Abstract
Grp170 and Hsp110 proteins constitute two evolutionary distinct branches of the Hsp70 family that share the ability to function as nucleotide exchange factors (NEFs) for canonical Hsp70s. Although the NEF mechanism of the cytoplasmic Hsp110s is well understood, little is known regarding the mechanism used by Grp170s in the endoplasmic reticulum. In this study, we compare the yeast Grp170 Lhs1 with the yeast Hsp110 Sse1. We find that residues important for Sse1 NEF activity are conserved in Lhs1 and that mutations in these residues in Lhs1 compromise NEF activity. As previously reported for Sse1, Lhs1 requires ATP to trigger nucleotide exchange in its cognate Hsp70 partner Kar2. Using site-specific cross-linking, we show that the nucleotide-binding domain (NBD) of Lhs1 interacts with the NBD of Kar2 face to face, and that Lhs1 contacts the side of the Kar2 NBD via its protruding C-terminal α-helical domain. To directly address the mechanism of nucleotide exchange, we have compared the hydrogen-exchange characteristics of a yeast Hsp70 NBD (Ssa1) in complex with either Sse1 or Lhs1. We find that Lhs1 and Sse1 induce very similar changes in the conformational dynamics in the Hsp70. Thus, our findings demonstrate that despite some differences between Hsp110 and Grp170 proteins, they use a similar mechanism to trigger nucleotide exchange.
Keywords: Chaperone Chaperonin, Endoplasmic Reticulum (ER), Enzyme Mechanisms, Heat Shock Protein, Protein Folding, Protein-Protein Interactions, Grp170, Hsp110, Hsp70 Chaperone, Nucleotide Exchange Factor
Introduction
The Hsp70 chaperones are essential components of the cellular machinery that controls the conformational states of proteins. They are involved in diverse functions, including folding of proteins, transport of proteins across membranes, and regulation of signal transduction components (1, 2). The common activity of Hsp70 chaperones that underlies these diverse functions is the transient binding to peptide segments of protein substrates. The association of Hsp70 with substrates is controlled by its ATPase cycle. When ATP is bound to the N-terminal nucleotide-binding domain (NBD),4 interdomain communication ensures that the C-terminal substrate-binding domain (SBD) exhibits low affinity for substrates, which consequently bind and release with high rates. Hydrolysis of ATP converts Hsp70 to the ADP-bound conformation, which traps the substrate with higher affinity. Exchange of ADP for ATP and concomitant substrate release completes this chaperone cycle.
The chaperone cycle of Hsp70s is regulated by cofactors that facilitate either ATP hydrolysis or exchange of ADP for ATP. ATP hydrolysis is stimulated by J-domain-harboring co-chaperones of the Hsp40 class that provide substrate specificity to the Hsp70 chaperone by delivering clients. Release of substrate from Hsp70 is facilitated by nucleotide-exchange factors (NEFs) that accelerate nucleotide dissociation, thereby allowing ATP to rebind. NEFs function by associating with and stabilizing specific conformations of the Hsp70 NBD that exhibit low affinity for nucleotide. Rebinding of ATP to the Hsp70 results in dissociation of the NEF.
Several structurally unrelated proteins function as NEFs for Hsp70s. Recent structural and biochemical analysis of the NEFs GrpE, Bag1, Bag2, HspBP1, and Hsp110 have revealed that each of these NEFs uses a unique Hsp70 NBD interaction interface (3–9). However, despite unique modes of binding, the actual mechanisms used to trigger nucleotide release fall into two principle classes. GrpE, Bag1, Bag2, and Hsp110 all promote nucleotide release by stabilizing similar Hsp70 NBD conformations that involve tilting NBD lobe II outwards. This tilting opens the NBD structure and facilitates nucleotide exchange (3–5, 7–9). In contrast, HspBP1/Fes1 associates with lobe II subdomain IIB and elicits a near global loss of tertiary structure of the NBD and thereby promotes nucleotide exchange (6, 9).
The eukaryotic Hsp110 proteins have recently been characterized as NEFs for Hsp70s in the cytosol (10–12). The Hsp110s are members of the Hsp70 superfamily of proteins and share overall structure with canonical Hsp70s but differ by an extended SBD with an acidic loop region inserted between the terminal strands of their β-sheet subdomain as well as by an extended flexible C terminus (13, 14). The yeast Saccharomyces cerevisiae harbors two highly homologous members of the Hsp110 family, Sse1 and Sse2 (15).
Sse1-ATP has been crystallized both alone and in complex with mammalian Hsp70s (7, 8, 13). The crystal structures together with biochemical analysis (9) of the interaction with its cognate Hsp70 Ssa1 have revealed how Hsp110 and the Hsp70 partners interact in the nucleotide exchange reaction. Sse1 has to bind nucleotide to fold into a conformation that is capable of associating with the Hsp70 NBD (26). In the complex formed, the NBDs of Hsp70 and Hsp110 interact face to face in such a way that lobe I of the Hsp70 NBD contacts lobe II of the Hsp110 NBD. A second contact is formed between the side of Hsp70 NBD lobe II and the Hsp110 α-helical SBD. As an outcome Hsp110 embraces the Hsp70 NBD. The multiple site binding results in a tilting of Hsp70 NBD lobe II leading to an opening of the Hsp70 structure.
In the endoplasmic reticulum, Sil1 and Grp170s are NEFs that function together with the Hsp70 chaperone BiP/Kar2 (16, 17). Sil1 is a homologue of HspBP1, whereas the Grp170s are members of the Hsp70 superfamily. Grp170s carry extensions of their primary sequence compared with Hsp70s including a predicted loop structure in their SBD and an extended C terminus (14). In yeast, the single representative of the Grp170s is termed Lhs1 (18). Cells carrying lhs1 inactivating mutations are viable but display partial defects in Hsp70-dependent translocation and an induction of the unfolded protein response (UPR) (18–20). ER folding stress is sensed by the kinase Ire1, which then triggers the UPR (21, 22), resulting in up-regulation of a number of genes including SIL1 and LHS1 (18, 20, 23). Cells carrying double lhs1 sil1 mutations are not viable suggesting a strict requirement for NEF activity for proper Hsp70 function in the ER (23).
Currently, it is not clear how Grp170s interact with Hsp70s and by what mechanism they induce nucleotide exchange. Given that both the cytoplasmic Hsp110s and the ER-localized Grp170s are members of the Hsp70 superfamily it is tempting to speculate that they utilize a common NEF mechanism. However, available phylogenetic comparisons do not suggest that Grp170 and Hsp110 are more related to each other than they are to other members of the Hsp70 superfamily (14). Moreover, their ATPase activities are oppositely regulated upon interaction with their cognate Hsp70s; Lhs1 displays stimulation of its ATPase activity when interacting with Kar2 (17), whereas the ATPase activity of Sse1 is dramatically reduced upon interaction with Ssa1 (10). Hence, it is not clear whether Hsp110s and Grp170s utilize common or distinct mechanisms to trigger nucleotide exchange of their respective Hsp70s. In this study, we employed a combination of genetic, biochemical, and biophysical approaches to show that Hsp110s and Grp170s employ a similar mechanism to trigger nucleotide exchange in Hsp70s.
