Abstract
Colicin M inhibits Escherichia coli peptidoglycan synthesis through cleavage of its lipid-linked precursors. It has a compact structure, whereas other related toxins are organized in three independent domains, each devoted to a particular function: translocation through the outer membrane, receptor binding, and toxicity, from the N to the C termini, respectively. To establish whether colicin M displays such an organization despite its structural characteristics, protein dissection experiments were performed, which allowed us to delineate an independent toxicity domain encompassing exactly the C-terminal region conserved among colicin M-like proteins and covering about half of colicin M (residues 124–271). Surprisingly, the in vitro activity of the isolated domain was 45-fold higher than that of the full-length protein, suggesting a mechanism by which the toxicity of this domain is revealed following primary protein maturation. In vivo, the isolated toxicity domain appeared as toxic as the full-length protein under conditions where the reception and translocation steps were by-passed. Contrary to the full-length colicin M, the isolated domain did not require the presence of the periplasmic FkpA protein to be toxic under these conditions, demonstrating that FkpA is involved in the maturation process. Mutational analysis further identified five residues that are essential for cytotoxicity as well as in vitro lipid II-degrading activity: Asp-229, His-235, Asp-226, Tyr-228, and Arg-236. Most of these residues are surface-exposed and located relatively close to each other, hence suggesting they belong to the colicin M active site.
Keywords: Bacterial Toxins, Cell Wall, Membrane Lipids, Membrane Proteins, Phosphodiesterases, Colicin M, Lipid II, Peptidoglycan
Introduction
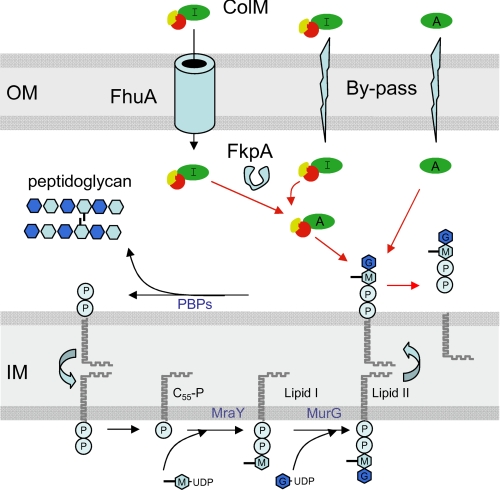
Colicins are plasmid-encoded toxins, secreted in the growth medium by certain strains of Escherichia coli to kill susceptible E. coli strains and closely related bacteria (1–3). Most colicins characterized to date possess either a pore-forming activity that causes depolarization of the cytoplasmic membrane or a nuclease activity that specifically targets rRNA, tRNA, or chromosomal DNA (3). Colicin M (ColM)2 is unique among the colicins, because it is the only one known to interfere with the biosynthesis of peptidoglycan (4–6). In vitro and in vivo analyses recently allowed us to elucidate the mode of action of ColM, i.e. to demonstrate that ColM interfered with the peptidoglycan pathway by catalyzing the hydrolysis of the lipid intermediates I and II (6) (see Fig. 1). The site of cleavage was unambiguously located between the C55 and pyrophosphoryl groups releasing undecaprenol (C55-OH) and either 1-pyrophospho-MurNAc-pentapeptide or 1-pyrophospho-MurNAc(pentapeptide)-GlcNAc (see Fig. 1 and supplemental Fig. S1).
FIGURE 1.
Synthesis of peptidoglycan lipid-linked intermediates and mode of action of ColM. The inner (IM) and outer (OM) membranes are depicted by the gray boxes. The MraY enzyme catalyzes the transfer of the phospho-MurNAc-pentapeptide from the nucleotide precursor onto the carrier lipid undecaprenyl phosphate (C55-P), yielding lipid I; subsequently, the MurG enzyme adds the GlcNAc moiety, yielding lipid II, which is translocated from the inner side of the membrane to the outer side (38, 39). Thereafter, the disaccharide-pentapeptide motif is polymerized and incorporated into the peptidoglycan through the action of the penicillin-binding proteins (PBPs) releasing the lipid carrier in a pyrophosphate form that will be recycled (40, 41). Incoming ColM was shown to cleave the lipid-linked intermediates between the C55 and pyrophosphoryl-disaccharide-peptide motifs (6). In the present report, an independent ColM toxicity domain (green) was identified (residues 124–271), which was active in vivo under conditions where the FhuA receptor was by-passed. In contrast to the full-length ColM, the independent toxicity domain did not require the periplasmic FkpA protein to be toxic. These data suggest that ColM exists in two active (A) and inactive (I) conformations, FkpA being involved in the activation process. The reception and translocation domains of ColM are represented in red and yellow, respectively.
ColM enters in susceptible cells by using the outer membrane protein FhuA as the receptor and the TonB/ExbB/ExbD machinery for its translocation (7–9). Mutants affected in the latter proteins are fully resistant to ColM. Recently, Hullmann et al. (10) reported that the cytotoxicity of the imported ColM was dependent on the presence of a specific periplasmic chaperone, FkpA. Incubation of denatured ColM with purified FkpA made it regain its cytotoxic activity, suggesting that ColM was denatured upon uptake through the outer membrane and then refolded by FkpA before exhibiting its deleterious effects (10). Colicinogenic cells are always protected against the toxin they produce by concomitant expression of a specific immunity protein (3). The ColM immunity protein, Cmi (also named ImM), is anchored to the cytoplasmic membrane via a putative α-helix located at its N-terminal end, whereas most of the polypeptide extends in the periplasmic space, where it likely inhibits incoming ColM by an unknown mechanism (11, 12). Interestingly, ColM should enter the cell from the outside to be bactericidal, explaining why it can be readily overproduced by import-deficient strains in the absence of Cmi. ColM most likely exerts its activity at the periplasmic face of the inner membrane, where it has access to lipid II (Fig. 1).
Most of the colicins that have been structurally characterized to date share a common three-domain organization, each of these domains being devoted to a particular function: the N-terminal domain is required for translocation across the outer membrane and into the periplasm, the central domain is involved in receptor binding, and the C-terminal domain displays the cytotoxic activity (3). This canonical organization and the fact that natural or recombinant chimeric colicins are functional suggest that their evolution is based on the exchange and mixing of similar domains originating from heterologous toxins (13–16). ColM, which is composed of only 271 amino acids, is the smallest colicin known to date. Biochemical data suggest that ColM, despite its low size, adopts a similar three-domain organization. A typical TonB box, that is characteristic of class B colicins, is located in the N-terminal region of the protein, and its mutation abolishes the uptake of ColM but not its capacity to bind the FhuA receptor (8). Similarly, mutations targeting the C-terminal domain affected the cytotoxicity but not the uptake (7–9). The three-dimensional structure of ColM, recently determined at a resolution of 1.7 Å, showed a unique fold with no similarity with other colicins or even known structures (17). Quite strikingly, unlike most other colicins, ColM displayed a compact structure in which distinct domains were difficult to differentiate. Genes encoding proteins that exhibit similarity with the C-terminal region of ColM were recently identified in the genomes of some pathogenic species of Pseudomonas and Burkholderia genera (Fig. 2) (18–20). Interestingly, no significant homology was observed in their respective N-terminal regions. The ColM-like proteins from Pseudomonas aeruginosa, P. syringae, and P. fluorescens were expressed and purified from E. coli cells and then demonstrated to also exhibit enzymatic properties of degradation of the peptidoglycan lipid intermediates (18). ColM and these few homologues identified so far thus constitute a novel and unique family of bacteriocins interfering with the pathway for peptidoglycan biosynthesis by enzymatic (phosphodiesterase type) degradation of its precursors. We here report a detailed analysis of the structural organization and mechanism of action of this class of enzymes. In particular, the catalytic domain of ColM was more precisely characterized by using complementary approaches of site-directed mutagenesis, protein dissection experiments, and in vivo activity assays in different genetic backgrounds.
FIGURE 2.
