by ND-96. B, ND-96 was followed by 50 nm α-CTX MII. C, 10 μm α-CTX LtIA was followed by 50 nm α-CTX MII.
Mutations in the Ligand-binding Site of the β Subunit Affect Blocking by LtIA
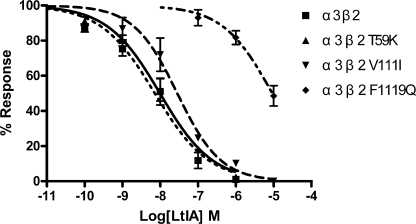
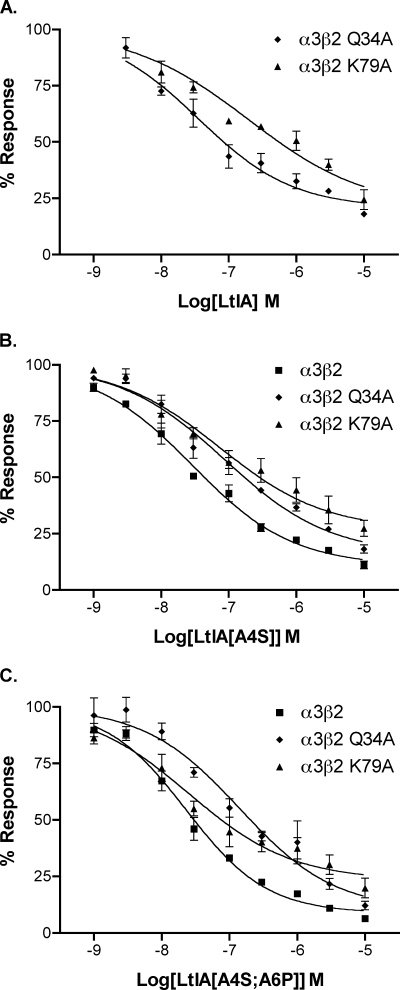
LtIA is 3 orders of magnitude more potent on α3β2 versus α3β4 nAChRs. Mutations within the nAChR β2 subunit ligand-binding interface were used to determine toxin potency differences for α3β2 versus α3β4 nAChRs. Point mutants were constructed in which β4 nAChR residues were substituted for β2 subunit residues that toxin-receptor docking studies have suggested are positioned to interact with α-conotoxins. nAChR β2 mutants T59K, V111I, and F119Q were used to examine the concentration response of α-CTX LtIA (Fig. 5 and Table 3). The toxin had little activity when tested on α3β2 F119Q. In contrast, LtIA potently blocked ACh-evoked currents of α3β2 T59K and V111I nAChRs. The ratios of mutant IC50/α3β2 IC50 were 0.7, 2.9, and 938 for α3β2 T59K, V111I, and F119Q, respectively. Thus, activity was modestly changed by Ile for Val substitution at β2 nAChR subunit position 111 and changed by >900-fold by Gln substitution for Phe at β2 nAChR subunit position 119.
FIGURE 5.
α-CTX LtIA concentration-response on wild type α3β2 nAChRs and their mutant receptors. All three mutant receptors exhibited similar sensitivity to acetylcholine relative to their wild type α3β2 counterpart. Values are means ± S.E. from 5 to 7 separate oocytes.
TABLE 3.
IC50 and Hill slope values for block of rat α3β2 nAChRs mutants by α-CTX LtIA
| Subtypes | IC50a | Ratiob | Hill slope |
|---|---|---|---|
| nm | |||
| α3β2 | 9.79 (7.3–13.2) | 1 | 0.63 (0.52–0.73) |
| α3β2(T59K) | 7.2 (4.8–10.7) | 0.7 | 0.61 (0.47–0.75) |
| α3β2(V111I) | 28.2 (19–42) | 2.9 | 0.76 (0.55–0.96) |
| α3β2(F119Q) | 9190 (5500–15,500) | 939 | 0.67 (0.41–0.92) |
a Numbers in parentheses are 95% confidence intervals.
b Ratio of nAChR subtype IC50/α3β2 IC50.
Circular Dichroism Analysis
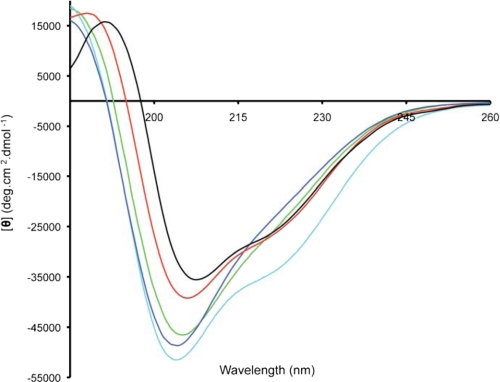
Globular LtIA was analyzed by circular dichroism spectroscopy to obtain information on any secondary structural elements present. The peptide was dissolved in 20 mm sodium phosphate buffer, pH 7, and CD spectra were obtained at two different peptide concentrations. Spectra obtained at 52 and 30 μm LtIA overlaid well suggesting that there was no concentration-dependent aggregation. The spectra of all three LtIA peptides, as well as α-conotoxins MII and Vc1.1, are shown in Fig. 6. All peptides show local minima at ∼205 nm and a positive band at ∼187 nm, which are characteristic of α-helical structure. Furthermore, minima at 220 nm are more pronounced for LtIA(A4S/A6P), MII, and Vc1.1 consistent with more regular helical structures in these Ser-Xaa-Pro-containing α-conotoxins.
