Abstract
Two distinct adenosine deaminases, ADA1 and ADA2, are found in humans. ADA1 has an important role in lymphocyte function and inherited mutations in ADA1 result in severe combined immunodeficiency. The recently isolated ADA2 belongs to the novel family of adenosine deaminase growth factors (ADGFs), which play an important role in tissue development. The crystal structures of ADA2 and ADA2 bound to a transition state analogue presented here reveal the structural basis of the catalytic/signaling activity of ADGF/ADA2 proteins. In addition to the catalytic domain, the structures discovered two ADGF/ADA2-specific domains of novel folds that mediate the protein dimerization and binding to the cell surface receptors. This complex architecture is in sharp contrast with that of monomeric single domain ADA1. An extensive glycosylation and the presence of a conserved disulfide bond and a signal peptide in ADA2 strongly suggest that ADA2, in contrast to ADA1, is specifically designed to act in the extracellular environment. The comparison of catalytic sites of ADA2 and ADA1 demonstrates large differences in the arrangement of the substrate-binding pockets. These structural differences explain the substrate and inhibitor specificity of adenosine deaminases and provide the basis for a rational design of ADA2-targeting drugs to modulate the immune system responses in pathophysiological conditions.
Keywords: Diseases/Cancer/Leukemia, Enzymes/Catalysis, Hormones/Growth Factors, Nucleoside, Protein/Ligand Binding, Protein/Structure, Protein/Zinc, Adenosine Deaminase
Introduction
Adenosine is an important signaling molecule. The concentration of this metabolite in tissues and blood is normally low. However, its local concentration can increase significantly as a result of cell damage or stress conditions such as inflammation, ischemia, and hypoxia (1). Extracellular adenosine can bind and activate four adenosine receptors (ADRs),3 A1, A2A, A2B, and A3, which are expressed in different combinations on the surface of almost all cell types, and the receptor activation triggers multiple intracellular processes either leading to cell activation or resulting in suppression of cell function and cell death (1).
Adenosine deaminases (ADAs) decrease the local concentration of adenosine by catalyzing the deamination of adenosine and 2′-deoxyadenosine to inosine and deoxyinosine, respectively. Two types of enzymes with the adenosine deaminase activity are known, the ADA1 family proteins and adenosine deaminase growth factors (ADGF) or ADA2-like proteins (2). ADA1, ADGF/ADA2, and the recently discovered ADA-like (ADAL) or ADA3 proteins (the activity of which have not yet been characterized) display a limited sequence homology in the region of the catalytic domain, thus constituting three distantly related branches of an ADA superfamily (2). ADA1 proteins are found in both prokaryotes and eukaryotes, whereas ADGF/ADA2 are observed only in eukaryotes and primarily in multicellular organisms (2). All three types of ADA are present in humans, where they are known as ADA1, ADA2, and ADA3 (2). In flies, ADGF/ADA2 is the only active adenosine deaminative enzyme (3), whereas in rodents, there is only a gene for ADA1 (2), suggesting that these proteins have overlapping functions but that the presence of the two in a given organism is beneficial for its survival.
The function of ADA1 is mainly to reduce the intracellular level of adenosine, which is toxic for cells, and more specifically, for lymphocytes. The apoptotic death of lymphocytes triggered by adenosine and its derivatives is thought to be one of the main reasons why the absence of functional ADA1 results in severe combined immunodeficiency in humans (4). In contrast, temporary inhibition of ADA1 with the specific inhibitor deoxycoformycin decreases the cancer cell proliferation in patients with leukemia (5). The discovery of the central role of ADA1 in severe combined immunodeficiency and blood cancers has stimulated intense structural studies of mammalian ADA1 (6–9), whereas recognizing ADA1 as an important drug target against malaria stimulated structural studies of ADA1 from Plasmodium (10). Both mammalian and protozoan ADA1 are single α/β-TIM barrel domain proteins, containing a catalytic zinc ion in the active site (see “Results and Discussion” and Figs. 1C, 2, 3, and 5A). Transition state analogues including coformycin, deoxycoformycin (the United States Food and Drug Administration (FDA)-approved anticancer drug, pentostatin), and 6-hydroxy-7,8-dihydro purine nucleoside (HDPR) display particularly high affinity to human ADA1; however, they also inhibit human ADA2, and (+)-erythro-9-(2-hydroxy-3-nonyl)adenine (EHNA) is the only known inhibitor that binds to human ADA1 proteins but not to ADA2 (11). Although ADA1 does not exhibit any signal sequence normally required for protein secretion by cells, small quantities of the human enzyme are detected in extracellular fluids and found in association with cell surface receptors (12, 13).
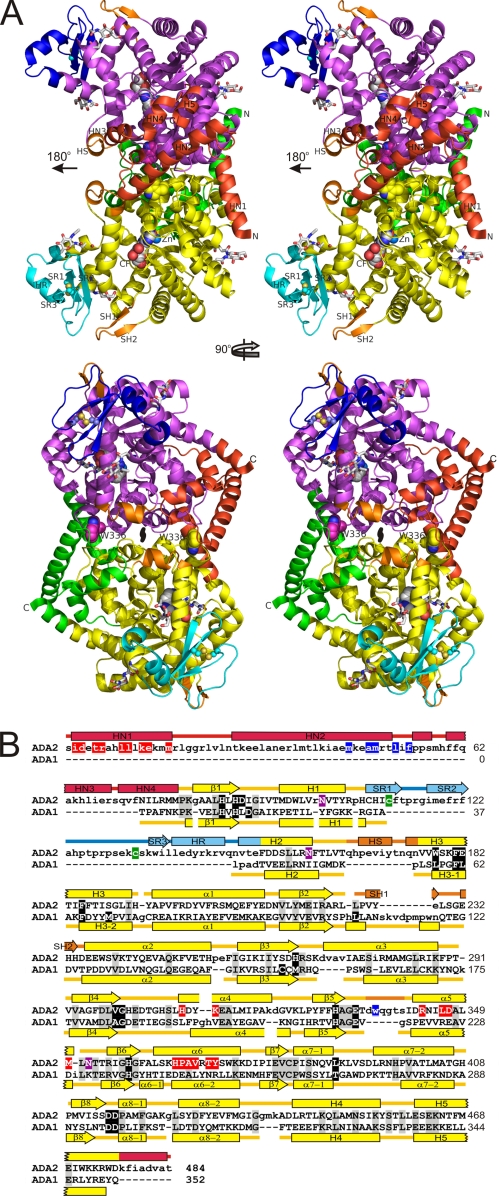
FIGURE 1.
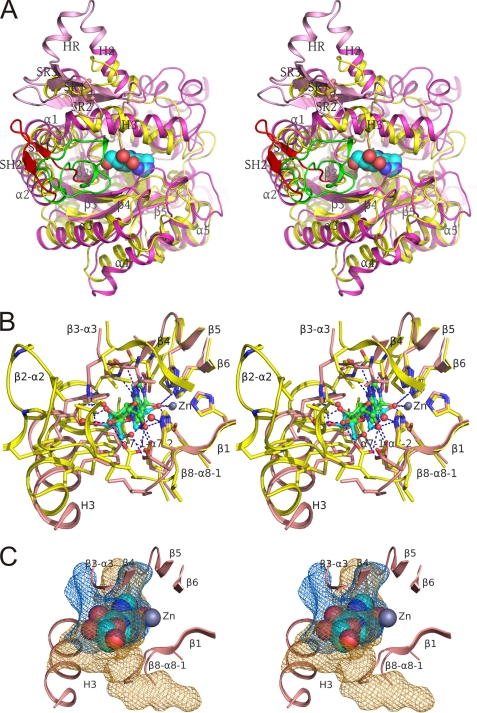
Structure of human ADA2. A, ribbon diagram of the ADA2 dimer (stereo view). “Butterfly” view of the molecule (top) and molecule rotated 90° around vertical axis toward the viewer (below). The ADA, dimerization, and PRB domains are shown in yellow, green, and cyan in one subunit and in magenta, red, and blue in the second subunit, respectively. Small unique elements are painted in orange. Catalytic Zn2+, coformycin molecules bound in the active sites, and Trp-336 are shown as spheres. Asparagine-linked N-acetyl glucosamine as well as the disulfide in PRB domains are shown with sticks. Oxygen, nitrogen, and sulfur atoms are painted in red, blue, and yellow, respectively, whereas carbon atoms are colored in white, green, yellow, or magenta. Secondary structure elements unique for ADA2 segments are labeled. The local two-fold symmetry axis is indicated. B, structural alignment of human ADA2 and mouse ADA1 (PDB accession code 2ADA). Structurally equivalent and nonequivalent residues are shown in upper and lower cases, respectively. Amino acid identities are indicated by background shading in gray. Active site residues are shown on the black background. Residues involved in the helix anchor and tryptophan catch intersubunit contacts are shown on red and blue backgrounds, respectively. The disulfide bond-linked cysteines and glycosylated asparagines are indicated by background shading in green and magenta, respectively. Secondary structure (rectangle, α-helix; arrow, β-strand; line, coil) is shown above and below the amino acid sequence of ADA2 and ADA1, respectively. The ADA domains of ADA2 and ADA1 are painted in yellow; ADA2 dimerization and PRB domains and small unique elements are painted in red, blue, and orange, respectively.
