Abstract
TRPC4 and TRPC5 are two closely related members of the mammalian transient receptor potential cation channel family that have been implicated in important physiological functions, such as growth cone guidance and smooth muscle contraction. To further unravel the role of TRPC4 and TRPC5 in these processes in vivo, detailed information about the molecular composition of native channel complexes and their association with cellular signaling networks is needed. We therefore searched a human aortic cDNA library for novel TRPC4-interacting proteins using a modified yeast two-hybrid assay. This screen identified SESTD1, a previously uncharacterized protein containing a lipid-binding SEC14-like domain as well as spectrin-type cytoskeleton interaction domains. SESTD1 was found to associate with TRPC4 and TRPC5 via the channel's calmodulin- and inositol 1,4,5-trisphosphate receptor-binding domain. In functional studies, we demonstrate that SESTD1 binds several phospholipid species in vitro and is essential for efficient receptor-mediated activation of TRPC5. Notably, phospholipid binding to SESTD1 was Ca2+-dependent. Because TRPC4 and -5 conduct Ca2+, SESTD1-channel signaling may be bidirectional and also couple TRPC activity to lipid signaling through SESTD1. The modulation of TRPC channel function by specific lipid-binding proteins, such as SESTD1, adds another facet to the complex regulation of these channels complementary to the previously described effects of direct channel-phospholipid interaction.
Keywords: Calcium, Channels/Ion, Lipid/Phospholipid, Membrane/Channels, Membrane/Trafficking, Signal Transduction/Calcium
Introduction
Seven homologous channel proteins belong to the TRPC3 family of nonselective cation channels. Within the TRPC family, TRPC4 and TRPC5 represent a structurally distinct subgroup that is characterized by its ability to form homo- and heteromultimeric channels with each other as well as with TRPC1 (1).
Moreover, TRPC4 and TRPC5 are functionally set apart from other TRPCs by their unique activation mechanism that is dependent on phospholipase C activity but does not involve the phospholipid hydrolysis product diacylglycerol (2). Further signaling pathways, including Ca2+ store depletion and diverse lipid-channel interactions, were also shown to control channel function (3–7). Nevertheless, the exact principles that govern TRPC4 and -5 gating have not been finally clarified and probably depend on the cellular context.
Studies in knock-out mice showed that TRPC4 plays an important role in vascular physiology. TRPC4−/− mice have markedly reduced store- and receptor-induced endothelial Ca2+ entry and impaired endothelium-dependent vasorelaxation (8). Moreover, TRPC4 knock-out strongly reduced acetylcholine-activated non-selective cation currents in visceral smooth muscle cells that are involved in the regulation of gastric motility (9). Similarly, functional expression of TRPC5 has been found in several vascular tissues, including human saphenous vein (10) and rabbit arteriolar smooth muscle cells (11). In addition, both TRPC4 and TRPC5 are strongly expressed in neuronal tissue and have been assigned functions in neurotransmitter release (12) and regulation of neurite growth, respectively (13, 14). Most recently TRPC5 has also been found to control innate fear behavior in mice (15).
Despite their physiological importance, the molecular composition of endogenous TRPC4 and -5 channel complexes is only poorly understood. Experiments in heterologous expression systems suggest that TRPC4 and TRPC5 can form functionally diverse channel complexes dependent on the co-assembly with other TRPC subunits, in particular TRPC1 (16). In addition to subunit heteromerization, several other accessory proteins can interact with TRPC4 and -5 and modulate plasma membrane targeting, gating, and downstream signaling of the channel complexes. Recently, the endoplasmic reticulum Ca2+ sensor STIM1 was demonstrated to co-immunoprecipitate with both channels, and siRNA knockdown of STIM1 prevented receptor-mediated activation (17). Also a direct interaction with the inositol 1,4,5-trisphosphate receptor has been proposed to link endoplasmic reticulum signaling with channel activity (18).
Besides a close association with the endoplasmic reticulum, several lines of evidence indicate a similar structural relationship with the cytoskeleton. Binding of TRPC4 to spectrin was shown to regulate basal and EGF-induced membrane insertion of the channel (19). Furthermore, interaction with adaptor proteins, such as protein 4.1 and NHERF/EBP50, was necessary for store-dependent activation of TRPC4 in endothelial cells and membrane localization in HEK cells (20, 21). Likewise, TRPC5 was found in a proteomics screen to interact with several cytoskeletal components (22). Interestingly, proper localization of TRPC5 in neuronal cells was mediated via binding to the microtubule-destabilizing protein stathmin (13).
Although it becomes increasingly evident that TRPC4 and -5 as well as other TRP homologs are embedded in supramolecular signaling complexes (“signalplexes”) (23), it remains to be elucidated how the different proteins work together to determine channel function. Moreover, the auxiliary proteins that co-localize and regulate TRPCs in diverse signalplexes are just beginning to emerge.
Our experiments focused on the identification of components of TRPC4 and -5 channels in vascular cells. SESTD1, a previously uncharacterized lipid-binding protein strongly expressed in aorta, was found to interact with both TRPC proteins and modulate receptor-activated TRPC5 currents in a recombinant cell line. Determination of whether SESTD1 is involved in phospholipid regulation of TRPC channels in vivo and contributes to tissue-specific function awaits further studies.
EXPERIMENTAL PROCEDURES
Yeast Two-hybrid Screening
In order to identify novel TRPC4-interacting proteins, we screened a human aortic cDNA library (Clontech) using the MATCHMAKER 3 system (Clontech). The TRPC4 yeast two-hybrid bait was constructed by integrating a sequence corresponding to the C terminus of murine TRPC4α (amino acids 615–974) in frame into pGBKT7 (Clontech). Because TRPC channels are thought to consist of tetramers, a mutated GCN4-leucine zipper (24, 25) was inserted between the C terminus and the GAL4-DNA binding domain. The amino acid sequence of the zipper and flanking sequences was GGGSG SRMKQ IEDKL EEILS KLYHI ENELA RIKKL LGERG GSGSA AA. The TRPC5 bait was constructed the same way by inserting the coding sequence of amino acids 619–975 of mTRPC5 into pGBKT7. For control experiments, the C termini of the channels were replaced by enhanced GFP. To map the SESTD1 interaction site on TRPC4, monomeric truncation bait constructs without the zipper were generated.