EXPERIMENTAL PROCEDURES
Yeast Strains and Plasmids
CAY1171 is a derivative of strain Y04922 (EUROSCARF, Germany) generated by introducing the complementing LHS1-plasmid pCA716 and deleting SIL1 using an hphMX4 cassette PCR amplified from pAG32 (24) with primers, GGTCCGGATTCTTCCCATAATTTTGAGCGCCCTATCTTCGCAGCTGAAGCTTCGTACGC and CTATGAGCCATGGGGTTGCCAAAGATCAAGTGTCTGCTGTCGATAAGGCATAGGCCACTAGTGGATCTG. CAY1172 was constructed by replacing IRE1 with a PCR-generated LEU2 fragment (primers, ATGCGTCTACTTCGAAGAAACATGTTAGTATTGACACTGCTCGTTTGTGTGAGAAATATCTTGACCGCAG and GGTGAAGTAATCGTAAAATCCATCGGGTACCGGCCCCATTAGTTCTGCTATATCTACCCTATGAACATATTCC). Plasmids pCA715 (HIS3 LHS1) and pCA716 (URA3 LHS1) contain similar LHS1 locus fragments amplified from genomic DNA using primers, GGCGGGGATCCTGCCCTTGTTTGTTCATAAGTC and GGCGGACTAGTAAGAAGTAACAAGTGGTTAACT, and BamHI/SpeI-cloned into pCA503 (9) and pRS316 (25), respectively. Mutations in LHS1 were introduced in pCA715 using oligonucleotide-based site-directed mutagenesis (lhs1–2, N608Y/E611A, GTTACTTGCAAGAACGTATAAATTTTCTTG; lhs1–3, A325T/N326A, CTTGCCTCAGAAGCGGTGCTTAAAATTAAC; lhs1–4, N415V/N417S, TCATCAGCAGAGACAACTCTCAACAC).
Plasmid pCA708 is a derivative of the His6-Smt3 vector pCA528 (26) and contains the Kar2 NBD coding sequence (amino acid residues 43–426) that was PCR amplified using primers, CCAGTGGGTCTCAGGTGGTGCCGATGATGTAGAAAACTACG and GGCGGGGGATCCttaGGATAAGACACCAGCTTGAAC, and cloned using BsaI/BamHI. Plasmid pCA709 contains LHS1 (from amino acid residue 21) and was cloned similarly using primers CCAGTGGGTCTCAGGTGGTGCCGTTTTAGGTGTTGATTACG and GGCCGGGGATCCTATAATTCATCATGCAAAATGTC. Plasmid pCA717 expresses Lhs1 carrying a C-terminal Strep-Tag II and was constructed by ligating a NdeI/BamHI-restricted PCR product (primers, CCGCCCCATATGGGTGCCGTTTTAGGTGTTGATTACG and CCGCCCGGATCCTACTTCTCGAACTGCGGGTGGCTCCACGCTGATAATTCATCATGCAAAATGTCTTCC) into pET24a (Novagen).
Protein Expression and Purification
Lhs1 (pCA709) and Sse1 and the NBDs of Kar2 (pCA708) and Ssa1 were expressed with an Ulp1 cleavable N-terminal His6-Smt3 tag and purified using Ni-IDA matrix (for details, see Ref. 26). During Ulp1 cleavage of His6-Smt3-Lhs1 1.75 m urea was added to the buffer. Proteins were made nucleotide-free by dialysis against buffer containing 50 mm EDTA for 24 h and then dialyzed extensively against EDTA-free buffer. Complexes of the Ssa1 NBD and Lhs1 or Sse1 for hydrogen amide exchange (HX) experiments were purified using a double tag strategy (26). Briefly, crude lysates from cells expressing His6-Smt3-Ssa1 and Lhs1 (pCA717) or Sse1 carrying a Strep-Tag II at their C termini were mixed and formed complexes were isolated by serial Ni-IDA and StrepTactin (IBA, GmbH, Göttingen, Germany) chromatography.
Nucleotide Release Assays
Nucleotide release was measured using the fluorescently labeled analogue of ADP, N8-(4-N′-methylanthraniloylaminobutyl)-8-aminoadenosine 5′-diphosphate (MABA-ADP; TriLink Technologies Inc., San Diego, CA), and the stopped flow instrumentation SX-18MV from Applied Photophysics (Surrey, UK). The method has been described elsewhere (26, 27).
Interaction Experiments
To assess complex formation between Lhs1 and Kar2 NBD, 30 μm Lhs1, supplemented with and without a 2-fold excess of ATP, was incubated with 30 μm His6-Smt3-Kar2 NBD (made nucleotide free using EDTA containing buffer) for 10 min at 30 °C in LWB150 buffer (40 mm Hepes-KOH, pH 7.4, 150 mm KCl, 5 mm MgCl2, 5% (v/v) glycerol) + 0.1% (v/v) Triton X-100. Alternatively, 39 μm Kar1 NBD was incubated with 13 μm Lhs1 wild-type or Lhs1 mutant proteins in the presence of 30 μm ATP. Co2+-NTA-Sepharose was added and proteins were allowed to bind for 20 min at 4 °C. After washing two times with LWB150 and two times with LWB500 (40 mm Hepes-KOH, pH 7.4, 500 mm KCl, 5 mm MgCl2, 5% (v/v) glycerol) + 0.1% (v/v) Triton X-100, proteins were eluted by digestion with Ulp1 protease for 10 min at 30 °C and analyzed on 10% SDS-PAGE that was stained with Coomassie Brilliant Blue.
Cross-linking Experiments
Complexes for cross-linking with bis(maleimido)hexane (BMH) (Thermo Scientific) were obtained by incubating 6 μm Lhs1/Sse1 with equimolar concentrations of Kar2/Ssa1 NBDs overnight at 4 °C in the presence of 1 mm ATP and 6 μm tris(2-carboxyethyl)phosphine. Cross-linking was carried out on ice by addition of 74 μm BMH and the reactions were stopped after 60 min by addition of sample buffer containing 50 mm dithiothreitol. The reactions were analyzed on 10% SDS-PAGE stained with Coomassie Brilliant Blue.