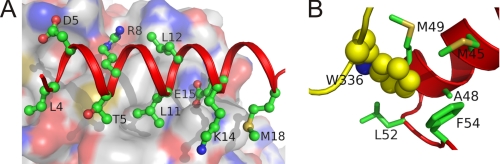
FIGURE 2.
ADA2 dimerization contacts: HN1 helix anchor (A) and Trp-336 catch (B). Helix HN1 (A) and helix HN2 and loop HN2-HN3 (B) of one subunit are shown as a cartoon in red. The second subunit is represented by molecular surface colored by type of element (A) or schematic (loop β5-α5) in yellow (B). Interactive side chains in the first subunit are shown as balls and sticks. Trp-336 in the second subunit is shown as spheres. Oxygen and nitrogen atoms are painted in red and yellow, respectively, whereas carbon atoms are colored in green, yellow, or white.
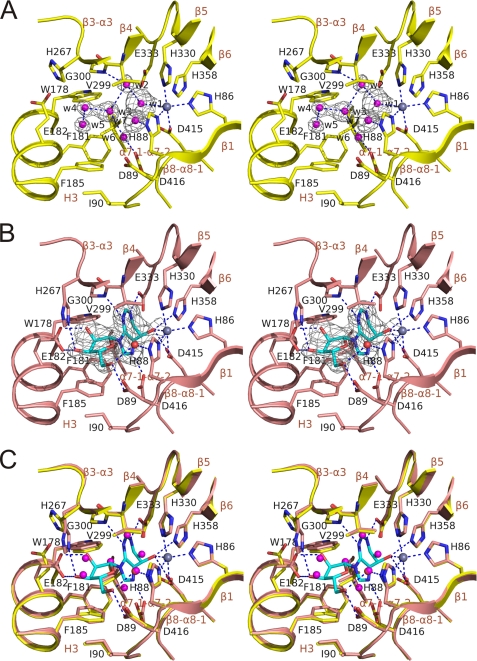
FIGURE 3.
Active site of ADA2 with and without coformycin (stereo views). A, ligand-free active site. B, ADA2 complexed with coformycin. C, superposition of active site residues of nonligated ADA2 (A) and ADA2 complexed with coformycin (B). The main chain is shown as a schematic painted in yellow and pink for nonligated and complexed ADA2, respectively. Side chains of the active site residues are shown with sticks and colored by the type of element. Oxygen and nitrogen atoms are also shown in red and blue, respectively. Carbon atoms in the nonligated and complexed ADA2 and coformycin are shown in yellow, pink, and cyan, respectively. Active site water molecules are shown as spheres and painted in magenta and red in the structures of ligand-free ADA2 and ADA2-CF complex, respectively. Zn2+ ion is shown as a gray sphere. (2|Fo| − |Fc|, αc) density for water molecules in A and coformycin in B is shown as a gray mesh at a contour level of 1 σ. Dashed lines represent coordinating interactions with zinc as well as hydrogen bonds between coformycin, active site water molecules, and ADA2. Active site residues, water molecules, and elements of secondary structure are labeled.
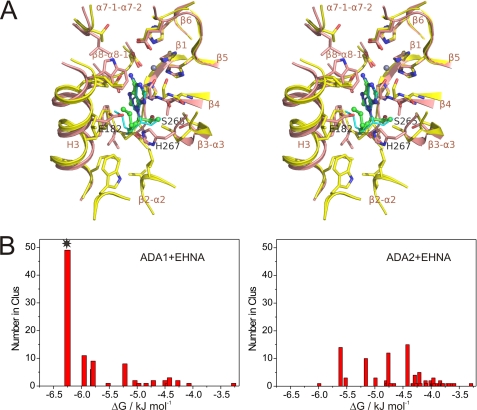
FIGURE 5.
Structural basis of selective binding of EHNA to ADA1 but not ADA2. A, structural superposition of the active sites of crystal structures of human ADA2 and bovine ADA1-EHNA complex (PDB accession code 2Z7G) and the low energy ADA1-EHNA complex obtained by docking of EHNA to the ADA1 structure (see bellow). The main chain is shown as a cartoon painted in pink and yellow for ADA2 and ADA1, respectively. Side chains of the active site residues are shown with sticks and colored by type of element. Carbon atoms in ADA2, ADA1, EHNA (crystal), and EHNA (docking experiment) are shown in pink, yellow, green, and cyan, respectively. Atoms of oxygen and nitrogen are painted in red and blue, respectively, in all structures. Zinc atoms are shown as spheres in gray. Secondary structure elements and Glu-182, His-267, and Ser-265 are labeled. Structures were superimposed by distance minimization between Cα atoms of the invariant active site residues and zinc ions. B, the binding energy statistic histograms of complexes between EHNA and bovine ADA1 (left) and human ADA2 (right) obtained from docking experiments using AutoDock 4 and AutoDockTools version 1.4.5. Note that although the majority of the predicted complexes between ADA1 and EHNA (marked by a star) have similar low energy structures that are very close to the crystal structure of the bovine ADA1-EHNA complex, no dominant low energy complexes are predicted for ADA2. Number in Clus, number in clusters.
The ADGF/ADA2 family proteins (3, 14, 15) were first described as growth factors in insects (16, 17) and later were found to possess the ADA activity, which was shown to be essential for the growth factor activity (3, 18, 19). The physiological role is best described for the Drosophila homologue, ADGF-A (3, 20). ADGF-A was shown to be crucial for fly survival, and the knock-out of the corresponding gene results in abnormal larval development and multiple melanocytic tumors. ADGF-A was found to have an autocrine ADA-dependent mutagenic activity toward macrophage-like hemocytes, suggesting its role in the function of the innate immune system in invertebrates. The knock-out of ADGF/ADA2 homologues in frogs causes a reduction in the body size and abnormalities of body axis in frog embryos, which illustrates the important role of ADGF/ADA2 proteins in the embryogenesis of amphibians (19). Unlike ADA1, all ADGF/ADA2 proteins possess secretion signal sequences and are found in the extracellular space (2). ADGF/ADA2 proteins contain considerably longer sequences than those of ADA1 proteins. One of the characteristic features of ADGF/ADA2 is an 80–100-amino acid extension at the N terminus that was suggested to be responsible for their growth factor activity (3, 14, 15).