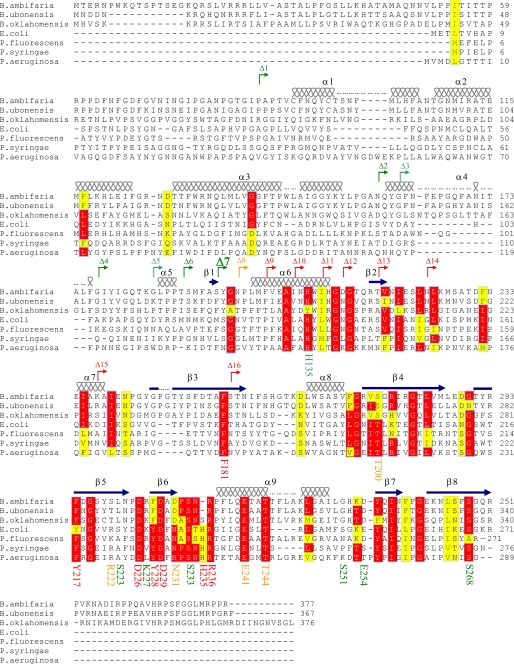
Sequence alignment of ColM with its homologues. Amino acid sequences of E. coli ColM (accession number P05820) and its homologues from Burkholderia ambifaria MC40–6, Burkholderia ubonensis Bu, Burkholderia oklahomensis Eo147, Pseudomonas fluorescens, Pseudomonas syringae pv. tomato DC3000 and Pseudomonas aeruginosa EXA13 were aligned using the ClustalW program. Based on the known three-dimensional structure of ColM (17), secondary structural elements are indicated at the top of the sequences, the wavy lines designating α-helices and the arrows designating β-strands. The numbers of these structural elements are given according to the structure. The conserved residues are color-coded in red for identical residues and yellow for similar ones. The starting points of each N-terminal deletion constructs (from Δ1 toward Δ16) are indicated by broken arrows. The truncations indicated in green exhibit high in vitro activities, whereas those in red had no activity (ColM-Δ8 had intermediate activity). The targeted residues for mutagenesis are also indicated by their respective numbers below the sequences. According to the ability of the corresponding mutants to inhibit cell growth, the residues were classified as non-important (green), important (orange), and essential residues (red).
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Growth Conditions
The E. coli strains DH5α (Invitrogen), XL1-blue (Stratagene), C43(DE3) (Avidis), and BW25113 (and its derivatives) were used as the hosts for propagation of plasmids, mutagenesis, production of proteins, and by-pass experiments, respectively. The E. coli BW25113 ΔfhuA::CmR, carrying a deletion of the chromosomal fhuA gene, was previously described (6). The E. coli BW25113 ΔfkpA::KanR, carrying a deletion of the chromosomal fkpA gene, was from the Keio collection (21). The pTO4 plasmid carrying the cma gene encoding ColM and the cmi immunity gene has been previously described (22). Plasmids pMLD189 and pMLD190 encoding ColM with a His6 tag at the N- and C-terminal extremities, respectively, have been described previously (6). The plasmid vector pTrc99A was from Amersham Biosciences, and the construction of pET2130 has been previously described (18). The pET2430 vector, a pET24d (Novagen) derivative, was constructed by removal (filling-in) of the unique BglII site of pET24d and replacement of the BamHI-HindIII polylinker by that from the Qiagen pQE30 vector. Similarly, the pET2460 vector derived from pET24d by removal of the BglII site and replacement of the NcoI-HindIII polylinker by that from the Qiagen pQE60 vector. The pET2130 and pET2430 plasmids allow expression of proteins with a His6 tag at the N-terminal extremity, whereas pET2460 allows expression of proteins with a His6 tag at the C-terminal extremity. Cells were grown in 2YT medium (1.6% bactotrypton, 1.0% bactoyeast extract, 0.5% NaCl, pH 7.0) (23) at 37 °C. Ampicillin, kanamycin, and chloramphenicol were used at 100, 35, and 25 μg/ml, respectively. Growth was monitored at 600 nm with a Shimadzu UV-1601 spectrophotometer.
General DNA Techniques and E. coli Cell Transformation
PCR amplification of genes was performed in a Thermocycler 60 apparatus (Bio-Med) using the Expand-Fidelity polymerase (Roche Applied Science). DNA fragments were purified using the Wizard PCR Preps DNA purification kit (Promega). Standard procedures for DNA digestion, ligation, agarose gel electrophoresis, and plasmid isolations were used (24). E. coli cells were transformed with plasmid DNA by the method of Dagert and Ehrlich (25) or by electroporation.
Construction of Expression Plasmids
Truncated forms of the cma gene encoding ColM variants, lacking between 32 and 181 amino acids from the N-terminal domain, were amplified by PCR from the pTO4 plasmid (22). Forward primers (Δx-O1) containing a BamHI site (in bold) 5′ to the new initiation codon (supplemental Table S1) and reverse primer Δ-O2 containing a HindIII site (in bold) 3′ to the gene, respectively, were used. In all cases, the amplified material was digested by BamHI and HindIII and inserted between the same sites of either the pET2130 (ApR) or pET2430 (KanR) vectors, generating plasmids that express the truncated ColM proteins with a Met-His6-Gly-Ser N-terminal tag. The sixteen resulting plasmids were then transformed into the E. coli C43(DE3) strain for expression experiments. To optimize expression of the full-length ColM, inserts from the pMLD189 and pMLD190 plasmids were recovered and cloned into the pET2430 and pET2460 vectors, generating the plasmids pMLD237 and pMLD238, respectively. A plasmid allowing expression of the ColM immunity protein (Cmi) was constructed as follows: the cmi gene was amplified by PCR from the pTO4 plasmid using primers containing a BspHI site (in bold) 5′ to the initiation codon (underlined), 5′-GCGCTCATGAAAGTAATTAGCATGAAATTTATTTTTATTTTAACG-3′, and a PstI site (in bold) 3′ to the gene, 5′-CCGGCTGCAGCCTTTATAACAGTGTATTAGTCATTCGC-3′, respectively. The amplified material was digested by BspHI and PstI and inserted between the compatible NcoI and PstI sites of the pTrc99A vector, generating the plasmid pMLD232 allowing expression of the wild-type cmi gene under the control of the strong isopropyl-β-d-thiogalactopyranoside-inducible trc promoter. In all cases, DNA sequencing was performed to control that the sequence of the cloned fragments was correct (Eurofins-MWG).
Site-directed Mutagenesis
Site-directed mutagenesis of the C-terminally His6-tagged ColM enzyme was performed directly on the expression plasmid pMLD238 (pET2460::cma) by using the QuikChange II XL site-directed mutagenesis kit from Stratagene. The couples of oligonucleotides used for introduction of these mutations in the gene sequence are shown in supplemental Table S1. Mutations introduced in the cma gene sequence were checked by DNA sequencing.
Preparation of Crude Extracts and Purification of Truncated and/or Mutated ColM Enzymes
The E. coli strain C43(DE3), which presents the double advantage of being naturally resistant to ColM, due to its lack of a functional FhuA protein, and suitable for gene expression from pET vectors, was used as the host strain for ColM production. Cells carrying expression plasmids were grown at 37 °C in 2YT medium containing ampicillin or kanamycin (200-ml cultures). When the optical density of the culture reached 0.8, isopropyl 1-thio-β-d-galactopyranoside was added at a final concentration of 1 mm, and growth was continued for 3 h at 37 °C. Cells were harvested and washed with 40 ml of cold 20 mm Tris-HCl buffer (pH 7.4) containing 10 mm 2-mercaptoethanol, 200 mm NaCl, and 10% glycerol (buffer A). The cell pellet was suspended in 6 ml of the same buffer, and cells were disrupted by sonication in the cold (Bioblock Vibracell sonicator, model 72412). The resulting suspension was either centrifuged at 4 °C for 30 min at 200,000 × g in a TL100 Beckman centrifuge and the supernatant (∼40 mg of proteins) stored at −20 °C before purification of the His6-tagged protein as described below, or centrifuged at 4 °C for 10 min at 7,000 × g when the protein was present as an insoluble form. In this case, the pellet was dispersed in 2 ml of 8 m urea buffer for 1 h at 4 °C and centrifuged again. The proteins were finally refolded by a 50-fold dilution of the supernatant in buffer A before purification.
The purification of the His6-tagged proteins was performed following the manufacturer's recommendations (Qiagen): native or refolded protein extracts obtained as described above were incubated for 1 h at 4 °C with nickel-nitrilotriacetic acid (Ni2+-NTA)-agarose pre-equilibrated in buffer A containing 10 mm imidazole. The resin was then washed extensively with buffer A containing 20 mm imidazole. The proteins of interest were eluted with 200 mm imidazole and either dialyzed overnight against 100 volumes of buffer A or desalted on PD-10 columns (Amersham Biosciences). The final preparations were stored at −20 °C. SDS-PAGE analysis of proteins was performed as described by Laemmli and Favre (26). Protein concentrations were determined by using the Bradford procedure (27), with bovine serum albumin as the standard, and by quantitative amino acid analysis with a Hitachi L8800 analyzer (ScienceTec) after hydrolysis of samples in 6 m HCl at 105 °C for 24 h.