Hydrogen-Deuterium Exchange Experiments and Mass Spectrometry
HX experiments were performed in a similar manner as described previously (9, 28, 29). Briefly, 100–350 pmol of purified monomeric Ssa1 NBD, or Ssa1 NBD in complex with Lhs1 or Sse1 were preincubated for 3 min at 30 °C and diluted 20-fold into D2O-based buffer (25 mm Hepes-KOH, pH 7.6, 50 mm KCl, 5 mm MgCl2) to initiate amide proton-deuterium exchange. The exchange reaction was stopped at defined times by addition of 1 volume of ice-cold quench buffer (0.4 m potassium phosphate buffer, pH 2.2). Quenched samples were immediately injected into the high pressure liquid chromatography setup, subjected to online peptic digest, and analyzed on an electrospray ionization quadrupole time-of-flight mass spectrometer (QSTAR Pulsar; Applied Biosystems) as described (29, 30). The data processing was performed according to Ref. 29. D2O buffer for the HX experiment was prepared by using 99.85% D2O (Euriso-top), lyophilized, and redissolved five times in equal D2O volumes.
Sequence Analysis
The primary sequence of Lhs1 and Lhs1 orthologues from Kluyveromyces lactis, Ashbya gossyppi, and Candida albicans were analyzed using PELE protein structure prediction. ClustalW (31) was used to globally align the sequences as well as for local alignments of sequences corresponding to SBDα and SBDβ (defined by secondary structure prediction and the fact that the sequences follow NBD sequences). Manual motif screening of Lhs1 and Sse1 ClustalW alignments identified the conserved pairs Ala325/Asn326, Asn415/Asn417, and Asn608/Glu609.
RESULTS
Lhs1 Shares Subdomain Organization with Sse1
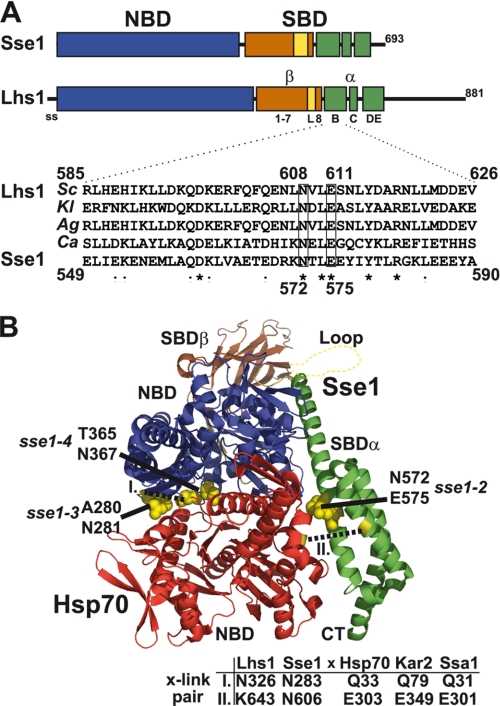
We wanted to perform biochemical analysis of Lhs1 and were therefore in need of a model that defined the subdomain organization of Lhs1. Initially, we attempted to use global sequence alignment methodology to gain information regarding potential sequence conservation between Hsp110s and Grp170s. Although sequence alignment programs could successfully align the more conserved NBD sequences of Hsp70s, Hsp110s, and Grp170s, they failed to properly align sequences derived from the poorly conserved SBDs. We hypothesized that Grp170s shared SBD subdomain structure with Hsp110s and tested this hypothesis by employing local instead of global sequence alignments of the SBD sequences of Lhs1 and Sse1 (see “Experimental Procedures”). Moreover, we applied secondary structure predictions to define SBDα and SBDβ. The result from this analysis is consistent with Lhs1 sharing subdomain organization with Sse1 (Figs. 1A and supplemental S1). Specifically, we find that Lhs1 carries a putative flexible loop structure right before the last β-sheet of SBDβ (Fig. 1A, L), a motif shared with Hsp110s that is not present in Hsp70s. Moreover, the sequence directly following the SBDβ appears to constitute the three α-helices B, C, and DE that are present in Hsp70s and Hsp110s.
FIGURE 1.
Lhs1 shares subdomain organization with Sse1. A, model for Lhs1 domain (NBD, SBD) and subdomain organization compiled from sequence alignment, local homology searches, and secondary structure prediction of Sse1 and fungal Lhs1 homologues. Indicated are the signal sequence (ss, amino acid residues 1–20), NBD (21–433, blue box), SBDβ sheet 1–7 (441–547, brown box), SBDβ loop (L, 548–564, yellow box), SBDβ sheet 8 (565–577, brown box), SBDα helix B (582–627, green box), SBDα helix C (635–650, green box), and SBDα helix DE (663–706, green box). Lhs1 has an extended C terminus with no significant homology to Sse1-(708–881). Homologous domains and subdomains of Sse1 are according to Ref. 13. Inset displays a sequence alignment of SBDα helix B from four fungal Lhs1 (S. cerevisiae, SC; K. lactis, KL; A. gossypii, Ag; C. albicans, Ca) and Sse1. Lhs1 Asn608 and Glu611 (boxed) exhibit sequence conservation with corresponding Sse1 residues (Asn572 and Glu575) that contact the Hsp70 NBD and are required for NEF activity (8). B, structure of a complex between Sse1 and the Hsp70 NBD (8). Sse1 is colored as in A and the Hsp70 NBD is colored red. Sse1 residues Thr280/Asn281, Thr365/Asn367, and Asn572/Glu575 that contact the Hsp70 NBD are marked (yellow). Residues used for pairwise cross-linking in Fig. 4 are marked (yellow, dotted lines) and corresponding positions in Lhs1/Sse1 and Hsp70/Kar2/Ssa1 are as indicated (table).
We asked if residues involved in the nucleotide exchange activity of Sse1 also are conserved in Lhs1. Polier et al. (8) documents three pairs of Sse1 residues involved in the interaction between Sse1 and Hsp70. Two of these pairs are localized in the NBD and one pair in the α-helix B of the SBDα (Fig. 1B). Sse1 NBD residue pairs Ala280/Asn281 and Thr365/Asn367 correspond to Ala326/Asn327 and Asn415/Asn417, respectively, in the Lhs1 sequence. Note that Sse1 Thr365 is not perfectly conserved with Lhs1 Asn415. However, in other fungal homologues, for example, such as C. albicans Lhs1, position 415 carries a threonine residue (data not shown). The Sse1 SBDα residue pair Asn572/Glu575 also appears to be conserved in fungal Lhs1 homologues despite the considerable variation in the α-helix B sequence (Fig. 1A, Asn608/Glu611). Our analysis suggests that Lhs1 carries corresponding residues at positions known to be important for Sse1 NEF activity.