Although the ADA2 activity in human plasma was discovered quite some time ago, the protein was purified only recently, and it was found to be encoded by the cat eye syndrome critical region gene 1 (CECR1), a gene located on chromosome 22 (21, 22). Human ADA2 and ADA1 have different catalytic and biochemical properties: ADA2 has 100-fold lower Km than ADA1, an acidic optimum pH for ADA activity, and forms a homodimer (21). ADA2 likely plays a role in development because the excess of ADA2 production due to a partial triplication of 22q11.2 chromosome results in several developmental abnormalities associated with cat eye syndrome (23). Our recent study4 showed that human ADA2 is specifically secreted by antigen-presenting cells and induces differentiation of monocytes to macrophages as well as stimulating the proliferation of macrophages and CD4+ T cells. This cytokine-like growth factor activity of ADA2 requires binding to cell surface receptors. We found that ADA2 binds to different types of cells via proteoglycans and more specifically to T cells via an unknown receptor. As we also found that ADA2 can bind to and modulate the affinity of ADRs, we hypothesized that the ADA2 binding to T cells is mediated by ADRs or an ADR-proteoglycan complex. Previously, the binding to the cell surface via unknown receptors was found for insects (17, 18) and proposed for amphibian homologues (19), suggesting that this might be a common signaling mechanism for ADGF/ADA2 proteins that is complementary to or cooperating with the signaling via the enzymatic activity.
In this study, we have determined the high resolution crystal structures of ADA2 and ADA2 complexed with a transition state analogue. In addition to the catalytic domain, the structures discovered two ADGF/ADA2-specific domains of novel folds that mediate the protein dimerization and binding to the cell surface receptors. Structures of the apoenzyme and enzyme complexed with coformycin allowed for the dissecting of the molecular details of the catalytic mechanism of ADGF/ADA2 proteins. Despite the structural conservation of the catalytic domain, a unique arrangement of the substrate-binding pocket was revealed, providing an explanation to the substrate and inhibitor specificity of ADGF/ADA2 proteins as well as providing the basis for the design of the first specific inhibitor for human ADA2.
EXPERIMENTAL PROCEDURES
ADA2 Expression and Purification
Total RNA was extracted from THP1 cells using RNAgents kit (Promega), and cDNA was prepared using random hexamers and SuperScript II reverse transcription-polymerase (Invitrogen). The CECR1 gene was amplified using forward and reverse primers (supplemental Table 1S) and inserted into the TOPO vector (Invitrogen). For the protein expression, the CECR1 gene was further modified with a biotinylation site and a His6 tag sequence on its C terminus and a leader BiP sequence on the N terminus (supplemental Fig. 1S). The construct was then subcloned into a plasmid pRMHa3 (24) for ADA2 expression in insect cells. S2 Drosophila cells were co-transfected with pRMHa3-ADA2 plasmid and pS2neo plasmids carrying the antibiotic resistance gene through the use of a calcium phosphate transfection kit (Invitrogen). Stable transformants were selected using 1.5 mg/ml G418 of antibiotic (Invitrogen). To overexpress ADA2, the cells were cultured for 10 days in Express Five SFM medium (Invitrogen) in the presence of 0.5 mm of CuSO4. The supernatant was then collected after centrifugation at 6,000 × g for 20 min, diluted with an equal volume of 40 mm Tris-HCl buffer (pH 6.8), and then applied to a column with heparin-Sepharose gel (GE Healthcare). The protein was eluted with 20 mm potassium phosphate buffer (pH 7.6) containing 500 mm NaCl and applied directly to a HiTrap chelating HP column charged with Ni2+. ADA2 was eluted using a 5–300 mm imidazole gradient. The fractions containing ADA2 activity were pooled, concentrated using Amicon Ultra concentrators (Millipore), and further purified on a Superdex 200 column (GE Healthcare) equilibrated either with phosphate-buffered saline or with 20 mm Tris-HCl (pH 6.8) buffer.
Crystallization and Structure Determination
Crystallization was performed by the hanging drop vapor diffusion method at 293 K. Large plate-like crystals were obtained in drops with 40% 2-methyl-2,4-pentanediol and 5% polyethylene glycol 8000 in 0.1 m sodium cacodylate buffer, pH 6.04, in the absence or presence of 2 mm coformycin (a kind gift from Dr. Bertil Fredholm, Karolinska Institute, Stockholm, Sweden). Crystals of both apoADA2 and ADA2 complexed with coformycin belong to space group P1 with unit cell dimensions around a = 63 Å, b = 73 Å, c = 81 Å, α = 113°, β = 94°, γ = 92° (see Table 1), and one copy of the ADA2 dimer per asymmetric unit. To obtain a derivative for heavy metal phasing, crystals were incubated with precipitant solution containing 5 mm K2Pt(NO2)4 for 12 h. Diffraction data were collected under liquid nitrogen cryoconditions at 100 K. Crystals were flash-cooled by rapidly dipping them in liquid nitrogen. X-ray diffraction data were collected on beam-line ID29 European Synchrotron Radiation Facility (ESRF), France, using an ADSC Q315R CCD-based detector (Area Detector Systems Corp.). Data were collected, processed, and reduced using MOSFLM and SCALA (25). Using PHENIX (26), heavy atom parameters were obtained and refined, and initial phases were calculated. The initial model was constructed using PHENIX and O (27). Positional, bulk solvent, and isotropic B factor refinement was performed using REFMAC5 (28). Noncrystallographic symmetry restraints were applied during the early cycles of modeling and refinement. Progress of refinement and selection of refinement schemes were monitored by the Rfree for a test set comprising ∼5% of the data. Data collection and refinement statistics are given in Table 1.
TABLE 1.
Data and refinement statistics
Values for high resolution shells are in parentheses.
| ADA2, native | ADA2 + coformycin | ADA2, platinum, peak | |
|---|---|---|---|
| Data collection | |||
| Space group | P1 | P1 | P1 |
| Unit cell a, b, c (Å); α, β, γ (°) | 63.21, 73.10, 80.66; 113.4, 94.2, 92.2 | 63.25, 72.99, 80.5; 113.6, 94.9, 91.5 | 62.61, 73.91, 79.37; 113.4, 93.1, 92.6 |
| Wavelength (Å) | 0.900 | 1.009 | 1.0715 |
| Resolution range (Å) | 73–1.9 (2.0–1.9) | 67–2.5 (2.64–2.50) | 72–3.5 (3.69–3.50) |
| No. of unique reflections | 101215 (31991) | 43967 (6368) | 15907 (2346) |
| Multiplicity | 2.1 (2.2) | 3.9 (3.9) | 6.3 (6.5) |
| Completeness | 97.4 (96.9) | 96.8 (95.8) | 97.1 (98.2) |
| Rmergea | 0.1 (0.43) | 0.069 (0.32) | 0.07 (0.18) |
| <I/s(I)> | 8.8 (1.9) | 16.8 (3.1) | 18.7 (6.7) |
| Refinement | ADA2 (Apo) | ADA2+coformycin | |
| Resolution (Å) | 40-2.0 | 67-2.5 | |
| No. of reflections | |||
| Total | 86867 | 43967 | |
| Work set | 82591 | 41746 | |
| Test set | 4276 | 2221 | |
| Rworkb | 0.188 | 0.192 | |
| Rfreec | 0.233 | 0.225 | |
| No. of atoms | |||
| Protein | 7946 | 7981 | |
| Solvent | 800 | 242 | |
| Average B-factor (Å2) | 27.5 | 33.0 | |
| r.m.s.d.d stereochemistry | |||
| Bonds (Å) | 0.013 | 0.009 | |
| Angles (°) | 1.325 | 1.225 | |
| r.m.s.d.e B-factors (Å2) | |||
| Main chain | 0.660 | 0.474 | |
| Side chain | 2.192 | 1.531 | |
| Coordinate error (Å) | 0.106 | 0.179 | |
| Ramachandran plot | |||
| Residues in most favored regions (%) | 92.2 | 92.2 | |
| Residues in additional allowed regions (%) | 7.3 | 7.3 | |
| Residues in disallowed regions (%), name | 0.5, His-358, Asp-415 | 0.5, His-358, Asp-415 | |
aRmerge = ΣhΣi|I(h)i − (I)|/ΣhΣi I(h)i, where I(h) is the intensity of a reflection h, Σh is the sum over all reflections and Σi is the sum over i measurements of reflection h.
bRwork = Σ|Fo − Fc|/ΣFo where Fo and Fc are the observed and calculated structure factors, respectively.
c Rfree is calculated for a test set of reflections randomly excluded from refinement.
d r.m.s.d. stereochemistry is the deviation from ideal values.
e r.m.s.d. B-factors are deviation between bonded atoms. The coordinate error was estimated based on maximum likelihood.
