Abstract
Drugs for cancer therapy belong to different categories of chemical substances. The cellular targets for the therapeutic efficacy are often not unambiguously identified. Here, we describe the process of ribosome biogenesis as a target of a large variety of chemotherapeutic drugs. We determined the inhibitory concentration of 36 chemotherapeutic drugs for transcription and processing of ribosomal RNA by in vivo labeling experiments. Inhibitory drug concentrations were correlated to the loss of nucleolar integrity. The synergism of drugs inhibiting ribosomal RNA synthesis at different levels was studied. Drugs inhibited ribosomal RNA synthesis either at the level of (i) rRNA transcription (e.g. oxaliplatin, doxorubicin, mitoxantrone, methotrexate), (ii) early rRNA processing (e.g. camptothecin, flavopiridol, roscovitine), or (iii) late rRNA processing (e.g. 5-fluorouracil, MG-132, homoharringtonine). Blockage of rRNA transcription or early rRNA processing steps caused nucleolar disintegration, whereas blockage of late rRNA processing steps left the nucleolus intact. Flavopiridol and 5-fluorouracil showed a strong synergism for inhibition of rRNA processing. We conclude that inhibition of ribosome biogenesis by chemotherapeutic drugs potentially may contribute to the efficacy of therapeutic regimens.
Keywords: Cell/Cycle, RNA/Nuclear RNA, RNA/Precursors, RNA/Ribosomal Processing, RNA/Ribosomal RNA, RNA/Ribosomes, Tumor/Suppressor, Chemotherapeutic Drugs
Introduction
Chemotherapeutic drugs (hereinafter drugs) are used for the treatment of neoplastic diseases for more than 50 years. The mode of action and specifically the therapeutic relevant targets of many drugs, however, are often less defined. Recent studies revealed that some drugs like 5-fluorouracil (5-FU),4 which were first assumed to interfere with DNA metabolism actually act mainly on RNA metabolism (1–9). In fact an increasing number of analyses identifies RNA metabolism as an important target of cancer drugs.
In a hallmark study, Rubbi and Milner (10) showed that accumulation of the tumor suppressor p53 in UV- or drug-damaged cells occurs only if nucleolar functions are affected. Local, severe UV irradiation of the nucleoplasm could not stabilize p53 accumulation. In contrast, UV damage in the nucleolus induced a strong p53 response suggesting that the major sensor controlling the stability and degradation of p53 is located in the nucleolus, the place of ribosome biogenesis.
The stability of the p53 protein is controlled by the ubiquitin ligase Mdm2, which targets p53 to the proteasome for degradation. Strikingly, several ribosomal proteins, including L5, L11, L23, and S7 proteins can bind and inactivate Mdm2 (11–14). Conditional knockdown of these ribosomal protein genes prevents Mdm2 inactivation and p53 stabilization in 5-FU-treated cells (15), consistent with the assumption that destruction of nucleolar functions by 5-FU inhibits ribosome biogenesis and results in liberation of ribosomal proteins followed by Mdm2 inactivation and p53 stabilization. The inhibition of rRNA transcription by knockout of the gene for the RNA polymerase I (Pol I) transcription factor TIF-1A (16), by blockage of the transcription factor UBF after microinjection of specific monoclonal antibodies (10), or by low concentrations (<5 nanomolar) of actinomycin D (10), blocked transcriptional activity of Pol I and consistently led to stabilization of p53. Likewise, the inhibition of specific processing steps of rRNA, e.g. after knockdown of genes required for maturation of 18 S and 28 S rRNA results in p53 stabilization (17–21). Thus, functional ribosome biogenesis is an essential prerequisite for inactivation of p53 in proliferating cells. Inhibition of this process at the level of rRNA transcription or rRNA maturation consistently leads to p53 accumulation, regardless of which level of the rRNA maturation processes is affected.
Stabilization of p53 is a consistent response of cells treated with classical chemotherapeutic drugs. The cellular signaling pathways triggering the increase in p53 protein levels are often not illuminated. We asked therefore, whether inhibition of ribosome biogenesis could be the basis for p53 stabilization in response to chemotherapy. To answer this question we studied the potency of 36 drugs of different chemical categories to interfere with ribosome biogenesis at the levels of transcription and processing of rRNA.
EXPERIMENTAL PROCEDURES
Tissue Culture
Human 2fTGH fibrosarcoma cells were cultured in Dulbecco's modified Eagle (DME) complete medium (Invitrogen) with 10% fetal bovine serum (PAA Laboratories) at 37 °C and 8% CO2. Cells were incubated with 36 different drugs at increasing concentrations. All drugs were stored according to the manufacturers' instructions and freshly dissolved as stock solutions in the solvents listed below. Prior to incubation, drugs were diluted in phosphate-buffered saline (PBS) and DME complete medium, resulting in 1-ml medium samples with defined drug concentrations.