Residues Conserved between Lhs1 and Sse1 Are Important for NEF Activity
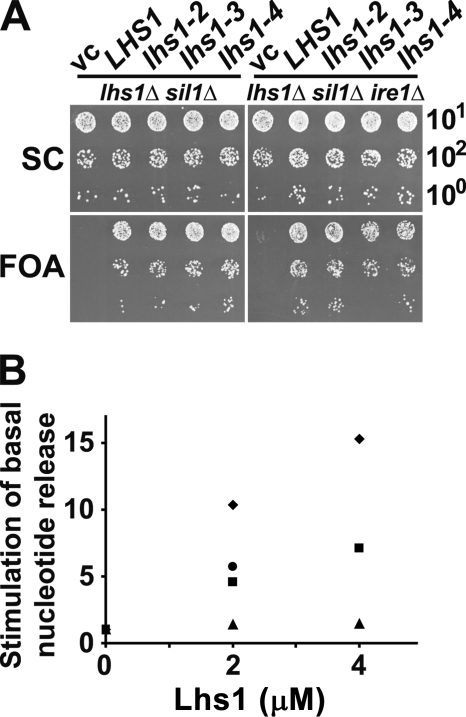
We tested if the identified residues in Lhs1 were important for NEF activity by mutational analysis. First, we introduced mutations in the LHS1 coding sequence on a yeast single copy plasmid in the context of the native promoter and terminator sequences. All three alleles (lhs1–2, N608Y/E611A, SBDα; lhs1–3 A326T/N327A, NBD; and lhs1–4, N415V/N417S, NBD) carry corresponding substitution mutations used in the previous functional analysis of Sse1 (see supplemental Fig. S1 and Ref. 8). We tested if the alleles could complement the lhs1Δ sil1Δ non-growth phenotype of strain CAY1171. This strain carries a complementing URA3-marked Lhs1-expressing plasmid that can be counterselected on 5-fluoroorotic acid (FOA) containing medium. Hence, cells will only grow on 5-FOA medium if they are supplied with a plasmid expressing at least partially active Lhs1. We introduced single copy plasmids harboring lhs1–2 (N608Y/E611A), lhs1–3 (A326T/N327A), and lhs1–4 (N415V/N417S) in the context of the endogenous LHS1 locus and found that all three alleles complemented the phenotype (Fig. 2A, left panels). However, lhs1–3 cells appeared to undergo a phase of adaptation right after they lost expression of wild-type LHS1 because they transiently formed unevenly sized colonies when they were transferred to selective FOA medium (data not shown). We reasoned that this weak phenotype might represent cellular adaptation to low Lhs1 NEF activity via the UPR regulation. Importantly, the LHS1 promoter itself is under UPR control (18, 20), which potentially allows lhs1 alleles encoding low activity to up-regulate their own expression. We tested this possibility by performing the complementation analysis of lhs1–2, lhs1–3, and lhs1–4 in lhs1Δ sil1Δ ire1Δ cells that lack a functional UPR pathway (22, 23). Both lhs1–2 and lhs1–4 complemented the non-growth phenotype fully, whereas lhs1–3 cells exhibited a lower plating efficiency on selective FOA medium compared with nonselective SC medium (Fig. 2A, right panels). Upon closer inspection we observed that lhs1–3 cells exhibit a slow-growth phenotype (data not shown) that explains why FOAR cells were underrepresented in the plasmid shuffle experiment, manifested as a lower plating efficiency on FOA medium.
FIGURE 2.
Residues conserved between Lhs1 and Sse1 are important for NEF activity. A, 10-fold serial dilutions of yeast strains CAY1171 (lhs1Δ sil1Δ) and CAY1172 (lhs1Δ sil1Δ ire1Δ) transformed to His+ using a vector control (vc), pCA715 (LHS1), or pCA715 carrying lhs1–2, lhs1–3, or lhs1–4 alleles. Cells were spotted onto control synthetic complete medium (SC) and medium containing FOA to counterselect against pCA716 (URA3 LHS1) that is present in the strains. Plates were incubated for 48 h at 30 °C. B, fold-stimulation of the basal rate of MABA-ADP dissociation from Kar2 NBD (2 μm) at the indicated concentrations of Lhs1 (♦) and derived mutants Lhs1–2 (■), Lhs1–3 (▲), and Lhs1–4 (●). Note that the data points for 4 μm Lhs1–2 and Lhs1–4 overlap. Proteins were preincubated with 3 mm ATP before measuring the release rates at 30 °C with a stopped flow instrument.
Combined Lhs1 mutations such as lhs1–2,3 and lhs1–2,4 also complemented the lhs1Δ sil1Δ non-growth phenotype (data not shown). This observation contrasts the predicted analogous sse1 mutations, which exhibited non-complementation (8), suggesting either that the residues are not essential for NEF activity of Lhs1 or that little residual NEF activity is sufficient to maintain cellular growth.
Next, we assessed the impact of the mutations on Lhs1 NEF activity in vitro using MABA-ADP release assays (Fig. 2B). In these assays preloaded complexes consisting of the fluorescent nucleotide derivative MABA-ADP and the NBD of Kar2 (2 μm) were rapidly mixed with Lhs1 and dissociation of MABA-ADP was followed by a decrease of its fluorescent signal in stopped flow measurements. Both Lhs1–2 and Lhs1–4 exhibited a low activity, corresponding to approximately half of the wild-type Lhs1 activity. The activity impairment of Lhs1–3 was even more pronounced; the protein exhibited only barely detectable activity under identical conditions. The low activity of Lhs1–3 in vitro correlates with the inability of the mutant to fully complement the in vivo phenotype. The residual activity is apparently sufficient to support growth of lhs1Δ sil1Δ ire1Δ cells, which might either mean that the in vitro assay underestimates the proteins activity in vivo, or that very little NEF activity is required for growth. In any case, we and others have previously observed similar partial complementation of growth phenotypes when testing sse1 NEF mutants in vivo (8, 9). All mutant Lhs1 proteins appeared to be properly folded as judged by CD measurements (data not shown).
Our in vivo and in vitro mutational analysis suggests that Lhs1 depends on similar residues as Sse1 for its NEF activity. The residual NEF activity of Lhs1–2 (in SBDα) and Lhs1–4 (in NBD) is expected if Lhs1, like Sse1, employs a large surface with multiple contact points to interact with the Hsp70 (8).