The full-length ColM proteins (wild-type and mutated variants) were expressed and purified to near homogeneity from the soluble fraction (data not shown). Only two mutants, F181A and Y217A, behaved differently as compared with the wild-type enzyme. They were quantitatively normally expressed, but only a small part was recovered in the soluble fraction, the rest forming inclusion bodies. F181A and Y217A proteins were purified from the soluble fraction but formed SDS-resistant aggregates, as judged by SDS-PAGE (supplemental Fig. S2), confirming a strong propensity to aggregate.
Protease Treatment and Polypeptide Sequencing
Purified ColM was incubated with sequencing grade trypsin, proteinase K, or chymotrypsin (Roche Applied Science) at a 10:1 or 100:1 ColM:protease molar ratio, for 30 min at room temperature. Digestions were stopped by the addition of 10% (v/v) trichloroacetic acid, whereupon protein precipitation occurred. Proteins were resuspended in SDS-loading buffer and heated (100 °C, 3 min) prior to analysis by Coomassie Blue-stained SDS-PAGE. For sequencing, protein samples were electroblotted onto polyvinylidene difluoride membranes (Pall Life Sciences) and visualized by Ponceau Red staining. Appropriate bands were excised and subjected to Edman degradation (Protein sequencing facility, University of Paris-Sud, Institut de Biochimie et Biophysique Moléculaire et Cellulaire, UMR 8619).
Native proteolysis experiments were used to address whether the mutated ColM proteins were native-like folded. All the mutants except F181A and Y217A behaved as the wild-type ColM in terms of susceptibility to chymotrypsin (similar patterns of characteristic fragments and kinetics of their release) (supplemental Fig. S2).
Fluorescence Spectroscopy
Fluorescence emission spectra were recorded with a Cary Eclipse spectrophotometer (Varian, Les Ulis, France). Spectra were recorded in buffer A (purification buffer) at a protein concentration of 1 μm by using an excitation wavelength of 295 nm and an emission wavelength comprised between 300 and 450 nm. The wild-type ColM had a fluorescence emission spectrum displaying a λmax at 330 nm, i.e. characteristic of a folded protein with tryptophan side chains buried in the protein core. The spectra of the mutants were identical to those obtained with the wild-type protein except in two cases, mutants F181A and Y217A. The latter mutants still exhibited an emission maximum at 330 nm but displayed an additional peak of emission at 350 nm that is indicative of solvent exposure of tryptophan residues.
Hydrolase Activity Assays
The enzymatic activity of mutated and/or truncated ColM forms was tested in a reaction mixture (10 μl) containing 100 mm Tris-HCl, pH 7.5, 20 mm MgCl2, 150 mm NaCl, 12 μm of [14C]radiolabeled lipid II (140 Bq), and 0.2% n-dodecyl-β-d-maltoside. The reaction was initiated by addition of the purified protein (in 5 μl of buffer A) and incubated for 1 h at 37 °C with shaking (Thermomixer, Eppendorf). The reaction was stopped by heating at 100 °C for 1 min and analyzed by TLC on LK6D silica gel plates (Whatman) using 1-propanol/ammonium hydroxide/water (6:3:1, v/v) as a mobile phase. The radioactive spots were quantified with a radioactivity scanner (model Multi-Tracemaster LB285, Berthold France). Under these conditions, the radiolabeled substrate (lipid II) and product (1-pyrophospho-MurNAc(pentapeptide)-GlcNAc) migrated with Rf values of 0.7 and 0.3, respectively. In all cases, the enzyme concentration was chosen so that substrate consumption was <30%, the linearity being ensured within this interval (supplemental Fig. S3).
For the determination of the kinetic constants, assay conditions were as described above, except that the concentration of lipid II varied from 6 to 200 μm. Data were fitted to the equation, v = VmaxS/(Km + S), by the Levenberg-Marquardt method (28). The MDFitt software developed by M. Desmadril (UMR 8619, CNRS, Orsay, France) was used for this purpose.
Determination of ColM Cytotoxicity (Standard in Vivo Assay)
The cytotoxic activity of mutated and/or truncated ColM forms was tested on 2YT-agar plates overlaid with 5 ml of soft nutrient agar containing susceptible BW25113 bacteria (100 μl of a liquid culture at A600 nm = 1.4). Serial 2-fold dilutions of pure ColM-containing samples were prepared in buffer A, and 3-μl aliquots were spotted on the overlay. Plates were incubated overnight at 37 °C, and the activity of ColM was expressed as the minimal amount of protein still yielding a clear halo indicative of growth inhibition (supplemental Fig. S4).
By-pass Experiments
These experiments were carried out essentially as described by Braun et al. (29, 30). Cells were grown in 2YT medium at 37 °C until the optical density reached 1.4. Cells (2 ml) were centrifuged, washed twice with 2 ml of ice-cold M9-salts solution (6 g of Na2HPO4, 3 g of KH2PO4, 0.5 g of NaCl, and 1 g of NH4Cl per liter), and finally resuspended in 1 ml of freshly prepared ice-cold 40 mm Tris-HCl buffer (pH 7.0) containing 10 mm EGTA (pH 7.0), and 48% sucrose. The cell suspension was incubated on ice for 30 min. Thereafter, 50 μl of cell suspension was rapidly diluted with 0.5 ml of ice-cold M9-salts solution in the presence or not of a 10-μl sample of ColM or derivatives at various concentrations. The suspension was kept on ice for 30 min before being appropriately diluted in 2YT medium and plated on 2YT-agar plates to determine the number of surviving cells.
Chemicals
[14C]Lipid II labeled in the GlcNAc moiety was prepared as previously described (6). The lipid II used in this work was C55-PP-MurNAc(l-Ala-γ-d-Glu-meso-A2pm-d-Ala-d-Ala)-GlcNAc, where A2pm represents diaminopimelic acid. N-Dodecyl-β-d-maltoside was purchased from Fluka, isopropyl 1-thio-β-d-galactopyranoside from Eurogentec, and Ni2+-NTA-agarose from Qiagen. Antibiotics and reagents were from Sigma. Synthesis of oligonucleotides and DNA sequencing were done by Eurofins MWG.
RESULTS
Design and Purification of ColM Mutants
ColM specifically degrades cell-wall peptidoglycan lipid precursors by cleaving the bond between undecaprenol and the pyrophospho-MurNAc-peptide motif. Neither the primary sequence of ColM (no similarity with known phosphatases) nor its three-dimensional structure has revealed the position of putative active site residues (6, 17). About 15 years ago, Pilsl et al. already reported the isolation of a few inactive point mutants of ColM, which had retained their capacity to enter the periplasm of susceptible cells (8). All these mutations were mapped in the C-terminal region of the protein that supposedly contained the active site. Some of them, G193S, L196F, G197S/D/V, and G252D affected glycine and leucine residues that are not typical catalytic residues and should be structurally important instead. Four other substitutions, however, concerned serine (S223L, S233A/T, and S251A) and aspartate (D226N) residues that could potentially be involved in the catalytic process and/or substrate binding. Interestingly, the Leu-196, Gly-197, Asp-226, and Ser-233 residues appeared as conserved in the recently identified ColM-like bacteriocins from Pseudomonas and Burkholderia species, whereas the Gly-193, Ser-223, Ser-251, and Gly-252 residues were not (Fig. 2) (17, 18).
On the basis of these preliminary data and sequence alignment of the different ColM homologues (Fig. 2), a series of putative active site residues was identified, whose participation in the ColM catalytic process was then questioned using a site-directed mutagenesis approach. We targeted all conserved charged residues that could potentially bind the electronegative pyrophosphate group, either directly or through coordination of the Mg2+ metal ion that is required for activity (6). Conserved aromatic residues as well as histidine, threonine, and serine residues that could act as nucleophile and or acid/base catalysts were also substituted. A total of twenty residues was subjected to mutagenesis and individually replaced by alanine (Table 1 and Fig. 2). Mutagenesis was performed directly on the pMLD238 plasmid that expresses ColM with a His6 tag at the C-terminal extremity under the control of the strong isopropyl 1-thio-β-d-galactopyranoside-inducible T7 promoter. All the mutated plasmids were obtained excepting the one encoding the Y255A mutant (all attempts to get the corresponding PCR product were unsuccessful). The wild-type and the different mutant proteins were expressed in the ColM-resistant E. coli strain C43(DE3) and subsequently purified to near homogeneity from the soluble fraction. Only two mutants, F181A and Y217A, were found to have a strong propensity to aggregate (see “Experimental Procedures”). To address whether the mutated variants were native-like folded, we analyzed their fluorescence emission spectrum as well as their proteolysis susceptibility. All the mutants behaved similarly to the wild-type protein except in two cases, mutants F181A and Y217A (see “Experimental Procedures” and supplemental Fig. S2). These data suggested that the overall structure of ColM was not affected by the selected substitutions, with the exception of the F181A and Y217A replacements that likely yielded altered structures.