FIGURE 6.
CD spectra of LtIA globular isomer (green), (A4S/A6P) (cyan), LtIA(A4S) (blue), MII (red), and Vc1. 1 (black). Spectra were recorded with concentrations of 30–50 μm.
NMR Studies
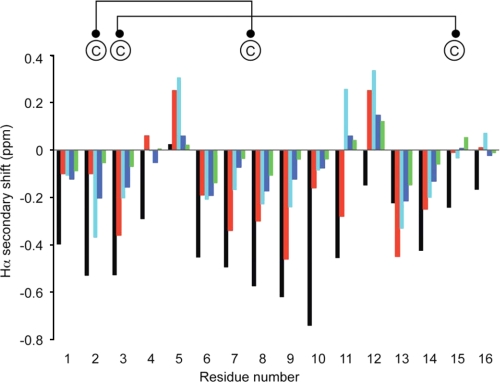
NMR spectroscopy was employed to further analyze the structure of LtIA. A series of NH-NHi + 1, Hα-NHi + 1, and Hβ-NHi + 1 connectivities obtained from the NOESY spectrum was used in conjunction with cross-peaks in the total correlation spectroscopy spectrum to obtain sequence-specific assignments. Differences between the observed Hα chemical shifts and random coil shifts, referred to as secondary shifts, which are generally characteristic of secondary structure elements (36), were calculated and are shown in Fig. 7. A series of small negative secondary shifts for residues 6–9 suggest that LtIA has a propensity for helical structure but is not as well structured as Vc1.1 or MII (29, 37), consistent with the findings from the CD spectral data. NMR spectra for LtIA at 280 K were consistent with those observed at higher temperatures. The double mutant of LtIA containing the Ser-Xaa-Pro motif has secondary shifts for residues 6–9 that are more negative than the wild type LtIA, indicating that the helical structure is stabilized in the central portion of this mutant (Fig. 7). Interestingly, LtIA(A4S) appears to have a lower helical propensity than the wild type peptide, indicating that the proline is important for stabilization of the helix.
FIGURE 7.
Secondary Hα NMR chemical shifts for LtIA (green bars), LtIA(A4S) (blue), LtIA(A4S/A6P) (cyan bars), Vc1.1 (black bars), and MII (red bars). Secondary shifts were calculated by subtracting the random coil chemical shifts (3) from the measured Hα chemical shifts. The disulfide connectivity is shown on the top of the diagram. Data for Vc1.1 and MII are from Refs. 29 and 37.
The apparently less well defined structure of LtIA relative to most α-conotoxins may explain the relative lack of nuclear Overhauser effects in the NOESY spectrum. Given this lack of nuclear Overhauser effects, no attempt was made to formally calculate a three-dimensional structure using restrained molecular dynamics as has been done for other conotoxins. Thus, the functional and structural studies show that LtIA binds to the α3β2 nAChR subtype despite possessing a less defined two-turn helical motif. This is an interesting observation as the helical region has been implicated as important for the activity of other α-conotoxins such as MII, GIC, PIA, and BuIA, which also bind to this receptor subtype (38).
Molecular Modeling of α3β2 nAChR and Docking Simulation of α-CTX LtIA and MII
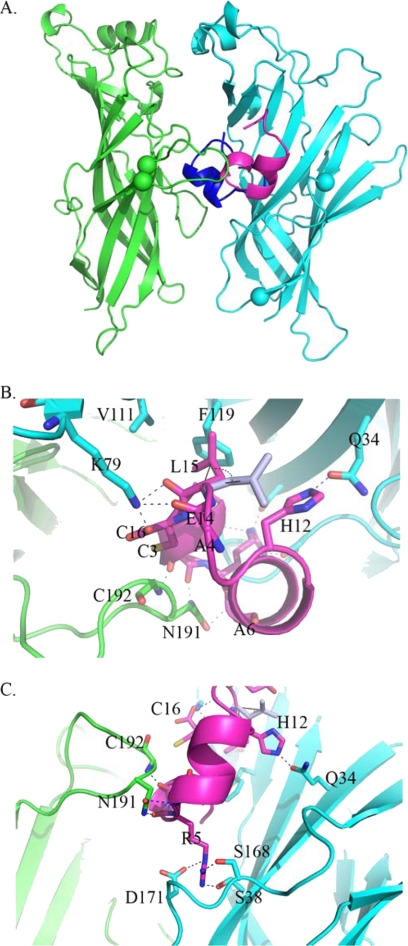
Homology modeling was used to study structure-function relationships of LtIA in the absence of experimental three-dimensional structures. We generated structural models for the extracellular domain of the α3β2 nAChR using the co-crystal structures of α1 nAChR-α-bungarotoxin (21) and of α-CTX LtIA and analogs using AChBP-TxIA(A10L) (25) as templates. The docking simulation of α-CTX LtIA with the α3β2 nAChR model predicted a novel binding mode (Fig. 8), with only the N terminus of LtIA found to overlap with the C terminus of α-CTX TxIA(A10L) (25), ImI (39), or PnIA(A10L/D14K) (38) in AChBP crystal structures. With respect to the TxIA(A10L) complex, α-CTX LtIA adopts an orientation that is rotated 180° around N and C termini along the y axis, with its α-helical backbone facing out the binding pocket of the α3β2 nAChR. The interface of α-CTX