After transformation of yeast, clones surviving on −Trp/−Leu/−His/−Ade agar plates were tested for β-galactosidase activity. Prey plasmids from positive clones were recovered and sequenced. One clone contained the full-length human SESTD1 cDNA (GenBankTM accession number NM_178123). The specificity of the yeast two-hybrid interaction was verified by co-transformation of the SESTD1 prey construct with the enhanced GFP control vector as bait.
Glutathione S-Transferase (GST) Pull-down
Constructs for recombinant expression of GST fusion proteins in bacteria were generated by inserting SESTD1 cDNA fragments coding for the Sec 14 (amino acids 1–192), Spec 1 (amino acids 193–406), or Spec 2 (amino acids 407–696) domain in pGEX-5X-3 (Amersham Biosciences). GST fusion proteins were recombinantly expressed in One Shot BL21 chemically competent Escherichia coli (Invitrogen). Fusion proteins were purified from bacteria using glutathione-Sepharose beads (Amersham Biosciences) and directly used for pull-down experiments.
Lysates (50 μg of total protein) of HEK293 cells transiently transfected with mTRPC4α, mTRPC4β (26), or mTRPC5 (27) were incubated with equal amounts of glutathione Sepharose-bound GST or GST fusion proteins and incubated for 2 h at room temperature. After washing, samples were eluted with NuPAGE LDS sample buffer (Invitrogen), heat-denatured, centrifuged, and analyzed by Western blot.
Co-immunoprecipitation
Plasmids coding for FLAG-tagged mTRPC4β and HA-tagged SESTD1 were constructed using pcDNA3.1-nFLAG-DEST (Invitrogen) and pCMV-HA (Invitrogen), respectively.
HM1 cells were co-transfected with FLAG-tagged mTRPC4, GFP-tagged mTRPC5 (28), and either HA-tagged SESTD1 or pcDNA3.1 as a control. 24 h after transfection, cells were washed with phosphate-buffered saline and scraped into lysis buffer (1 mm EDTA, 150 mm NaCl, 50 mm Tris-HCl, 1% Triton X-100, pH 7.4, supplemented with Complete protease inhibitor mix (Roche Applied Science). After centrifugation (15 min, 16,000 × g) supernatants were incubated with anti-TRPC4 or anti-GFP antibodies at 4 °C overnight.
Samples were then incubated with protein A-Sepharose or protein G-Sepharose (Amersham Biosciences). After washing with lysis buffer, precipitated proteins were eluted with NuPAGE LDS sample buffer (Invitrogen), heat-denatured, centrifuged, and analyzed by Western blot.
SDS-PAGE and Western Blots
Before electrophoresis, protein content of probes was quantified with the BCA protein assay kit (Pierce). Samples prepared in NuPAGE LDS sample buffer (Invitrogen) were supplemented with 5% β-mercaptoethanol, heat-denatured (95 °C, 3 min), centrifuged (16,000 × g, 2 min), and loaded on 4–12% BisTris-HCl gradient gels (Invitrogen). Electrophoresis was carried out in NuPAGE MOPS SDS running buffer (Invitrogen). Proteins were transferred to nitrocellulose membranes using NuPAGE transfer buffer (Invitrogen) and a Novex Xcell 2 blot module (Invitrogen) or the iBlot gel transfer system (Invitrogen) according to the manufacturer's instructions. To verify transfer, the blotted proteins were stained with 0.1% Ponceau S solution (Sigma). The membrane was blocked with 50% Odyssey blocking buffer (LI-COR, Lincoln, NE) and 50% TBST (500 mm NaCl, 20 mm Tris-HCl, 0.6% Tween 20) before incubation with primary (1 h, room temperature) and secondary fluorescence-labeled or horseradish peroxidase-conjugated antibodies (45 min, room temperature). Proteins were analyzed using the Odyssey infrared imaging system (LI-COR).
Quantitative Reverse Transcription-PCR
Total RNA of 47 human tissues from at least five different donors and of 19 cell lines or primary cell types were isolated, and 1 μg of each total RNA was reverse-transcribed into cDNA using TaqMan reverse transcription reagents (Applied Biosystems, Foster City, CA), Oligo(dT) (2.5 μm), and random hexamer primers (2.5 μm) in 50 μl of reaction volume.
Specific probe/primer sets for analysis of human SESTD1 and the reference gene RPL37a were designed using Primer Express software (Applied Biosystems) and checked against the mammalian expressed sequences.
The following primers were used: RPL37a-forward, 5′-ACAGCGGAAGTGGTATTGTACGT-3′; RPL37a-reverse, 5′-GGCACTGTGGTTCCTGCAT-3′; SESTD1-forward, 5′-CAACATCCCTAATAAGCCATCCA-3′; SESTD1-reverse, 5′-GCAAAATCTCTTACAAGCAGCTCTT-3′. The RPL37a TaqMan probe was 5′-CAGGCACCGCCAGCCACTGTCT-3′ labeled with VIC/TAMRA. The SESTD1 TaqMan probe was 5′-TGCTGAGACAAATCTTGGGCAACACCA-3′ labeled with FAM/TAMRA.
Real-time PCR was carried out with 5 μl of 10-fold diluted cDNA in a 20-μl final reaction volume in 384-well plates. Each experiment was done in duplicate with a duplex reaction with RPL37a as a reference gene. Concentrations of TaqMan probes, target primers, and reference primers used were 200, 900, and 50 nm, respectively. Real-time PCR was carried out on the ABI PRISM 7900 Sequence Detection System using TaqMan Universal PCR Master Mix (Applied Biosystems). PCR cycling conditions were as follows: AmpliTaq Gold enzyme activation for 10 min at 95 °C, followed by 40 cycles of PCR amplification (denaturation step, 15 s at 95 °C; annealing/extension step, 1 min at 60 °C).
The relative abundance of the gene of interest was calculated relative to the expression of RPL37a as follows, relative expression = 2−(ΔCt) × 1000, where ΔCt is the difference between the Ct values of SESTD1 and RPL37a. SESTD1 is considered to be significantly expressed if the Ct value is below 35 and ΔCt between duplicates is less than 1.