Nucleotide Is Required for NEF Activity of Lhs1
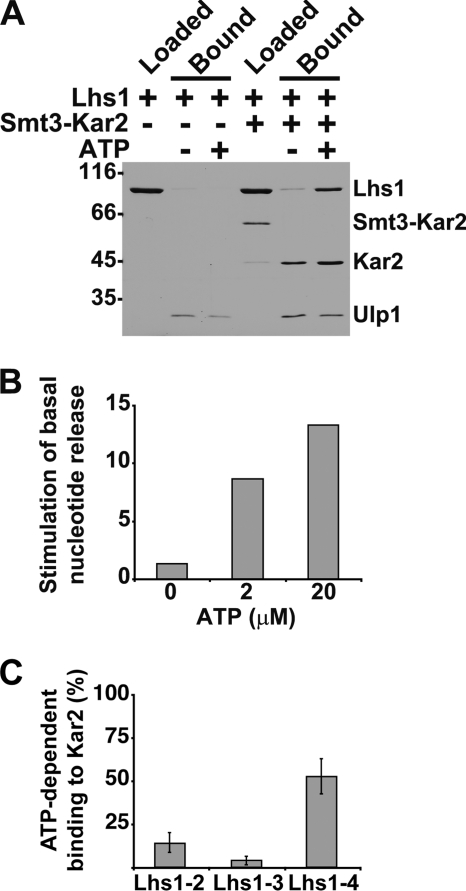
In contrast to other characterized NEFs, Sse1 requires ATP binding to reach an active conformation that is capable of interacting with Hsp70 and trigger nucleotide exchange. We wondered also if Lhs1 required nucleotide for functional interaction with Kar2 and investigated this possibility by testing if Lhs1 could form a complex with Kar2 in the absence or presence of ATP. The NBD of Kar2 was incubated with nucleotide-free Lhs1 in the presence or absence of added ATP and immobilized onto cobalt affinity resin by means of an N-terminal His6-Smt3 tag. After washing, Kar2 and bound Lhs1 were eluted by proteolytic severing of the His6-Smt3 tag and analyzed by SDS-PAGE. In the absence of nucleotide, little or no Lhs1 was bound to Kar2, whereas the presence of ATP resulted in formation of substantial amounts of complex (Fig. 3A).
FIGURE 3.
Lhs1 requires ATP to associate with Kar2 and trigger nucleotide exchange. A, Lhs1 and His6-Smt3-Kar2 were incubated in the presence and absence of ATP and loaded onto cobalt-NTA matrix. After washing, bound protein was eluted by proteolytic cleavage of the His6-Smt3 using Ulp1 protease and analyzed on SDS-PAGE. B, fold-stimulation of the basal rate of MABA-ADP dissociation from Kar2 NBD (2 μm) induced by Lhs1 (2 μm) preincubated at the indicated concentrations of ATP. The release rates were measured with a stopped flow instrument. C, ATP-dependent interaction between Lhs1–2, Lhs1–3, and Lhs1–4 and Kar2. Quantitation of pulldown assays (see A) given as percent of wild-type Lhs1 bound to the Kar2 NBD. Error bars indicate the standard error (n = 4).
To directly assess if nucleotide is required for NEF activity of Lhs1, we measured MABA-ADP release from Kar2 under nucleotide-depleted conditions (no excess nucleotide to compete with the released MABA-ADP in the assay). In the absence of added nucleotide, 2 μm Lhs1 could only accelerate the basal release rate of Kar2 (2 μm) 1.4-fold, whereas preincubation of Lhs1 with 2 μm ATP resulted in 9-fold acceleration (Fig. 3B). Preincubation with 20 μm ATP further enhanced the activity to 13-fold acceleration of the basal release rate. The interaction experiments together with the activity determinations indicate that Lhs1 requires nucleotide to interact with Kar2 and trigger nucleotide release. During review of this manuscript a parallel study was published that also reported on an ATP requirement for Lhs1 NEF activity (32).
Lhs1-Kar2 Association Depends on Residues Conserved between Lhs1 and Sse1
We wanted to test if the residues in Lhs1 that are important for NEF activity (Asn608/Glu611, SBDα; Ala326/Asn326, NBD; and Asn415/Asn417, NBD) also are important for association of Lhs1 and Kar2. We undertook quantitative pulldown assays, where the Kar2 NBD was immobilized onto cobalt affinity resin by means of an N-terminal and cleavable His6-Smt3 tag (see above). We applied Lhs1 and our Lhs1 with amino acid substitutions that impair NEF activity to Kar2 and assayed the interaction after washing. Lhs1–2 (N608Y/E611A, SBDα) and Lhs1–3 (A326T/N327A, NBD) exhibited approximately 15 and 4% ATP-dependent Kar2 binding compared with wild-type Lhs1 (Fig. 3C). Lhs1–4 (N415V/N417S, NBD) exhibited about 53% binding. In the case of Lhs1–3 and Lhs1–4 the Kar2 binding correlates directly with their determined NEF activity; Lhs1–3 is a poor binder and exhibits very little NEF activity and Lhs1–4 exhibits both substantial NEF activity and binding. Interestingly, Lhs1–2 displays NEF activity at the level of Lhs1–4 but a significantly lower level of complex formation. Hence, this observation implies that Hsp70 complex stability and NEF activity might not be directly coupled. A similar phenomenon has been reported previously for the Sse1-Hsp70 interaction (8).
Lhs1 and Sse1 Contact the Hsp70 NBD Similarly
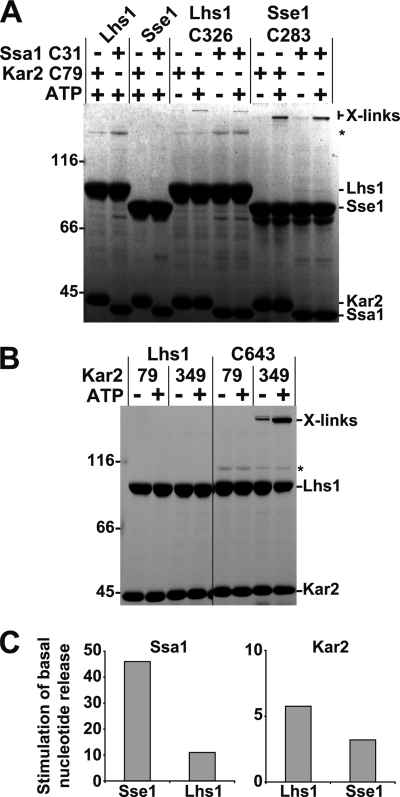
We used site-specific cross-linking to investigate if Lhs1 and Kar2 form a complex with architecture reminiscent of the Sse1-Ssa1 complex. We have previously established that a cysteine residue replacing Asn283 in NBD lobe II of Sse1 forms a cross-link to a cysteine replacing Gln31 of the Ssa1 NBD using bismaleimide cross-linkers (9). This cross-link provides evidence that the NBD of Sse1 interacts face to face with the NBD of Ssa1, an observation consistent with recently published crystal structures (7, 8). We introduced cysteines at corresponding positions in Lhs1 (N326C) and the NBD of Kar2 (Q79C) and tested their ability to cross-link using BMH (10 Å length). Because both Lhs1 and Sse1 require ATP to form complexes with Hsp70s, the proteins were mixed with BMH in the presence and absence of ATP to control for cross-linking specificity. We observed that Lhs1-N326C and Sse1-N283C formed ATP-dependent cross-links to both Kar2-Q79C and Ssa1-Q31C (Fig. 4A), indicating that the NBDs of Lhs1 and Kar2 and Sse1 and Ssa1 are oriented similarly in their respective complexes. Furthermore, the data suggest that key interacting residues in Lhs1 and Sse1 and their cognate Hsp70s are conserved.