LtIA and the β2 subunit is characterized by two hydrogen bonds between β2-Gln-34 and LtIA-His-12, β2-Lys-79, LtIA-Glu-14, Leu-15, and/or Cys-16, and a strong hydrophobic interaction between LtIA-Leu-15 and β2-Phe-119 (Fig. 8B and Table 4). This hydrophobic interaction induced a significant re-orientation of the LtIA-Leu-15 side chain upon energy minimization of the docked structure, highlighting the importance of β2-Phe-119 in coordinating LtIA and further implicating its role in stabilizing the conformation of α3β2-LtIA complex. The unique orientation of α-CTX LtIA is further sustained by Arg-5, which is likely to form hydrogen bonds with Ser-38, Ser-168, and Asp-171 from the β2 F-loop (Fig. 8C and Table 4), an interaction that is not seen in any of the previously described α-CTX complexes. This unusual anchor interaction of LtIA-Arg-5 with the β2 F-loop appears to prevent LtIA from binding deeper into the binding pocket. This shallow binding mode of LtIA supports its fast koff value.
FIGURE 8.
Homology modeling and docking simulations for α3β2 (green/cyan), LtIA (magenta), and MII (blue). The interacting residues are presented as sticks, and the potential hydrogen bonds are indicated by blue dashed lines. C-loop and F-loop are located between the two green and two cyan spheres, respectively. The superposition of LtIA and MII (A) reveals a novel binding mode for LtIA that partially overlaps with that of MII. A noteworthy finding (B) in the LtIA binding is that LtIA-Leu-15 side chain swings to form an energetically favorable hydrophobic interaction with β2-Phe-119 after energy minimization. C, most prominent interaction of LtIA binding is formed between LtIA-Arg-5 and the β2 F-loop, which acts as an anchor, preventing it from binding deeper into the active site as seen in other α-CTX complexes.
TABLE 4.
Potential hydrogen bonds and hydrophobic interactions between LtIA and α3β2
| α3β2 nAChR | LtIA | Distance |
|---|---|---|
| Å | ||
| Potential hydrogen bonds | ||
| β2_K79_NZ | Glu-14_O | 2.8a |
| β2_K79_NZ | Leu-15_O | 2.5a |
| β2_K79_NZ | Cys-16_OXT | 1.6a |
| β2_D171_OD2 | Arg-5_NE | 2.8 |
| β2_S168_OG | Arg-5_NH1 | 1.8 |
| β2_S38_OG | Arg-5_NH2 | 1.9 |
| β2_Q34_OE1 | His-12_NE2 | 2.0 |
| α3_C192_N | Cys-3_O | 1.9 |
| α3_N191_ND2 | Ala-4_O | 1.8 |
| α3_N191_OD1 | Ala-6_N | 2.7 |
| Hydrophobic interactions | ||
| β2_V111 | Leu-15 | ∼4.5 |
| β2_F119 | Leu-15 | ∼3 |
a The possible H bonds (the closer interaction most likely dominates) are shown.
Mutation of the β2 Subunit and LtIA
The LtIA/receptor modeling analysis (assuming a helical structure for bound toxin) predicted that LtIA interacts with nAChR subunit β2-Gln-34 and Lys-79. To experimentally examine these predictions, the β2 subunit was mutated to express α3β2,Q34A and α3β2,K79A in oocytes for assessment of the functional block by LtIA. The β2 subunit Gln to Ala mutation reduced LtIA potency by ∼3.5-fold, whereas the Lys to Ala mutation reduced potency by 20-fold, supporting the predictions of the model (see Fig. 8 and Table 5). We next created toxin mutations to examine the effect the Ala-Arg-Ala sequence of LtIA that replaces the highly conserved Ser-Xaa-Pro motif of other α-conotoxins LtIA, A4S, and LtIA(A4S/A6P). The mutant peptides were then tested on wild type and α3β2, Q34A, and α3β2, K79A mutant receptors Results are shown in Fig. 9, B and C, and Table 5. In contrast to the native LtIA, the double mutant LtIA, which contains the highly conserved Ser-Xaa-Pro of other conotoxins, does not appear to interact with β2-Lys-79.
TABLE 5.