Cell Culture, Transfection, and Cell Line Generation
Cells were grown at 37 °C in a humidified atmosphere (5 or 7% CO2) under standard cell culture conditions. HEK293 cells were maintained in Iscove medium (Biochrom, Berlin, Germany) supplemented with 10% fetal bovine serum (Biochrom) and 2 mm glutamine (Invitrogen). HEK293 cells stably expressing the human muscarinic receptor type 1 (HM1 cells) (29) were maintained in Dulbecco's modified Eagle's medium/nutrient F-12 with GlutaMAX I (Invitrogen) supplemented with 10% fetal bovine serum (PAA, Pasching, Austria), 1 mm glutamine, and 400 μg/ml geneticine (Invitrogen). 50 μg/ml zeocin (Invitrogen) was added for selection and culture of clones stably expressing mTRPC5-YFP (HM1-C5 cells).
Primary human aortic smooth muscle cells, coronary artery smooth muscle cells, aortic artery endothelial cells, microvascular endothelial cells (Cambrex, East Rutherford, USA), and the A7r5 cell line (ATCC, Manassas, VA) were cultured according to the provider's instructions.
Cells were transfected in growth medium using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. For siRNA experiments, HM1-C5 cells were transfected at 30–50% confluence with either nonspecific SilencerR negative control number 2 (Ambion, Austin, TX), liposomes only (mock) or with a specific SESTD1 siRNA pool consisting of an equimolar mixture of three siGENOME human SESTD1 duplexes (Dharmacon, Lafayette, LA).
Transfection efficiency was determined either by Western blot, enhanced GFP controls, or siGLO red transfection indicator (Dharmacon). Transfection rates of 80% and more were defined as mandatory for functional experiments with cell populations.
In order to generate a cell line with robust TRPC5 currents, we utilized HM1 cells that overexpress the human muscarinic m1 receptor (29). A mTRPC5-YFP fusion construct was derived from mTRPC5-GFP (28) by replacing enhanced GFP with YFP and transfer into pcDNA3.1(−)zeo (Invitrogen). The stable HM1-C5 cell line was generated by clonal selection starting 24 h post-transfection. Clones were functionally characterized by fluorometric Ca2+ measurements and patch clamp experiments, and one clone was selected for further studies.
Electrophysiological Techniques
Ion currents were measured with the whole-cell patch clamp technique (30). Heat-polished patch pipettes with resistances of 2–4 megaohms were pulled from borosilicate glass capillaries (Hilgenberg, Malsfeld, Germany) using a DMZ-Universal puller (Zeitz-Instruments, Munich, Germany) and filled with standard intracellular solution (120 mm CsOH, 120 mm gluconic acid, 2 mm MgCl2, 3 mm CaCl2, 5 mm Cs4-BAPTA, 10 mm HEPES, pH 7.4).
Cells grown on poly-l-lysine-coated coverslips were continuously superfused with extracellular solution (140 mm NaCl, 5.4 mm KCl, 2 mm CaCl2, 1 mm MgCl2, 10 mm glucose, 10 mm HEPES, pH 7.4), and agonists were applied using an ALA BPS-8 perfusion system (ALA Scientific Instruments, Westbury, NY). Whole-cell recordings were performed with an EPC-10 amplifier and Pulse software (HEKA, Lambrecht, Germany). Cells were held at −70 mV, and current-voltage (I-V) relationships were routinely measured every 3 s by applying voltage ramps (180 ms) from −100 mV to +80 mV.
Data were acquired at 6.67 kHz and filtered with 2.22 kHz. A series resistance of <10 megaohms was accepted and routinely compensated by 50–70%. Liquid junction potentials (∼12 mV) were not corrected. All experiments were performed at room temperature.
Measurement of [Ca2+]i
Fluo-4 Measurements
HM1 or HM1-C5 cells grown to an almost confluent monolayer on black poly-d-lysine-coated 96-well plates (Greiner, Frickenhausen, Germany) were washed with standard extracellular solution (135 mm NaCl, 1 mm MgCl2, 5.4 mm KCl, 2 mm CaCl2, 10 mm HEPES, 10 mm glucose, pH 7.35) and stained (30 min, room temperature) with dye solution (2 μm fluo-4/AM, 0.02% Pluronic F127, 0.1% bovine serum albumin in standard extracellular solution). Cells were washed and incubated with either standard extracellular solution or Ca2+-free standard extracellular solution (135 mm NaCl, 1 mm MgCl2, 5.4 mm KCl, 0.5 mm EGTA, 10 mm HEPES, 10 mm glucose, pH 7.35). Fluo-4 fluorescence was excited at 488 nm with an argon laser and measured using a fluorometric imaging plate reader (Molecular Devices, Sunnyvale, CA).
Fura-2 Measurements
HM1-C5 cells were seeded in black poly-l-lysine-coated glass bottom 96-well plates (Sensoplates; Greiner) and grown overnight to an almost confluent monolayer. They were loaded in standard extracellular solution supplemented with 2 μm fura-2/AM (30 min, 37 °C) and allowed to de-esterify (15 min, 37 °C). Subsequently, cells were either incubated with standard extracellular solution or Ca2+-free standard extracellular solution. Intracellular calcium signals were ratiometrically measured in a FLEX station (Molecular Devices). Fluorescence was excited, alternating at 340 and 380 nm, long pass-filtered at 495 nm, and captured at 4-s intervals. The F340/F380 ratio was calculated using SoftMax Pro software (Molecular Devices). Fluorescence ratios were plotted versus time, and aggregate Ca2+ changes were derived from the area under the curve with SigmaPlot 9 (Systat Software Inc., Chicago, IL). All fluorometric [Ca2+]i measurements were performed at room temperature.
Phospholipid Overlay Assay
PIP strips (Echelon, Salt Lake City, UT) were blocked with either TBST (150 mm NaCl, 10 mm Tris-HCl, 0.1% Tween 20, pH 8), TBST with 2.5 μm free Ca2+ (150 mm NaCl, 10 mm Tris-HCl, 1 mm EGTA, 1 mm MgCl2, 1 mm CaCl2, 0.1% Tween 20, pH 8), or TBST with 0.06 μm free Ca2+ (150 mm NaCl, 10 mm Tris-HCl, 1 mm EGTA, 1 mm MgCl2, 0.9 mm CaCl2, 0.1% Tween 20, pH 8) and 3% essentially fatty acid-free bovine serum albumin (Sigma). Free Ca2+ concentrations were calculated using Cabuf software (G. Droogmans; available on the Internet).