Docking
Docking of a pentasaccharide model ligand to protein structures was performed with AutoDock 4 (29) essentially as described in Ref. 30. The ligand consisted of three N-sulfo-d-glucosamine 6-O-sulfate (GlcNS6S) residues separated by l-iduronate 2-O-sulfate (IdoA2S) residues (OMe-GlcNS6S-IdoA2S-GlcNS6S-IdoA2S-GlcNS6S-OMe). Because the IdoA2S residue can adopt different ring conformations, two model conformations were used, one in which all IdoA2S residues were in the 1C4 ring form and another in which the 2S0 ring form was adopted. The model ligand had fixed glycosidic torsion angles, taken from a heparin NMR structure (Protein Data Bank (PDB) accession number 1HPN). Flexibility was allowed for all exocyclic torsion angles.
ADA2 Binding to Glycosaminoglycans
Approximately 0.2 μg of radiolabeled glycosaminoglycans (GAGs) (31) were incubated with indicated concentrations of ADA2 in 200 μl of phosphate-buffered saline, 0.1% bovine serum albumin for 2 h at 37 °C. Unbound GAG was removed by filtration through a nitrocellulose filter, whereas protein-bound GAG was trapped on the filter surface. The amount of protein-bound GAG was determined by counting the radioactivity associated with the filter using scintillation counting.
Mutagenesis
Site-directed mutagenesis was carried out by using the QuikChange site-directed mutagenesis kit (Stratagene). The Arg-8–Lys-14 deletion and point mutations W336G, C111G, and W136G were introduced in the pRMHa3-iCECR1 plasmids using the oligonucleotides R8-K14, W336G, C111G, and W136G, respectively (supplemental Table 1S).
RESULTS AND DISCUSSION
Architecture
To elucidate the structural basis of the catalytic and growth factor activities of ADGF/ADA2 proteins, we determined high resolution crystal structures of the human ADA2 apoenzyme and of the enzyme complexed with a transition state analogue, coformycin (CF). The crystal structure of the secreted apoenzyme was solved using the platinum single anomalous diffraction method and refined to a final resolution of 2.0 Å; this structure was later used as a search model to solve the structure of the enzyme complexed with CF that was subsequently refined to a final resolution of 2.5 Å (Table 1). Asymmetric units of both apo-complexed and CF-complexed enzyme crystals contain two nearly identical monomers related by a local dyad axis (Fig. 1A). The monomers display large interactive surfaces (∼4700 Å2) dominated by hydrophobic interactions (∼2800 Å2), strongly suggesting that ADA2 is a functional homodimer. This is in agreement with our gel filtration (see Fig. 6C) and cross-linking (supplemental Fig. 3S) analysis of purified ADA2 as well as gel filtration studies of partially purified ADA2 from human plasma (21).
FIGURE 6.
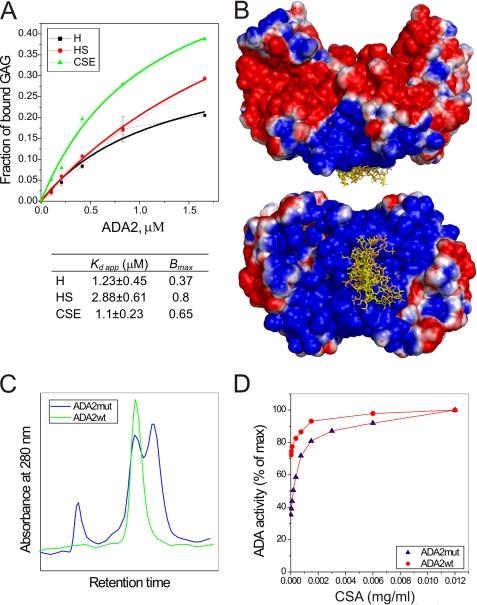
ADA2 interaction with GAGs. A, equilibrium binding of ADA2 to heparin (black), HS (red), and chondroitin sulfate E (CSE) (green). Approximately 0.2 μg of intact 3H-labeled GAGs were incubated with increasing concentrations of ADA2 for 2 h at 37 °C. Error bars indicate S.D. B, heparin-binding site on ADA2 as determined by docking model ligands. A pentasaccharide ligand consisting of 3 GlcNS6S residues separated by IdoA2S residues was used in the docking study. In one version (yellow), all IdoA2S residues are in the 1C4 ring conformation, whereas in another (orange), these residues are in 2S0 ring conformation. The four lowest energy complexes between ADA2 and each of the ligands are shown overlaid. Solvent-accessible surface of ADA2 is shown with the electrostatic potential of the ADA2 molecule (from −1 to 1 kiloteslas) mapped onto it. Note that ligands bind in the positively charged area located in the region between two subunits of the ADA2 dimer. C and D, a point mutation in the intersubunit space of ADA2 facilitates dissociation of ADA2 into monomers and decreases the ADA activity. C, T336G mutant of ADA2 (ADA2mut) dissociates into monomers under denaturing conditions. Elution profiles of gel filtration of wild type and mutant ADA2 on a Superdex 200 column in the presence of 2 m urea are shown in green and blue, respectively. D, binding of GAGs to ADA2 increases the ADA activity. The activity of ADA2 wild type and ADA2 T336G mutant has been determined in the presence of increasing concentrations of chondroitin sulfate A (CSA).
The core structure of the subunit is formed by an ADA1-like, adenosine deaminase (ADA) domain comprising more than 70% of the amino acid sequence of the protein. Despite a low sequence homology between the ADA domain and ADA1 proteins (18–21% identical residues), the overall structure of the ADA domain is similar to known ADA1 structures, showing the highest similarity to ADA1 from Plasmodium vivax (PDB accession number 2PGF). 342 Cα in these structures are superimposed with r.m.s.d. of 2.8 Å. As in ADA1 proteins, the ADA domain of human ADA2 is composed of an eight-stranded, parallel β-sheet that closes into a barrel and is surrounded by classical α/β-TIM barrel motif helices and five additional, specific for ADA1 helices located between β1 and α1 (H1, H2, and H3) and at the C terminus (H4 and H5) (Fig. 1). Loops between β-strands and α-helices constitute most of the essential features of the active site (see below).
ADGF family proteins contain ∼80-, ∼50-, and ∼10-amino-acid long sequences at the N terminus, between H1 and H2 helices of the ADA domain, and at the C terminus, respectively, which are absent in ADA1 proteins (Fig. 1B and supplemental Fig. 2S). In human ADA2, these sequences comprise two additional domains.
The N-terminal extension forms N-terminal α-helices HN1, HN2, HN3, and HN4, whereas the C-terminal extension elongates the C-terminal helix H5, which together with the N-terminal helices forms a unique helix-turn-helix arrangement (Fig. 1). Because ADA domains form nearly as many interactions with these helices as the helices between each other, this structural element resembles more a second α-helical layer wrapping the surface of ADA domains than a separate domain. Nevertheless, we will call it “dimerization domain” based on its clear functional role in connecting subunits of the dimer; the dimerization domain mediates more than 60% of the total and 70% of the nonpolar intersubunit interactions.
Helix HN1 projects as a finger from its own subunit and almost entirely interacts with the ADA domain of the neighboring subunit (Fig. 2A). This “helix anchor” provides the major contact between subunits in the dimer that contributes ∼40% of the hydrophobic interactive area. Helix HN1 docks to the surface created by helices α5 and α6. Two highly conserved charged residues of helix HN1, Arg-8 and Glu-15, are engaged in ionic interactions with the Asp-347 and His-365 of the neighboring subunit, respectively. Hydrophobic Ile-4, Leu-11, Leu-12, and Met-18 as well as parts of aliphatic chains of polar Thr-7 and Lys-14 form hydrophobic contacts with residues of the neighboring subunit.