IC50 values of native and mutant LtIA on native and mutant nAChRs
Numbers are IC50 values in nanomolar. Numbers in parentheses indicate 95% confidence intervals.
| Peptide | α3β2 | α3β2, Q34A | α3β2, K79A |
|---|---|---|---|
| LtIA | 9.79 (7.3–13.2) | 35.7 (19.7–64.4) | 195 (121–315) |
| LtIA(A4S) | 29.1 (20.7–40.8) | 87.3 (48.1–158) | 73.4 (26.7–202) |
| LtIA(A4S/A6P) | 23.1 (17.3–30.9) | 142 (57.0–352) | 25.4 (11.8–54.8) |
FIGURE 9.
Concentration response of LtIA and LtIA analogs. Each α-conotoxin was tested on nAChRs expressed in Xenopus oocytes including α3β2 (see Fig. 5 for LtIA), α3β2 with β2 Gln-34 mutated to Ala, and α3β2 with β2 Lys-79 mutated to Ala. A, LtIA; B, LtIA with Ala-4 mutated to Ser; C, LtIA with Ala-4 mutated to Ser and Ala-6 mutated to Pro. Results are summarized in Table 5. Values are mean ± S.E. from 3 to 4 separate oocytes.
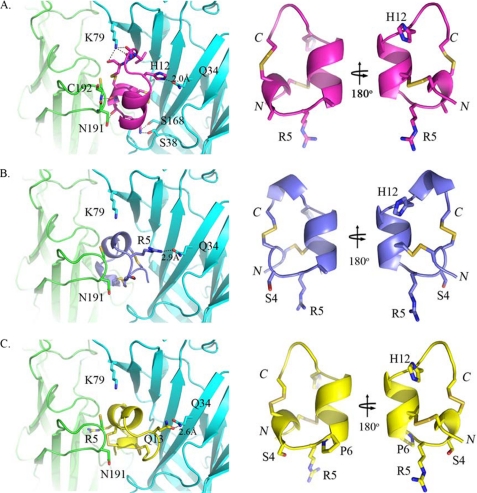
Interestingly, the conserved Ser-Xaa-Pro motif is replaced with Ala-Arg-Ala in LtIA, where both Ala main chains form hydrogen bonds with Asn-191 from the α3 C-loop (Fig. 8C). The Ala substitutions seem to be essential for binding in the traditional orientation as Ser and Pro at positions 4 and 6, respectively, may contribute to a loss of hydrogen bonding and a resulting steric clash with α3-Asn-191. To confirm this hypothesis, we created homology models for LtIA(A4S) and LtIA(A4S/A6P) using TxIA(A10L) as the template and performed docking simulations to the α3β2 model (Fig 10). In contrast to the native LtIA, both analogs exhibited different binding orientations and lost the intermolecular interactions via Arg-5 and from β2-Lys-79, and yet they retained interactions with β2-Gln-34. This explains why the β2-K79A does not have significant impact to both analogs compared with that with β2-Q34A (Table 5). Furthermore, an additional main chain-main chain interaction between two highly conserved residues, LtIA-Cys-3 and α3-Cys-192, was also identified. A docking model of α-conotoxin MII bound to the α3β2 nAChR was also constructed. Comparison of MII and LtIA binding revealed that the C terminus of MII overlapped the N terminus of LtIA, suggesting LtIA would be a competitive inhibitor of MII binding consistent with our experimental findings (Fig. 4).
FIGURE 10.
Homology models (right panel) and docking simulations (left panel) for LtIA (magenta) and its analogs LtIA(A4S) (marine blue) and LtIA(A4S/A6P) (yellow) to α3β2. The homology models are shown in the identical orientation with the N and C termini labeled in italic. The docking complexes on the left panel are also presented in the same orientation with key interacting residues shown as sticks and the potential hydrogen bonds indicated as black dashed lines with distances (Å) labeled. The docking complexes of both LtIA analogs (B and C, left panel) showed loss of interactions via LtIA-Arg-5 and β2-Lys-79 and rearrangements of hydrogen bonds with β2-Gln-34 compared with that with LtIA (A, left panel). The superposition of all three peptides showed no significant alteration in the backbone (root mean square of <0.5 Å), and thus the differences in binding orientation might be due to the steric clash with α3-Asn-191 observed in modeling when A6 was mutated to Pro and a slight shift of α-helix (B; right panel) from the N to C termini when A4 was mutated to Ser alone.
DISCUSSION
This study describes the synthesis, pharmacological characterization, structure determination, and receptor interactions of the first α-conotoxin from C. litteratus. The peptide, α-CTX LtIA, is highly unusual in that it lacks the conserved Ser-Xaa-Pro motif found in the first Cys loop of all previously reported α4/7 conotoxins that target neuronal nAChRs (see Table 1). In LtIA, Ala residues replace these conserved Ser and Pro residues found in other conotoxins. Interestingly, substitution of Ala for Ser-4 in synthetic analogs of another potent α4/7 conotoxin, α-CTX MII (see Table 1), shifts selectivity to favor binding to α6/α3β2β3 versus α3β2 nAChRs (39, 40). In contrast, LtIA is more potent on α3β2 than α6/α3β2β3 nAChRs (Table 2). In addition, whereas substitution of Ala for Pro in MII dramatically reduces activity at α3β2 nAChRs by 2000-fold (40), α-CTX LtIA has high affinity for α3β2 nAChRs (450-fold higher than the MII (P6A) analog). Thus, the structural and functional studies on LtIA reported here provide a foundation for important mechanistic insights into the interactions of the α-conotoxin class of toxins with nAChRs.
