Abstract
Pili are surface-exposed virulence factors involved in bacterial adhesion to host cells. The Streptococcus pneumoniae pilus is composed of three structural proteins, RrgA, RrgB, and RrgC and three transpeptidase enzymes, sortases SrtC-1, SrtC-2, and SrtC-3. To gain insights into the mechanism of pilus formation we have exploited biochemical approaches using recombinant proteins expressed in Escherichia coli. Using site-directed mutagenesis, mass spectrometry, limited proteolysis, and thermal stability measurements, we have identified isopeptide bonds in RrgB and RrgC and demonstrate their role in protein stabilization. Co-expression in E. coli of RrgB together with RrgC and SrtC-1 leads to the formation of a covalent RrgB-RrgC complex. Inactivation of SrtC-1 by mutation of the active site cysteine impairs RrgB-RrgC complex formation, indicating that the association between RrgB and RrgC is specifically catalyzed by SrtC-1. Mass spectrometry analyses performed on purified samples of the RrgB-RrgC complex show that the complex has 1:1 stoichiometry. The deletion of the IPQTG RrgB sorting signal, but not the corresponding sequence in RrgC, abolishes complex formation, indicating that SrtC-1 recognizes exclusively the sorting motif of RrgB. Finally, we show that the intramolecular bonds that stabilize RrgB may play a role in its efficient recognition by SrtC-1. The development of a methodology to generate covalent pilin complexes in vitro, facilitating the study of sortase specificity and the importance of isopeptide bond formation for pilus biogenesis, provide key information toward the understanding of this complex macromolecular process.
Keywords: Cell/Wall/Bacteria, Methods/Mass Spectrometry, Methods/Site-directed Mutagenesis, Protein/Assembly, Protein/Self-Assembly, Pilus, Pneumococcus, Sortase
Introduction
Attachment of pathogenic bacteria to host cells is mediated by bacterial surface-exposed adhesins which, in some instances, are located at the tip of a filamentous surface appendages known as pili. Pili of the Gram-negative bacteria have been extensively described (1). Despite the diversity in their assembly pathways, they share a common structural feature involving the noncovalent association of pilin subunits. Even though a first description of Gram-positive pili was published in 1968 (2), the first studies regarding their assembly mechanism were published more recently for the Corynebacteria diphtheriae pilus (3). Since then, pili have been identified in many other Gram-positive bacteria, including Actinomyces, Enterococcus, Bacillus, and several Streptococcal species, S. pyogenes, S. agalactiae, and S. pneumoniae (4–10). In contrast to Gram-negative bacteria, both major pilin polymerization and minor pilin association to the Gram-positive pilus backbone occur through the formation of covalent bonds catalyzed by specialized transpeptidases. These membrane-associated sortase enzymes recognize a LPXTG-like sequence (sorting motif) within their protein substrate (11, 12). The enzymatic reaction involves a nucleophilic cysteine residue, which is essential for the covalent intermolecular association between pilus subunits (13–15). In addition, intramolecular bonds within pilin subunits, formed between lysine and asparagine side chains, have been identified in the high resolution structures of major pilins Spy0128 from S. pyogenes (16) and SpaA from C. diphtheriae (17) as well as in the main adhesin of the Streptococcus pneumoniae pilus, RrgA (18). The role of these unusual intramolecular cross-links in pilin subunit stabilization has been shown by site-specific mutagenesis and both proteolytic and thermal stability studies (19, 20). Notably, Lys-Asn intramolecular bonds were also identified in the Cna collagen-binding domains A and B (CnaA and CnaB, respectively) of Staphylococcus aureus (20–22) as well as the N2CnaB domain of the S. agalactiae minor pilin subunit GBS52 (23) and the CnaA-similar ACE protein from Enterococcus faecalis (24). The CnaB domains are repetitive regions whose fold is reminiscent of the IgG fold (22).
S. pneumoniae is an important human pathogen causing a large number of respiratory tract infections, such as pneumonia and sinusitis, but also invasive diseases like septicemia and meningitis. Among the surface-exposed virulence factors, the pneumococcal pilus has been recently shown to play a role in host cell adhesion (10). Pilus formation requires the expression of seven genes encoded by the rlrA pathogenicity islet, including a RofA-like transcriptional regulator (RlrA), three sortases (SrtC-1, SrtC-2, and SrtC-3), and three structural proteins (RrgA, RrgB, and RrgC) (10). Electron microscopy observations of immunogold-labeled pneumococcus allowed investigations on the structural composition of the pilus and showed that RrgB is the major pilin subunit forming the elongated fiber shaft, but data concerning the precise localization of the minor RrgA and RrgC pilin subunits remained controversial (10, 25–27). Recently, an elegant structural analysis of the native pneumococcal pilus using a combination of electron microscopy techniques provided evidence that the RrgB-shaft is composed of monomeric covalently linked subunits oriented head-to-tail (28). Furthermore, RrgA and RrgC do not localize together in the pili, but are present at the distal and the proximal end, respectively, in accordance with the host adhesion function of RrgA and the putative cell wall anchoring role of RrgC (28).
Investigation of the roles and relative contributions of the three sortases to the pilus formation process by genetic mutational approaches revealed that although deletion of individual sortases does not compromise RrgB polymerization, indicating a redundancy in sortase functionality, deletion of all three sortases abolishes RrgB fiber formation (27, 29). The role played by the three different sortases in incorporation of the minor pilin subunits onto the fiber remains largely unresolved: both SrtC-1 and SrtC-2 have been suggested as playing a role in incorporation of RrgA, and both SrtC-1 and SrtC-3 have been proposed as participating in association of RrgC to the RrgB backbone (27, 29).
Here, we provide insights into the molecular mechanims of S. pneumoniae pilus biogenesis and of the specificity of cognate sortases by combining biochemical and biophysical approaches. In this work, we identify the intramolecular bonds present in RrgB and RrgC as well as their stabilization properties. We address the question of sortase specificity by employing a recombinant protein expression platform in which pilins and sortases are co-expressed. We show that co-expression of RrgB, together with RrgC and SrtC-1, leads to the formation of a covalent RrgB-RrgC complex whose association is abrogated when SrtC-1 carries a mutation in the nucleophilic cysteine. The deletion of the IPQTG sequence of RrgB, but not of the corresponding sequence in RrgC, impairs complex formation, indicating that SrtC-1 recognizes the sorting motif of RrgB prior to the final transpeptidation reaction with an amine group of RrgC. Finally, we provide evidence that the intramolecular bonds stabilizing RrgB may play a role in its recognition by SrtC-1 and subsequent association to RrgC.