Calculation of Clinical Relevant Concentrations
Standard clinical therapy protocols were used to translate commonly used clinical doses into concentrations used in our experiments. We are aware that this is a very simplified calculation in respect to the often very different pharmacokinetic behavior of many substances in patients. Nevertheless it will allow the comparison of ranges. The clinical doses are typically indicated as [mg/m2]. To determine roughly the equivalent chemical concentration in [mol/liter], we considered a standard patient having 70 kg distributed over 2 m2. We calculated the maximal chemical concentration of drug applied per day, using the molar mass of each drug in [g/mol] and 70 liter-volumes/patient, which is equivalent to 70 kg body weight (see Table 1).
TABLE 1.
Effect of cytostatic drugs on rRNA synthesis and nucleolar integrity
Substances are divided into nine classes. The mode and the efficiency of rRNA synthesis inhibition are depicted together with the potential of drug-mediated nucleolar phenotype induction. Transcription: inhibition of the 47 S rRNA; early processing: inhibition of 32 S rRNA precursor; late processing: inhibition of mature 28 S and 18 S rRNA. “Caps,” “spots,” and “necklace” indicate nucleolar protein translocation phenotypes. The inhibitory effect of drugs on rRNA synthesis was rated as follows: (−) little or no effect <50%; (+) 50–90% inhibition; (++) >90% inhibition; (+++) >90% inhibition occurs with 4 or less doublings of concentrations. The concentrations relevant to inhibit rRNA synthesis with 50 and 80% (IC50/IC80) efficiency are shown and compared to the mean clinical concentrations applied per kg body mass, roughly calculated as described under “Experimental Procedures.”
In Vivo Labeling of RNA and rRNA Analysis
For metabolic labeling, 2 × 105 2fTGH cells were grown in multiwell plates with DME/10% FBS complete medium for 24 h. Subsequently, cells were treated with increasing concentrations of drugs, freshly dissolved in DME/10% FBS complete medium, for 2 h. For phosphate depletion, DME/10% FBS complete medium was replaced by phosphate-free DME/10% dialyzed FBS (Invitrogen), and cells were incubated in the presence of unchanged drug concentrations for 1 h. Medium was then replaced by DME/10%-dialyzed FBS medium containing 15 μCi/ml [32P]orthophosphate (Hartmann), and phosphate-depleted cells were labeled for 1 h. Medium was again replaced by DME/10% FBS complete, drug-containing medium, and total RNA was isolated after 3 h using the PeqLab Gold total RNA kit (PeqLab). RNA concentration was determined using a Biophotometer (Eppendorf), and 1 μg of total RNA was separated on a 1% agarose-formaldehyde gel. After electrophoresis, 28 S rRNA amounts were controlled under UV light, and gels were placed on Whatman paper and dried for 2 h at 80 °C under vacuum suction. Dried agarose gels were exposed to regular x-ray films (Kodak), and rRNA was visualized by autoradiography. A PhosphorImager (Fuji) was used for the quantification of signal intensities.
Immunofluorescence Microscopy
Cells were grown on coverslips with DME/10% FBS complete medium for 24 h, and then incubated with several defined concentrations of drugs freshly dissolved in DME/10% FBS complete medium for 6h. Cells were washed in PBS and fixed with warm 3.7% paraformaldehyde for 4 min, permeabilized with PBS/Tween 0.04% for 7 min, and unspecific binding was blocked with PBS/10% FBS for 2 h. Nucleophosmin (NPM), Pescadillo1 (Pes1), and Fibrillarin (Fib) were detected with a 1:2000 dilution of anti-NPM (Sigma, B0556), a 1:1000 dilution of 8E9 anti-Pes1 hybridoma supernatant (18), and a 1:500 dilution of anti-Fib (Abcam, ab5821), respectively. Primary antibodies were incubated overnight at 4 °C in a humidified chamber. After washing with PBS/Tween 0.04%, Cy3- (Jackson) or Alexa Flour 488- (Invitrogen) labeled secondary antibodies were incubated at room temperature for 2 h in a humidified chamber. Cells were washed with PBS/Tween 0.04% again, and nuclei were counterstained with 0.1 μg/ml DAPI (Sigma) for 2 min. Prior to microscopy, cells were treated with Vectashield mounting medium (Vectalabs). Digital images were acquired using the Openlab acquisition software (Improvision) and a microscope (model Axiovert 200 m; Carl Zeiss MicroImaging, Inc.) with a 100× (1.30) plan oil objective connected to a 5 charge-coupled device camera (model ORCA-479; Hamamatsu). Exposure times: Cy3 10–150 ms, Alexa Flour 488: 50–700 ms, DAPI: 5–12 ms.
Western Blot Analysis
Cells were washed in PBS and lysed in warm SDS-loading buffer (50 mm Tris-HCl, 100 mm dithiothreitol, 2% SDS, 0.1% bromphenol blue, and 10% glycerol). Total cell lysates were sonicated, heated, separated on 10–12% SDS-PAGE gel, and blotted on nitrocellulose membranes (GE Healthcare). Immunodetection was performed with anti-Pes1 (8E9) and anti-p53 (DO-1 Santa Cruz Biotechnology, sc-126) antibodies, horseradish peroxidase-labeled secondary antibodies.