TABLE 1.
In vitro and in vivo activity of the ColM mutants
| Protein | Plasmid | Enzymatic activitya | Cytotoxicityb |
|---|---|---|---|
| pmol.min−1.mg−1 | ng | ||
| WT | pMLD238 | 630 ± 57 (100%) | 0.4 |
| E254A | pTT935 | 2100 ± 150 (330%) | 0.4 |
| S223A | pTT635 | 930 ± 70 (150%) | 0.6 |
| S268A | pTT788 | 540 ± 81 (86%) | 0.8 |
| S251A | pTT913 | 510 ± 83 (81%) | 0.4 |
| K227A | pTT908 | 320 ± 43 (51%) | 0.5 |
| S233A | pTT793 | 280 ± 39 (44%) | 0.8 |
| H135A | pTT621 | 190 ± 65 (30%) | 0.8 |
| N231A | pTT927 | 77 ± 5 (12%) | 4 |
| T244A | pTT931 | 76 ± 6 (12%) | 4 |
| T200A | pTT904 | 49 ± 5 (7.8%) | 14 |
| E241A | pTT653 | 36 ± 10 (5.7%) | 2 |
| R222A | pTT627 | 14 ± 5 (2.2%) | 5 |
| D229A | pTT641 | 12 ± 5 (1.9%) | NDc |
| H235A | pTT645 | 9 ± 4 (1.4%) | ND |
| D226A | pTT629 | 2 ± 0.9 (0.3%) | ND |
| R236A | pTT946 | ND | ND |
| Y228A | pTT919 | ND | ND |
| F181A | pTT897 | ND | ND |
| Y217A | pTT914 | ND | ND |
a The enzymatic activity was measured in vitro in the presence of 12 μm of [14C]lipid II and appropriate amounts of enzyme. Values represent the mean ± S.D. of triplicate determinations.
b The cytotoxicity of ColM was measured by spotting various amounts of pure protein samples (serial 2-fold dilutions) on a lawn of susceptible BW25113 strain. The cytotoxicity was expressed as the minimal amount of protein required to obtain a clear halo indicative of growth inhibition (see supplemental Fig. S4). All assays were performed at least in triplicate.
c ND, no detectable activity (up to 10 μg of protein tested).
Cytotoxicity of ColM Mutants
The various ColM mutants were tested for their cytotoxicity by spotting serial 2-fold dilutions of pure protein samples onto a lawn of the susceptible E. coli strain BW25113. The minimal amount of toxin required to get a clear halo indicative of total growth inhibition was determined in each case (Table 1). Based on their ability and relative efficiencies to inhibit cell growth, the ColM mutants were classified in three categories: (i) seven mutants (D229A, H235A, D226A, R236A, Y228A, F181A, and Y217A) appeared totally unable to inhibit cell growth even when tested undiluted (∼10 μg of protein spotted); (ii) five mutants (N231A, T244A, T200A, E241A, and R222A) were significantly less active than the wild-type protein but still exhibited a strong toxicity (2–14 ng of protein required to inhibit growth, versus 0.4 ng for the wild-type ColM); and finally (iii), seven mutants (E254A, S223A, S268A, S251A, K227A, S233A, and H135A) appeared as active as the wild-type protein or slightly less active (inhibition observed with <1 ng of protein).
In Vitro Enzymatic Activity and Correlation with Cytotoxicity
The ability of the mutant proteins to hydrolyze lipid II in vitro was tested in the standard conditions (Table 1). We first determined whether the amino acid substitutions that had caused a total loss of cytotoxicity (mutants of the first category) also had a similar effect on the in vitro enzyme activity. Four of the seven mutants (R236A, Y228A, F181A, and Y217A) were totally inactive, in perfect agreement with the in vivo data. The lack of activity of the F181A and Y217A mutants was not really surprising, because it should likely be correlated with their structural alteration (see above). These two substitutions may therefore not be relevant in the search for specific active site residues. The activity of the other three mutants from the same category (D229A, H235A, and D226A) was also strongly diminished, representing at most 2% of the wild-type activity. The second category of mutants displayed in vitro activities in the 2–12% range as compared with wild-type ColM, that here also correlated well with their respective cytotoxic effects. Mutants N231A and T244A, which were 10-fold less active than the wild-type protein in vivo, also displayed 12% of its activity in vitro. Similarly, the mutant T200A, which required 35-fold more protein to inhibit cell growth, displayed an 8% residual activity in vitro. The mutants E241A and R222A, which demanded 5- and 12-fold more protein in vivo, displayed only 5 and 2% of in vitro activity, respectively. Finally, the third category of mutants, whose in vivo cytotoxicity levels were quite similar to that of the wild-type toxin, displayed an in vitro enzyme activity either moderately lower (>30% of relative activity) or even higher (334%) than the wild-type level.
Pilsl et al. had earlier isolated an S223L mutant and reported that its cytotoxic activity was totally abolished (8). We here observed that the replacement of the same serine residue by alanine did not affect its cytotoxic and enzymatic activities at all. This discrepancy can be explained by the nature of the residue of substitution (leucine) used by Pilsl et al. The bulky leucine residue might have affected the protein conformation. Therefore, serine 223 does not seem to be essential per se. More surprisingly, Pilsl et al. had found that mutations S233A or S251A caused a 1000-fold decrease of cytotoxicity (8). Here we showed that the same mutations did not significantly alter the cytotoxic properties of ColM and caused only moderate 66 and 20% decreases, respectively, of its in vitro enzyme activity. On one hand, the fact that the Ser-223 and Ser-251 residues are not conserved in the ColM-like homologues supports our activity data indicating that these residues are not essential for ColM activity. On the other hand, the conserved Ser-233 residue seems to be somewhat important but clearly not as much as Pilsl and coworkers had suggested. In fact, among the substitutions that have been analyzed in common by both these authors and us, only the substitution D226A yielded similar results in terms of cytotoxicity. It should be mentioned that, to estimate the toxicity of their mutants, Pilsl et al. used a highly susceptible strain that overexpressed the different components of the ColM uptake machinery, namely FhuA, TonB, ExbB, and ExbD, which likely raised the amount of incoming ColM molecules (8). For some reasons, the use of this strain might exacerbate small differences in uptake capacity or activity between the wild-type ColM and its derivatives. Moreover, these authors used ColM-containing crude cell extracts to test the activity of their mutants, which might engender differences in efficacies with respect to purified proteins. Unfortunately, Pilsl and coworkers could not address the effect of their mutations toward the enzymatic activity, as the function of ColM has only been recently elucidated (6).
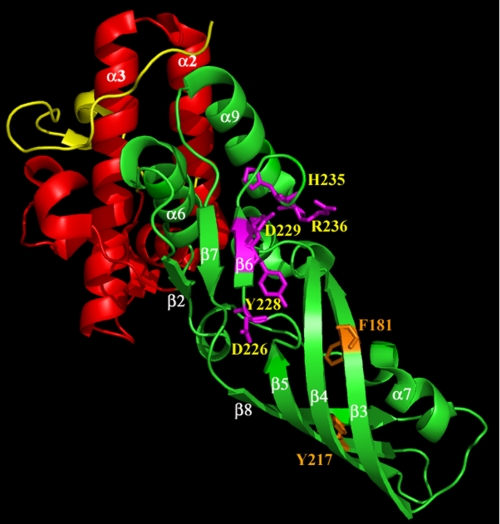
From this analysis, residues Asp-226, Tyr-228, Asp-229, His-235, and Arg-236 likely belong to the active site, as judged by the particularly strong effect their substitution had on ColM activity. Among them, the two aspartate and arginine residues appeared as conserved in all the ColM-like bacteriocins (Fig. 2). The aromatic tyrosine residue was conserved in toxins from Pseudomonas species but was replaced by a phenylalanine residue in the putative toxins from Burkholderia, and the histidine residue was only conserved in the Pseudomonas homologues. Importantly, these five residues formed a cluster at the surface of ColM with most of their side chains being protruding toward the exterior (Fig. 3) (17). Interestingly, the Phe-181 and Tyr-217 residues, the substitution of which caused more likely a major structural default, were located in the open β-barrel formed by β3, β4, β5, and β8 strands (Fig. 3). Their side chains, which are oriented toward the interior of this structure, may therefore be important for the stabilization of this sub-domain that constitutes the continuation of the sub-domain containing the active site residues.
FIGURE 3.