EXPERIMENTAL PROCEDURES
Cloning Procedures
Sequences corresponding to coding regions of RrgB (30–633) and RrgC (22–368), lacking signal peptides, transmembrane anchors, and C-terminal tails, were amplified and cloned into a pLIM vector using the LIC method (P'X Therapeutics, Grenoble). Primers (listed in supplemental Table S1) and S. pneumoniae TIGR4 chromosomal DNA were used for PCR amplification. DNA fragments were also cloned into pETDuet and pACYCDuet bi-cistronic plasmids (Novagen), leading to constructs expressing combinations of proteins carrying histidine tags, S-tags, or devoid of tags. Resulting plasmids were then singly transformed or co-transformed into Escherichia coli STAR (Invitrogen) to obtain strains expressing all protein combinations (supplemental Table S2).
Site-directed Mutagenesis
Mutations were introduced by PCR-based site-directed mutagenesis (Stratagene QuikChange® II XL site-directed mutagenesis kit). Mutant plasmids were verified by DNA sequencing (Cogenics, Grenoble) and transformed into E. coli STAR for protein expression.
Expression and Purification of RrgB, RrgC, and Mutants
For both RrgB and RrgC, protein expression was induced in Terrific Broth with 0.5 mm IPTG2 at 37 °C during 3 h. Cells were harvested by centrifugation and lysed by sonication in 50 mm HEPES, pH 7.5, 200 mm NaCl, 20 mm imidazole (buffer A), and a protease inhibitor mixture (Complete EDTA-free, Roche). The lysates were clarified by centrifugation and applied onto HisTrapTM HP columns (Amersham Biosciences) pre-equilibrated in buffer A. Protein elution was performed using linear gradients of 20–500 mm imidazole over 20 column volumes. Eluted proteins were dialyzed and purified by gel filtration chromatography. These proteins were used to immunize mice to produce polyclonal antibodies.
Co-expression of Pilin Subunits, Sortase, and Complex Purification
Bacterial cultures of Duet vectors transformed into E. coli STAR cells were performed, and protein expression was induced in 4 ml of Terrific Broth with 0.5 mm IPTG at 37 °C during 3 h. Cells were harvested by centrifugation and chemically lysed for 20 min in 1 ml of BugBuster® 1× (Novagen) diluted in buffer A and supplemented with 2 μl of r-lyzozyme (Novagen) and 0.8 μl benzonase nuclease (Novagen). The lysates were clarified by centrifugation and applied onto His MultiTrapTM HP (GE Healthcare) pre-equilibrated in buffer A. Protein were eluted in 50 mm HEPES, pH 7.5, 200 mm NaCl, 500 mm imidazole.
Purification of the RrgB-RrgC Complex
E. coli strain co-transformed with pETDuet-RrgB and pACYCDuet-RrgC-SrtC-1 vectors was cultured in 2 liters of Terrific Broth, and protein expression was induced with 0.5 mm IPTG at 37 °C during 3 h. Cells were harvested by centrifugation and lysed by sonication in buffer A containing protease inhibitor mixture (Complete EDTA-free, Roche). The lysates were clarified by centrifugation and applied onto a NiNTA resin column (Qiagen) pre-equilibrated in buffer A. Protein elution was performed using imidazole steps of 60, 100, and 300 mm. Fractions containing the RrgB-RrgC complex were pooled, dialyzed against 50 mm MES pH 6.0, 50 mm NaCl, and loaded onto an ion-exchange RQ-1 ml column (GE Healthcare) before elution by a linear gradient of NaCl (50–250 mm). A gel filtration chromatography S200 (GE Healthcare) equilibrated in 50 mm MES pH 6.0, 150 mm NaCl was the final purification step. The purification yield of the RrgB-RrgC complex was ∼500 μg per liter of culture.
Western Blotting
Samples were loaded onto 4–12% Criterion XT Precast gels (Bio-Rad) and were run and subsequently electrotransferred in Trans-Blot Transfer Medium (Bio-Rad) for 30 min at 100 V. Incubation times of 1 h were successively performed using the appropriate antibody (polyclonal mouse, diluted 1:5 000) and anti-mouse horseradish peroxidase conjugate (Sigma) was diluted 1:120,000 before detection with a chemiluminescent substrate (Pierce).
Thermal Shift Assay
TSA experiments were carried out using an IQ5 96-well format real-time PCR instrument (Bio-Rad). Briefly, 25 ng of recombinant RrgC or RrgB proteins, native or mutant, were mixed with 2 μl of 5000× Sypro Orange (Molecular Probes) previously diluted 1:100 in water. Samples were heat-denatured from 20 to 95 °C at a rate of 1 °C per minute. Protein thermal unfolding curves were monitored by detection of changes in fluorescence of the SyproOrange. The inflection point of the fluorescence versus temperature curves was identified by plotting the first derivative over the temperature and the minima were referred to as the melting temperatures (Tm). The fluorescence of buffers and salt solutions were checked as controls.
Mass Spectrometry Analyses
Accurate molecular masses of proteins were determined by ESI-TOF mass spectrometry performed on a LC/MSD-TOF mass spectrometry system (Applied Biosystems) in 95% acetonitrile, 5% water, 0.03% formic acid. 3 μl of each sample were diluted in 97 μl of formic acid 0.1% and 100 μl of each dilution were desalted on line. LC/MS-MS experiments were carried out on the RrgB-RrgC complex. The protein complex band was excised from the gel and subsequently washed with 50 mm acetonitrile/ammonium bicarbonate 50/50 (v/v) for 30 min, before dehydratation with acetonitrile. Subsequently, the sample was treated with 7% H2O2. After drying was complete, the band was rehydrated in 15 μl of digestion buffer (50 mm ammonium bicarbonate, pH 8.1) containing 150 ng of trypsin and incubated at 4 °C for 15 min. 30 μl of digestion buffer were then added, and the digestion reaction was carried out at 37 °C overnight. Peptides were extracted from the gel by diffusion for 15 min with agitation, followed by three sequential 5 min sonication steps in 50% acetonitrile, 5% formic acid, and 100% acetonitrile. Digestion and extraction solutions were pooled and dried under vacuum. Peptide mixtures were re-dissolved in 25 μl of water/acetonitrile 95/5 (v/v) containing 0.2% trifluoroacetic acid prior to LCMS/MS analysis. All experiments were performed in a 96-well system coordinated by an EVO15 (Tecan) robot.
Proteolysis Assay and N-terminal Sequencing
RrgC, RrgB, and mutants were diluted in a buffer composed of 50 mm HEPES pH 7.5 and 200 mm NaCl. Subsequently, 20 μg of each Rrg protein was mixed with 50 ng of trypsin. The reaction was carried out at room temperature for 16 h. The samples were analyzed by SDS-PAGE. The proteins were either stained with a Coomassie Blue solution or transferred to a Problot membrane (Applied Biosystems) and stained according to supplier's instructions. The protein bands were cut from the membrane and sequenced using automated Edman degradation on an Applied Biosystem gas-phase sequencer model 477A with on-line analysis of the phenyl thiohydantoin derivatives.