Helix HN1 is followed by a sharp twist and 5-residue loop, positioned at the right angle relative to the helix. The twist and loop as well as helices HN2, HN3, and HN4 pack on the surface of the ADA domain of own subunit. The N-terminal part of helix HN2 and helix HN4 form coil-coil-like interactions with the ADGF family-specific C-terminal end of helix HN5.
Helices HN2 and HN3 are connected with a long loop containing a Pro-Pro-Ser twist in the middle. The loop residues have the lowest B factors in the dimerization domain, indicating the high conformational stability of this segment. The conserved hydrophobic residues of loop HN2-HN3 (Leu-52 and Phe-52) and helix HN2 (Met-45, Ala-48, and Met-49) form a hydrophobic binding pocket, accommodated by the highly conserved Trp-336 from the neighboring subunit (Fig. 2B and supplemental Fig. 2S). This tryptophan-mediated binding, which we will further refer to as “tryptophan catch,” is the second largest after the anchoring helix contact between subunits in the ADA2 dimer, providing ∼20% nonpolar intersubunit surface. Trp-336 is located in the sequence inserted between strand β5 and helix α5, which is unique for ADGF/ADA2 proteins (Fig. 1B).
A short helix HS further extends the second layer of helices wrapping the ADA domain (Fig. 1A). The helix HS and adjacent loops are formed by the ADGF/ADA2-specific ∼10-residue insertion between helices H2 and H3 (Fig. 1B and supplemental Fig. 2S). Helices HS from two subunits of the dimer are closely positioned, but they do not interact with each other.
The ∼50-amino acid insertion between H1 and H2 helices of the ADA domain folds into an additional “putative receptor-binding” (PRB) domain, sitting on the top of the ADA domain above the H3 helix at the edge of the active site (Figs. 1 and 4). The PRB domain has an α- and β-fold and is composed of a three-stranded antiparallel β-sheet SR1-SR2-SR3, surrounded by the HR and partially H2 α-helices on one side and the proline-rich SR2-SR3 loop on the other side. The SR2-SR3 loop is linked to the SR1 strand with a disulfide bond between Cys-111 and Cys-133. This disulfide bond is highly conserved for ADGF/ADA2 proteins from multicellular organisms (supplemental Fig. 2S). A C111G mutation completely abolishes the ADA2 secretion, whereas in vitro reduction of the disulfide bond with 1,4-dithiothreitol leads to massive aggregation of the protein (data not shown), suggesting that the disulfide bond is structurally important.
FIGURE 4.
Comparison of the active site in ADA2 and ADA1. A, superposition of structures of human ADA2 and mouse ADA1 (PDB accession code 2ADA) (cartoon diagrams, stereo view). ADA2 is painted in magenta except for the putative receptor-binding domain (pink) and SH1-SH2 hairpin (red). ADA1 is painted in yellow except for loop β2-α2 (green). The Cys-111–Cys-133 disulfide bond is shown with sticks. The coformycin molecule in the ADA2 catalytic site is shown as spheres. B, superposition of active site residues in human ADA2 and mouse ADA1 complexed with coformycin and HDPR, respectively (stereo view). The main chain is shown as a schematic painted in pink and yellow for ADA2 and ADA1, respectively. Side chains of the active site residues are shown with sticks and colored by the type of element. Carbon atoms in ADA2, ADA1, coformycin, and HDPR are shown in pink, yellow, cyan, and green, respectively. Atoms of oxygen and nitrogen are painted in red and blue, respectively, in all structures. Zinc atom and water molecules in contact with ligands are shown as spheres in gray and red, respectively. ADA2 is shown in the same orientation as in Fig. 3. Dashed lines represent hydrogen bonds between ligands and proteins and zinc coordination by ligands. C, the catalytic site of ADA2 is more open than that in ADA1. Cavities in the active sites of ADA2 and ADA1 (which were superimposed and displayed in the same orientation as that in B) are shown with mesh in orange and blue colors, respectively. For clarity only the ribbon diagrams of the ADA2 active site and ADA2 complexed with the coformycin molecule are shown. In A–C, structures were superimposed by distance minimization between Cα atoms of invariant catalytic residues.
The PRB domain is apparently not involved in the catalytic function of ADA2. Its good exposure on the surface of the molecule suggests that it might be responsible for the binding of ADA2 to specific receptors. The PRB domains are conveniently located on the same side of the ADA2 dimer to bind dimeric receptors or induce receptor dimerization, a common mechanism for signal transmission into the interior of the cell. Intriguingly, the functional adenosine receptors, which we show to bind ADA2,4 are dimers (32). The DALI search in the Protein Data Bank revealed no structures similar to the structure of the PRB domain. Interestingly, the PRB domain possesses structural features typical for monomers of chemokines. Having a similar size, they also possess an α-helix/three-stranded antiparallel sheet fold, contain proline-rich loops, and are stabilized by a disulfide bond (33).
Fourier difference maps revealed an extra electron density close to asparagines 101, 159, and 352 in each ADA2 subunit, which allowed modeling of 1–2 N-acetyl-glucosamine residues of Asn-linked oligosaccharide chains (Fig. 1). Two N-glycans are situated close to the PRB domain (Asn-101 and Asn-159), whereas the third one is located on the opposite side of the molecule (Asn-352). Oligosaccharide chains located on three different faces of the ADA2 molecule may protect the enzyme against proteolysis in the extracellular space. The extensive glycosylation, the presence of a conserved disulfide bond in the PRB domain, and a signal peptide strongly suggest that ADA2, in contrast to ADA1, is specifically designed for the secretion to the extracellular environment.
Active Site
The (|Fo| − |Fc|, αc) difference electron density map revealed a strong density peak indicative of a metal-binding site. An independent analysis of ADA2 crystals with x-ray absorption spectroscopy suggested the presence of zinc in the protein. Therefore, a zinc ion was included in the structure refinement, which after completion attained a B factor of the modeled atom consistent with bound zinc. The discovery of the zinc ion confirms the earlier suggestion that ADGF/ADA2 as well as ADA1 family proteins are zinc-dependent hydrolases (6, 14).
The ADA domain contains a deep oblong-shaped active site cavity lined by the C-terminal segments and connecting loops of the β-barrel strands, acting as a “floor” and “walls” and capped from above with a “ceiling” made of helices H3 and α7 and loop α7-1-α7-2 (Figs. 1 and 4). The zinc ion sits in the deepest part of the active site cavity in the center of the C-terminal end of the barrel. It is coordinated to 4 invariant residues: His-86 and His-88 in the β1 strand, His-330 in the β5 strand, and Asp-415 in the β1-α8-1 loop (Fig. 3A).
CF binds in the cavity above the zinc ion (Fig. 3B). The purine-like unit of CF lying perpendicular to the β-barrel is sandwiched between loops connecting β-barrel strands 1, 4, 5, and 6 and hydrophobic parts of helix H3 and loop α7-1-α7-2. The ribose group is located closer to the opening of the binding cavity interacting with loops β1-α1, β3-α3, β4-α4, and helix H3.
CF is a powerful transition state inhibitor, mimicking the tetrahedral intermediate at the C6 position of adenine. The 8-hydroxyl group of CF occupies the transition state position of the attacking hydroxyl ion. It lies at nearly optimal distance to fill the fifth coordination site of the zinc ion. The resulting coordination geometry of the bound zinc is distorted trigonal bipyramidal with atoms Nϵ2 of His-86 and His-88 and the O8 atom of CF occupying the equatorial positions and atoms Nϵ2 of His-330 and Oδ2 of Asp-415 occupying the axial positions (supplemental Table 2S). This binding geometry is very similar to that observed for the zinc coordination in ADA1 proteins (6, 8, 10) (Fig. 4B and supplemental Table 2S).