RESULTS
Analysis of rRNA Transcription and Processing
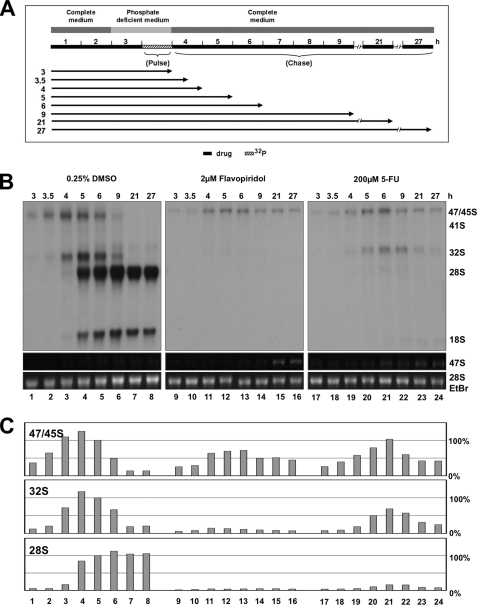
The impact of chemotherapeutic drugs on ribosome biogenesis was studied in the human sarcoma cell line 2fTGH. To measure the impact of drugs on transcription and processing of ribosomal RNA (rRNA) simultaneously, cells were metabolically labeled with [32P]orthophosphate along the protocol depicted in Fig. 1A. The primary 47 S rRNA transcript is first processed into intermediate products and finally into the mature 18 S, 5.8 S, and 28 S rRNAs (Fig. 1B). This process becomes apparent, if labeled total RNA is separated by gel electrophoresis and visualized by autoradiography. The amount of label incorporated into individual fragments was determined for the primary 47 S transcript, the 32 S major intermediate form, and for the mature 18 S and 28 S rRNAs by phosphorimager analysis (Fig. 1C). Precursor and mature rRNAs were sufficiently labeled when cells were pulsed for 1 h followed by a 3 h chase (supplemental Fig. S1, lane 5). Reduced incorporation rates into the 47 S rRNA were interpreted as inhibition of rRNA transcription (Fig. 1C, lane 2). Reduced amounts of label in the 32 S rRNA and rRNAs downstream thereof were interpreted as inhibition of early rRNA processing steps (lane 3) and reduction of only 28 S and 18 S rRNA as inhibition of late rRNA processing steps (lane 4).
FIGURE 1.
Analysis of rRNA transcription and processing. A, inhibition of rRNA transcription and processing by cytotoxic drugs in metabolic labeling experiments. Cells were cultured with cytostatic drugs, phosphate-depleted, and labeled with [32P]orthophosphate as indicated. B, schematic of rRNA processing in mammalian cells. ETS, external transcribed spacer; ITS, internal transcribed spacer. C, total RNA was isolated, separated by agarose-formaldehyde gel electrophoresis and transferred to a Whatman paper. The signal intensities of all detectable rRNA forms were quantified by phosphorimager analysis. Lane 1 represents a schematic pattern of rRNA of cells with unaffected rRNA synthesis, lanes 2–4 rRNAs of cells after inhibition of rRNA transcription, early rRNA processing, or late rRNA processing.
Inhibition of rRNA Transcription by Chemotherapeutic Drugs
Cisplatin, oxaliplatin, doxorubicin, and mitoxantrone strongly inhibited transcription of rRNA genes. Labeling of the 47 S rRNA precursor was entirely blocked by these substances. The signals for the intermediate and mature rRNAs disappeared concomitantly, indicating that rRNA processing was not directly affected (Fig. 2, A–D).
FIGURE 2.
Cytostatic drugs inhibit rRNA transcription and processing. A–D, cytostatic drugs cisplatin, oxaliplatin, doxorubicin, and mitoxantrone inhibited transcription of rRNA genes. Specific inhibition was demonstrated by a fast and complete decrease of the 47 S/45 S rRNA signal within a small range of concentrations. The signals of the intermediate and mature rRNA forms downstream of the 47 S/45 S transcript decreased concomitantly indicating that rRNA processing is not primarily affected. E–H, cytostatic drugs DRB, roscovitine, MG-132, and homoharringtonine inhibited rRNA processing at various levels. DRB and roscovitine inhibited the occurrence of the 32 S rRNA indicative for inhibition of early processing steps. MG-132 and homoharringtonine inhibited the occurrence of the 18 S and 28 S rRNAs indicative for inhibition of late rRNA processing steps. Green bars indicate mean body concentrations for clinical applications. Ethidium bromide (EtBr)-stained 28 S rRNA served as loading control. Control 1, water; control 2, solvent with highest concentration; control 2*, 1% ethanol; control 3, 0.125% ethanol. I, quantification of signals by phosphorimager, controls were set as 100%.