RESULTS
Identification of the Residues Involved in the Formation of Two Intramolecular Bonds in the RrgC Minor Pilin
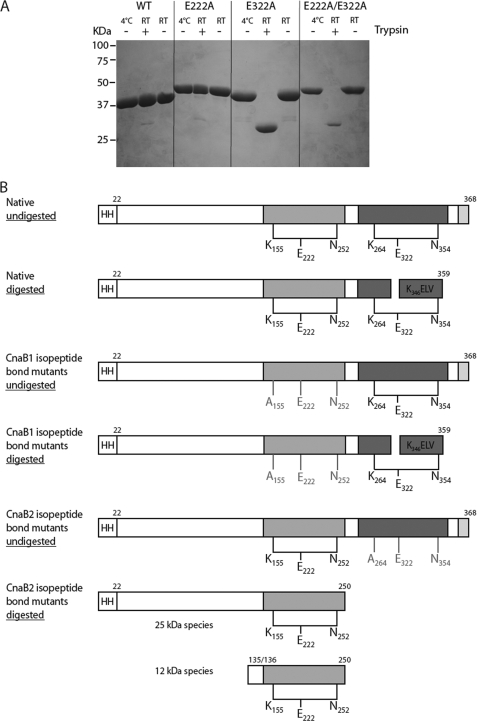
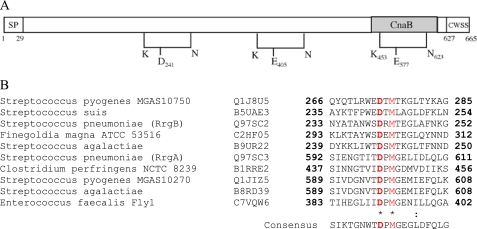
Pfam analysis of the RrgC amino acid sequence predicted the presence of a signal peptide, the cell wall sorting signal (CWSS) and two CnaB domains, CnaB1 and CnaB2 (Fig. 1A). The crystal structure of the pneumococcal pilin adhesin RrgA reveals the presence of two intramolecular Lys-Asn cross-links in an IgG-fold domain and in a CnaB domain (18). To test for the presence of such bonds in RrgC, we first searched its sequence for the presence of the putative conserved residues involved in isopeptide bond formation through multiple sequence alignments (Fig. 1B). This analysis clearly highlights a pattern of highly conserved lysine and glutamate residues which, in the crystal structures of GBS52 (23) and RrgA (18), are involved in the formation of the isopeptide bonds. The lysine and glutamate residues are included in KVD and GXYXLXEXAPXGY (E-box) motifs, respectively, while the asparagine residue is located ∼25–30 residues downstream from the glutamate. We thus predicted that the intramolecular isopeptide bond in the CnaB1 domain of RrgC could be formed by Lys-155, Glu-222, and Asn-252, while the CnaB2 domain could harbor a bond formed by Lys-264, Glu-322, and Asn-354 (Fig. 1B).
FIGURE 1.
Intramolecular bonds in RrgC. A, predicted topology of RrgC. SP, signal peptide; the CWSS comprises a LPXTG-like motif followed by a hydrophobic membrane-spanning domain and a cytoplasmic positively charged tail. B, sequence alignments of pilin subunits from S. pneumoniae (RrgA, RrgC, and RrgB), S. agalactiae (GBS52 and GBS104), B. cereus (BcpB), E. faecalis (EpbB and EpbC), and C. diphteriae (SpaG). The CnaB domain is highlighted in gray; lysine, asparagine, and glutamate residues involved in the intramolecular bond are shown in red, and the consensus motifs including the Lys and the Glu residues are indicated in blue.
A His-tagged recombinant form of RrgC (residues 22–368) was produced and mass spectrometry confirmed the presence of two isopeptide bonds in the native form (Table 1). The observed molecular mass (40,948 Da) was consistent with the loss of two NH3 units (when compared with the calculated mass, 40,983 Da). Each bond-forming residue was mutated into alanine; single mutants as well as the double E222A/E322A mutant were produced and purified as the native molecule. Mass spectrometry measurements confirmed the loss of one NH3 unit in each single mutant and the loss of two NH3 units in the double mutant, supporting the role of Lys-155, Glu-222, Asn-252, Lys-264, Glu-322, and Asn-354 in the formation of the isopeptide bonds in RrgC.
TABLE 1.
ESI-TOF mass spectral analyses of the native and mutant proteins of RrgC
| RrgC | Maveragea |
Δ(Mexpected − Mobserved)b | NH3 units lost | |
|---|---|---|---|---|
| Expected | Observed by ESI-TOF | |||
| Da | Da | Da | ||
| Native | 40,983.07 | 40,948.19 | 34.88 | 2 |
| K155A | 40,925.98 | 40,907.71 | 18.27 | 1 |
| E222A | 40,925.04 | 40,906.89 | 18.15 | 1 |
| N252A | 40,940.05 | 40,921.88 | 18.17 | 1 |
| K264A | 40,925.98 | 40,907.79 | 18.19 | 1 |
| E322A | 40,925.04 | 40,907.09 | 17.95 | 1 |
| N354A | 40,940.05 | 40,921.83 | 18.22 | 1 |
| E222A/E322A | 40,867.00 | 40,867.49 | −0.49 | 0 |
a Average molecular mass.
b Difference between expected and observed molecular mass.
The Two Internal Covalent Bonds Stabilize RrgC
Both native and mutant RrgC forms were tested for resistance to proteolytic digestion (Fig. 2A). Neither the native nor the CnaB1-E222A mutant seemed to be affected by trypsin digestion while the CnaB2-E322A and the double E222A-E322A mutant proteins were degraded into a proteolytic product of ∼30kDa (Fig. 2A, lanes 8 and 11). It is interesting to note that in the absence of the protease, all three mutant proteins, lacking either one or two isopeptide bonds, migrated on SDS-PAGE slightly slower than the native protein, suggesting that the intramolecular bonds could aid in the formation of a more compact protein fold (Fig. 2A).
FIGURE 2.
Proteolysis stability of RrgC. A, trypsin digestion products were separated on 12.5% SDS-PAGE. The molecular weight markers are shown on the left side of the gel. Whether trypsin was added or not is noted on top of the gel as well as the reaction temperatures. B, summary of the trypsin digestion data.
Further characterization of the proteolytic products were carried out by mass spectrometry and N-terminal sequencing (Table 2 and Fig. 2B). In the native RrgC protein as well as in the three mutant variants which lack the CnaB1 isopeptide bond, an internal tryptic cleavage occurred in the CnaB2 domain at position 346KELVT without dissociation of the polypeptide chains (Fig. 2A), indicating that the CnaB2 intramolecular bond is capable of holding these two chains together (Fig. 2B). In the mutants lacking the CnaB2 isopeptide bond, proteolysis induces the total digestion of the CnaB2 domain (25-kDa species) and of the RrgC N-terminal region (12-kDa species) suggesting that the isopeptide bond is key for the stabilization of this domain but also influences the overall stability of RrgC (Fig. 2C).