FIGURE 4.
Lhs1 and Sse1 interact with Hsp70s in a similar manner. A, Lhs1, Sse1, or derivatives carrying introduced cysteine residues (Lhs1-C326 and Sse2-C283) were preincubated in the presence or absence of 1 mm ATP with the NBDs of Kar2 and Ssa1 carrying introduced cysteine residues (Kar2-C79 and Ssa1-C31). After cross-linking with BMH, cross-linked products were detected by SDS-PAGE analysis. The asterisk (*) refers to background protein bands that are not products of cross-linking. B, control Lhs1 and Lhs1 carrying a cysteine at position 643 (C643) was tested for cross-linking (as in A) to Kar2 NBDs carrying cysteines in place of residues 79 or 349. The asterisk (*) refers to background protein bands that are not products of cross-linking. C, fold-stimulation of the basal rate of MABA-ADP dissociation from the Kar2 and Ssa1 NBDs (0.5 μm) induced by Lhs1 or Sse1 (1 μm). Lhs1 and Sse1 were preincubated with 0.5 mm ATP before measuring the release rates with a stopped flow instrument.
Next, we wanted to investigate if Lhs1 SBDα contacts the side of the Kar2 NBD lobe II in a fashion similar to how Sse1 contacts Ssa1 (see Fig. 1B). Aided by the recently published crystal structures of complexes between Sse1 and Hsp70s (7, 8), we introduced cysteines positioned in Lhs1 SBDα (K643C) and at the side of Kar2 NBD lobe II (E349C) so that they potentially could be cross-linked using BMH. Again, we took advantage of the ATP requirement for efficient complex formation between Lhs1 and Kar2 to control for specificity and found that BMH cross-links Lhs1-K643C efficiently to Kar2-E349C in an ATP-dependent manner (Fig. 4B). Cross-links were not detected between Lhs1-K643C and Kar2-Q79C (Fig. 4B), providing another specificity control. Taken together, our cross-linking data indicate that Lhs1 interacts with Kar2 NBD using two contact surfaces similar to the architecture of the Sse1-Ssa1 complex.
Lhs1 and Sse1 Trigger Nucleotide Exchange in Both Kar2 and Ssa1
Because Lhs1 and Sse1 interacted in the cross-linking experiments with both the cognate and noncognate Hsp70s, we wished to test if they catalyze nucleotide exchange from both Ssa1 and Kar2. Using MABA-ADP release assays we found that 1 μm Lhs1 accelerated the basal nucleotide release rate from its cognate partner Kar2 6- and 11-fold from non-cognate Ssa1, both at 0.5 μm (Fig. 4C). Sse1 accelerated the basal release rate of cognate Ssa1 46-fold and of Kar2 3-fold under identical experimental conditions. Although both Lhs1 and Sse1 function less well with the non-cognate Hsp70s than the cognate interaction partners, we find that the basic features required for functional interaction with Lhs1 and Sse1 are conserved in both Kar2 and Ssa1.
Lhs1 and Sse1 Employ the Same Mechanism to Trigger Nucleotide Exchange in Hsp70s
The similarity in the interactions of Lhs1 and Sse1 with Hsp70s raises the possibility that the two NEFs employ the same mechanism to trigger nucleotide exchange in Hsp70s. We have previously devised a methodology that allows us to directly probe for NEF-induced conformational changes in the Hsp70 NBD using HX combined with mass spectrometry (9). Briefly, by measuring HX protection or deprotection of the Ssa1 NBD at a peptide level we can monitor conformational changes induced by the NEFs upon interaction with the Hsp70 NBD.
We wished to compare the specific patterns of HX protection and deprotection that Lhs1 and Sse1 induce in the NBD of Ssa1. The Ssa1 NBD was chosen for analysis because we have previously annotated all of its peptides and established the effect that several NEFs, including Sse1, induce on this NBD (9). Furthermore, our cross-linking analysis and activity assays demonstrate that Lhs1 functionally interacts with this particular Hsp70.
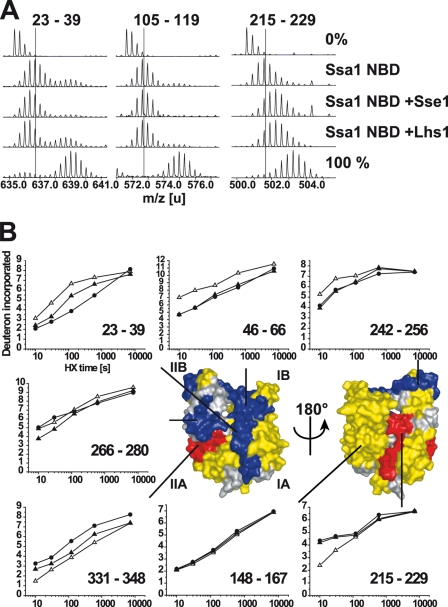
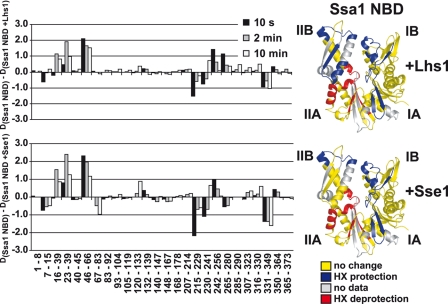
We purified nucleotide-free monomeric Ssa1 NBD and the NBD in complex with either Lhs1 or Sse1 (see “Experimental Procedures”) and subjected these proteins to HX in heavy water buffer for defined durations. Online pepsin digestion followed by mass spectrometry allowed us to identify Ssa1 NBD peptides and the extent of HX of each peptide at the different time points (Fig. 5A). Upon comparison of the HX behavior of each peptide derived from either monomeric Ssa1 NBD or the NBD in complex with either Lhs1 or Sse1 we found mainly three classes of peptides: (i) peptides that exhibited HX protection induced by both Lhs1 and Sse1 (Fig. 5A, left panel, peptide 23–39), (ii) peptides that were not affected by Lhs1 and Sse1 (Fig. 5A, middle panel, peptide 105–119), and (iii) peptides that exhibited HX deprotection induced by Lhs1 and Sse1 (Fig. 5A, right panel, peptide 215–229). Very few peptides exhibited different HX behavior induced by Lhs1 and Sse1. We mapped the changes onto a structural model of the Ssa1 NBD (9) derived from the crystallized bovine Hsc70 NBD (33) (Fig. 5B). Strikingly, for nearly all peptides analyzed, Lhs1 and Sse1 induced the same general pattern of HX protection and deprotection (Figs. 5B, and 6, and supplemental S2). Specifically, Lhs1 or Sse1 induced protection in subdomain I and deprotection of peptides in subdomain II, whereby deprotection was localized to the interface of subdomains IIA and IIB. The induced HX changes were more pronounced for Sse1 than for Lhs1, likely reflecting a higher affinity of Sse1 over Lhs1 for the Ssa1 NBD. One segment in subdomain IIB spanned by peptide 265/266–280 (Figs. 5B and 6), exhibited protection induced by Lhs1, whereas Sse1 induced a slight deprotection of the same segment. This is the only segment that exhibits differences in HX effects induced by Lhs1 and Sse1.