In detail, the drugs were titrated from nanomolar to micromolar concentrations. Half-maximal inhibition of rRNA synthesis was detectable at concentrations >50 μm for cisplatin, >3 μm for oxaliplatin, >0.4 μm for doxorubicin, and >0.8 μm for mitoxantrone. The strong inhibitory effect of oxaliplatin, doxorubicin, and mitoxantrone was achieved at concentrations at or below clinical relevance, whereas cisplatin was effective only at higher concentrations (Fig. 2, A–D; green bars). All four substances induced a sharp drop in the synthesis of the 47 S rRNA precursor at increasing concentrations.
Inhibition of rRNA Processing by Chemotherapeutic Drugs
DRB (5, 6-dichloro-1-β-d-ribofuranosylbenzimidazole), roscovitine, MG-132, and homoharringtonine blocked processing of rRNA at different levels (Fig. 2, E–H). DRB and roscovitine, inhibitors of cellular kinases, blocked early rRNA processing steps. DRB and roscovitine inhibited the occurrence of the 32 S rRNA at concentrations of >12 μm and >6 μm, respectively (Fig. 2, E and F), whereas the signal intensity of the primary 47 S rRNA was only marginally or not affected. The proteasome inhibitor MG-132 blocked processing steps for maturation of 18 S and 28 S rRNA at concentrations >6 μm, and at higher concentrations for synthesis of the 47 S rRNA (Fig. 2G). A similar inhibitory profile was observed for homoharringtonine, an inhibitor of translation (Fig. 2H).
All together, the impact of 36 substances on rRNA synthesis was studied (Table 1). Ten substances inhibited rRNA transcription, four early rRNA processing steps, and seven late rRNA processing steps (for further details, see supplemental Fig. S2). We conclude that chemotherapeutic drugs can inhibit ribosome biogenesis at the levels of (i) rRNA transcription, (ii) early rRNA processing, and (iii) late rRNA processing.
Does Inhibition of rRNA Processing Feedback to rRNA Transcription?
Short pulse labeling for 15–60 min in the presence and absence of flavopiridol or 5-FU revealed a similar labeling index for the 47 S rRNA, whereas labeling of 47 S rRNA was entirely blocked by doxorubicin (supplemental Figs. S3 and S4). To rule out that the block in rRNA processing by flavopiridol and 5-FU can feedback and block the Pol I transcriptional machinery, as recently reported for demethylated ribosomal gene loci (22), long term kinetic experiments were performed. Cells were pulse-labeled for 1 h and chased for 21 and 27 h. After this period, label was detectable in the mature 18 S and 28 S rRNAs in control cells, but no longer in the 47 S rRNA precursor (Fig. 3). In contrast, label in the 47 S rRNA precursor was still present in flavopiridol and 5-FU-treated cells after a chase of 27 h. This observation suggests a processing block. Importantly, we also noticed an accumulation of the 47 S rRNA precursor in the EtBr-stained gel 21 and 27 h after chase. The increase of steady-state 47 S rRNA levels is a clear indication for ongoing Pol I transcription in flavopiridol- and 5-FU-treated cells.
FIGURE 3.
Inhibition of rRNA processing does inhibit production of 47 S rRNA precursor. A, cells were pulse-labeled for 1 h and chased for various periods of time as indicated. B, autoradiography of labeled rRNAs separated by gel electrophoresis. 47 S rRNA precursor and 28 S rRNA stained by EtBr. C, relative signal intensities determined by phosphorimager. Signals in lane 5 were set as 100%.
Inhibition of rRNA Transcription and Early rRNA Processing Steps, but Not of Late rRNA Processing Steps, Coincides with the Loss of Nucleolar Integrity
Various substances have been reported to affect ribosome biogenesis and nucleolar integrity (23, 24). After defining three different levels for inhibition of ribosome biogenesis by chemotherapeutic drugs, we asked to which extent does inhibition of rRNA synthesis at different levels contribute to disintegration of nucleolar structures? For this purpose, we treated cells with methotrexate, flavopiridol, and 5-fluorouracil, which inhibit ribosome biogenesis at the level of transcription, early rRNA processing, and late rRNA processing, respectively (Fig. 4, A–C). For each substance the critical concentration range for inhibition was determined (blue box) and applied for experiments, in which the translocation of NPM from the nucleolus into the nucleoplasm was studied (Fig. 4, right panels). Control cells show a preferred localization of NPM in the nucleolus, which steadily diminished, if cells were treated with increasing concentrations of methotrexate or flavopiridol (Fig. 4, A and B). Completion of the nucleoplasmic translocation of NPM coincided with either the complete inhibition of rRNA transcription or the complete inhibition of the occurrence of the 32 S rRNA precursor. In contrast, the complete inhibition of late processing steps by 5-fluorouracil did not provoke the translocation of NPM into the nucleoplasm (Fig. 4C). For further results see also supplemental Fig. S5.
FIGURE 4.
Inhibition of rRNA transcription and early rRNA processing, but not late rRNA processing induces nucleoplasmic translocation of NPM. A, methotrexate inhibited transcription of rRNA; B, flavopiridol early rRNA processing steps; C, 5-fluorouracil late rRNA processing steps. A panel of concentrations with increasing inhibitory activity (blue box) was analyzed in parallel for NPM translocation to the nucleoplasm and the ratio of 32 S/28 S rRNA.