TABLE 2.
Molecular masses and N-terminal sequencing of the trypsin digestion of wild type and mutant RrgC variants
The calculated masses take into account the presence of one or two isopeptide bonds, depending on the RrgC variant considered.
| Undigested |
Digested |
||||
|---|---|---|---|---|---|
| Calculated mass | Measured mass | N-terminal sequence | Measured mass | N-terminal sequence | |
| Native | 40,949.07 | 40,948.19 | GSSHHHH | 40,009.86 | GSSHHHH |
| KELVTVV | |||||
| K155A | 40,908.98 | 40,907.71 | GSSHHHH | 39,969.72 | GSSHHHH |
| KELVTVV | |||||
| E222A | 40,908.04 | 40,906.89 | GSSHHHH | 39,968.94 | GSSHHHH |
| KELVTVV | |||||
| VEPLAGY | |||||
| ELVTVVK | |||||
| N252A | 40,923.05 | 40,921.88 | GSSHHHH | 39,983.95 | GSSHHHH |
| KELVTVV | |||||
| K264A | 40,908.98 | 40,907.79 | GSSHHHH | 28,551.07 | GSSHHHH |
| 12,893.41 | NDa | ||||
| 12,765.19 | NDa | ||||
| E322A | 40,908.04 | 40,907.09 | GSSHHHH | 28,551.21 | GSSHHHH |
| 12,893.43 | NDa | ||||
| 12,765.23 | NDa | ||||
| N354A | 40,923.05 | 40,921.83 | GSSHHHH | 28,551.32 | GSSHHHH |
| 12,893.37 | NDa | ||||
| 12,765.24 | NDa | ||||
a ND, not determined.
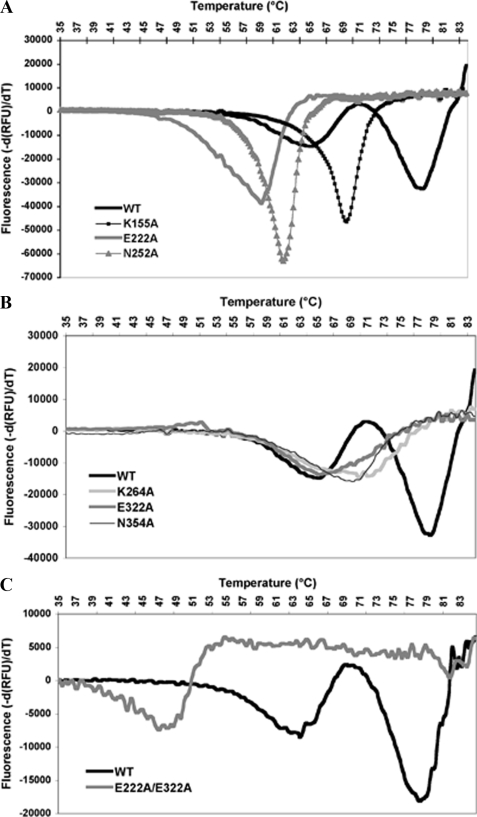
To further investigate the role played by the isopeptide bonds in RrgC stability, we performed thermal shift analyses of the different RrgC variants (Fig. 3). Native RrgC showed two distinct melting temperature (Tm) values of 63.7 °C (±0.6) and 77.7 °C (±0.5). The CnaB1 bond mutants, namely K155A, E222A, and N252A, each presented an unique Tm value of 68.3 °C (±0.6), 57.4 °C (±1.3), and 61 °C (±1), respectively (Fig. 3A). The proteins mutated in the CnaB2 bond, namely K264A, E322A, and N354A also showed unique Tm values of 66, 65, and 67 °C, respectively (Fig. 3B). The double mutant E222A-E322A, deleted in both isopeptide bonds showed a drastically diminished Tm value of 47.9 °C (±2.1) (Fig. 3C). Taken together, these data show that the isopeptide bonds influence the stability of RrgC.
FIGURE 3.
Thermal shift assay of RrgC and variants. The wild-type curve has been included in each figure to facilitate the comparison with the different RrgC variants. A, WT, and CnaB1 mutants. B, WT and CnaB2 mutants. C, WT and the double glutamate mutant.
Identification of Residues Involved in the Formation of Three Intramolecular Isopeptide Bonds in the Major RrgB Pilin
We investigated the presence of intramolecular bonds in RrgB by using the same approach as for RrgC. A His-tagged recombinant form of RrgB (residues 30–633) was produced and purified from E. coli cells. Mass spectrometry provided evidence of the presence of three isopeptide bonds since the molecular mass observed (67,400 Da) is consistent with the loss of three NH3 units (when compared with the expected mass, 67,453 Da) (Table 3).
TABLE 3.
ESI-TOF mass spectral analyses of the native and mutant proteins of RrgB
| RrgB | Maveragea |
Δ(Mexpected − Mobserved)b | NH3 units lost | |
|---|---|---|---|---|
| Expected | Observed by ESI-TOF | |||
| Da | Da | Da | ||
| Native | 67,453.11 | 67,400.51 | 52.60 | 3 |
| D241A | 67,409.10 | 67,375.51 | 33.59 | 2 |
| E405A | 67,395.07 | 67,361.38 | 33.69 | 2 |
| E577A | 67,395.07 | 67,361.84 | 33.23 | 2 |
| D241A/E405A/E577A | 67,293.03 | 67,293.78 | −0.75 | 0 |
aAverage molecular mass.
b Difference between expected and observed molecular mass.
In contrast to RrgC, the topology of RrgB presents a single predicted CnaB domain at the C terminus (Fig. 4A). The alignment of this RrgB domain with other pilins carrying CnaB predicted domains allowed us to identify that a potential isopeptide bond could involve residues Lys-453, Glu-577, and Asn-623, with the lysine and glutamate residues being included in KVD-like and GXYXLXEXAPXGY (E-box) motifs (Figs. 1B and 4A). The E405, also present in an E-box motif, could play a role in the formation of another intramolecular bond in RrgB (Fig. 4A). Because no additional glutamate residue could be identified to explain the formation of the third bond, we looked for an aspartate residue which might play such a role, as observed in the RrgA crystal structure (18). Sequence alignments of RrgA and RrgB homologs suggested that Asp-241, defined in a D-box, might be involved in the formation of an intramolecular bond in RrgB (Fig. 4B). To assess the role of these various acidic residues in isopeptide bond formation, they were replaced by Ala either individually or together. Mass spectrometry confirmed that all single mutants lacked one NH3 unit, indicating that each mutated residue is involved in the formation of an isopeptide bond (Table 3). No isopeptide bonds were detected in the triple mutant (Table 3).
FIGURE 4.