A GST-SESTD1 fusion construct was prepared by inserting full-length human SESTD1 in pGEX-4T-1 (Amersham Biosciences). The fusion protein was expressed and purified as described above. GST-SESTD1 was eluted from glutathione-Sepharose beads with elution buffer (20 mm glutathione, 50 mm Tris-HCl, pH 8) and dialyzed against 50 mm Tris-HCl buffer (pH 8). PIP strips were incubated with 500 ng/ml purified GST or GST-SESTD1 protein (4 h, room temperature) in the respective Ca2+-containing or Ca2+-free blocking buffer. Subsequently, strips were washed with blocking buffer and incubated at 4 °C overnight with anti-GST antibody followed by secondary horseradish peroxidase-conjugated anti-rabbit antibody. Lumi-LightPLUS Western blotting substrate (Roche Applied Science) was added, and the chemiluminescent signals were analyzed with a Lumi imager (Roche Applied Science).
Materials
An affinity-purified rabbit polyclonal antibody directed against the epitope KRQQLRHPEMVTTES (amino acids 682–696) of human SESTD1 was custom-made by Eurogentec (Seraing, Belgium). Other primary antibodies used were rat anti-HA and mouse anti-GFP (Roche Applied Science), mouse anti-GAPDH (Chemicon, Wiesbaden, Germany), rabbit anti-GST (Sigma), and rabbit anti-TRPC4 and anti-TRPC5 (Alomone, Jerusalem, Israel). Secondary antibodies used were Alexa Fluor 680 goat anti-rabbit, Alexa Fluor 680 goat anti-rat, and Alexa Fluor 680 rabbit anti-mouse (Invitrogen) and goat anti-rabbit horseradish peroxidase-conjugated (Pierce). siRNAs against human SESTD1 (siGENOME duplexes D-018379-01, D-018379-03, and D-018379-04) were purchased from Dharmacon.
Fluo-4/AM, fura-2/AM, Cs4-BAPTA, and Pluronic F127 were from Invitrogen, Tris-HCl was from Invitrogen, MgCl2 was from Merck, and agarose was from Bio-Rad. All other chemicals were from Sigma.
Statistics
Averaged data are expressed as means ± S.E. For statistical analysis, analysis of variance was performed with Origin 6.0 software (Microcal Software Inc., Northampton, MA). p values less than 0.05 were considered as statistically significant.
RESULTS
To further define components and associated proteins of TRPC4 channels in smooth muscle, we screened a human aortic cDNA library for interaction with the channel's C terminus in yeast. Because TRPC channels are probably tetramers (31), we utilized a modified bait vector containing a tetramerization domain (24, 25) to aid proper assembly of the channel fragments. In this screen, several proteins, including the previously described α-spectrin, were identified as putative TRPC4 binding partners.
Sequence analysis revealed that one of the potential TRPC4-interacting proteins was SESTD1, a protein containing a SEC14-like lipid binding domain as well as two spectrin domains (Spec1 and -2), which typically mediate interaction with the cytoskeleton. Considering the dependence of TRPC4 activity on phospholipid signaling and its close link to cytoskeletal structures, SESTD1 seemed a promising candidate for a new TRPC4-regulating protein.
A directed yeast two-hybrid assay with consecutively shortened parts of the TRPC4 tail showed that binding to SESTD1 was mediated by a short 29-amino acid sequence. This sequence is highly conserved in TRPC5 and largely overlaps with the previously described calmodulin- and inositol 1,4,5-trisphosphate receptor-binding (CIRB) domain (amino acids 695–724 of mTRPC4) (Fig. 1). We hypothesized that TRPC5 also interacts with SESTD1. To confirm binding of SESTD1 to TRPC4 and TRPC5, GST pull-down experiments were performed. These experiments were also designed to define the TRPC binding region of SESTD1. Therefore, three GST fusion proteins were constructed that contained one of the identified structural domains of SESTD1 (SEC14-like, Spec1, and Spec2) each. The results shown in Fig. 2 demonstrate that the Spec1 domain of SESTD1 bound to full-length TRPC4α and the short TRPC4β isoform as well as TRPC5. In contrast, no interaction of the channels with Spec2 could be detected. Binding to the SEC14-like domain could not be tested in this assay because the SEC14-GST fusion protein was toxic to E. coli. However, a directed yeast two-hybrid assay using the SEC14 domain did not show any binding to TRPC4, whereas binding to Spec1 was confirmed (not shown). Thus, SESTD1 interacts via its Spec1 domain with the CIRB domains of TRPC4 and TRPC5.
FIGURE 1.
Mapping of the SESTD1-binding site on the mTRPC4α C terminus. A, illustration of truncation mutants of the mTRPC4α C terminus used in yeast two-hybrid assays to identify the SESTD1-binding site. B, yeast colonies co-transformed with the given truncation mutants as baits and full-length human SESTD1 as prey were plated on selective −Trp/−Leu/−Ade/−His agar plates. Growth (light color) indicates protein-protein interaction. C, alignment of the identified SESTD1 binding site in mTRPC4α (amino acids (aa) 700–728) with mTRPC5 shows that this region is highly conserved between the two proteins (identical and diverse amino acids are denoted by the red and blue bars, respectively). The binding site of mTRPC4α is completely conserved in mTRPC4β. wt, wild type.
FIGURE 2.
Binding of the SESTD1 spectrin 1 domain to TRPC4 and TRPC5. GST fusion proteins of the spectrin 1 or 2 domains of SESTD1 (GST-Spec1 or -2) were used to pull down full-length mTRPC4α (A), mTRPC4β (B), or mTRPC5 (C) from lysates of HEK293 cells transiently transfected with the respective cDNAs. Immunoprecipitates were Western blotted and stained with anti-TRPC4 (A and B) or anti-TRPC5 (C). 10% of the lysate input was run as expression control. GST alone was used to verify specificity of the pull-down assays.
To verify the significance of the interaction in intact cells, we attempted co-immunoprecipitation of SESTD1 with TRPC4β and TRPC5 proteins. For this purpose, HM1 cells were first transfected with HA-tagged SESTD1, and the protein was detected on Western blots using an HA antibody or a direct anti-SESTD1 antibody. Both antibodies recognized an ∼80-kDa protein in accordance with the predicted molecular mass of SESTD1 (Fig. 3A). After co-transfection with tagged channel constructs, SESTD1 could be identified in TRPC4 and -5 immunoprecipitates (Fig. 3B), confirming its association with the TRPC channel complexes.
FIGURE 3.