Three-dimensional structure of ColM. Based together on structural data from Zeth et al. (17) and biochemical data from this study, the three functional domains are illustrated as follows: the translocation domain in yellow (residues 1–35), the receptor-binding domain in red (residues 36–123), and the toxicity domain in green (124–271). The residues whose substitution caused a total loss of cytotoxicity are indicated as sticks. Residues that likely belong to the active site (Asp-226, Tyr-228, Asp-229, His-235, and Arg-236) are indicated in magenta, and structurally important residues (Phe-181 and Tyr-217) are in orange. The figure was prepared with the atomic coordinates 3DA4 deposited by Zeth et al. (17) by using PyMOL (DeLano Scientific).
Construction and Purification of Truncated Forms of ColM
Although most colicins are organized in three distinct regions with each a specialized function, the compact structure of ColM did not allow distinguishing easily such a structural organization. Protease accessibility experiments were performed to investigate whether ColM could display linker regions between putative domains. ColM was hydrolyzed by trypsin and proteinase K into numerous fragments; chymotrypsin treatment, however, gave an interesting pattern. The latter protease, which is specific of aromatic residues, had 26 potential cleavage sites distributed all along the ColM sequence. Digestion of purified C-terminally His6-tagged ColM (30.5 kDa) by chymotrypsin released four fragments of ∼11, 12, 19, and 21 kDa in equivalent amounts, as judged by SDS-PAGE analysis (supplemental Fig. S2). These data and subsequent N-terminal sequence analyses showed that the cleavage had occurred after either of the two tyrosine residues Tyr-99 and Tyr-112, at a similar frequency. Interestingly, these cleavage sites were very close, i.e. only a few residues upstream from the well conserved C-terminal domain that started at residue 124 (Fig. 2). Attempts to purify the His6-tagged (C-terminal) fragments apart from the N-terminal ones on Ni2+-NTA-agarose failed, suggesting that the two generated fragments remained tightly associated following chymotrypsin treatment. As reported below, these protein fragments were subsequently individually expressed and purified by using protein engineering experiments.
To determine which part of the C-terminal region constituted the catalytic domain and whether the activity domain was independent or not from the N-terminal (translocation) and central (reception) domains, a large series of truncated ColM derivatives (ColM-Δ) were generated (Fig. 2 and Table 2), which carried increasing deletions of the N-terminal region of the protein. Truncated forms of the cma gene (truncation at the 5′-end) were amplified by PCR and subsequently cloned in-frame with a N-terminal His6 tag (Met-His6-Gly-Ser extension) into plasmid vector pET2430. The ColM-Δ1 derivative was deleted of the 32 first residues, which form an unstructured region containing the characteristic “TonB box.” The latter sequence is recognized by the TonB protein involved in translocation of ColM through the outer membrane. Constructs ColM-Δ3 and ColM-Δ5 were designed according to the aforementioned protease accessibility data; they started with 100DAYH and 113DYRS sequences, respectively. The other constructs were chosen with respect to secondary structural elements. The construct ColM-Δ2 started with the 97SVYD sequence localized just upstream from helix α4. The other derivatives were chosen downstream and along the protein sequence to get gradually shorter derivatives (from ColM-Δ4 to ColM-Δ16) (Fig. 2 and Table 2).
TABLE 2.
In vitro activity analysis of ColM-truncated derivatives
| Protein | Plasmid | N-terminal residuea | Number of residuesb | Molecular weightb | Enzymatic activity |
|---|---|---|---|---|---|
| Da | nmol.min−1.mg−1 | ||||
| ColM WT | pMLD237 | Met-1 | 271 | 29,483 | 0.6 |
| ColM-Δ1 | pMLD262 | Ala-33 | 239 | 26,263 | 32 |
| ColM-Δ2 | pMLD263 | Ser-97 | 175 | 19,204 | 17 |
| ColM-Δ3 | pMLD264 | Asp-100 | 172 | 18,854 | 48 |
| ColM-Δ4 | pMLD265 | Ala-105 | 167 | 18,221 | 13 |
| ColM-Δ5 | pMLD257 | Asp-113 | 159 | 17,378 | 55 |
| ColM-Δ6 | pMLD258 | Met-117 | 155 | 16,856 | 30 |
| ColM-Δ7 | pMLD259 | Met-122 | 150 | 16,223 | 25 |
| ColM-Δ8 | pMLD250 | Asn-125 | 147 | 15,948 | 0.4 |
| ColM-Δ9 | pMLD266 | Pro-129 | 143 | 15,533 | NDc |
| ColM-Δ10 | pMLD267 | Leu-133 | 139 | 15,152 | ND |
| ColM-Δ11 | pMLD273 | Leu-137 | 135 | 14,668 | ND |
| ColM-Δ12 | pMLD274 | Asn-140 | 132 | 14,311 | ND |
| ColM-Δ13 | pMLD275 | Ser-145 | 127 | 13,784 | ND |
| ColM-Δ14 | pMLD276 | Gly-152 | 120 | 13,072 | ND |
| ColM-Δ15 | pMLD277 | Asp-165 | 107 | 11,620 | ND |
| ColM-Δ16 | pMLD278 | Thr-182 | 90 | 9,843 | ND |
a ColM and all the truncated variants were expressed with a N-terminal His6 tag (Met-His6-Gly-Ser extension). Sequences of ColM fused to the tag extended from the amino acid residue indicated (three-letter abbreviation followed by its position in wild-type ColM sequence, see Fig. 2) to the last residue of the protein (Arg-271).
b The N-terminal His6 tag is not included.
c ND, no detectable activity (up to 10 μg of protein tested).
All these plasmids were transformed in C43(DE3), and attempts to express and purify the truncated derivatives were performed. In contrast to the full-length ColM and with the exception of ColM-Δ1, all the truncated derivatives formed inclusion bodies and were recovered essentially in the particulate fraction. The ColM-Δ1 protein was also partially detected in the soluble fraction and thus purified from it. For other ColM-Δ derivatives, various conditions of expression (temperature, host, and chaperones) were tested to produce soluble polypeptides, but none was really satisfactory. This prompted us to perform a solubilization/renaturation procedure. Protein aggregates were thus dissolved in 8 m urea buffer and then renaturated by 50-fold dilution in non-urea buffer (see “Experimental Procedures”). This approach allowed us to get soluble and stable polypeptides, as judged by their quantitative recovery in the supernatant after ultracentrifugation. Subsequently, the different polypeptides were purified to homogeneity by affinity chromatography on Ni2+-NTA agarose (supplemental Fig. S5). The fluorescence emission spectrum of the purified polypeptides displayed a maximum at 330 nm, as the full-length protein, suggesting that the various ColM derivatives were structured.
ColM Possesses an Independent Catalytic Domain
As expected, due to the lack of the N-terminal region that is involved in ColM uptake, none of the truncated derivatives could inhibit the growth of susceptible E. coli cells. The truncated proteins were then tested for their ability to hydrolyze lipid II in vitro. All derivatives with sizes between 26.3 kDa (ColM-Δ1) and 16.2 kDa (ColM-Δ7) exhibited an in vitro activity in the range of 13–55 nmol·min−1·mg−1 (Table 2). Quite surprisingly, the activity of these truncated forms was much higher (20- to 92-fold) than that reproducibly observed for the full-length protein (∼0.6 nmol·min−1·mg−1). ColM-Δ8 (15.9 kDa), which only differed from ColM-Δ7 by the removal of three additional residues, exhibited a residual activity lower than that of the full-length toxin. None of the shorter variants (from ColM-Δ9 to ColM-Δ16) showed detectable lipid II-degrading activity, even after prolonged incubation or in the presence of higher amounts of proteins. It is noteworthy to mention that ColM-Δ7 started at residue 122, i.e. two residues upstream from the well conserved region. The slightly shortest ColM-Δ8 protein, the activity of which was dramatically decreased as compared with the Δ7 construct, lacked the first residue of this conserved region. The four-residue sequence 122MSGN125 could therefore be considered as the N-terminal limit of the minimal fragment required for full expression of ColM catalytic activity. The fact that the Gly-124 residue, that is present within this small region, was highly conserved in the sequences of other members of this bacteriocin family (Fig. 2) was consistent with our findings, suggesting that this particular borderline sequence was needed for a proper conformation of the activity domain. Based on the three-dimensional structure of ColM, Zeth et al. hypothesized that the activity domain should begin around residue 140 (17). Our data show that the minimal size required for activity is by 18 residues larger. The ColM-Δ7 construct was thus used in subsequent experiments as the toxicity domain.
Comparison of the Full-length ColM with the Catalytic Domain
As mentioned above, the in vitro activities of purified ColM-Δ1- to ColM-Δ7-truncated derivatives were much higher than that of the full-length protein. To address whether this gain of activity was a consequence of the denaturation/renaturation treatment undergone by these proteins, wild-type ColM was subjected to the same treatment. However, following renaturation, the urea-treated ColM exhibited the same activity as the non-treated sample. The position of the His6 tag (C- and N-terminal extremities in the full-length ColM and the ColM-Δ constructs, respectively) could not be incriminated as we had previously shown that the specific activity of ColM did not vary with the position of the tag (6). Together, these data suggest that the gain of activity observed with the isolated C-terminal catalytic domain was a consequence of its partition from the other domains of the protein that are required for its internalization.