Intramolecular bonds in RrgB. A, topology of RrgB indicating the positions of the intramolecular bonds. The predicted CnaB domain is comprised between residues 547 and 605. The lysine and asparagine residues forming the bond-containing Glu-405 and Asp-241 have not been identified. SP, signal peptide; the cell wall sorting signal (CWSS) comprises a LPXTG-like motif followed by a hydrophobic membrane-spanning domain and a cytoplasmic positively charged tail. B, sequences alignment showing the D-box motif.
The Three Internal Covalent Bonds Stabilize RrgB
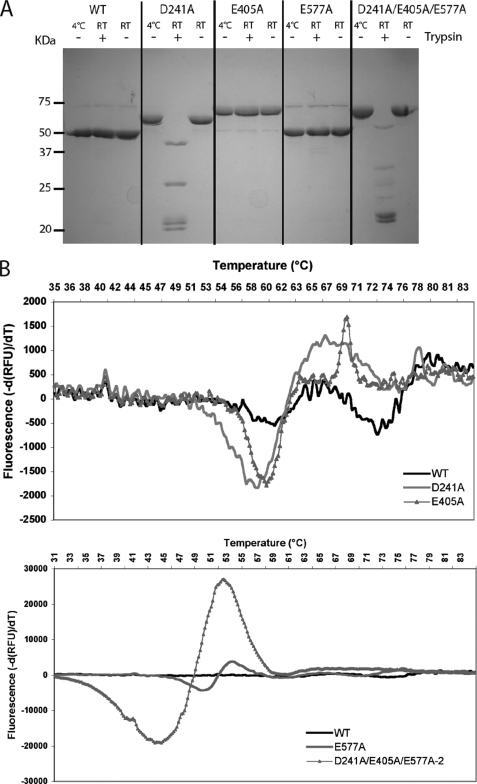
We investigated the role played by the three isopeptide bonds in RrgB stabilization by analyzing the proteolytic susceptibility and the thermal stability of the different RrgB variants (Fig. 5). Wild-type (WT) and RrgB variants were tested for resistance to proteolytic digestion (Fig. 5A). Neither native, E405A nor E577A mutant proteins were affected by trypsin digestion while the D241A and the triple mutant D241A/E505A/E577A were cleaved in multiple fragments ranging from 20 to 50 kDa (Fig. 5A, lanes 5 and 14). It is interesting to note that in the absence of the protease, the D241A, E405A, and the triple mutant proteins migrate on SDS-PAGE slightly slower than the native protein, indicating that, as in the case of RrgC, isopeptide bond formation enhances protein compactness (Fig. 5A).
FIGURE 5.
Proteolysis and thermal stability of RrgB and variants. A, trypsin digestion products were separated on 12.5% SDS-PAGE. The molecular weight markers are shown on the left side of the gel. Whether trypsin was added or not is noted on top of the gel as well as the reaction temperatures. B, wild-type curve has been included in each figure to facilitate the comparison with the different RrgB variants.
No significant thermal denaturation of the native RrgB was observed, an indication of the stable fold of the protein (Fig. 5B) while the D241A and E405A mutants displayed Tm values of 58.5 °C (±0.5) and 58.9 °C (±2.5), respectively (Fig. 5B). The E577A mutant presents an even more pronounced defect in stability as a three-state unfolding pattern is observed, characterized by the Tm values of 48.6 °C (±2.1), 59.2 °C (±2.0), and 68.8 °C (±1.9) (Fig. 5B). The triple RrgB mutant that does not contain any intramolecular bonds has a deeply decreased thermal stability reflected by the single low Tm value of 43.1 °C (±1.1) (Fig. 5B).
SrtC-1 Covalently Associates RrgB and RrgC
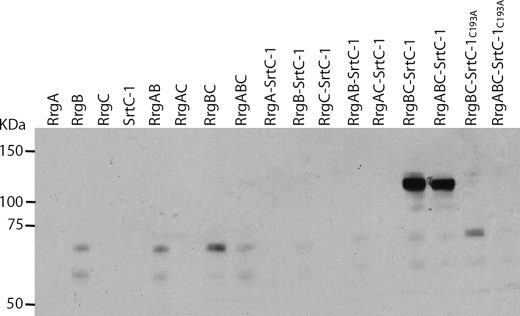
SrtC-1 is the main sortase responsible for RrgB polymerization (13). To determine if SrtC-1 displays restrictive specificity toward RrgB or is also able to recognize RrgC and RrgA, we designed a co-expression system which allows testing of sortase-pilin interactions (Fig. 6). Genes encoding the soluble forms of RrgA, RrgB, RrgC and SrtC-1 proteins were cloned in the bi-cistronic plasmids pETDuet and pACYCDuet, and the corresponding proteins were expressed in E. coli (Fig. 6). The co-expression products were purified with HisTrapTMHP columns to which only His-tagged RrgA and RrgC, associated or not to RrgB, could be retained; eluted proteins were separated onto gradient polyacrylamide gels and immunoblotted with mouse polyclonal antibodies specific for RrgB (Fig. 7). As expected, no RrgB was immunodetected in the eluted fractions of strains expressing only RrgA or RrgC and SrtC-1 (Fig. 7, lanes 1, 3, 4). Despite the absence of a His tag on RrgB, two weak RrgB bands (60 and 70 kDa) are detected in the RrgB-containing eluted fractions which could be due to nonspecific binding to the HisTrap column (Fig. 7, lanes 2, 5, 7, 8, 16). However, strains that co-express SrtC-1, RrgB, and RrgC, present an intense band which migrates with a molecular apparent mass of 120 kDa and is recognized by both anti-RrgB (Fig. 7, lane 14) and anti-RrgC antibodies (data not shown). This finding suggests that SrtC-1 is able to covalently link the pilin subunits RrgB and RrgC. This same product was identified when RrgA was also co-expressed (Fig. 7, lane 15), although this pilin subunit is not involved in the formation of the RrgB-RrgC complex. In contrast, co-expression of SrtC-1 with RrgA and RrgB did not yield a complex between the two pilins (Fig. 7, lane 12) demonstrating a substrate specificity of sortase SrtC-1 for the association of RrgB with RrgC exclusively. To verify the catalytic nature of RrgB-RrgC complex formation, we mutated the SrtC-1 active site cysteine 193 into alanine and performed co-expression experiments as described above. No RrgB-RrgC complex could be identified, indicating that the association between RrgB and RrgC is specifically catalyzed by SrtC-1 (Fig. 7, lanes 16 and 17).
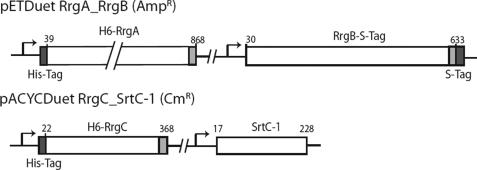
FIGURE 6.
Constructs for the co-expression platform. Schematic representation of the pETDuet RrgA-RrgB and pACYCDuet RrgC-SrtC-1 constructs.