Given the divergent results between α-CTX LtIA and α-CTX MII, we examined whether the two peptides might overlap in their respective binding sites. Both α-CTX LtIA and α-CTX MII potently block α3β2 nAChRs (Figs. 2 and 3); however, α-CTX LtIA differs from α-CTX MII in that the former has a relatively fast off-rate. We utilized this finding to perform a competition experiment to assess whether pre-block of the α3β2 nAChR with a high concentration of α-CTX LtIA could prevent block by subsequent addition of α-CTX MII. The outcome was assessed by measurement of the kinetics of recovery from toxin block. Pre-application of α-CTX LtIA followed by application of α-CTX MII resulted in block of the nAChR with rapid recovery upon toxin washout. This profile is similar to that seen with a block by α-CTX LtIA alone, although not to block by α-CTX MII alone, thus supporting the notion that the two peptides have overlapping binding sites (Fig. 4).
To more rigorously examine the nAChR-binding site, we exploited the different binding selectivity of α-CTX LtIA for α3β2 versus α3β4 nAChRs. α-CTX LtIA blocks α3β4 nAChRs with an IC50 value that is >1000 higher than that of α3β2 nAChRs. Although there is no NMR or crystal structure of the α3β2 nAChR, the structure of the molluscan AChBP serves as a surrogate (22). The AChBP is a homolog of the extracellular ligand-binding domain of the nAChR, and the AChBP crystal structure sheds significant light on the three-dimensional structure of the ACh-binding site. Furthermore, α-conotoxins have been co-crystallized with the AChBP (41, 42), providing insight into their binding modes. The highly conserved first Cys loop of the α-conotoxins contains a small α-helix important for AChBP and nAChR binding. Thus, the unusual first Cys loop of α-CTX LtIA is of significant interest. Molecular surface analysis and modeling of the α3β2 nAChR and docking of previously characterized α-conotoxins provide evidence of a small cleft above the nAChR β2 subunit β9β10 hairpin as a site for α-conotoxin binding (15). Residues that rim this cleft include Thr-59, Val-111, and Phe-119 (16). Mutation of the nAChR β2 subunit Thr-59 to Lys has a 4-fold effect on α-CTX-MII block (43) and substantially affects the kinetics of block by α-CTX BuIA (44). However, Ala substitution of Val-111 and Ala replacement of Phe-119 have little or no effect on the IC50 value of α-CTX MII (16). In contrast, although nAChR β2 subunit mutations T59K and V111I had no effect or a small effect (2.9-fold for V111I) on α-CTX LtIA binding, the F119Q mutation reduced LtIA block by over 900-fold (Fig. 4 and Table 3). Thus, although the mutation of the nAChR β2 subunit Phe-119 did not affect the IC50 value of block by MII, suggesting that MII does not have close interactions with Phe-119, changing Phe-119 to the Gln present in the homologous position of the nAChR β4 subunit dramatically affected block by α-CTX LtIA. These results are consistent with the 3 orders of magnitude higher IC50 value of α-CTX LtIA for α3β4 versus α3β2 nAChRs (Fig. 3 and Table 2).
Analysis of LtIA by CD and NMR spectroscopy suggested that it has a helical propensity characteristic of most other α-conotoxins, albeit less prominently than Ser-Xaa-Pro containing α-conotoxins. It seems likely that LtIA is relatively flexible in solution, a suggestion supported by the lack of classic helical nuclear Overhauser effects in the NOESY spectrum. Such flexibility is consistent with the absence of the conserved Ser and Pro residues that likely have a structure-defining role in other α-conotoxins. This suggestion is supported by the stabilization of the helical structure in the central portion of the peptide when the Ser and Pro residues are introduced (LtIA(A4S/A6P)). The helical propensity of LtIA suggests that it might be able to adopt a well defined helical conformation on receptor binding. Modeling assumed a helical conformation for the peptides and predicted interaction with nAChR β2 subunit residues Gln-34 and Lys-79. Mutation of these receptor residues decreased potency of LtIA consistent with the model. Peptide analogs of LtIA were also synthesized to introduce the Pro and/or Ser residues found in other α-conotoxins (Table 1). Interestingly, the doubly substituted LtIA(A4S/A6P) no longer showed a drop in potency at the α3β2-Lys-79 mutant (Table 5). Thus, although these findings do not prove LtIA adopts a helical conformation when bound, they are consistent with the atypical α-conotoxin LtIA having a distinct binding interaction at α3β2 nAChRs. Although conotoxins are generally regarded as rather rigid peptides, precedent exists for multiple conformations for the globular isomer of other conotoxins, as exemplified for the 4/4 α-conotoxin BuIA (45). Without the conserved Ser-Xaa-Pro motif, it appears that the 4/7 conotoxin framework becomes more flexible, thus providing new opportunities for tuning α-conotoxin selectivity.