CF forms nine hydrogen bonds with 8 residues of ADA2 (Fig. 3B). The purine-like ring is hydrogen-bonded to 5 residues (Gly-300, Glu-333, His-358, Asp-415, and Asp-416), occupying invariant positions in the ADA superfamily alignment. The main chain nitrogen of Gly-300 and side chains of Asp-416 and Glu-333 make hydrogen bonds with all three nitrogen atoms of the ring, whereas side chains of Asp-415 and His-358 form hydrogen bonds with the 8-hydroxyl group. These residues almost perfectly superimpose with the corresponding residues in ADA1 proteins (Figs. 4B and 5). Hence, as it is illustrated by the structural superposition of complexes ADA2-CF and mouse ADA1-HDPR, in Fig. 5B, the purine(-like) rings of substrates are bound in similar orientations. In contrast, all 3 residues engaged in hydrogen bonds with the ribose group (Asp-89, Glu-182, and His-286) have no analogs in ADA1 proteins, and the mechanism of binding the ribose is different. Glu-182 in helix H3 accepts hydrogen bonds from 2′- and 3′-hydroxyl groups. His-286 in strand β3 donates a hydrogen bond to the 3′O. Asp-89 in loop β1-α1 accepts a hydrogen bond from the 5′-hydroxyl group, which in turn accepts a hydrogen bond from structured water interacting with the zinc-coordinating His-88. The ribose in CF is rotated by ∼35° around the N3-1′C bond, and its 5′C-5′OH group is rotated by ∼120° around the 4′C-5′C bond relative to the positions of these groups in HDPR.
Although β-α-loops are predominantly engaged in polar contacts with CF, the majority of hydrophobic interactions between ADA2 and CF are mediated by helix H3 and loop α7-1-α7-2 (Fig. 3B). Large aromatic residues (Trp-178, Phe-181, and Phe-185) in helix H3 together with Leu-389 in loop α7-1-α7-2 and Ile-90 in loop β1-α1 create a continuous hydrophobic surface capping both the purine-like ring and the ribose of CF. Together with Val-299, interacting with the opposite side of CF, these residues form a hydrophobic binding subpocket.
In contrast to catalytic parts of active sites, hydrophobic binding subpockets in ADA2 and ADA1 have surprisingly different structures (Fig. 4). Only 4 ligand-binding hydrophobic residues are structurally equivalent, and only 2 of these are identical in ADA2 and mouse ADA1. This dissimilarity arises from differences both in the structure of hydrophobic ligand-binding segments and in the distribution of hydrophobic side chains.
Helix H3 carries the majority of the active site hydrophobic residues in proteins of both families. In ADA2, it is situated up to 4.5 Å further away from the purine-like ring of the ligand in the direction perpendicular to the plane of the ring and shifted by about 3–4 Å in the direction parallel to the ring toward the ribose relative to its position in ADA1. Differences in the position of helix H3 are reflected in the elevated r.m.s.d. value for the corresponding Cα atoms (3.76 Å). However, the most striking difference between ADA2 and ADA1 proteins is observed in the conformation of loop β2-α2 In ADA1 proteins; this highly hydrophobic loop is coiled to form a high wall at one side of the active site cavity. In ADA2, this loop is more than half a size shorter than that in mammalian ADA1. Instead of packing close to the cavity as in ADA1 proteins, it forms a hairpin projecting away from the active site in a direction perpendicular to the β-barrel. Both differences in the orientation of helix H5 and conformation of loop β2-α2 make the active site in ADA2 considerably more open than ADA1 proteins (Fig. 4C).
The binding site in ADA2 contains only half the number of hydrophobic residues than that in mouse ADA1. All these missing residues are located at the entrance of the binding site in mouse ADA1 where they predominantly interact with the ribose group of the ligand. In contrast, ADA2 contains two charged (Glu-182 and His-267) and one polar (Ser-265) residues in this area. Hence, the entrance to the active site in ADA2 is not only considerably more open but also much more hydrophilic than that in ADA1.
The CF binding causes surprisingly little change in the structure of the active site (Fig. 3C). The structure of the main chain remains practically unchanged. The greatest changes are observed in the structure of side chains unique to ADA2 charged residues Glu-182 and His-267. These side chains act as lids, partially enclosing CF in the active site. Upon the CF binding, side chains of Glu-182 and His-267 change rotamer configurations to be able to form hydrogen bonds with 2′- and 3′-hydroxyl groups of the substrate ribose. Side chains of other active site residues undergo only minor changes. This is in sharp contrast to the dramatic conformational changes in the structures of ADA1 proteins, which switch between open (ligand-free) and closed (complexed) conformations (8). Interestingly, superposition of 40 active site residues in ADA2 with corresponding residues in the open and closed conformations in ADA1 revealed significantly higher similarity to the open (r.m.s.d. = 1.17 Å) than the closed (r.m.s.d. = 1.6 Å) forms of ADA1. As both helix H3 and loop β4-α4 are involved in the intersubunit contacts associated with the tryptophan catch, it is logical to suggest that these unique intersubunit interactions for ADA2 may restrain the conformational changes in the binding site and hence arrest ADA2 in the open conformation.
CF replaces six active site water molecules (W1–6) (Fig. 3A). Waters 1–3 and 6 create a planar geometry and occupy or are located close to the position of the purine-like ring of CF. W4 and W5 are situated in the ribose-binding pocket in positions corresponding to the 2′- and 3′-hydroxyl groups of the ribose, respectively. W1 is situated at a 3 Å distance from the zinc ion. This molecule sits in a high density spherical peak in the (2|Fo| − |Fc|, αc) Fourier map. The position of W1 is unexpected. The structure of the apoform of bovine ADA1 also shows a water molecule close to the zinc ion. However, this water molecule coordinates to the active center of zinc with a 2.1 Å distance and is thought to represent an activated water or hydroxyl ion that plays a role in attacking nucleophile for adenosine deamination. The reason for this striking difference is not clear. It is possible that the observed density peak belongs to a relatively small unknown ion and not to a water molecule.
Catalytic Mechanism
The high similarity of the coordination structure of the zinc ion and hydrogen bonding of the purine(-like) ring in active sites of ADA2 and ADA1 proteins leaves little doubt that these enzymes share a similar enzymatic mechanism (6, 7). Side chains of His-86, His-88, His-330, and Asp-415 play a role in the coordination of the zinc active center, which acts as a powerful electrophile activating the attacking water to a hydroxide ion (mimicked by the 8-hydroxyl group of CF). Hydrogen bonding between side chains of His-358 and Asp-415 and the 8-hydroxyl group of CF suggests that these residues are involved in the stereo-specific positioning of the attacking water and stabilizing the sp3 configuration of the tetrahedral adduct at the C6 atom of adenosine. Interestingly, side chains of His-358 and Asp-415 in ADA2 and those of the corresponding residues in ADA1 proteins have similar high energy conformations (supplemental Table 1). Although the reason for this phenomenon is not clear, it indicates that the catalytic function of these residues is the same in both types of enzymes. By analogy with ADA1 enzymes, His-358 may also serve as a basis for abstracting protons from the attacking water, whereas the side chain of Glu-333, located in the hydrophobic environment, may serve as a proton donor to the N1 atom of adenosine, promoting the nucleophilic attack at the C6 atom by reducing the N1 to C6 double bond character.