The kinetic constants of the ColM-Δ7 enzyme toward lipid II were determined. Its apparent Km and kcat values were 41 μm and 2.2 min−1, respectively. In the case of the wild-type enzyme, these constants were 44 μm and 0.055 min−1, respectively (18). Because the reaction mechanism of ColM, in particular the number of bound enzyme species, is not known, the significance of these steady-state parameters is not straightforward (31, 32). Nevertheless, the fact that the specificity constant (kcat/Km) of the ColM-Δ7 enzyme (0.054 μm−1·min−1) is 45-fold higher than that of the wild-type enzyme (0.0012 μm−1·min−1) confirms that ColM-Δ7 is a better catalyst than the full-length enzyme. Furthermore, we wondered whether the point mutations we had previously introduced in the full-length protein could have a different impact on the activity of the isolated domain, the specific activity of which is much greater. Mutations R222A, S223A, D226A, D229A, H235A, and E241A were thus individually introduced into the ColM-Δ7 sequence, and the activity of the corresponding purified proteins were determined. Each of these mutations was shown to cause a decrease of activity to the same extent as in the full-length ColM (data not shown).
Cytotoxicity of the by-passed Catalytic Domain of ColM
The requirement for FhuA and the TonB/ExbB/ExbD machinery components for the energy-dependent translocation of ColM across the outer membrane can be by-passed by osmotic shock (7, 33). This procedure allows the introduction of proteins from the external milieu into the periplasm of Gram-negative bacteria while preserving cell integrity. We used this method to address the toxicity of ColM and its derivatives (point mutants and truncated forms) in different genetic backgrounds. Exponentially growing cells were suspended in a buffer of high osmolarity and then suddenly diluted in a low osmolarity buffer (assay volume, 0.5 ml), resulting in a hypo-osmotic shock. This shock was performed in the presence or not of ColM derivatives at various concentrations, and the number of surviving cells was determined. Results were reported as percentages of survivors observed following treatments in the presence of the toxin, compared with the same treatment without toxin.
We first tested the toxicity of the various ColM derivatives toward the E. coli strain BW25113 ΔfhuA that does not express the ColM receptor. As expected, when the full-length ColM was added during the hypo-osmotic shock, these cells that are normally fully resistant recovered sensitivity to the toxin (Table 3). At least 2 μg of ColM/ml was required to elicit a significant mortality (3.9% survivors) and, below this concentration, the number of survivors rapidly increased to the level obtained in the absence of toxin. This was consistent with previous data by Pilsl et al. who also reported that amounts of colicins used in by-pass experiments were much higher than those (∼10 ng/ml) needed to cause lysis of susceptible cells in normal conditions (6, 8). Interestingly, a higher amount of toxin did not significantly increase the mortality, indicating that a threshold of intracellular toxin had been reached. When inactive ColM mutant proteins D226A, D229A, H235A, Y228A, and R236A were tested, we found that 2 μg/ml of these purified proteins was able to kill a significant part of the cell population (31.9–67% survivors), although this effect was much lower than that obtained with the wild-type enzyme. Two mutants T200A and T244A from the second category were tested, which were found to have a stronger effect as compared with the fully inactive mutants (∼19% survivors). Also in these cases, increasing the amount of protein did not elicit an increase of mortality. We then tested the effect of ColM-Δ1 (central and catalytic domains) and ColM-Δ7 (catalytic domain) derivatives (2 μg/ml); both were found to be active in vivo as very low numbers of survivors were obtained: 0.2% and 1.3%, respectively (Table 3). Importantly, the effects of these various ColM derivatives were quite similar when wild-type BW25113 cells instead of BW25113 ΔfhuA cells were used for the by-pass assay (data not shown).
TABLE 3.
In vivo activity analysis of ColM and its derivatives (mutant and truncated forms) under by-pass conditions
For these experiments, exponentially growing cells of the indicated strains were washed and then incubated in a high osmolarity solution (48% sucrose) before being suddenly diluted in M9-salts solution (1/50, v/v) in the presence or not of 1 μg of ColM or its derivatives. After 30-min incubation in the cold, the number of surviving cells was determined by plating cell suspensions on 2YT-agar medium. Values represent the mean ± S.D. of at least triplicate experiments.
| BW25113 ΔfhuA (×106) | BW25113 ΔfkpA (×106) | BW25113 + pMLD232a (×106) | |
|---|---|---|---|
| Control (buffer) | 57.0 ± 20.0 (100%) | 21.0 ± 4.6 (100%) | 36.0 ± 9.4 (100%) |
| Full-length ColM | 2.2 ± 1.0 (3.9%) | 15.8 ± 3.5 (75.5%) | 26.7 ± 14.6 (74.3%) |
| ColM-Δ7 (catalytic domain) | 0.7 ± 0.3 (1.3%) | 0.35 ± 0.18 (1.7%) | 0.85 ± 0.85 (2.37%) |
| ColM-Δ1 (central + catalytic domains) | 0.12 ± 0.07 (0.2%) | 3.6 ± 0.7 (17.0%) | 10.6 ± 3.3 (29.5%) |
| ColM D226A | 29.9 ± 2.0 (52.5%) | NDb | ND |
| ColM D229A | 38.2 ± 8.9 (67.0%) | ND | ND |
| ColM H235A | 18.2 ± 2.4 (31.9%) | ND | ND |
| ColM Y228A | 19.7 ± 7.6 (34.5%) | ND | ND |
| ColM R236A | 26.8 ± 5.3 (47.0%) | ND | ND |
| ColM T200A | 11.2 ± 6.0 (19.7%) | ND | ND |
| ColM T244A | 10.6 ± 5.0 (18.7%) | ND | ND |
a The plasmid pMLD232 allows expression of the ColM immunity protein (Cmi). BW25113 cells bearing this plasmid become fully resistant to ColM under the standard in vivo assay.
b ND, not determined.
As expected, BW25113 cells expressing the wild-type Cmi protein (plasmid pMLD232, see “Experimental Procedures”) appeared as fully resistant to native ColM and all its derivatives in the standard in vivo assay (data not shown). We then addressed whether the Cmi immunity protein could still protect cells against by-passed molecules of the various ColM derivatives. These cells were not significantly affected by the full-length ColM protein under by-pass assay conditions (up to 170 μg of protein/ml was tested). This finding demonstrated that the toxin did not need to go through the natural entrance to be inactivated by Cmi, and thus that Cmi presumably did not proceed through an inhibition of ColM uptake. In contrast, BW25113(pMLD232) cells appeared as susceptible as BW25113 cells that do not express the Cmi protein to the by-passed catalytic domain ColM-Δ7 (Table 3). The ColM-Δ1 derivative was found to be partially inhibited by the Cmi protein (29.5% survivors), and higher amounts of this toxin did not greatly increase the mortality level. These data indicated that the ColM-Δ1 variant (27.2 kDa), which comprises both the central and catalytic domains, could still be inhibited by Cmi, although less efficiently than the full-length protein, whereas ColM-Δ7 (17.2 kDa), which only carries the catalytic domain, somehow escaped recognition by the immunity protein.
Because the whole translocation machinery can be by-passed by osmotic shock, we then wondered whether the FkpA chaperone protein, for which the exact role in the ColM pathway remains to be elucidated (10), could also be dispensable in these conditions. The effects of the ColM derivatives were thus tested toward the ColM-resistant strain BW25113 ΔfkpA. Full-length ColM protein did not display a significant activity against these cells under by-pass conditions. This result demonstrated that the FkpA chaperone was still required for expression of the ColM activity even though the latter toxin did get access to the periplasm without the need of its dedicated uptake machinery. This clearly indicated that the action of FkpA and ColM uptake were not coupled events, a finding that disagreed with the assumption that FkpA was required for the refolding of ColM molecules, which had been partially unfolded during import through the FhuA channel (10). Interestingly, BW25113 ΔfkpA cells remained fully susceptible toward the by-passed ColM-Δ7 derivative (1.7% survivors) and were comparatively less susceptible toward the by-passed ColM-Δ1 derivative (11.1% survivors). This demonstrated that the catalytic domain per se did not absolutely require FkpA to exert its toxic effects in vivo. It also suggested that larger ColM derivatives comprising the central domain needed FkpA to be fully active in vivo.