Molecular modeling of the α3β2 nAChR and docking simulation also suggested that the binding site of LtIA partially overlaps with that of other 4/7 α-CTXs (Fig. 8A) consistent with experimental findings (Fig. 4). This novel binding mode may be driven by several unique interactions that appear to explain LtIA-β2 specificity. The most prominent is the anchor interaction between LtIA-Arg-5 and the β2 F-loop, which is believed to be the key determinant for this shallow binding mode (Fig. 8C). Sequence analysis shows no conservation in the F-loop, indicating that this interaction is exclusive to the β2 subunit. Another obvious feature is an energetically favorable hydrophobic interaction between LtIA-Leu-15 and β2-Phe-119, which is achieved by re-orientation of LtIA-Leu-15 side chain after energy minimization. This finding is in agreement with our site-directed mutagenesis data that show a 900-fold increase in the IC50 value of LtIA when the hydrophobic interaction is disrupted by β2-F119Q (Fig. 5). In contrast, Val-111 and Thr-59 are >4 and >5 Å distant from LtIA, respectively, suggesting a weak or no interaction with LtIA, again consistent with our mutagenesis data. In the case of MII, although it is located in a close proximity to Phe-119, Val-111, and Thr-59, only Van der Waals interactions are observed. Furthermore, Ala residues in LtIA that replace the conserved Ser-Xaa-Pro motif allow the accommodation of the α3-Asn-191 side chain and potentially allow the formation of two new hydrogen bonds (Fig. 8C). The distinctive binding modes of LtIA(A4S) and LtIA(A4S/A6P) to α3β2 (Fig 10) and the functional studies (Fig. 9 and Table 5) highlight the role of the Ser-Xaa-Pro motif in establishing the binding mode of previously studied α-conotoxins. Intermolecular complementarity between ligand and receptor are essential for ligand binding. As hydrophobic interactions in the active site of nAChR have been postulated as the dominant contributors to α-CTX binding (40, 46), we anticipate that the decrease in hydrophobicity of LtIA due to Ala substitutions may be another key factor for this novel binding mode. Other interactions identified in our docking simulations, especially the interactions distributed from the α3 C-loop, are believed to be involved only in orienting LtIA to the correct position for binding. As most of the ligand-receptor interactions to LtIA appear to arise from the β2 subunit, it is suggested that the ligand selectivity of LtIA for the α3β2 nAChR is determined by the β2 subunit.
In summary, α-CTX LtIA is a novel α-conotoxin that potently blocks α3β2 nAChRs. Structural analysis indicates that the lack of Ser and Pro in the first loop results in a flexible structure that still retains elements of helicity in the solution. Although the bound LtIA structure remains undefined, the profound effect on the block by Gln-34, Lys-79, and Phe-119 mutations in β2 reveals that LtIA binds in a mode that is distinctly different from previously characterized α-conotoxins.
Acknowledgments
We thank Layla Azam, Baldomero Olivera, Doju Yoshikami, and David Adams for advice and help.
This work was supported, in whole or in part, by State High Tech Research and Development Project (863) of the Ministry of Science and Technology of China Grant 2007AA02Z114, Major New Drug Discovery Project 2009ZX09103-644, International Key Project of Science and Technology Collaboration Grant 2005DFA30600, National Natural Science Foundation of China Grant 30860368, Cultivation Fund of the Key Scientific and Technical Innovation Project, and Ministry of Education of China Grant 705043. This work was also supported by National Institutes of Health Grants MH53631 and GM48677 (to J. M. M.), and Australian Research Council grants (to P. F. A., D. J. C., and N. L. D.). A preliminary account of part of this work was presented in the patent literature (Chinese Patents CN ZL200410103563.X (November 5, 2008) and CN 200810182970.2-A (December 12, 2008)).
- nAChR
- nicotinic acetylcholine receptor
- AChBP
- acetylcholine-binding protein
- CTX
- conotoxin
- HPLC
- high pressure liquid chromatography
- ACh
- acetylcholine
- PDB
- Protein Data Bank
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- ESI-MS
- electrospray ionization-mass spectrometry
- NOESY
- nuclear Overhauser effect spectroscopy.