SESTD1 co-immunoprecipitates with mTRPC4β and mTRPC5. A, lysates of nontransfected HM1 cells (−) and cells transfected with HA-SESTD1 (+) were analyzed on Western blots using anti-HA antibodies and a polyclonal antibody against SESTD1. Both antibodies detected an ∼80-kDa protein in transfected cells, indicating that this protein is SESTD1. Anti-SESTD1 also detected an 80-kDa protein in nontransfected HM1 cells that we suggest is native SESTD1 (see also Fig. 7). In addition, the antibody recognized an unrelated ∼50-kDa protein. B, Western blot of anti-TRPC4 immunoprecipitates (P) and the corresponding lysates (L) from HM1 cells transfected with HA-tagged SESTD1 and FLAG-tagged mTRPC4β or HA-tagged SESTD1 alone (left). Shown are Western blots of anti-GFP immunoprecipitates (P) and the corresponding lysates (L) from HM1 cells transfected with HA-tagged SESTD1 and GFP-tagged mTRPC5 or HA-tagged SESTD1 alone (right). IP, immunoprecipitation; WB, Western blot.
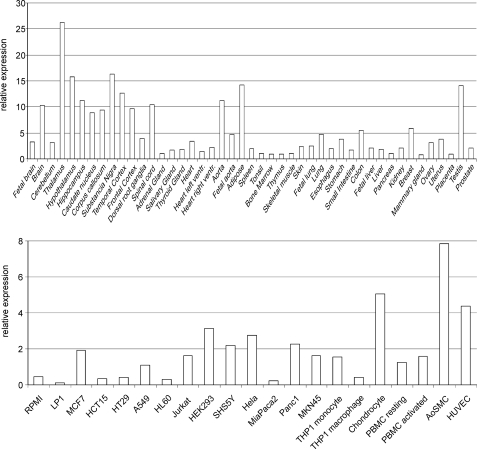
Before investigating possible functional consequences of the SESTD1-TRPC channel interaction, we wondered whether SESTD1 and TRPC4 and -5 are indeed co-expressed in vivo. To approach this question, we conducted an expression profiling of SESTD1. Using quantitative RT-PCR, we found that SESTD1 is broadly expressed in human tissues and cells at moderate to low levels. The highest abundance of SESTD1 mRNA was measured in thalamus, with expression levels reaching about 2.5% of that of the housekeeping gene RPL37a (Fig. 4). Interestingly, SESTD1 shows relatively high expression in brain and is also significantly expressed in vasculature, two tissues in which TRPC4 and -5 had previously been detected (8, 27, 32, 33). The presence of SESTD1 in several human vascular cell types and in rat brain was confirmed on the protein level by Western blot (Fig. 5). Unfortunately, however, we were unable to unequivocally isolate TRPC4/5-like currents in SESTD1-expressing cells derived from these tissues, hampering the study of native SESTD1 and TRPC protein interaction.
FIGURE 4.
Quantitative expression profiling of SESTD1 mRNA. SESTD1 mRNA expression was determined in different human tissues (top) or cell lines and selected primary cell types (bottom) using quantitative RT-PCR. Expression levels were normalized to the expression of the housekeeping gene RPL37a. Data shown are means of duplicates. Cell line designations are according to the ATCC and standard literature. Primary cells used were chondrocytes, peripheral blood mononuclear cells (PBMC), aortic smooth muscle cells (AoSMC), and human umbilical vein endothelial cells (HUVEC).
FIGURE 5.
Expression of SESTD1 in different rat and human tissues. Proteins from rat brain microsomes (brain), total cell lysates of the rat smooth muscle cell line A7r5, primary human aortic endothelial cells (HAEC), microvascular endothelial cells (HMVEC), coronary smooth muscle cells (CASMC), and aortic smooth muscle cells (AoSMC) were separated by SDS-PAGE, and SESTD1 was detected on Western blots using anti-SESTD1 antibody.
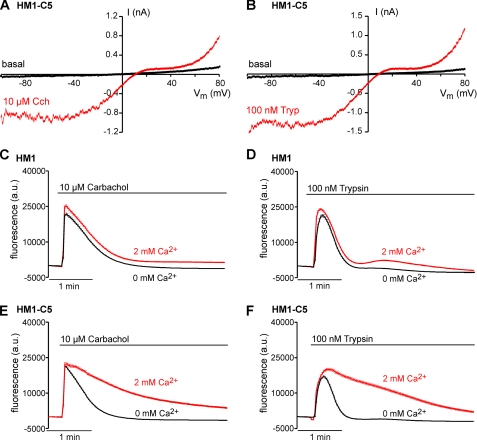
When testing SESTD1 antibodies, we noted a protein of the expected size of SESTD1 in nontransfected HM1 cells (see Fig. 3), suggesting that these cells contain SESTD1 endogenously. Therefore, we decided to use a stable HM1 cell line expressing mTRPC5-YFP (HM1-C5 cells) for further studies. In response to the muscarinic agonist carbachol and the protease-activated receptor agonist trypsin, HM1-C5 cells showed whole-cell currents undistinguishable from currents through wild-type mTRPC5 channels that lead to prolonged Ca2+ influx compared with the parental cell line (Fig. 6). An siRNA knockdown strategy was employed to test the effect of SESTD1 abolition on TRPC5. As illustrated in Fig. 7, transfection of specific siRNA against SESTD1 reduced SESTD1 protein by about 85%, whereas expression of an unrelated protein also recognized by anti-SESTD1 and expression of the housekeeping enzyme GAPDH were unchanged. Moreover, siRNA treatment had no effect on the expression of the TRPC5-YFP fusion protein.
FIGURE 6.
Functional characterization of TRPC5 currents and Ca2+ entry in HM1-C5 cells. Electrophysiological characterization of HM1-C5 cells by whole-cell patch clamp. The M1-agonist carbachol (10 μm) (A) and protease-activated receptor agonist trypsin (100 nm) (B) induced nonselective cation currents with double-rectifying I-V relationships characteristic for TRPC5. Time-dependent changes of [Ca2+]i in fluo-4-loaded cells were assayed using a fluorometric imaging plate reader. Ca2+ influx (2 mm extracellular Ca2+) or release from internal stores (0 mm extracellular Ca2+) in HM1 cells was evoked by application of 10 μm carbachol (C) or 100 nm trypsin (D). Data are means of 40–48 measurements. Ca2+ influx (2 mm extracellular Ca2+) or release from internal stores (0 mm extracellular Ca2+) in response to application of 10 μm carbachol (E) or 100 nm trypsin (F) was measured in HM1-C5 cells. Data are means of eight measurements.