FIGURE 7.
Formation of the covalent complex RrgB-RrgC by SrtC-1. Co-expression of RrgA, RrgB, RrgC, and SrtC-1 in E. coli. The purification was performed on HisTrapTMHP columns and only RrgA and RrgC are fused to His tag. The eluted proteins were separated on 4–12.5% SDS-PAGE, transferred on nitrocellulose membrane and immunodetected with an anti-RrgB polyclonal antibody.
SrtC-1 Recognizes the RrgB Sorting Signal
To further characterize the covalent complex, we purified it to homogeneity and performed mass spectrometry analysis, which yielded a mass of 106,565.51 Da. This value corresponds precisely to the sum of the mass of one His6-tagged RrgC subunit associated to an IPQT-cleaved-S-tag RrgB subunit. The purified RrgB-RrgC complex was submitted to MS/MS experiments and peptides from both proteins were identified (supplemental Table S3). To verify which cell wall sorting signal is recognized by SrtC-1, both RrgC and RrgB motifs were deleted, and the mutated proteins were tested in the SrtC-1 co-expression system (Fig. 8). The RrgB-RrgC complex could be successfully purified in the absence of the RrgC VPDTG motif, while complex production was totally abrogated when RrgB lacks its IPQTG motif (Fig. 8). This observation demonstrates that catalysis by SrtC-1 proceeds through recognition of the RrgB IPQTG motif.
FIGURE 8.
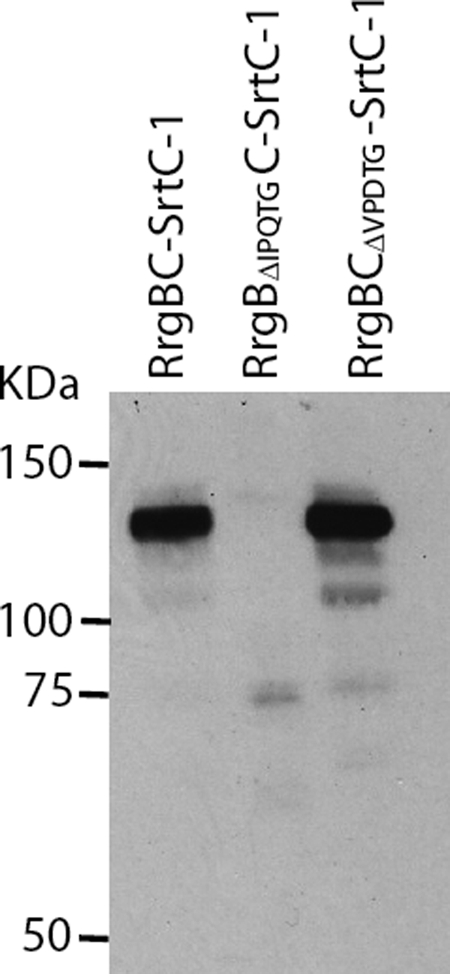
Substrate recognition specificity of SrtC-1. Both sorting signals in RrgB and RrgC were deleted, and each mutant was analyzed for its ability to be recognized and processed by SrtC-1. The purification of the co-expressed proteins, RrgB, RrgC, and SrtC-1 was performed on HisTrapTMHP columns. The eluted proteins were separated on 4–12.5% SDS-PAGE, transferred onto a nitrocellulose membrane, and immunodetected with an anti-RrgB polyclonal antibody.
Role of the Isopeptide Bonds in the RrgB-RrgC Complex Formation
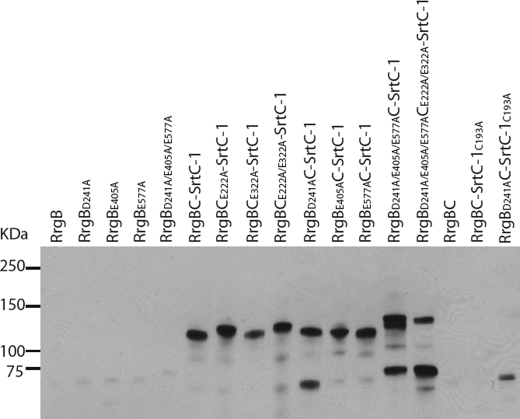
Isopeptide bonds stabilize RrgB and RrgC subunits, which are covalently associated by SrtC-1. Consequently, we investigated if the compact fold induced by the lysine-asparagine intramolecular bonds could influence the formation of the covalent RrgB-RrgC complex. The acidic residues shown to be involved in the formation and in the stabilization of the isopeptide bonds in both RrgB and RrgC were mutated into alanines in the co-expression system described above and the RrgB-RrgC complexes bearing the point mutations were purified and detected by the anti-RrgB antibodies as previously described (Fig. 9). The Glu-222 and Glu-322 mutations in RrgC and the single Asp-241, Glu-405, and Glu-577 mutations in RrgB did not affect complex formation (Fig. 9). However, when Asp-241 in RrgB was mutated into alanine, a significant quantity of monomeric RrgB was present in the HisTrapTMHP column eluate, indicating that a large amount of RrgB remained bound to RrgC through noncovalent interactions (Fig. 9, lane 10). The co-elution of the mutated RrgB variant is not due to artifactual binding to the resin as the recombinant RrgB mutants behave as the wild-type RrgB (Fig. 9). Furthermore, the noncovalent association of RrgB-D241A to RrgC is not dependent on the activity of the SrtC-1 as the RrgB-D241A is still co-eluted with RrgC and present in the elution fraction when the SrtC-1 is mutated in the catalytic Cys-193 (Fig. 9). When all three isopeptide bonds in RrgB were deleted, the RrgB-RrgC complex migrates with two apparent molecular masses of 140kDa and 150kDa (Fig. 9, lane 13). Finally, the absence of all the isopeptide bonds in both RrgC and RrgB lead to inefficient complex formation, since only a minor amount of RrgB-RrgC complex could be formed (Fig. 9, lane 14). In conclusion, the stabilization of the fold of both RrgB and RrgC by intramolecular bond formation plays a role in the efficient recognition of the pilin subunits by SrtC-1 to catalyze the formation of the RrgB-RrgC covalent complex.
FIGURE 9.
Role of the internal bonds in RrgB and RrgC in the covalent complex formation. The variants of RrgB and RrgC deleted in the internal bonds were introduced in the co-expression platform. The purification of the different RrgB forms (wild-type, single, and triple mutants) and of the co-expressed proteins, RrgB, RrgC, and SrtC-1 was performed on HisTrapTMHP columns. The eluted proteins were separated on 4–12.5% SDS-PAGE, transferred on nitrocellulose membrane, and immunodetected with an anti-RrgB polyclonal antibody.