Transcription and Early Processing Steps of rRNA Are Essential for Maintenance of Nucleolar Integrity
To verify whether inhibition of rRNA transcription and early processing steps, but not late processing steps, lead to disintegration of the nucleolar structure we extended the analysis to all 36 substances and the nucleolar factors fibrillarin (Fib) and pescadillo (Pes1), which are involved in early and late rRNA processing steps. Treatment of cells with substances that inhibit transcription and early processing of rRNA all resulted in translocation of NPM into the nucleoplasm (Fig. 5 and supplemental Figs. S6 and S7). The same substances affected also the localization of Pes1 and Fib, which translocated into nuclear spot and nucleolar cap structures (Fig. 5 and supplemental Fig. S6). Divergent from these phenotypes, the kinase inhibitors DRB, roscovitine, and flavopiridol, which all inhibited early rRNA processing steps, induced necklace structures for Fib (Fig. 5, F and G and supplemental Fig. S6H). Inhibitors of late rRNA processing steps, MG-123, homoharringtonine, cycloheximide, and 5-fluorouracil apparently did not induce an altered nucleolar localization of NPM, Pes1, and Fib (Fig. 5, H and I and supplemental Fig. S6, G and J). The nucleolar integrity was maintained for drugs without inhibitory activity on ribosome biogenesis (supplemental Fig. S7).
FIGURE 5.
Inhibition of rRNA transcription and early rRNA processing, but not late rRNA processing alters the nucleolar localization of NPM, Pes1 and Fib. Cells were treated with inhibitory concentrations of drugs for 6 h, fixed, and cellular localization of the nucleolar proteins nucleophosmin (NPM), pescadillo (Pes1), and fibrillarin (Fib) was determined after immunochemical staining with specific antibodies. Nucleoplasmic translocation of NPM is indicated by an arrow, nucleolar cap structures by arrowheads, and necklace structures by a star. PhC, phase contrast. Pictures for PhC, DAPI, and NPM are taken from the same cell.
Synergistic Inhibition of rRNA Processing by Flavopiridol and 5-Fluorouracil
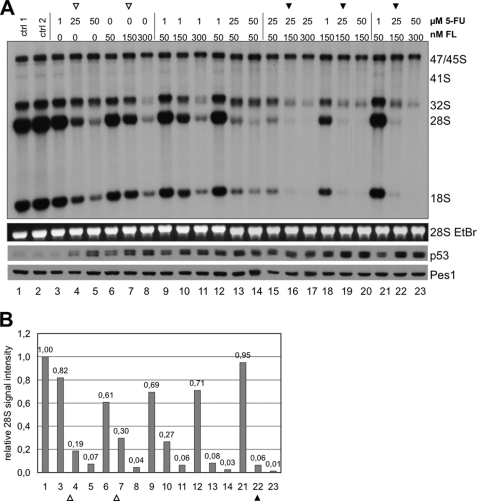
Having defined different inhibitory levels for production of rRNA, we asked, if chemotherapeutic drugs might synergize in their inhibitory activity. Therefore, we titrated increasing concentrations of flavopiridol and 5-FU, two drugs, inhibiting early and late processing steps of 28 S rRNA. The levels of 28 S rRNA were reduced to 0.19-fold by 25 μm 5-FU (Fig. 6, A and B, lane 4) and to 0.30-fold by 150 nm flavopiridol (Fig. 6, A and B, lane 7). The combination of both drugs reduced the levels of 28 S rRNA further to 0.06-fold (Fig. 6, A and B, lane 22), suggesting that both substances might act additively in inhibition of rRNA processing. The same additive effect was seen when cells were treated with 50 μm 5-FU (lane 5), 300 nm flavopiridol (lane 8), or a combination of both (lane 23). The result indicates that 5-FU and flavopiridol block two different maturation steps for 28 S rRNA, which are uncoupled from each other. This may establish the basis for a combinatorial analysis of more substances presented in this study.
FIGURE 6.
5-Fluorouracil and flavopiridol have additive inhibitory effects on rRNA processing. A, cells were treated with 5-FU and FL alone or in combination for 6 h as indicated. Maturation of 28 S rRNA is additively inhibited by 5-FU and FL (filled arrowheads). Levels of p53 induction are determined by Western analysis. EtBr-stained 28 S rRNA and Pes1 served as loading controls. B, quantification of the label of 28 S rRNA signals of A reveals additive inhibition of 28 S rRNA maturation by 5-FU and FL. Signal intensities were determined by phosphorimager and plotted as relative signal intensity normalized to control lane 1.
The strength of inhibition of rRNA processing correlated with the level of p53 induction (Fig. 6A). It is important to note in this context that the accumulation of p53 did not inhibit transcription of rRNA genes and labeling of the 47 S rRNA precursor. An inhibitory effect of p53 on Pol I transcription has been suggested earlier (25, 26).