FIGURE 7.
Knockdown of SESTD1 expression by specific SESTD1 siRNA. Western blot of HM1-C5 cells transfected with liposomes only (mock), 20 nm nonspecific control siRNA, or a 20 nm concentration of a specific SESTD1 siRNA pool (see “Experimental Procedures”). Blots were incubated with anti-SESTD1, anti-GFP, and anti-GAPDH antibodies. SESTD1-specific siRNA decreased SESTD1 protein by 85.5 ± 5.5% (n = 4, compared with mock-transfected cells) or 82.3 ± 5.3% (n = 4, compared with cells treated with nonspecific, non-silencing siRNA), whereas expression of TRPC5-YFP, an unrelated protein recognized by anti-SESTD1, as well as GAPDH expression were unchanged.
Fura-2 measurements of carbachol- and trypsin-induced Ca2+ transients revealed that TRPC5-mediated Ca2+ influx was significantly reduced in HM1-C5 cells treated with SESTD1-specific siRNA compared with controls treated with unspecific siRNA or mock-transfected cells (Fig. 8). Specifically, evaluation of the data at t > 120 s, when contribution of Ca2+ release was negligible, showed a significant reduction of the fluorescence ratios in SESTD1 siRNA-treated cells. Furthermore, total Ca2+ changes as determined from the area under the fluorescence curves were also decreased in SESTD1 knockdown cells. In the SESTD1 siRNA group, the mean area under the curve after carbachol stimulation amounted to 150.5 ± 6.4 arbitary units (n = 15) compared with 289.4 ± 9.8 arbitary units in the mock-treated and 277.1 ± 13.4 arbitary units in the control siRNA-treated group (both n = 15; p < 0.001 versus mock or control siRNA, analysis of variance).
FIGURE 8.
TRPC5 activity is reduced in HM1-C5 cells transfected with SESTD1 siRNA. Ratiometric measurements of [Ca2+]i in fura-2-loaded HM1-C5 cells transfected with 20 nm SESTD1 siRNA, nonspecific control siRNA, or liposomes only (Mock). Ca2+ release from internal stores was activated in Ca2+-free buffer by application of 10 μm carbachol (A) or 100 nm trypsin (B). Carbachol-induced (C) or trypsin-induced (D) TRPC5-mediated Ca2+ influx was assessed in standard Ca2+-containing buffer (2 mm). Shown are means ± S.E. of three independent experiments (each performed with n = 4–6 wells/experimental condition). Statistical analysis shows a selective reduction of agonist-induced TRPC5-mediated Ca2+ influx in SESTD1 siRNA-treated cells after termination of Ca2+ release (t = 121 s) as well as at the end of the experiment. (***, p < 0.001; *, p < 0.05 versus unselective siRNA; ***, p < 0.001 versus mock; analysis of variance).
Trypsin-induced Ca2+ transients were similarly reduced by SESTD1 down-regulation. The area under the curve derived from SESTD1 siRNA-transfected cells amounted to 138 ± 8.3 arbitary units (n = 15) versus 281.1 ± 15.6 arbitary units (n = 15) in the mock and 230.8 ± 16.9 arbitary units (n = 15) in the control siRNA group, respectively (p < 0.001 versus mock or control siRNA, analysis of variance).
In contrast, there was no significant difference between the groups when carbachol- or trypsin-induced Ca2+ release in Ca2+-free solution was compared. This indicates that the receptor signaling pathways upstream of TRPC5 were not compromised by the siRNA treatment.
Previous studies (19) have shown that TRPC4 interacted with spectrin repeats 19–21 of the cytoskeletal protein αII-spectrin via a sequence adjacent to the SESTD1-Spec1 binding site identified in this study. It is conceivable that SESTD1 utilizes a similar regulation mechanism and that its spectrin domains serve as linkers of TRPC4 and -5 to the cytoskeleton. Nevertheless, SESTD1 features another distinct structural motif, the SEC14 domain, that has been implicated in phospholipid transport in yeast and, for instance, intracellular protein targeting in mammalian cells (34). It is, likewise, possible that this domain of SESTD1 is involved in transport of the channel proteins or that it controls channel function by modifying the local lipid environment. Therefore, it was important to test the functionality of the SESTD1 SEC14 domain. To this end, we assessed the ability of SESTD1 to bind different phospholipids using an overlay assay. Fig. 9 shows that SESTD1 bound phosphatidylinositol monophosphates, phosphatidylinositol diphosphates, and phosphatidic acid, whereas no binding to less polar lipids, including phosphatidylcholine, phosphatidylserine, and phosphatidylinositol, was observed. Interestingly, binding to phosphatidylinositol diphosphates was Ca2+-dependent and was strongly reduced in buffer containing a Ca2+ concentration typical for resting cells. Thus, it is possible that Ca2+ influx through TRPC4 or -5 strengthens phospholipid binding of SESTD1. These experiments put the TRPC-SESTD1 interaction in a new perspective, suggesting a two-way functional interdependence of the associated proteins.
FIGURE 9.
Selective Ca2+-dependent binding of SESTD1 to phospholipids. PIP strips containing various immobilized phospholipids (left) were probed with GST-SESTD1 or GST in buffer containing 60 nm or 2.5 μm free Ca2+. Bound proteins were detected with anti-GST antibodies and visualized using horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence. LPA, lysophosphatidic acid; LPC, lysophosphatidylcholine; PI, phosphatidylinositol; PE, phosphatidylethanolamine; PC, phosphatidylcholine; S1P, sphingosine-1-phosphate; PS, phosphatidylserine; PA, phosphatidic acid.
DISCUSSION
In this work, we identified SESTD1 as a novel TRPC4- and TRPC5-interacting protein. SESTD1 belongs to a large class of mammalian proteins that feature a Sec14-like lipid binding domain. Definition of the Sec14 domain is based on the protein Sec14p that is essential for phospholipid transfer and secretory function in yeast (35, 36). However, the function of Sec14 domains in mammalian phospholipid transport is less clear, and no functional information was available for SESTD1.
The second structural feature of SESTD1 consists of two C-terminal spectrin repeats. Spec1 was responsible for binding to TRPC4 and TRPC5. The binding site on mTRPC4 was mapped to the proximal C terminus comprising amino acids 700–728.