DISCUSSION
The recent discovery of pili expressed at the surface of Gram-positive bacteria allowed the publication of large amounts of data concerning pilus assembly, at the genetic and biochemical levels. However, the molecular details of substrate recognition and sortase catalysis remain largely unraveled. The pneumococcal pilus is composed of a fiber formed by RrgB, to which two minor pilins, RrgA and RrgC, are associated. Three sortases are involved in pilus assembly, SrtC-1, SrtC-2, and SrtC-3. One major challenge is to define the specificity of the sorting signal recognition and the molecular basis of the transpeptidation reaction of the pneumococcal-pilus-associated sortases.
The recent crystal structure of the RrgA adhesin showed the presence of two intramolecular bonds (18). In this work, we identified and characterized the intramolecular bonds in RrgC and RrgB. Sequence alignment analyses and mutagenesis data show that the presence of an E-box, containing a glutamate residue, is associated to the formation of the isopeptide bonds. The E-box motif had been described as playing a key role in the incorporation of minor pilin subunits (30), and here we present experimental evidence that the role of the E-box is related to the stabilization of the intramolecular bonds (see also below). We also describe a new D-box motif in which the aspartate residue plays a role in the formation of the isopeptide bond. The lysine residue forming the intramolecular bond is contained in a small conserved sequence motif, KVD. Recombinant RrgB contains three intramolecular isopeptide bonds even though sequence analysis revealed a potential fourth site involving residues Lys-41, Asn-184, and Glu-143. It cannot be excluded that this supplementary bond is formed in the native protein. The intramolecular bonds in the CnaB domains of RrgC and RrgB are most probably located in the same position as in the analog domains GBS52-N2 (23) and RrgA-D4 (18) : this is suggested by the structural alignments of GBS52-N2 and RrgA-D4 with the RrgC and RrgB CnaB sequences, highlighting the conservation of the lysine and glutamate residues participating in the intramolecular bonds, with the secondary structures elements (supplemental Fig. S1).
The role of the isopeptide bonds in RrgC and RrgB in protein stability was also investigated. The thermostability assay used in this study is based on the modification of the fluorescence signal emitted by the SyproOrange probe upon its binding to hydrophobic residues that are exposed while the protein denatures. Consequently, the protein thermal unfolding assay is performed by monitoring of a mixture of protein with the probe over a range of temperatures. The reported temperature values correspond to the melting temperatures (Tm) calculated as the inflection point of the fluorescence versus temperature; they correspond to the mid-point temperature values determined in circular dichroïsm experiments. TSA studies thus are good indicators of the multidomain organization of a protein; if distinct domains unfold independently, this could give rise to multiple Tm curves, as observed for RrgB.
Both thermostability and protease sensitivity data show that the absence of intramolecular bonds in mutated variants of RrgC and RrgB destabilizes the proteins. However, how these intramolecular bonds cause the protein to be more stable and compact remains an open question. We propose the following hypothesis. The CnaB domains resemble the IgG-like structure with a four- and a three-β-stranded barrel, but exhibit a novel IgG reverse fold (23). The Lys-Asn bond cross-links the three-β-stranded sheet while the Glu residue points from the four-β-stranded sheet (supplemental Fig. S2). Consequently, the intramolecular bonds act as locks which maintain a globally closed conformation of the β-barrel, as observed for the disulfide bonds in IgG, thus leading to an increased compactness and stability of the proteins.
We observed that point mutations of the Lys and Asn residues and of the catalytic Glu or Asp residues impair the isopeptide bond formation. Rearrangements induced by such mutations have been observed recently by Kang and Baker (19) in the S. pyogenes major pilin. The crystal structure of the E117A variant showed that the side chains of Lys-36 and Asn-168 can adopt alternative orientations due probably to the inappropriate positioning of ionic bonding partners. However, this remains a local effect which does not cause major protein conformational changes. We believe that a similar situation occurs in the mutants described in this work.
Our results also show that the pilin intramolecular bonds influence the formation of the RrgB-RrgC complex. Indeed, a significant amount of the D241A RrgB mutant monomer was co-eluted with RrgC showing that both pilin subunits interact in a stable noncovalent complex whose formation is independent on the enzymatic activity of the SrtC-1. The structural changes caused by the absence of this single intramolecular isopeptide bond in RrgB reduce the ability of SrtC-1 to covalently form RrgB-RrgC. These findings suggest that SrtC-1, in addition to recognizing the IPQTG motif of RrgB, also interacts with other region(s) of the pilin, which could be less accessible when the Asp-241 intramolecular bond does not stabilize the RrgB structure. This result is in accordance with what was observed for BcpA, the shaft forming pilin of B. cereus, which contains three intramolecular bonds, one of which plays an important role in pilus formation (31).
To address the question of the pilus biogenesis mechanisms at the molecular level, we used biochemical approaches that mimic the assembly reactions in vitro. We developed an expression platform dedicated to the production of recombinant pilins and sortases, in different combinations, in order to reconstitute the pneumococcal pilus assembly. The co-expression in E. coli of RrgB, RrgC, and SrtC-1 led to the formation of a covalent RrgB-RrgC complex which was catalyzed by SrtC-1. Following cleavage of the IPQTG sorting signal of RrgB, SrtC-1 forms an amide bond between the C-terminal threonine and an NH2 group on the RrgC subunit. The RrgB-RrgC complex is the first sortase-product between two different pneumococcal pilins obtained in in vitro conditions. In this work, we do not provide accurate quantitative data about the sortase enzymatic efficiency as the overexpression is performed in an heterologous host. However, an estimation of the pneumococcal sortase efficiency in E. coli can be estimated to be around 1% based on the purification of 500 μg of RrgB-RrgC complex from about 30–50 mg of RrgB, RrgC, and SrtC-1 expressed.
The number of pilus-associated sortases is variable: one sortase is present in Bacillus and Corynebacteria species, two in GAS and GBS species and three in S. pneumoniae. Consequently, it can be assumed that one pilus-associated sortase may recognize different sorting motifs and/or accept different nucleophilic amine groups. One example of such redundancy concerns SrtD of B. cereus which recognizes both BcpA and BcpB (31, 32). We describe a different situation in S. pneumoniae: SrtC-1 only recognizes and cleaves the IPQTG sorting signal of RrgB but catalyzes the formation of an amide bond with either another subunit of RrgB, leading to formation of the fiber polymer (13), or with a RrgC monomer, to form the heterodimeric RrgB-RrgC complex (this work). The mechanism at the basis of the balance between both reactions remains to be described. The in vitro RrgB-RrgC complex formation is relevant in regard to the current knowledge of pilus biogenesis: RrgC is associated to the pilus fiber (26, 28) and SrtC-1 has been proposed to incorporate RrgC in the native pilus as observed by mutational genetics in the TIGR4 strain (27). Consequently, we propose that the complex RrgB-RrgC whose association is catalyzed by SrtC-1, may represent a molecular unit onto which pilus elongation and/or its cell wall insertion may take place.