DISCUSSION
The identification of the cellular targets of chemotherapeutic drugs is an important challenge for the improvement of therapy regimens in the future. Previous molecular and biochemical approaches have characterized a number of potential molecular targets, but were faced with the limitation that these analyses are usually not comprehensive. In addition, many drugs have an impact on a variety of cellular structures and enzymatic activities and affect cellular processes such as ribosome biogenesis at multiple levels and often indirectly. Moreover, if a cellular target has been identified for a specific drug at the molecular level the relevance of this target for the therapeutic use could often not ultimately be determined. The lack of a general overview for a cellular system has recently been overcome in yeast: the yeast fitness data base describes the impact of chemotherapeutic drugs on cell viability dependent on the genetic background (27, 28). The growth of a library of Saccharomyces cerevisiae haplo-deficient strains was studied in response to different kinds of stresses by chemotherapeutic drugs, including 14 drugs investigated in our study. The rational of this approach is the identification of cellular targets (genes) or pathways with high sensitivity to a specific drug. The results obtained in yeast are useful for the identification of cellular targets in mammalian cells, because many cellular processes of DNA, RNA, and protein metabolism, as well as the factors involved are conserved between yeast and mammals. The identification of pathways affected in yeast should help to clarify the relevant pathways in mammals.
5-FU is a potent chemotherapeutic drug described as an inhibitor of the enzyme thymidylate synthetase. Inhibition of this enzyme leads to a depletion of dTTP pools accompanied by a misincorporation of deoxyuridine into newly synthesized DNA and irreversible DNA damage. The effects of 5-FU on DNA synthesis have been characterized in detail (29–32). However, mounting evidence indicates that 5-FU also has important effects on RNA metabolism that contribute significantly to the toxicity of the drug. Incorporation of 5-FU into RNA inhibits rRNA processing, post-transcriptional modification of tRNA, rRNA, and snRNA as well as mRNA splicing (2–9). 5-FU-marked RNAs inhibit pseudouridylation, the most abundant post-transcriptional modification of noncoding RNA, and are target of exosome-mediated degradation (33). Interestingly, haplo-insufficiency of several components of the exosome, namely MTR3, Rrp4, Rrp6, Rrp42, and Rrp46, make yeast strains extremely sensitive to 5-FU (27), suggesting that exosome-linked processes are the major cellular targets of 5-FU in terms of its growth inhibitory and cytotoxic behavior in yeast. This is in line with the observations that yeast cells with a functional knock-out of the exosome accumulate huge amounts of unprocessed rRNAs in response to 5-FU (34), and that 5-FU enhances exosome-dependent accumulation of polyadenylated ribosomal RNA in a discrete domain within the nucleolus (2). Because inhibition of rRNA processing triggers inhibition of p53 degradation, the cellular response to 5-FU consistently involves the stabilization of p53. Disruption of p53 in vitro and in vivo renders cells strikingly resistant to the effects of 5-FU (1).
Whereas there is ample evidence in the literature for targeting of RNA metabolism by 5-FU, this question has not fully been addressed for substances like oxaliplatin, doxorubicin, mitoxanthrone, methotrexate, or actinomycin D, which all inhibited the synthesis of the 47 S rRNA precursor. Several of these substances have been studied in haplo-insufficient yeast strains (27, 28). Unexpectedly, yeast strains haplo-insufficient for genes of RNA metabolism including several subunits of the exosome, but also essential factors for the formation of 90 S preribosome as well as nucleolar RNA helicases, displayed a particular high sensitivity to these drugs. Again, this observation identified the nucleolus and ribosome biogenesis as the cellular process with the highest sensitivity toward chemotherapeutic drugs. But how can the exosome be a critical factor for substances, if the production of the 47 S rRNA precursor is blocked? A recent study showed that actinomycin D, a global inhibitor of transcription, does not block cellular transcription entirely. In exosome-depleted cells, high levels of upstream promoter transcripts have been reported to accumulate in the presence of Actinomycin D (35). Thus, an imbalance of the RNAs produced could render cells particular dependent on exosome function after treatment with actinomycin D.
Besides rRNA transcription, and late rRNA processing steps, chemotherapeutic drugs could also inhibit the early rRNA processing steps required for the production of the 32 S rRNA precursor. Several kinase inhibitors could specifically block the processing of the internal transcribed sequence 1 (ITS1, see Fig. 1) without having a detectable effect upstream on the production of the 47 S rRNA. The targets of these inhibitors involve several cyclin-dependent kinases, which are critical for RNA polymerase II-dependent gene expression. Thus, the effect of these inhibitors on early rRNA processing could be indirect. However, inhibition of protein synthesis by cycloheximide (supplemental Fig. S2I) had only a little effect on rRNA transcription and early rRNA processing steps, supporting the notion that short-lived proteins are not critical downstream targets of the applied kinase inhibitors. The kinase inhibitors used in our study have not yet been tested in the yeast fitness data base. However, treatment of haplo-insufficient yeast strains with 5-FU or methotrexate identified the kinases CMK1 and YCK2 (25). Mammalian homologues of CMK1 and YCK2 are glycogen synthase kinase 3 (GSK3) and casein kinase 1, respectively. Interestingly, roscovitine and flavopiridol are currently used in clinical trials as CDK/GSK3 inhibitors for the treatment of renal cell carcinoma (36) and high risk chronic lymphatic leukemia patients (37).