DISCUSSION
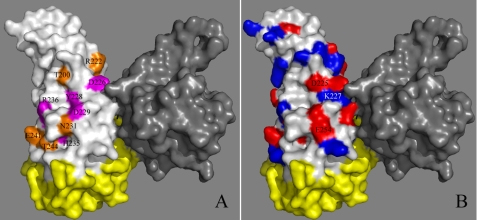
In the present work, residues essential for the ColM activity that were identified are likely involved in the binding and/or the catalytic process. These residues, namely Asp-226, Tyr-228, Asp-229, His-235, and Arg-236, were all conserved in the confirmed ColM-like toxins from Pseudomonas species, whereas Tyr-228 was replaced by a phenylalanine residue and His-235 was missing in the putative toxins from Burkholderia species (Fig. 2). They formed a linear patch at the surface of ColM, distributed within and apart of the small β6 sheet (17) (Figs. 3 and 4). The side chains of the two aspartate residues as well as those of the arginine and tyrosine residues were surface-accessible, whereas that of the histidine residue was rather embedded. These side chains were positioned relatively far from each other and did not lie within hydrogen bonding distances. Therefore, these residues constituted an atypical, particularly elongated active site, which might be relevant when considering the large size of the lipid II substrate. In addition to these five critical residues, five other conserved residues (Thr-200, Arg-222, Asn-231, Glu-241, and Thr-244) were identified for which mutation significantly reduced the toxin activity, although to a much lower extent. Interestingly, these residues were also mostly surface-accessible and surrounded the central patch formed by the first group of essential residues (Fig. 4). These residues might thus participate in the formation of the active site as well. Among the essential residues, the well exposed aspartate residues could be involved in the coordination of a Mg2+ ion, which together with the arginine residue could then bind the negatively charged pyrophosphate group.
FIGURE 4.
Surface representation of the ColM structure. Because ColM was crystallized as a dimer, one protomer is indicated in gray, the other protomer is illustrated as follows: the N-terminal and central domains (residues 1–123) are shown in yellow, and the toxicity domain is indicated in white. A, the data of the mutational analysis were mapped onto the structure of ColM. The amino acids whose mutation resulted in a total loss of cytotoxicity are in pink, and the amino acids with a less substantial effect in orange. B, ColM is in the same orientation as in panel A except the surface of the toxicity domain is colored on the basis of electrostatic potential, with negative charges in red and positive charges in blue.
Because little is known about the mechanism of ColM, it is thus quite difficult to assign specific functions to the essential residues we have identified. Some speculations could be made, however, based on analogies with known phosphatases. Most dephosphorylation reactions occur as a two-step mechanism, yielding a transition state phosphoenzyme intermediate. In this case, a nucleophilic attack of the phosphoester bond by a catalytic residue would lead to the formation of an unstable ColM-pyrophospho-sugar-peptide covalent intermediate and the release of undecaprenol. This step would be followed by a second nucleophilic attack by an activated water molecule, releasing the pyrophospho-sugar-peptide product. The aspartate and histidine residues are potential candidates for the nucleophilic substitution. By analogy, the acid phosphatases proceed through a phosphohistidine intermediate (34), whereas the sarcoplasmic reticulum ATPases involve a carboxyl-phosphate intermediate. Catalytic mechanisms involving a serine (alkaline phosphatase) or a cysteine protein-tyrosine phosphatases as the nucleophilic amino acid could be excluded, because none of the serine residues were found to be essential for ColM activity, and there is no cysteine present in the catalytic domain. Such a catalytic mechanism could also require a proton donor to give a proton to the isoprenol leaving group; here again a histidine residue could likely be involved. Alternatively, the catalysis could occur as a one-step mechanism. In this case, the substrate would be positioned in the active site so that a direct nucleophilic attack by a water molecule could occur. Ser/Thr phosphatases are metalloenzymes with bimetallic centers, which typically use such a single displacement mechanism (35).
Based on structural and biochemical data, Zeth et al. tentatively divided ColM into three functional domains (17). Such an assignment was difficult, because no domain was really structurally independent. The N-terminal, translocation domain was thought to contain the first 35 residues corresponding to an unstructured and flexible random coil domain wrapping around the central domain. The receptor-binding domain was delineated from residue 36 to 140, and the C-terminal activity domain was defined from residue 141 to 271. In the present study, we showed that an active and independent catalytic domain can be produced, which included at least residues 122–271. Interestingly, this domain encompassed exactly the region that is conserved in ColM-like toxins from Pseudomonas species (18); therefore, we considered this region as constitutive of the catalytic domain. In our model, the helix α6 was attributed to the catalytic domain, whereas Zeth et al. considered it as constitutive of the central domain (17). Of course, we cannot exclude the possibility that this helix may also contribute to the folding of the central domain. The interface between the central and C-terminal domains is formed by two small β-sheets (β1 and β2) connected by hydrogen bonds together with a hydrophobic core formed by four long helices (α2, α3, α6, and α9), each domain contributing to two helices (Fig. 5). Interestingly, when the α2 and α3 helices were removed to delineate the catalytic domain, the resulting truncated constructs were found to aggregate, being consistent with the solvent exposure of the hydrophobic interface. That these derivatives were recovered soluble and stable after renaturation likely indicated that the hydrophobic interface may not be exposed anymore. Interestingly, the isolated catalytic domain was found to exhibit a much greater (45-fold) in vitro activity toward lipid II than the full-length protein. This activity enhancement also suggested that a conformational change between the full-length protein and the catalytic domain (ColM-Δ7 derivative) had occurred, the latter being in an “active” conformation. These data may be relevant to the mechanism of action of ColM. The toxin may be synthesized in the cytoplasm of producer cells in an “inactive” form, which would explain why cleavage of lipid I and II does not occur in the cytoplasm. After secretion, the toxin would acquire its active conformation only once it has penetrated the periplasm of targeted cells. This change of conformation may involve the import machinery and/or the FkpA chaperone, which to date are the only known components from the targeted cells required for ColM activity. Here we showed that the FkpA chaperone cannot be by-passed when using the native ColM (Fig. 1). This result strongly suggested that the native toxin was produced in an “inactive” conformation, which requires FkpA to become active. In contrast, FkpA was dispensable for ColM-Δ7 derivative activity in vivo (Fig. 1), suggesting that this truncated ColM variant is already in the active conformation, which is correlated with an increase of specific activity in vitro. Therefore, the partition of the central domain from the activity domain would likely mimic what happens in vivo. FkpA comprises two independent chaperone (N-terminal) and cis/trans-peptidyl-prolyl isomerase (C-terminal) domains (36). The analysis of ColM-resistant FkpA mutants showed that the domain displaying the cis/trans-isomerase activity was required for expression of ColM activity (10, 29). Indeed, three mutants out of four carried a transposon insertion in the C-terminal domain and in vitro renaturation of ColM by FkpA was shown to be inhibited by the immunosuppressant ligand FK506 that specifically binds to the same region (10). Interestingly, ColM is particularly rich in proline residues, especially in the N-terminal and central domains. In the three-dimensional structure of native ColM, only one proline residue (Pro-107) was in a cis configuration, and this residue was localized at the surface of the protein. Because ColM has to encounter FkpA to be active, we can reasonably presume that Pro-107 is targeted and that a cis to trans isomerization of the corresponding peptidyl-prolyl bond then provides a potential mechanism switching ColM from an inactive toward an active conformation. Interestingly, this proline residue is located between the two chymotrypsin cleavage sites and in proximity to the starting point of the activity domain. Unfortunately, preincubation of native ColM with purified FkpA did not improve its toxicity toward the ΔfkpA strain under the by-pass conditions (data not shown), that would have supported our assumption. Nevertheless, according to the cis/trans isomerization hypothesis, the fact that purified preparations of native ColM exhibit an in vitro activity (although lower than that of the active domain) could reflect the coexistence of both cis (inactive) and trans (active) conformations in these preparations, with the cis conformation being in a large excess. In that case, a low proportion of trans molecules would have been generated spontaneously or following contact with FkpA molecules (during ColM overexpression and/or preparation of crude cell extracts). To further analyze this point, the ColM protein was recently expressed in and purified from the BW25113 ΔfkpA strain. The in vitro enzymatic activity of this preparation equaled that purified from the wild-type strain BW25113 (data not shown), demonstrating that the presence of FkpA did not influence the intrinsic activity of ColM molecules produced in E. coli cells. We can alternatively envisage that ColM molecules are all in a cis conformation, which exhibits intrinsically low activity.
FIGURE 5.