Supplementary Material
Acknowledgments
We thank Izabel Bérard and Eric Forest from the IBS mass spectroscopy facility (LSMP, IBS) for analyses, Alexandra Kraut and Jérôme Garin (Lab. d'Etude de la Dynamique des Protéomes, EDyP Grenoble) for access to the LC-MS/MS platform and experiments, and Jean-Pierre Andrieu for (LEM, IBS) for N-terminal sequencing.
This work was supported in part by EC Grant LSHM-CT-2004-512138 and ANR Grant 05-JC-JC-0049.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Tables S1–S3.
- IPTG
- isopropyl-1-thio-β-d-galactopyranoside
- MES
- 4-morpholineethanesulfonic acid
- WT
- wild type
- CWSS
- cell wall sorting signal.
REFERENCES
- 1.Proft T., Baker E. N. (2009) Cell Mol. Life Sci. 66, 613–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yanagawa R., Otsuki K., Tokui T. (1968) Jpn J. Vet. Res. 16, 31–37 [PubMed] [Google Scholar]
- 3.Ton-That H., Schneewind O. (2003) Mol. Microbiol. 50, 1429–1438 [DOI] [PubMed] [Google Scholar]
- 4.Mishra A., Das A, Cisar J. O., Ton-That H. (2007) J. Bacteriol. 189, 3156–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kline K. A., Kau A. L., Chen S. L., Lim A., Pinkner J. S., Rosch J., Nallapareddy S. R., Murray B. E., Henriques-Normark B., Beatty W., Caparon M. G., Hultgren S. J. (2009) J. Bacteriol., 191, 3237–3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Budzik J. M., Marraffini L. A., Schneewind O. (2007) Mol. Microbiol. 66, 495–510 [DOI] [PubMed] [Google Scholar]
- 7.Mora M., Bensi G., Capo S., Falugi F., Zingaretti C., Manetti A. G., Maggi T., Taddei A. R., Grandi G., Telford J. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15641–15646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosini R., Rinaudo C. D., Soriani M., Lauer P., Mora M., Maione D., Taddei A., Santi I., Ghezzo C., Brettoni C., Buccato S., Margarit I., Grandi G., Telford J. L. (2006) Mol. Microbiol. 61, 126–141 [DOI] [PubMed] [Google Scholar]
- 9.Dramsi S., Caliot E., Bonne I., Guadagnini S., Prévost M. C., Kojadinovic M., Lalioui L., Poyart C., Trieu-Cuot P. (2006) Mol. Microbiol. 60, 1401–1413 [DOI] [PubMed] [Google Scholar]
- 10.Barocchi M. A., Ries J., Zogaj X., Hemsley C., Albiger B., Kanth A., Dahlberg S., Fernebro J., Moschioni M., Masignani V., Hultenby K., Taddei A. R., Beiter K., Wartha F., von Euler A., Covacci A., Holden D. W., Normark S., Rappuoli R., Henriques-Normark B. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2857–2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dramsi S., Magnet S., Davison S., Arthur M. (2008) FEMS Microbiol. Rev. 32, 307–320 [DOI] [PubMed] [Google Scholar]
- 12.Maresso A. W., Schneewind O. (2008) Pharmacol. Rev. 60, 128–141 [DOI] [PubMed] [Google Scholar]
- 13.Manzano C., Contreras-Martel C., El Mortaji L., Izoré T., Fenel D., Vernet T., Schoehn G., Di Guilmi A. M., Dessen A. (2008) Structure 16, 1838–1848 [DOI] [PubMed] [Google Scholar]
- 14.Manzano C., Izoré T., Job V., Di Guilmi A. M., Dessen A. (2009) Biochemistry 48, 10549–10557 [DOI] [PubMed] [Google Scholar]
- 15.Neiers F., Madhurantakam C., Fälker S., Manzano C., Dessen A., Normark S., Henriques-Normark B., Achour A. (2009) J. Mol. Biol. 393, 704–716 [DOI] [PubMed] [Google Scholar]
- 16.Kang H. J., Coulibaly F., Clow F., Proft T., Baker E. N. (2007) Science 318, 1625–1628 [DOI] [PubMed] [Google Scholar]
- 17.Kang H. J., Paterson N. G., Gaspar A. H., Ton-That H., Baker E. N. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 16967–16971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Izoré T., Contreras-Martel C., El Mortaji L., Manzano C., Terrasse R., Vernet T., Di Guilmi A. M., Dessen A. (2010) Structure 18, 106–115 [DOI] [PubMed] [Google Scholar]
- 19.Kang H. J., Baker E. N. (2009) J. Biol. Chem. 284, 20729–20737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Symersky J., Patti J. M., Carson M., House-Pompeo K., Teale M., Moore D., Jin L., Schneider A., DeLucas L. J., Höök M., Narayana S. V. (1997) Nat. Struct. Biol. 4, 833–838 [DOI] [PubMed] [Google Scholar]
- 21.Zong Y., Xu Y., Liang X., Keene D. R., Höök A., Gurusiddappa S., Höök M., Narayana S. V. (2005) EMBO J. 24, 4224–4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deivanayagam C. C., Rich R. L., Carson M., Owens R. T., Danthuluri S., Bice T., Höök M., Narayana S. V. (2000) Structure 8, 67–78 [DOI] [PubMed] [Google Scholar]
- 23.Krishnan V., Gaspar A. H., Ye N., Mandlik A., Ton-That H., Narayana S. V. (2007) Structure 15, 893–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Q., Ponnuraj K., Xu Y., Ganesh V. K., Sillanpää J., Murray B. E., Narayana S. V., Höök M. (2007) J. Biol. Chem. 282, 19629–19637 [DOI] [PubMed] [Google Scholar]
- 25.LeMieux J., Hava D. L., Basset A., Camilli A. (2006) Infect Immun. 74, 2453–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hilleringmann M., Giusti F., Baudner B. C., Masignani V., Covacci A., Rappuoli R., Barocchi M. A., Ferlenghi I. (2008) PLoS Pathog. 4(3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fälker S., Nelson A. L., Morfeldt E., Jonas K., Hultenby K., Ries J., Melefors O., Normark S., Henriques-Normark B. (2008) Mol Microbiol. 70, 595–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hilleringmann M., Ringler P., Müller S. A., DeAngelis G., Rappuoli R., Ferlenghi I., Engel A. (2009) EMBO J. 28, 3921–3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LeMieux J., Woody S., Camilli A. (2008) J Bacteriol. 190, 6002–60013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ton-That H., Marraffini L. A., Schneewind O. (2004) Mol. Microbiol. 53, 251–261 [DOI] [PubMed] [Google Scholar]
- 31.Budzik J. M., Marraffini L. A., Souda P., Whitelegge J. P., Faull K. F., Schneewind O. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10215–10220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Budzik J. M., Oh S. Y., Schneewind O. (2009) J. Biol. Chem. 284, 12989–12997 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.