In our study, we identified different cellular levels of rRNA synthesis as targets for inhibition by chemotherapeutic drugs. This raised the question, whether combinations of drugs were able to inhibit rRNA synthesis synergistically. We tested two drugs, which affected rRNA processing at different levels. Flavopiridol inhibited the formation of the 32 S rRNA precursor, while 5-FU specifically inhibited the formation of the mature 28 S and 18 S rRNAs. The flow diagram in Fig. 1 suggests that the formation of the 32 S rRNA is a major intermediate form of the processing cascade and appears to be a prerequisite for the formation of the 28 S rRNA. Surprisingly, flavopiridol and 5-FU inhibited the formation of the 28 S rRNA additively, suggesting that the flowchart in Fig. 1C does not necessarily reflect the quantitative order of processing steps in vivo. The flowchart may depict the stability of processing intermediates rather than their quantitative importance for the entire rRNA processing process. Otherwise, a plausible explanation for the synergism is difficult to achieve. An explanation could be that flavopiridol is inhibitory for the proper 3′-end processing of the 28 S rRNA while 5-FU inhibits the proper formation of the 5′-end of 28 S rRNA. Alternatively, the drugs act additively simply because neither drug gives absolute inhibition at the concentration tested, and the fractional decrease from one drug is maintained in the presence of the other. Thus, our assay does not only identify the cellular levels for action of drugs, but also may allow the testing for their synergism.
In conclusion, we have identified a large panel of chemotherapeutic drugs with an inhibitory effect on ribosome biogenesis. These drugs inhibit either the production (transcription) of the 47 S rRNA precursor or different processing steps further downstream. The question, whether the drugs used in our study act as direct inhibitors of distinct steps of ribosome biogenesis has not been addressed. Many effects are probably indirect.
Combinations of drugs, like oxaliplatin/5-FU, methotrexate/5-FU, are currently used in protocols for the treatment of various forms of cancer. Whether these drugs also act synergistically in therapeutic regimens in vivo is unknown and has to be clarified. The utilization of the yeast fitness data base identified the RNA metabolism, and in particular rRNA synthesis, as a major target of many chemotherapeutic drugs. If this holds true for cancer cells, the question arises: how important is the genotoxic activity of many drugs for their therapeutic efficacy? If the therapeutic efficacy is achieved mainly via the RNA metabolism, as already proven for 5-FU, the screen for new substances with less genotoxic activity can even be considered for inhibition of the RNA metabolism, with a focus on rRNA synthesis.
Supplementary Material
Acknowledgment
We thank Dr. Rob Chapman for reading the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB684 and SFB-Transregio 5), Fonds der Chemischen Industrie, and German José Carreras Leukemia Foundation (DJCLS, Project F09/03).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
- 5-FU
- 5-fluorouracil
- PBS
- phosphate-buffered saline
- FBS
- fetal bovine serum
- NPM
- nucleophosmin
- DAPI
- 4′,6-diamidino-2-phenylindole
- DME
- Dulbecco's modified Eagle's
- DRB
- 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole.
REFERENCES
- 1.Bunz F., Hwang P. M., Torrance C., Waldman T., Zhang Y., Dillehay L., Williams J., Lengauer C., Kinzler K. W., Vogelstein B. (1999) J. Clin. Invest. 104, 263–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fang F., Hoskins J., Butler J. S. (2004) Mol. Cell. Biol. 24, 10766–10776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghoshal K., Jacob S. T. (1994) Cancer Res. 54, 632–636 [PubMed] [Google Scholar]
- 4.Lenz H. J., Manno D. J., Danenberg K. D., Danenberg P. V. (1994) J. Biol. Chem. 269, 31962–31968 [PubMed] [Google Scholar]
- 5.Longley D. B., Harkin D. P., Johnston P. G. (2003) Nat. Rev. Cancer 3, 330–338 [DOI] [PubMed] [Google Scholar]
- 6.Parker W. B., Cheng Y. C. (1990) Pharmacol. Ther. 48, 381–395 [DOI] [PubMed] [Google Scholar]