FIGURE 5.
HX properties of the Ssa1 NBD in complex with Lhs1. A, mass spectra of representative peptides from the monomeric Ssa1 NBD or Ssa1 NBD in complex with Lhs1 or Sse1 incubated in H2O (0%), or for 10 s in D2O buffer. Control spectra (100%) of the same peptides from fully deuterated Ssa1. The lines indicate the centroids of the peptide ions for monomeric Ssa1 NBD after 10 s in D2O buffer. B, kinetics of deuteron incorporation during HX into selected segments of the Ssa1 NBD in its monomeric form (▵) and when in complex with Lhs1 (▴) or Sse1 (●) from data set one (see supplemental Fig. S2, A and B). The structural representation of the Ssa1 NBD (modeled onto the structure of Hsc70 NBD (33)) is colored to display segments in which Lhs1 and Sse1 induce average protection/deprotection effects at 10 s, 2 min, and 10 min of ≥0.5 Da and 5% of total possible exchange compared with the monomeric NBD in duplicate data sets (see supplemental Fig. S2); HX protection (blue), deprotection (red), no significant HX differences (yellow), or no data (gray) are as indicated.
FIGURE 6.
Lhs1 and Sse1 use the same NEF mechanism. Difference in deuteron incorporation between monomeric Ssa1 NBD and NBD in complex with Lhs1 (upper left panel) or Sse1 (lower left panel). The data were resolved to individual non-redundant peptic peptides as indicated by the start and end residue numbers of the corresponding segments. The data presented are from data set two (see supplemental Fig. S2, C and D). Structural representations of the Ssa1 NBD are colored to display the Lhs1 (upper right panel) and Sse1 (lower right panel) induced changes as in Fig. 5B.
Taken together, the HX data for the interaction of Lhs1 with the Ssa1 NBD are consistent with the mechanism we previously derived for interaction of Sse1 with Ssa1 using HX and cross-linking (9). Two recently published crystal structures of the Sse1-Hsp70 complex show how Sse1 contacts the Hsp70 NBD and induces outward rotation of subdomain II (7, 8). Hence, our HX data indicate that Lhs1 and Sse1 employ a very similar mechanism to trigger nucleotide exchange from Hsp70s.
DISCUSSION
We have defined how the yeast Grp170 Lhs1 binds Hsp70s and how this interaction induces nucleotide release. Initially the published data on Lhs1 suggested some mechanistic differences to the Sse1/Ssa1 system (see below). Our study, however, demonstrates that the ER-localized Lhs1 shares both binding interface and NEF mechanism with the cytoplasmic Hsp110 Sse1. The main contact between Lhs1 and Kar2 is mediated via their NBDs so that they orient to face each other and Lhs1 NBD lobe II contacts lobe I of Kar2 (see Fig. 1B). A second contact between the protruding SBDα of Lhs1 and the side of Kar2 NBD lobe II induces tilting of lobe II thereby opening the Kar2 NBD. The resulting conformation is incompatible with high nucleotide binding affinity.
Our understanding of how Lhs1 interacts with Hsp70s is based on a series of experiments directly comparing the well characterized Sse1 with Lhs1. Key observations include sequence conservation between the NEFs, the finding that Lhs1 and Sse1 can be cross-linked to the Hsp70 NBD at similar residues, and that both NEFs induce near identical conformational dynamics of an Hsp70 NBD as judged from HX experiments. Given the similarities we conclude that Lhs1 employs the same interaction mode and NEF mechanism as Sse1.
Our data indicate that there are small differences in the Hsp70-interaction interface of Lhs1 and Sse1. For example, the interaction between Sse1 SBDα and the side of Ssa1 NBD subdomain IIB is critical for NEF activity because substitution of the interacting Sse1 residues almost abolishes the activity in vitro and results in inefficient complementation in vivo (8). When we introduced the corresponding substitutions in Lhs1 (lhs1–2), the complex formation and NEF activity were reduced, but approximately half the activity remained and the allele complemented in vivo. In addition, combining the mutations, which abolishes the NEF activity of Sse1 (8), was in the case of Lhs1 tolerated in vivo. Although the extensive interaction interface between Hsp70 and NEF may account for some of the resilience observed in Lhs1, our findings raise the possibility that Lhs1 might employ additional residues to contact the side of the Hsp70 NBD. Indeed, we observe a peptide located nearby (residues 266–280) that exhibits protection from HX when the Ssa1 NBD is in complex with Lhs1 but not when in complex with Sse1. Hence, this peptide defines a potential contact point for Lhs1 that is not used by Sse1.
Curiously, the ATPase activities of Lhs1 and Sse1 offer examples of similarities and differences between the two proteins. In this study we document that ATP binding of Lhs1 is a prerequisite for NEF interaction with the Kar2 NBD. A parallel study has addressed the same issue and provides further evidence for the finding (32). Also Sse1 requires nucleotide for its NEF activity (10, 26) and we have previously documented that the molecular basis for this is a two-step folding event upon which the protein acquires the active conformation (26). Apparently, Lhs1 undergoes a similar conformational activation event upon ATP binding. Hydrolysis of the bound ATP does not play a role for the NEF activity of the two proteins (12, 17). Despite this fact, both Lhs1 and Sse1 hydrolyze the ATP and their ATPase activities are affected differently by complex formation with Hsp70s. When Lhs1 forms a complex with Kar2, its ATPase rate is stimulated (17), whereas Hsp70 potently inhibits the ATP hydrolysis of Sse1 (10). At this point, there is no explanation for this apparent difference between Lhs1 and Sse1.
Our finding that the Grp170 Lhs1 and the Hsp110 Sse1 employ a similar mechanism to trigger NEF in Hsp70s addresses a controversy in the field. One opinion has been that Hsp110s and Grp170s share a common origin and therefore are expected to employ a similar mechanism (see for example, Ref. 34). However, because experimental evidence or even rigorous sequence alignments have been missing, another idea has been that Grp170s and Hsp110s are more different (see for example, Ref. 14). In such a scenario the NEF mechanisms of the two protein families might differ. Our experimental analysis of Lhs1 supports the notion that Grp170s and Hsp110s are quite similar. Because both Lhs1 and Sse1 require nucleotide for NEF activity it is likely that this feature is mechanistically and evolutionarily linked to the NEF activity.