REFERENCES
- 1.Lester H. A., Dibas M. I., Dahan D. S., Leite J. F., Dougherty D. A. (2004) Trends Neurosci. 27, 329–336 [DOI] [PubMed] [Google Scholar]
- 2.Wonnacott S., Barik J., Dickinson J., Jones I. W. (2006) J. Mol. Neurosci. 30, 137–140 [DOI] [PubMed] [Google Scholar]
- 3.Dani J. A., Bertrand D. (2007) Annu. Rev. Pharmacol. Toxicol. 47, 699–729 [DOI] [PubMed] [Google Scholar]
- 4.Gotti C., Zoli M., Clementi F. (2006) Trends Pharmacol. Sci. 27, 482–491 [DOI] [PubMed] [Google Scholar]
- 5.Gotti C., Riganti L., Vailati S., Clementi F. (2006) Curr. Pharm. Des. 12, 407–428 [DOI] [PubMed] [Google Scholar]
- 6.Olivera B. M., Quik M., Vincler M., McIntosh J. M. (2008) Channels 2, 143–152 [DOI] [PubMed] [Google Scholar]
- 7.Armishaw C. J., Alewood P. F. (2005) Curr. Protein Pept. Sci. 6, 221–240 [DOI] [PubMed] [Google Scholar]
- 8.Craik D. J., Adams D. J. (2007) ACS Chem. Biol. 2, 457–468 [DOI] [PubMed] [Google Scholar]
- 9.Zhangsun D., Luo S., Wu Y., Zhu X., Hu Y., Xie L. (2006) Chem. Biol. Drug Des. 68, 256–265 [DOI] [PubMed] [Google Scholar]
- 10.Luo S., Zhangsun D., Zhang B., Chen X., Feng J. (2006) Peptides 27, 2640–2646 [DOI] [PubMed] [Google Scholar]
- 11.Terlau H., Olivera B. M. (2004) Physiol. Rev. 84, 41–68 [DOI] [PubMed] [Google Scholar]
- 12.Olivera B. M., Teichert R. W. (2007) Mol. Interv. 7, 251–260 [DOI] [PubMed] [Google Scholar]
- 13.Dutertre S., Lewis R. J. (2004) Eur. J. Biochem. 271, 2327–2334 [DOI] [PubMed] [Google Scholar]
- 14.Ellison M., Olivera B. M. (2007) Chem. Rec. 7, 341–353 [DOI] [PubMed] [Google Scholar]
- 15.Dutertre S., Nicke A., Tyndall J. D., Lewis R. J. (2004) J. Mol. Recognit. 17, 339–347 [DOI] [PubMed] [Google Scholar]
- 16.Dutertre S., Nicke A., Lewis R. J. (2005) J. Biol. Chem. 280, 30460–30468 [DOI] [PubMed] [Google Scholar]
- 17.Luo S., Zhangsun D., Zhang B., Quan Y., Wu Y. (2006) J. Pept. Sci. 12, 693–704 [DOI] [PubMed] [Google Scholar]
- 18.Peng C., Han Y., Sanders T., Chew G., Liu J., Hawrot E., Chi C., Wang C. (2008) Peptides 29, 1700–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dowell C., Olivera B. M., Garrett J. E., Staheli S. T., Watkins M., Kuryatov A., Yoshikami D., Lindstrom J. M., McIntosh J. M. (2003) J. Neurosci. 23, 8445–8452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cartier G. E., Yoshikami D., Gray W. R., Luo S., Olivera B. M., McIntosh J. M. (1996) J. Biol. Chem. 271, 7522–7528 [DOI] [PubMed] [Google Scholar]
- 21.Dellisanti C. D., Yao Y., Stroud J. C., Wang Z. Z., Chen L. (2007) Nat. Neurosci. 10, 953–962 [DOI] [PubMed] [Google Scholar]
- 22.Brejc K., van Dijk W. J., Klaassen R. V., Schuurmans M., van Der Oost J., Smit A. B., Sixma T. K. (2001) Nature 411, 269–276 [DOI] [PubMed] [Google Scholar]
- 23.Fiser A., Sali A. (2003) Methods Enzymol. 374, 461–491 [DOI] [PubMed] [Google Scholar]
- 24.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 25.Dutertre S., Ulens C., Büttner R., Fish A., van Elk R., Kendel Y., Hopping G., Alewood P. F., Schroeder C., Nicke A., Smit A. B., Sixma T. K., Lewis R. J. (2007) EMBO J. 26, 3858–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 27.Bowie J. U., Lüthy R., Eisenberg D. (1991) Science 253, 164–170 [DOI] [PubMed] [Google Scholar]
- 28.Laskowski R. A., Watson J. D., Thornton J. M. (2005) Nucleic Acids Res. 33, W89–W93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hill J. M., Oomen C. J., Miranda L. P., Bingham J. P., Alewood P. F., Craik D. J. (1998) Biochemistry 37, 15621–15630 [DOI] [PubMed] [Google Scholar]
- 30.Bourne Y., Talley T. T., Hansen S. B., Taylor P., Marchot P. (2005) EMBO J. 24, 1512–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ritchie D. W., Kemp G. J. (2000) Proteins Struct. Funct. Genet. 39, 178–194 [PubMed] [Google Scholar]
- 32.Vriend G. (1990) J. Mol. Graph. 8, 52–56, 29 [DOI] [PubMed] [Google Scholar]
- 33.Gehrmann J., Alewood P. F., Craik D. J. (1998) J. Mol. Biol. 278, 401–415 [DOI] [PubMed] [Google Scholar]