The high structural similarity of the catalytic region of the active site in ADA2 and ADA1 is consistent with the determination of close values of kcat for these enzymes (21). At the same time, the dissimilar structure of the substrate-binding hydrophobic subpocket in ADA2 and ADA1 explains the large differences in the affinity of these proteins to their substrates and inhibitors. Thus, human ADA2 has 100 times higher value of Km for adenosine deamination than that of mammalian ADA1 (21). The affinity to adenosine decreases for ADGF/ADA2 proteins from lower to higher species. Interestingly, this affinity loss strongly correlates with the hydrophobicity of the mouth of the binding pocket. Positions of charged Glu-182 and His-267 and polar Ser-265 in human ADA2 (Km ∼2 mm (21)), in ADGF/ADA2 proteins from the salivary glands of the flesh fly, Sarcophaga peregrina (Km ∼15 μm (18)), and the blood-sucking insect, Lutzomyia longipalpis (Km ∼50 μm (14)) are occupied by Met, Val, and Ala and Gly, Ile, and Ala, respectively. ADGF/ADA2 proteins from frog and chicken, which have intermediate values of Km (∼250 (19) and 150 μm (34), respectively), share most of the ligand-binding residues with the human homologue; however, instead of Ser-265 in the human enzyme, they contain the more hydrophobic Thr and Ala residues. The hydrophobic-to-polar mutation, adjusting the activity of ADGF/ADA2 proteins from higher species to higher levels of adenosine, might have occurred in parallel with acquiring the ability to act locally at specific physiological conditions. This is consistent with our findings suggesting a highly specialized function of human ADA24.
Differences in the structure of the hydrophobic subpocket of ADA2 and ADA1 also determine the selective inhibition of ADA1, but not ADA2, by EHNA. In the ADA1-EHNA complex (9), the aliphatic chain of EHNA is exposed to the highly hydrophobic environment of the large binding subpocket of this protein (Fig. 5A). The orientation of the ligand is well predicted by docking experiments (Fig. 5B). In ADA2, the corresponding part of the binding pocket is highly hydrophilic. The superposition of ADA2 and the ADA1-EHNA complex suggests that the aliphatic chain of EHNA would collide with Glu-182 and form close contacts with His-267 and Ser-265 (Fig. 5A). This polar environment would greatly weaken the affinity for EHNA, which is supported by the failure to reveal high affinity ADA2-EHNA complexes in the docking experiments (Fig. 5B).
A Glycosaminoglycan-binding Site Is Formed Jointly by the Two Subunits of the Dimer
We showed that ADA2 binds to different types of cells via heparan sulfate (HS)- and chondroitin sulfate (CS)-containing proteoglycans.4 To study the affinity of ADA2 to different GAGs, we performed in vitro binding experiments with purified enzyme and radiolabeled heparin, HS, and CS E (Fig. 6A). All these GAGs strongly bind to ADA2. The interactions between ADA and CS E and heparin are tighter than those between ADA2 and the less sulfated HS, suggesting that the binding involves extensive ionic interactions.
To identify the binding site for GAGs, we performed docking with a model heparin pentasaccharide (30) to ADA2 using AutoDock 4. This ligand consists of 3 GlcNS6S residues separated by IdoA2S residues. To test for the effect of the flexible IdoA ring structure, the ligand was either kept in an all 1C4 ring conformation for both IdoAs or kept in a 2S0 conformation. The 10 lowest energy complexes were analyzed for each ligand. In all resulting complexes the ligand was bound to the same area within a large, highly positively charged surface around the interface between the subunits in the dimer, involving helices HN1, α6, and α7-2 of each subunit (Fig. 6B). The ligands form extensive interactions with both subunits, suggesting that the protein dimerization is likely to be essential for GAG-dependent protein localization on the cell surface.
ADA2 Dimerization Is Required for the Full Enzymatic Activity and Secretion on the Cell Surface
To investigate the role of intersubunit contacts in ADA2 activity, we have generated a mutant of ADA2 in which the tryptophan in position 336 (Fig. 1) was substituted with a glycine. This point mutation in ADA2 slightly affects the Km of the enzyme but significantly reduces the turnover rate (supplemental Fig. 4S), indicating that the dissociation of ADA2 subunits rather than a change in ADA2 conformation decreases ADA2 activity. Indeed, gel filtration of ADA2-W336G in the presence of low concentrations of urea shows that this mutant of ADA2 partly dissociates into monomers, whereas this did not occur in the case of the wild type enzyme (Fig. 6C). Interestingly, the addition of GAGs to the reaction mix increases the ADA activity of ADA2 (Fig. 6D). Moreover, this effect is more prominent in the case of mutant ADA2 (Fig. 6D and supplemental Fig. 4S). These results suggest that the dimerization of ADA2 is required for the full enzymatic activity and that the interaction of ADA2 with cell surface GAGs (see below) stabilizes the dimer. Deletion of amino acid residues in the region between Thr-7 and Glu-15 completely abolishes the release of ADA2 into the culture medium (data not shown), suggesting that subunit dimerization via the helix anchor is crucial for the secretion of this protein.
Conclusions
Despite the fact that ADGF/ADA2 proteins play such an important role in tissue development, very little is known about their structure and functional mechanism. In this study, we present the first high resolution structures of an ADGF, human ADA2, which constitutes the basis for understanding the catalytic/signaling activities of these enzyme growth factors.
The high resolution crystal structure of ADA2 (apoenzyme and in complex with coformycin) revealed a symmetric dimer of two identical subunits with a complex multidomain architecture. The unique N-terminal part of ADGF/ADA2 proteins, which was previously suggested to form a growth factor domain, was observed as an α-helical domain that mediates ADA2 dimerization. However, the structure revealed that an insertion sequence within the catalytic domain folds into a highly exposed independent domain sharing several structural properties with chemokines. This novel domain could be involved in the enzyme binding to cell surface receptors, such as human ADA2 T cell receptors, and hence might be responsible for the receptor-mediated growth factor activity of ADGF/ADA2 proteins, such as the cytokine activity of human ADA2.4 Although the human ADA2 T cell receptors are yet to be identified, our cellular and in vitro data demonstrate high affinity binding to different cell types via GAG chains of proteoglycans. The docking experiments using model substrates unambiguously identified a GAG-binding site in the highly positively charged region around the contact between ADA2 subunits. Consistent with this result, the analysis of the W336G mutant with decreased dimerization affinity suggests an important role of the ADA2 dimerization in the GAG binding. Many cytokines bind to GAG chains of proteoglycans, and this binding often promotes the high affinity binding to more specific protein receptors. We found that in addition to proteoglycans, ADA2 binds to ADRs,4 which function as dimers (32). Hence, it is plausible to speculate that ADA2 activates T cells by binding to certain ADR-GAG complexes similar to, for example, ternary complexes of fibroblast growth factor-fibroblast growth factor receptor-HS (35).
Although the overall structure of the catalytic domain and catalytic site of ADA2 and functionally related ADA1 proteins is similar, predicting essentially the same catalytic mechanism, the structure of their hydrophobic binding (specificity) subpocket is very different. The difference is larger for the ligand-bound forms than for apoenzymes, possibly due to restraints for conformational changes imposed by the dimerization contacts in ADA2. The observed structural differences provide an explanation for the previously documented large differences in the specificity of the enzymes to their substrates and inhibitors (11) and suggest a mechanism for the evolutionary decrease in the affinity of ADGF/ADA2 proteins for adenosine. It could be speculated that the evolutionary decrease in the affinity is the result of the specialization of ADGF/ADA2 from higher species to act in pathological conditions. Human ADA2, specifically secreting by antigen-presenting cells, may act locally in or navigate to sites with a high concentration of adenosine (e.g. sites of inflammation) by binding to specific cell surface receptors.
We speculate that the differences in the specificity pockets of human ADA2 and mammalian ADA1 could be exploited to generate the first specific inhibitor for ADA2. Such a compound will greatly simplify further functional studies of ADA2 and may be employed for the development of novel anticancer drugs avoiding the severe side effects (5) of the current drug, pentostatin (deoxycoformycin), which inhibits both enzymes. Our cellular studies4 show that both ADA1 and ADA2 stimulate monocyte-dependent proliferation of CD4+ T cells. However, we show that the unique role of ADA2 is to induce differentiation of monocytes into macrophages. Hence, although the drugs inhibiting the ADA1 function could be employed to treat leukemias (5), a specific inhibitor for ADA2 could be efficient to treat lymphomas or infectious mononucleosis where the activity of ADA2 is significantly elevated (36). At the same time, ADA2 and its mutants could be used as efficient immunostimulators by increasing the number of CD4+ T cells and macrophages in immunocompromised patients and, therefore, find their application in the treatment of tuberculosis and human immunodeficiency virus (HIV) infections.