The TRPC-SESTD1 interaction has striking similarities with the interaction of TRPC4 with αII- and βV-spectrin, an association crucial for regulated basal and EGF-induced plasma membrane localization of TRPC4 (19). Although αII- and βV-spectrin binding to TRPC4 was disrupted by deletion of a site (amino acids 730–758) just downstream of the SESTD1 binding site, it is possible that both binding sites are in fact continuous. In the intact channel protein, the minimal spectrin repeat binding peptide identified in our study may require the further downstream sequence described by Odell et al. (19) for proper folding or accessibility. Moreover, knockdown of αII-spectrin as well as SESTD1 led to impaired channel function. Thus, it is possible that both proteins are part of a common regulatory mechanism. It has been suggested that spectrin as well as the adaptor protein 4.1 stabilize TRPC4 in a macromolecular cytoskeleton-associated complex that is required for efficient transport of the channel to the cell membrane and/or correct association with other signaling components (19, 21). SESTD1 may act as another adaptor similar to protein 4.1 and link TRPC4 and TRPC5 to the cytoskeleton through its spectrin domains.
However, taking into account the proposed requirement of phosphatidylinositol 4,5-bisphosphate for vesicular insertion of TRPC5 into the plasma membrane (37) and the fact that SESTD1 directly binds phospholipids, another appealing hypothesis is that SESTD1 represents the primary docking protein directing membrane turnover and assembly of TRPC4 and TRPC5.
Besides regulating assembly or translocation of the channel proteins, SESTD1 may have more direct effects on TRPC function. In particular, it has been proposed that removal of calmodulin from the CIRB site contributed to TRPC4 activation (18). Because the SESTD1 binding site is almost identical to the CIRB site, competition of calmodulin by SESTD1 may be required for full activation of TRPC4 and -5.
Because we did not investigate the mechanism by which SESTD1 modulates TRPC5 currents in detail, additional experiments will be required to validate the above hypotheses and to explore possible alternative mechanisms of SESTD1-mediated TRPC channel regulation.
An important finding of this study is that the affinity of SESTD1 with selected phospholipids is Ca2+-dependent. Opening of TRPC4 and -5 significantly raises intracellular Ca2+ and, therefore, will increase binding of SESTD1 to phosphatidylinositol diphosphates. The impact of this switch on TRPC4 and -5 function is unclear, but a reinforced attachment of the channels to, for example, phosphatidylinositol 4,5-bisphosphate would be compatible with both the basic requirement of phosphatidylinositol 4,5-bisphosphate for TRPC5 activity (3) and the observed role of phosphatidylinositol 4,5-bisphosphate in TRPC5 translocation (37).
Considering its widespread expression, SESTD1 is likely to have other functions apart from TRPC channel regulation. In preliminary experiments, we observed discrete changes in cell morphology accompanied by a decreased plasma membrane localization of the adherens junction-associated protein β-catenin in SESTD1 siRNA-treated cells. Moreover, co-immunoprecipitation studies indicated an interaction of SESTD1 with β-catenin (see supplemental Fig. S1). β-Catenin has recently been found to co-assemble with TRPC4 in endothelial cells and to modulate TRPC4 activity in a cell contact-dependent manner (38). Although our findings have to be further corroborated, it is an intriguing possibility that SESTD1 is linked to β-catenin and its complex functional network.
Our study aimed at discovering novel TRPC channel-interacting proteins in vascular cells. Definition and reconstitution of vascular TRP channel complexes remains an important task because the properties of these channels have not been finally clarified. The significant uncertainty regarding the function and structure of, for example, endothelial TRPC channels is illustrated by data indicating that store-operated Ca2+ entry in certain endothelial cells is mediated by Orai1 channels rather than TRPC4 or TRPC1 (39). Nevertheless, there is convincing knock-out data demonstrating an involvement of TRPC4 in endothelial Ca2+ signaling (8). These issues continue to be considerable hurdles for the development of specific TRPC channel modulators that may have therapeutic value in the treatment of major cardiovascular diseases.
Based on its association with the C terminus of TRPC4 in yeast, we isolated SESTD1 from an aortic cDNA library. Subsequently, we confirmed expression of SESTD1 in primary human endothelial and smooth muscle cells by Western blot. Because we could not detect TRPC4 or TRPC5 proteins in the same preparations using commercially available antibodies, it will be critical for functional studies on native proteins to identify cell types that co-express both SESTD1 and TRPC4 or TRPC5. Although more work is needed to understand the physiological impact of the TRPC-SESTD1 interaction, the present findings represent a further step toward deciphering the complex associations that govern the function of TRPC4 and -5 in vivo.
Supplementary Material
Acknowledgments
We are grateful to Drs. Yasuo Mori and Michael Schäfer for TRPC channel cDNAs and Dr. David Clapham for HM1 cells. We also thank Dr. Florian Bundis for help with yeast two-hybrid assays and Drs. David Clapham, Grigory Krapivinsky, and Klaus Steinmeyer for comments on the manuscript.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- TRPC
- transient receptor potential canonical
- mTRPC
- murine TRPC
- GST
- glutathione S-transferase
- CIRB domain
- calmodulin- and inositol 1,4,5-trisphosphate receptor-binding domain
- siRNA
- small interfering RNA
- GFP
- green fluorescent protein
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- YFP
- yellow fluorescent protein
- HA
- hemagglutinin.