- 34.Schnölzer M., Alewood P., Jones A., Alewood D., Kent S. B. (1992) Int. J. Pept. Protein Res. 40, 180–193 [DOI] [PubMed] [Google Scholar]
- 35.McIntosh J. M., Azam L., Staheli S., Dowell C., Lindstrom J. M., Kuryatov A., Garrett J. E., Marks M. J., Whiteaker P. (2004) Mol. Pharmacol. 65, 944–952 [DOI] [PubMed] [Google Scholar]
- 36.Sönnichsen F. D., Van Eyk J. E., Hodges R. S., Sykes B. D. (1992) Biochemistry 31, 8790–8798 [DOI] [PubMed] [Google Scholar]
- 37.Clark R. J., Fischer H., Nevin S. T., Adams D. J., Craik D. J. (2006) J. Biol. Chem. 281, 23254–23263 [DOI] [PubMed] [Google Scholar]
- 38.Wishart D. S., Sykes B. D., Richards F. M. (1992) Biochemistry 31, 1647–1651 [DOI] [PubMed] [Google Scholar]
- 39.Ulens C., Hogg R. C., Celie P. H., Bertrand D., Tsetlin V., Smit A. B., Sixma T. K. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3615–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Celie P. H., Kasheverov I. E., Mordvintsev D. Y., Hogg R. C., van Nierop P., van Elk R., van Rossum-Fikkert S. E., Zhmak M. N., Bertrand D., Tsetlin V., Sixma T. K., Smit A. B. (2005) Nat. Struct. Mol. Biol. 12, 582–588 [DOI] [PubMed] [Google Scholar]
- 41.Hansen S. B., Sulzenbacher G., Huxford T., Marchot P., Taylor P., Bourne Y. (2005) EMBO J. 24, 3635–3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Celie P. H., Klaassen R. V., van Rossum-Fikkert S. E., van Elk R., van Nierop P., Smit A. B., Sixma T. K. (2005) J. Biol. Chem. 280, 26457–26466 [DOI] [PubMed] [Google Scholar]
- 43.Azam L., Yoshikami D., McIntosh J. M. (2008) J. Biol. Chem. 283, 11625–11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shiembob D. L., Roberts R. L., Luetje C. W., McIntosh J. M. (2006) Biochemistry 45, 11200–11207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin A. H., Brandstaetter H., Nevin S. T., Tan C. C., Clark R. J., Adams D. J., Alewood P. F., Craik D. J., Daly N. L. (2007) BMC Struct. Biol. 7, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quiram P. A., McIntosh J. M., Sine S. M. (2000) J. Biol. Chem. 275, 4889–4896 [DOI] [PubMed] [Google Scholar]
- 47.McIntosh J. M., Dowell C., Watkins M., Garrett J. E., Yoshikami D., Olivera B. M. (2002) J. Biol. Chem. 277, 33610–33615 [DOI] [PubMed] [Google Scholar]
- 48.Vincler M., Wittenauer S., Parker R., Ellison M., Olivera B. M., McIntosh J. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17880–17884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sandall D. W., Satkunanathan N., Keays D. A., Polidano M. A., Liping X., Pham V., Down J. G., Khalil Z., Livett B. G., Gayler K. R. (2003) Biochemistry 42, 6904–6911 [DOI] [PubMed] [Google Scholar]
- 50.Chi S. W., Kim D. H., Olivera B. M., McIntosh J. M., Han K. H. (2006) Biochem. Biophys. Res. Commun. 345, 248–254 [DOI] [PubMed] [Google Scholar]
- 51.Luo S., Nguyen T. A., Cartier G. E., Olivera B. M., Yoshikami D., McIntosh J. M. (1999) Biochemistry 38, 14542–14548 [DOI] [PubMed] [Google Scholar]
- 52.Loughnan M. L., Nicke A., Jones A., Adams D. J., Alewood P. F., Lewis R. J. (2004) J. Med. Chem. 47, 1234–1241 [DOI] [PubMed] [Google Scholar]
- 53.Hogg R. C., Miranda L. P., Craik D. J., Lewis R. J., Alewood P. F., Adams D. J. (1999) J. Biol. Chem. 274, 36559–36564 [DOI] [PubMed] [Google Scholar]
- 54.McIntosh J. M., Plazas P. V., Watkins M., Gomez-Casati M. E., Olivera B. M., Elgoyhen A. B. (2005) J. Biol. Chem. 280, 30107–30112 [DOI] [PubMed] [Google Scholar]
- 55.Loughnan M., Bond T., Atkins A., Cuevas J., Adams D. J., Broxton N. M., Livett B. G., Down J. G., Jones A., Alewood P. F., Lewis R. J. (1998) J. Biol. Chem. 273, 15667–15674 [DOI] [PubMed] [Google Scholar]
- 56.Luo S., Kulak J. M., Cartier G. E., Jacobsen R. B., Yoshikami D., Olivera B. M., McIntosh J. M. (1998) J. Neurosci. 18, 8571–8579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.López-Vera E., Aguilar M. B., Schiavon E., Marinzi C., Ortiz E., Restano, Cassulini R., Batista C. V., Possani L. D., Heimer de la Cotera E. P., Peri F., Becerril B., Wanke E. (2007) FEBS J. 274, 3972–3985 [DOI] [PubMed] [Google Scholar]