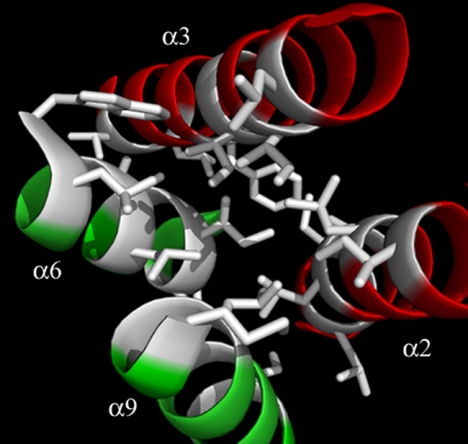
Representation of the interface between the central and toxicity domains of ColM. The ribbon diagram of the α-helices from the central domain (α2 and α3, in red) and from the toxicity domain (α6 and α9, in green) is shown. This four-helix bundle forms a hydrophobic core constitutive of the interface between the two domains. The side chains of hydrophobic residues are shown as sticks.
The recent identification of the mode of action of ColM and characterization of several homologues from Pseudomonas species exhibiting a similar activity of degradation of the peptidoglycan intermediates has opened the way to detailed investigations of the catalytic mechanism and antibacterial spectrum of action of the different members of this new family of bacteriocins. Because the lipid I and II precursors targeted by these enzymes are essential for growth and conserved in bacterial world, an exploitation of these bacteriocins as potential antibacterial agents could thus be envisaged. The present demonstration that the isolated catalytic domain of ColM exhibits a higher activity as compared with the full-length protein and could potentially by-pass the requirements for the outer-membrane receptor, translocation, and maturation steps of the latter form is interesting in this respect. Given that the structure of peptidoglycan and of its precursors somewhat varies in bacterial world, in particular in the composition and structure of the peptide moiety (37), further investigations of the substrate specificity of the ColM homologues and of the in vivo activity of full-length and isolated catalytic domains on a wide range of Gram-negative and Gram-positive bacterial species are needed.
Supplementary Material
Acknowledgments
We thank Philippe Bouloc for providing E. coli strains from the Keio collection and Michel Arthur for helpful discussion.
This work was supported by grants from the Agence Nationale de la Recherche (PEPGLYCOL project, ANR-07-MIME-020) and the CNRS (UMR 8619).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5 and Table S1.
- ColM
- colicin M
- MurNAc
- N-acetylmuramic acid
- C55-P
- undecaprenyl phosphate
- Ni2+-NTA
- nickel-nitrilotriacetic acid.
REFERENCES
- 1.Pugsley A. P. (1984) Microbiol. Sci. 1, 203–205 [PubMed] [Google Scholar]
- 2.Pugsley A. P. (1984) Microbiol. Sci. 1, 168–175 [PubMed] [Google Scholar]
- 3.Cascales E., Buchanan S. K., Duché D., Kleanthous C., Lloubès R., Postle K., Riley M., Slatin S., Cavard D. (2007) Microbiol. Mol. Biol. Rev. 71, 158–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harkness R. E., Braun V. (1989) J. Biol. Chem. 264, 6177–6182 [PubMed] [Google Scholar]
- 5.Schaller K., Höltje J. V., Braun V. (1982) J. Bacteriol. 152, 994–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El Ghachi M., Bouhss A., Barreteau H., Touzé T., Auger G., Blanot D., Mengin-Lecreulx D. (2006) J. Biol. Chem. 281, 22761–22772 [DOI] [PubMed] [Google Scholar]
- 7.Braun V., Schaller K., Wolff H. (1973) Biochim. Biophys. Acta 323, 87–97 [DOI] [PubMed] [Google Scholar]
- 8.Pilsl H., Glaser C., Gross P., Killmann H., Olschläger T., Braun V. (1993) Mol. Gen. Genet. 240, 103–112 [DOI] [PubMed] [Google Scholar]
- 9.Kock J., Olschlager T., Kamp R. M., Braun V. (1987) J. Bacteriol. 169, 3358–3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hullmann J., Patzer S. I., Römer C., Hantke K., Braun V. (2008) Mol. Microbiol. 69, 926–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ölschläger T., Turba A., Braun V. (1991) Mol. Microbiol. 5, 1105–1111 [DOI] [PubMed] [Google Scholar]
- 12.Olschläger T., Braun V. (1987) J. Bacteriol. 169, 4765–4769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braun V., Pilsl H., Gross P. (1994) Arch. Microbiol. 161, 199–206 [DOI] [PubMed] [Google Scholar]
- 14.Chavan M., Rafi H., Wertz J., Goldstone C., Riley M. A. (2005) J. Mol. Evol. 60, 546–556 [DOI] [PubMed] [Google Scholar]
- 15.Kageyama M., Kobayashi M., Sano Y., Masaki H. (1996) J. Bacteriol. 178, 103–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jakes K. S., Davis N. G., Zinder N. D. (1988) J. Bacteriol. 170, 4231–4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeth K., Römer C., Patzer S. I., Braun V. (2008) J. Biol. Chem. 283, 25324–25331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barreteau H., Bouhss A., Fourgeaud M., Mainardi J. L., Touzé T., Gérard F., Blanot D., Arthur M., Mengin-Lecreulx D. (2009) J. Bacteriol. 191, 3657–3664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kulasekara B. R., Kulasekara H. D., Wolfgang M. C., Stevens L., Frank D. W., Lory S. (2006) J. Bacteriol. 188, 4037–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mavrodi D. V., Mavrodi O. V., McSpadden-Gardener B. B., Landa B. B., Weller D. M., Thomashow L. S. (2002) Appl. Environ. Microbiol. 68, 5170–5176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., Mori H. (2006) Mol. Syst. Biol. 2, 2006.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olschläger T., Schramm E., Braun V. (1984) Mol. Gen. Genet. 196, 482–487 [DOI] [PubMed] [Google Scholar]
- 23.Miller J. H. (1972) Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, pp. 431–435 [Google Scholar]
- 24.Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 25.Dagert M., Ehrlich S. D. (1979) Gene 6, 23–28 [DOI] [PubMed] [Google Scholar]
- 26.Laemmli U. K., Favre M. (1973) J. Mol. Biol. 80, 575–599 [DOI] [PubMed] [Google Scholar]
- 27.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 28.Press W. H., Flannery B. P., Teukolsky S. A., Vetterling W. T. (1986) Numerical Recipes: The Art of Scientific Computing, pp. 521–528, Cambridge University Press, Cambridge, UK [Google Scholar]
- 29.Braun V., Frenz J., Hantke K., Schaller K. (1980) J. Bacteriol. 142, 162–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimizu K., Sekiguchi M. (1979) Mol. Gen. Genet. 168, 37–47 [DOI] [PubMed] [Google Scholar]
- 31.Fehrst A. (1977) Enzyme Structure and Mechanism, W. H. Freeman and Company, San Francisco, CA [Google Scholar]
- 32.Johnson K. A., Benkovic S. J. (1990) The Enzymes, 3rd Ed. (Sigman D. S., Boyer P. D. eds) vol. XIX, pp. 159–211, Academic Press, New York [Google Scholar]
- 33.Eick-Helmerich K., Braun V. (1989) J. Bacteriol. 171, 5117–5126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rigden D. J. (2008) Biochem. J. 409, 333–348 [DOI] [PubMed] [Google Scholar]
- 35.Barford D., Das A. K., Egloff M. P. (1998) Annu. Rev. Biophys. Biomol. Struct. 27, 133–164 [DOI] [PubMed] [Google Scholar]
- 36.Saul F. A., Arié J. P., Vulliez-le Normand B., Kahn R., Betton J. M., Bentley G. A. (2004) J. Mol. Biol. 335, 595–608 [DOI] [PubMed] [Google Scholar]
- 37.Vollmer W., Blanot D., de Pedro M. A. (2008) FEMS Microbiol. Rev. 32, 149–167 [DOI] [PubMed] [Google Scholar]
- 38.Bouhss A., Trunkfield A. E., Bugg T. D., Mengin-Lecreulx D. (2008) FEMS Microbiol. Rev. 32, 208–233 [DOI] [PubMed] [Google Scholar]
- 39.van Dam V., Sijbrandi R., Kol M., Swiezewska E., de Kruijff B., Breukink E. (2007) Mol. Microbiol. 64, 1105–1114 [DOI] [PubMed] [Google Scholar]
- 40.Sauvage E., Kerff F., Terrak M., Ayala J. A., Charlier P. (2008) FEMS Microbiol. Rev. 32, 234–258 [DOI] [PubMed] [Google Scholar]
- 41.Touzé T., Mengin-Lecreulx D. (February12, 2008, posting date) Undecaprenyl Phosphate Synthesis. EcoSal-Escherichia coli and Salmonella: Cellular and Molecular Biology (Böck A., Curtiss R., III, Kaper J. B., Karp P. D., Neidhardt F. C., Nyström T., Slauch J. M., Squires C. L. eds) Chapter 4.7.1.7, ASM Press, Washington, D. C. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.