- 7.Yu Y. T., Shu M. D., Steitz J. A. (1998) EMBO J. 17, 5783–5795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao X., Yu Y. T. (2004) RNA 10, 681–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao X., Yu Y. T. (2007) Nucleic Acids Res. 35, 550–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rubbi C. P., Milner J. (2003) EMBO J. 22, 6068–6077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dai M. S., Zeng S. X., Jin Y., Sun X. X., David L., Lu H. (2004) Mol. Cell. Biol. 24, 7654–7668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horn H. F., Vousden K. H. (2008) Oncogene 27, 5774–5784 [DOI] [PubMed] [Google Scholar]
- 13.Lindström M. S., Jin A., Deisenroth C., White Wolf G., Zhang Y. (2007) Mol. Cell. Biol. 27, 1056–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lohrum M. A., Ludwig R. L., Kubbutat M. H., Hanlon M., Vousden K. H. (2003) Cancer Cell 3, 577–587 [DOI] [PubMed] [Google Scholar]
- 15.Sun X. X., Dai M. S., Lu H. (2007) J. Biol. Chem. 282, 8052–8059 [DOI] [PubMed] [Google Scholar]
- 16.Yuan X., Zhou Y., Casanova E., Chai M., Kiss E., Gröne H. J., Schütz G., Grummt I. (2005) Mol. Cell 19, 77–87 [DOI] [PubMed] [Google Scholar]
- 17.Grimm T., Hölzel M., Rohrmoser M., Harasim T., Malamoussi A., Gruber-Eber A., Kremmer E., Eick D. (2006) Nucleic Acids Res. 34, 3030–3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hölzel M., Rohrmoser M., Schlee M., Grimm T., Harasim T., Malamoussi A., Gruber-Eber A., Kremmer E., Hiddemann W., Bornkamm G. W., Eick D. (2005) J. Cell Biol. 170, 367–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rohrmoser M., Hölzel M., Grimm T., Malamoussi A., Harasim T., Orban M., Pfisterer I., Gruber-Eber A., Kremmer E., Eick D. (2007) Mol. Cell. Biol. 27, 3682–3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlosser I., Hölzel M., Mürnseer M., Burtscher H., Weidle U. H., Eick D. (2003) Nucleic Acids Res. 31, 6148–6156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holzel M., Orban M., Hochstatter J., Rohrmoser M., Harasim T., Malamoussi A., Kremmer E., Laengst G., Eick D. (2010) J. Biol. Chem. 285, 6364–6370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gagnon-Kugler T., Langlois F., Stefanovsky V., Lessard F., Moss T. (2009) Mol. Cell 28, 414–425 [DOI] [PubMed] [Google Scholar]
- 23.Shav-Tal Y., Blechman J., Darzacq X., Montagna C., Dye B. T., Patton J. G., Singer R. H., Zipori D. (2005) Mol. Biol. Cell 16, 2395–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sirri V., Hernandez-Verdun D., Roussel P. (2002) J. Cell Biol. 156, 969–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Budde A., Grummt I. (1999) Oncogene 18, 1119–1124 [DOI] [PubMed] [Google Scholar]
- 26.Zhai W., Comai L. (2000) Mol. Cell. Biol. 20, 5930–5938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hillenmeyer M. E., Fung E., Wildenhain J., Pierce S. E., Hoon S., Lee W., Proctor M., St Onge R. P., Tyers M., Koller D., Altman R. B., Davis R. W., Nislow C., Giaever G. (2008) Science 320, 362–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lum P. Y., Armour C. D., Stepaniants S. B., Cavet G., Wolf M. K., Butler J. S., Hinshaw J. C., Garnier P., Prestwich G. D., Leonardson A., Garrett-Engele P., Rush C. M., Bard M., Schimmack G., Phillips J. W., Roberts C. J., Shoemaker D. D. (2004) Cell 116, 121–137 [DOI] [PubMed] [Google Scholar]
- 29.Dornfeld K., Johnson M. (2005) Nucleic Acids Res. 33, 6644–6653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kouzminova E. A., Kuzminov A. (2006) J. Mol. Biol. 355, 20–33 [DOI] [PubMed] [Google Scholar]
- 31.Ladner R. D. (2001) Curr. Protein Pept. Sci. 2, 361–370 [DOI] [PubMed] [Google Scholar]
- 32.Seiple L., Jaruga P., Dizdaroglu M., Stivers J. T. (2006) Nucleic Acids Res. 34, 140–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoskins J., Butler J. S. (2008) Genetics 179, 323–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carneiro T., Carvalho C., Braga J., Rino J., Milligan L., Tollervey D., Carmo- Fonseca M. (2007) Mol. Cell. Biol. 27, 4157–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Preker P., Nielsen J., Kammler S., Lykke-Andersen S., Christensen M. S., Mapendano C. K., Schierup M. H., Jensen T. H. (2008) Science 322, 1851–1854 [DOI] [PubMed] [Google Scholar]
- 36.Soos T. J., Meijer L., Nelson P. J. (2006) Drug News Perspect. 19, 325–328 [DOI] [PubMed] [Google Scholar]
- 37.Byrd J. C., Lin T. S., Dalton J. T., Wu D., Phelps M. A., Fischer B., Moran M., Blum K. A., Rovin B., Brooker-McEldowney M., Broering S., Schaaf L. J., Johnson A. J., Lucas D. M., Heerema N. A., Lozanski G., Young D. C., Suarez J. R., Colevas A. D., Grever M. R. (2007) Blood 109, 399–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.