REFERENCES
- 1.Hofmann T., Schaefer M., Schultz G., Gudermann T. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 7461–7466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schaefer M., Plant T. D., Obukhov A. G., Hofmann T., Gudermann T., Schultz G. (2000) J. Biol. Chem. 275, 17517–17526 [DOI] [PubMed] [Google Scholar]
- 3.Trebak M., Lemonnier L., DeHaven W. I., Wedel B. J., Bird G. S., Putney J. W., Jr. (2009) Pflugers Arch. 457, 757–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flemming P. K., Dedman A. M., Xu S. Z., Li J., Zeng F., Naylor J., Benham C. D., Bateson A. N., Muraki K., Beech D. J. (2006) J. Biol. Chem. 281, 4977–4982 [DOI] [PubMed] [Google Scholar]
- 5.Philipp S., Cavalié A., Freichel M., Wissenbach U., Zimmer S., Trost C., Marquart A., Murakami M., Flockerzi V. (1996) EMBO J. 15, 6166–6171 [PMC free article] [PubMed] [Google Scholar]
- 6.Philipp S., Hambrecht J., Braslavski L., Schroth G., Freichel M., Murakami M., Cavalié A., Flockerzi V. (1998) EMBO J. 17, 4274–4282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeng F., Xu S. Z., Jackson P. K., McHugh D., Kumar B., Fountain S. J., Beech D. J. (2004) J. Physiol. 559, 739–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freichel M., Suh S. H., Pfeifer A., Schweig U., Trost C., Weissgerber P., Biel M., Philipp S., Freise D., Droogmans G., Hofmann F., Flockerzi V., Nilius B. (2001) Nat. Cell Biol. 3, 121–127 [DOI] [PubMed] [Google Scholar]
- 9.Lee K. P., Jun J. Y., Chang I. Y., Suh S. H., So I., Kim K. W. (2005) Mol. Cells 20, 435–441 [PubMed] [Google Scholar]
- 10.Xu S. Z., Muraki K., Zeng F., Li J., Sukumar P., Shah S., Dedman A. M., Flemming P. K., McHugh D., Naylor J., Cheong A., Bateson A. N., Munsch C. M., Porter K. E., Beech D. J. (2006) Circ. Res. 98, 1381–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu S. Z., Boulay G., Flemming R., Beech D. J. (2006) Am. J. Physiol. Heart Circ. Physiol. 291, H2653–H2659 [DOI] [PubMed] [Google Scholar]
- 12.Munsch T., Freichel M., Flockerzi V., Pape H. C. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 16065–16070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greka A., Navarro B., Oancea E., Duggan A., Clapham D. E. (2003) Nat. Neurosci. 6, 837–845 [DOI] [PubMed] [Google Scholar]
- 14.Hui H., McHugh D., Hannan M., Zeng F., Xu S. Z., Khan S. U., Levenson R., Beech D. J., Weiss J. L. (2006) J. Physiol. 572, 165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riccio A., Li Y., Moon J., Kim K. S., Smith K. S., Rudolph U., Gapon S., Yao G. L., Tsvetkov E., Rodig S. J., Van't Veer A., Meloni E. G., Carlezon W. A., Jr., Bolshakov V. Y., Clapham D. E. (2009) Cell 137, 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strübing C., Krapivinsky G., Krapivinsky L., Clapham D. E. (2001) Neuron 29, 645–655 [DOI] [PubMed] [Google Scholar]
- 17.Yuan J. P., Zeng W., Huang G. N., Worley P. F., Muallem S. (2007) Nat. Cell Biol. 9, 636–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang J., Lin Y., Zhang Z., Tikunova S., Birnbaumer L., Zhu M. X. (2001) J. Biol. Chem. 276, 21303–21310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Odell A. F., Van Helden D. F., Scott J. L. (2008) J. Biol. Chem. 283, 4395–4407 [DOI] [PubMed] [Google Scholar]
- 20.Mery L., Strauss B., Dufour J. F., Krause K. H., Hoth M. (2002) J. Cell Sci. 115, 3497–3508 [DOI] [PubMed] [Google Scholar]
- 21.Cioffi D. L., Wu S., Alexeyev M., Goodman S. R., Zhu M. X., Stevens T. (2005) Circ. Res. 97, 1164–1172 [DOI] [PubMed] [Google Scholar]
- 22.Goel M., Sinkins W., Keightley A., Kinter M., Schilling W. P. (2005) Pflugers Arch. 451, 87–98 [DOI] [PubMed] [Google Scholar]
- 23.Montell C. (2005) Sci. STKE 2005, re3. [DOI] [PubMed] [Google Scholar]
- 24.Zerangue N., Malan M. J., Fried S. R., Dazin P. F., Jan Y. N., Jan L. Y., Schwappach B. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 2431–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harbury P. B., Zhang T., Kim P. S., Alber T. (1993) Science 262, 1401–1407 [DOI] [PubMed] [Google Scholar]
- 26.Schaefer M., Plant T. D., Stresow N., Albrecht N., Schultz G. (2002) J. Biol. Chem. 277, 3752–3759 [DOI] [PubMed] [Google Scholar]
- 27.Okada T., Shimizu S., Wakamori M., Maeda A., Kurosaki T., Takada N., Imoto K., Mori Y. (1998) J. Biol. Chem. 273, 10279–10287 [DOI] [PubMed] [Google Scholar]
- 28.Strübing C., Krapivinsky G., Krapivinsky L., Clapham D. E. (2003) J. Biol. Chem. 278, 39014–39019 [DOI] [PubMed] [Google Scholar]
- 29.Peralta E. G., Ashkenazi A., Winslow J. W., Ramachandran J., Capon D. J. (1988) Nature 334, 434–437 [DOI] [PubMed] [Google Scholar]
- 30.Hamill O. P., Marty A., Neher E., Sakmann B., Sigworth F. J. (1981) Pflugers Arch. 391, 85–100 [DOI] [PubMed] [Google Scholar]
- 31.Amiri H., Schultz G., Schaefer M. (2003) Cell Calcium 33, 463–470 [DOI] [PubMed] [Google Scholar]
- 32.Flemming R., Xu S. Z., Beech D. J. (2003) Br. J. Pharmacol. 139, 955–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riccio A., Medhurst A. D., Mattei C., Kelsell R. E., Calver A. R., Randall A. D., Benham C. D., Pangalos M. N. (2002) Brain Res. Mol. Brain Res. 109, 95–104 [DOI] [PubMed] [Google Scholar]
- 34.Saito K., Tautz L., Mustelin T. (2007) Biochim. Biophys. Acta 1771, 719–726 [DOI] [PubMed] [Google Scholar]
- 35.Bankaitis V. A., Phillips S., Yanagisawa L., Li X., Routt S., Xie Z. (2005) Adv. Enzyme Regul. 45, 155–170 [DOI] [PubMed] [Google Scholar]
- 36.Griac P., Holic R., Tahotna D. (2006) Biochem. Soc. Trans. 34, 377–380 [DOI] [PubMed] [Google Scholar]
- 37.Bezzerides V. J., Ramsey I. S., Kotecha S., Greka A., Clapham D. E. (2004) Nat. Cell Biol. 6, 709–720 [DOI] [PubMed] [Google Scholar]
- 38.Graziani A., Poteser M., Heupel W. M., Schleifer H., Krenn M., Drenckhahn D., Romanin C., Baumgartner W., Groschner K. (2010) J. Biol. Chem. 285, 4213–4223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abdullaev I. F., Bisaillon J. M., Potier M., Gonzalez J. C., Motiani R. K., Trebak M. (2008) Circ. Res. 103, 1289–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.