The recent development in the understanding of how NEFs promote nucleotide exchange from Hsp70s has previously allowed us to propose two general classes of nucleotide release mechanisms (9): (i) catalysis of nucleotide release through a global loss of tertiary structure of the Hsp70 NBD (HspBP1/Fes1) and (ii) catalysis of nucleotide release through opening of the nucleotide binding cleft by tilting lobe II. Our data regarding Lhs1 place the Grp170s in the second group together with the structurally related Hsp110s and the structurally unrelated GrpE and Bag1 proteins. During writing of this article the NEF mechanism for the mammalian Bag2 protein was published (5). The authors propose that Bag2 might have been mis-annotated as a classical Bag-family member and document that Bag2 induces outward tilting of Hsp70 NBD lobe II in a different manner than Bag1. Thus, according to our proposed classification, Bag2 also belongs to the same functional NEF group as GrpE, Bag1, Hsp110s, and now Grp170s. Apparently, the limited conformational repertoire of the Hsp70 NBD drives evolution to produce similar nucleotide release mechanisms despite different NEF-binding modes.
Supplementary Material
Acknowledgments
We thank Matthias P. Mayer for helpful discussions and the mass spectrometry facility of the ZMBH for support.
This work was supported in part by Deutsche Forschungsgemeinschaft Grant SFB638 and a grant from the Fonds der Chemischen Industrie (to B. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- NBD
- nucleotide-binding domain
- SBD
- substrate-binding domain
- NEF
- nucleotide exchange factor
- HX
- hydrogen amide exchange
- BMH
- bis(maleimido)hexane
- FOA
- 5-fluoroorotic acid
- UPR
- unfolded protein response
- ER
- endoplasmic reticulum
- MABA-ADP
- N8-(4-N′-methylanthraniloylaminobutyl)-8-aminoadenosine 5′-diphosphate.
REFERENCES
- 1.Mayer M. P., Bukau B. (2005) Cell. Mol. Life Sci. 62, 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Craig E. A. H. P. (2005) Protein Folding Handbook, pp. 490–515, Wiley, Weinheim, Germany [Google Scholar]
- 3.Harrison C. J., Hayer-Hartl M., Di Liberto M., Hartl F., Kuriyan J. (1997) Science 276, 431–435 [DOI] [PubMed] [Google Scholar]
- 4.Sondermann H., Scheufler C., Schneider C., Hohfeld J., Hartl F. U., Moarefi I. (2001) Science 291, 1553–1557 [DOI] [PubMed] [Google Scholar]
- 5.Xu Z., Page R. C., Gomes M. M., Kohli E., Nix J. C., Herr A. B., Patterson C., Misra S. (2008) Nat. Struct. Mol. Biol. 15, 1309–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shomura Y., Dragovic Z., Chang H. C., Tzvetkov N., Young J. C., Brodsky J. L., Guerriero V., Hartl F. U., Bracher A. (2005) Mol. Cell 17, 367–379 [DOI] [PubMed] [Google Scholar]
- 7.Schuermann J. P., Jiang J., Cuellar J., Llorca O., Wang L., Gimenez L. E., Jin S., Taylor A. B., Demeler B., Morano K. A., Hart P. J., Valpuesta J. M., Lafer E. M., Sousa R.(2008) Mol Cell 31, 232–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Polier S., Dragovic Z., Hartl F. U., Bracher A. (2008) Cell 133, 1068–1079 [DOI] [PubMed] [Google Scholar]
- 9.Andréasson C., Fiaux J., Rampelt H., Druffel-Augustin S., Bukau B. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 16519–16524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaner L., Sousa R., Morano K. A. (2006) Biochemistry 45, 15075–15084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dragovic Z., Broadley S. A., Shomura Y., Bracher A., Hartl F. U. (2006) EMBO J. 25, 2519–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raviol H., Sadlish H., Rodriguez F., Mayer M. P., Bukau B. (2006) EMBO J. 25, 2510–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Q., Hendrickson W. A. (2007) Cell 131, 106–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Easton D. P., Kaneko Y., Subjeck J. R. (2000) Cell Stress Chaperones 5, 276–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mukai H., Kuno T., Tanaka H., Hirata D., Miyakawa T., Tanaka C. (1993) Gene 132, 57–66 [DOI] [PubMed] [Google Scholar]
- 16.Chung K. T., Shen Y., Hendershot L. M. (2002) J. Biol. Chem. 277, 47557–47563 [DOI] [PubMed] [Google Scholar]
- 17.Steel G. J., Fullerton D. M., Tyson J. R., Stirling C. J. (2004) Science 303, 98–101 [DOI] [PubMed] [Google Scholar]
- 18.Craven R. A., Egerton M., Stirling C. J. (1996) EMBO J. 15, 2640–2650 [PMC free article] [PubMed] [Google Scholar]
- 19.Hamilton T. G., Flynn G. C. (1996) J. Biol. Chem. 271, 30610–30613 [DOI] [PubMed] [Google Scholar]
- 20.Baxter B. K., James P., Evans T., Craig E. A. (1996) Mol. Cell. Biol. 16, 6444–6456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mori K., Ma W., Gething M. J., Sambrook J. (1993) Cell 74, 743–756 [DOI] [PubMed] [Google Scholar]
- 22.Cox J. S., Shamu C. E., Walter P. (1993) Cell 73, 1197–1206 [DOI] [PubMed] [Google Scholar]
- 23.Tyson J. R., Stirling C. J. (2000) EMBO J. 19, 6440–6452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldstein A. L., McCusker J. H. (1999) Yeast 15, 1541–1553 [DOI] [PubMed] [Google Scholar]
- 25.Sikorski R. S., Hieter P. (1989) Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andréasson C., Fiaux J., Rampelt H., Mayer M. P., Bukau B. (2008) J. Biol. Chem. 283, 8877–8884 [DOI] [PubMed] [Google Scholar]
- 27.Theyssen H., Schuster H. P., Packschies L., Bukau B., Reinstein J. (1996) J. Mol. Biol. 263, 657–670 [DOI] [PubMed] [Google Scholar]
- 28.Rist W., Jørgensen T. J., Roepstorff P., Bukau B., Mayer M. P. (2003) J. Biol. Chem. 278, 51415–51421 [DOI] [PubMed] [Google Scholar]
- 29.Rist W., Graf C., Bukau B., Mayer M. P. (2006) J. Biol. Chem. 281, 16493–16501 [DOI] [PubMed] [Google Scholar]
- 30.Rist W., Rodriguez F., Jørgensen T. J., Mayer M. P. (2005) Protein Sci. 14, 626–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Keyzer J., Steel G. J., Hale S. J., Humphries D., Stirling C. J. (2009) J. Biol. Chem. 284, 31564–31571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flaherty K. M., DeLuca-Flaherty C., McKay D. B. (1990) Nature 346, 623–628 [DOI] [PubMed] [Google Scholar]
- 34.Shaner L., Morano K. A. (2007) Cell Stress Chaperones 12, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.