This work was supported by grants from the Swedish Research Council (K2008-58X-20689-01-3) and FORMAS (221-2007-1057) (to Ant. V. Z.), European Molecular Biology Organization Grant ALT-978-2005 and Human Frontier of Science Program (HFSP) Long Term Fellowship LT00395-2006-L (to And. V. Z.), and grants from INSERM (Avenir) and HFSP carrier development award (CDA0040-2005C) (to G. L.).
This article was selected as a Paper of the Week.
The atomic coordinates and structure factors (codes 3LGD and 3LGG) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1S–2S and supplemental Figs. 1S–4S.
A. Zavialov, E. Gracia, N. Glaichenhaus, R. Franco, G. Lauvau, and A. Zavialov, unpublished results.
- ADR
- adenosine receptor
- ADA
- adenosine deaminase
- ADGF
- adenosine deaminase growth factor
- CF
- coformycin
- CS
- chondroitin sulfate
- HS
- heparan sulfate
- EHNA
- (+)-erythro-9-(2-hydroxy-3-nonyl)adenine
- GAG
- glycosaminoglycan
- HDPR
- 6-hydroxy-7,8-dihydro purine nucleoside
- PRB
- putative receptor-binding
- r.m.s.d.
- root mean square deviation
- GlcNS6S
- N-sulfo-d-glucosamine 6-O-sulfate
- IdoA2S
- l-iduronate 2-O-sulfate.
REFERENCES
- 1.Haskó G., Linden J., Cronstein B., Pacher P. (2008) Nat. Rev. Drug. Discov. 7, 759–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maier S. A., Galellis J. R., McDermid H. E. (2005) J. Mol. Evol. 61, 776–794 [DOI] [PubMed] [Google Scholar]
- 3.Zurovec M., Dolezal T., Gazi M., Pavlova E., Bryant P. J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 4403–4408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hershfield M. S. (2005) Eur. J. Immunol. 35, 25–30 [DOI] [PubMed] [Google Scholar]
- 5.Dillman R. O. (2004) Expert. Rev. Anticancer Ther. 4, 27–36 [DOI] [PubMed] [Google Scholar]
- 6.Wilson D. K., Rudolph F. B., Quiocho F. A. (1991) Science 252, 1278–1284 [DOI] [PubMed] [Google Scholar]
- 7.Wilson D. K., Quiocho F. A. (1993) Biochemistry 32, 1689–1694 [DOI] [PubMed] [Google Scholar]
- 8.Kinoshita T., Nakanishi I., Terasaka T., Kuno M., Seki N., Warizaya M., Matsumura H., Inoue T., Takano K., Adachi H., Mori Y., Fujii T. (2005) Biochemistry 44, 10562–10569 [DOI] [PubMed] [Google Scholar]
- 9.Kinoshita T., Tada T., Nakanishi I. (2008) Biochem. Biophys. Res. Commun. 373, 53–57 [DOI] [PubMed] [Google Scholar]
- 10.Larson E. T., Deng W., Krumm B. E., Napuli A., Mueller N., Van Voorhis W. C., Buckner F. S., Fan E., Lauricella A., DeTitta G., Luft J., Zucker F., Hol W. G., Verlinde C. L., Merritt E. A. (2008) J. Mol. Biol. 381, 975–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niedzwicki J. G., Kouttab N. M., Mayer K. H., Carpenter C. C., Parks R. E., Jr., Abushanab E., Abernethy D. R. (1991) J. Acquir. Immune Defic. Syndr. 4, 178–182 [PubMed] [Google Scholar]
- 12.Weihofen W. A., Liu J., Reutter W., Saenger W., Fan H. (2004) J. Biol. Chem. 279, 43330–43335 [DOI] [PubMed] [Google Scholar]
- 13.Franco R., Pacheco R., Gatell J. M., Gallart T., Lluis C. (2007) Crit. Rev. Immunol. 27, 495–509 [DOI] [PubMed] [Google Scholar]
- 14.Charlab R., Valenzuela J. G., Andersen J., Ribeiro J. M. (2001) Gene 267, 13–22 [DOI] [PubMed] [Google Scholar]
- 15.Maier S. A., Podemski L., Graham S. W., McDermid H. E., Locke J. (2001) Gene 280, 27–36 [DOI] [PubMed] [Google Scholar]
- 16.Matsushita T., Fujii-Taira I., Tanaka Y., Homma K. J., Natori S. (2000) J. Biol. Chem. 275, 36934–36941 [DOI] [PubMed] [Google Scholar]
- 17.Homma K., Matsushita T., Natori S. (1996) J. Biol. Chem. 271, 13770–13775 [DOI] [PubMed] [Google Scholar]
- 18.Homma K. J., Tanaka Y., Matsushita T., Yokoyama K., Matsui H., Natori S. (2001) J. Biol. Chem. 276, 43761–43766 [DOI] [PubMed] [Google Scholar]
- 19.Iijima R., Kunieda T., Yamaguchi S., Kamigaki H., Fujii-Taira I., Sekimizu K., Kubo T., Natori S., Homma K. J. (2008) J. Biol. Chem. 283, 2255–2264 [DOI] [PubMed] [Google Scholar]
- 20.Dolezal T., Dolezelova E., Zurovec M., Bryant P. J. (2005) PLoS Biol. 3, e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zavialov A. V., Engström A. (2005) Biochem. J. 391, 51–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riazi M. A., Brinkman-Mills P., Nguyen T., Pan H., Phan S., Ying F., Roe B. A., Tochigi J., Shimizu Y., Minoshima S., Shimizu N., Buchwald M., McDermid H. E. (2000) Genomics 64, 277–285 [DOI] [PubMed] [Google Scholar]
- 23.Meins M., Burfeind P., Motsch S., Trappe R., Bartmus D., Langer S., Speicher M. R., Mühlendyck H., Bartels I., Zoll B. (2003) J. Med. Genet. 40, e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bunch T. A., Grinblat Y., Goldstein L. S. (1988) Nucleic Acids Res. 16, 1043–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 26.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 27.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 28.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 29.Huey R., Morris G. M., Olson A. J., Goodsell D. S. (2007) J. Comput Chem. 28, 1145–1152 [DOI] [PubMed] [Google Scholar]
- 30.Sachchidanand, Lequin O., Staunton D., Mulloy B., Forster M. J., Yoshida K., Campbell I. D. (2002) J. Biol. Chem. 277, 50629–50635 [DOI] [PubMed] [Google Scholar]
- 31.Petersen F., Brandt E., Lindahl U., Spillmann D. (1999) J. Biol. Chem. 274, 12376–12382 [DOI] [PubMed] [Google Scholar]
- 32.Canals M., Burgueño J., Marcellino D., Cabello N., Canela E. I., Mallol J., Agnati L., Ferré S., Bouvier M., Fuxe K., Ciruela F., Lluis C., Franco R. (2004) J. Neurochem. 88, 726–734 [DOI] [PubMed] [Google Scholar]
- 33.Fernandez E. J., Lolis E. (2002) Annu. Rev. Pharmacol. Toxicol. 42, 469–499 [DOI] [PubMed] [Google Scholar]
- 34.Iwaki-Egawa S., Namiki C., Watanabe Y. (2004) Comp Biochem. Physiol. B Biochem. Mol. Biol. 137, 247–254 [DOI] [PubMed] [Google Scholar]
- 35.Yayon A., Klagsbrun M., Esko J. D., Leder P., Ornitz D. M. (1991) Cell 64, 841–848 [DOI] [PubMed] [Google Scholar]
- 36.Ungerer J. P., Oosthuizen H. M., Bissbort S. H., Vermaak W. J. (1992) Clin. Chem. 38, 1322–1326 [PubMed] [Google Scholar]