Non-natural polymers have the capacity to recapitulate both the well-folded structures and resultant functionalities of biological polymers. Peptides composed of β3-amino acids, which have one additional backbone methylene unit compared to natural R-amino acids, can adopt stable helices in water1-3 and interact with cellular proteins and membranes.4-8 More sophisticated β-peptide functionality, however, must await the incorporation of higher order structure. Previous efforts toward this goal have generated constrained tertiary interactions using disulfide bonds,9 nucleobase pairing,10 or metal chelation.11 Quaternary structures lacking a well-defined stoichiometry have also been reported.12,13 These advances, however, did not provide structures that rival the stable, discrete, and compact assemblies of natural proteins. Here we report a β-dodecapeptide (Zwit-1F, Figure 1A) that self-assembles spontaneously in aqueous solution into a highly thermostable β-peptide octamer and determine its structure by X-ray crystallography.
Figure 1.
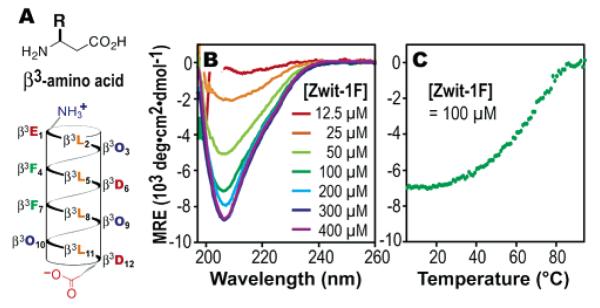
(A) Helical net cartoon of Zwit-1F with β-amino acids represented by single-letter codes. β3O signifies β3-homoornithine. Concentration (B) and temperature (C) dependent CD spectra of Zwit-1F at 25 °C in phosphate buffer (pH 7.1).
Peptides composed of β3-amino acids favor a 314 helical secondary structure characterized by a 14-membered hydrogen-bond ring between the amide at position i and the carbonyl of position i + 2, a left-handed helical twist, and three distinct faces.14,15 We recently reported a β-peptide pair (Acid-1F, Base-1F) that assembles in an equimolar ratio into a β-peptide quaternary structure, a cooperatively folded assembly of β-peptide subunits.16
Like Acid-1F and Base-1F, the design of Zwit-1F uses residues on each of the three 314-helical faces to control higher order structure. Residues along one helical face promote 314 helicity through salt-bridge formation, while those on a second use β3-homoleucines to drive oligomerization, akin to leucine zipper proteins (Figure 1A). While Acid-1F and Base-1F bear cross-complementary charges at positions 1 and 10 on the third face to drive hetero-oligomerization, Zwit1-F employs self-complementary charges to favor homo-oligomerization.
Circular dichroism (CD) spectroscopy indicates that Zwit-1F is minimally structured in buffered aqueous solution at low μM concentration but undergoes a concentration-dependent increase in 314-helical structure (Figure 1B), suggesting the formation of a well-structured oligomer. The thermally induced change in the CD spectrum of Zwit-1F is sigmoidal, indicating cooperativity in the unfolding process (Figure 1C). Moreover, the Zwit-1F oligomer is exceptionally stable; even at 25 μM the TM is approximately 72 °C, definitively within the range observed for well-folded small proteins.17 Data from sedimentation equilibrium experiments are consistent with an oligomer of discrete stoichiometry, fitting well to both hexamer and octamer models.18
To unambiguously define the oligomerization state of Zwit-1F and detail the interactions of its assembly, we determined its structure by X-ray crystallography. To aid phasing, we synthesized an analogue of Zwit-1F bearing β3-4-iodohomophenylalanine at position 4 (Zwit-1F*). Zwit-1F* exhibits concentration-dependent secondary structure and sedimentation equilibria comparable to Zwit-1F18 and crystallizes from similar conditions, allowing calculation of initial phases by single-wavelength anomalous scattering. The final model of Zwit-1F thus derived is refined against data to 1.45 Å resolution and contains two pairs of parallel 314 helices per asymmetric unit. Individually, these four crystallographically unique monomers exhibit metrics that deviate only slightly from an ideal 314 helix. The root-mean-square deviation between all monomers is 1.44 Å, and all possible i to i + 2 main-chain hydrogen bonds are observed, with the exception of the N-terminal residue in one monomer. The average backbone dihedral angles (φ = −135.5°, ψ = −126.4°, μ = 55.6°) correspond well to those expected for the 314 helix.19 Crystallographic symmetry relates two copies of the asymmetric unit, however, to bury 2421 Å2 of surface area and reveal the octameric structure of Zwit-1F (Figure 2A).
Figure 2.
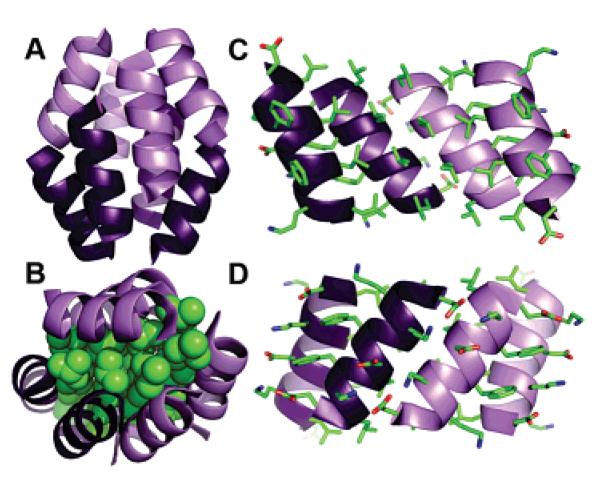
(A) Ribbon diagram of the Zwit-1F octamer, with parallel 314 helices in like shades. (B) Space-filling rendering of β3-homoleucine sidechains in green illustrate the well-packed hydrophobic core, while interior (C) and exterior (D) views of each hand detail the hydrophobic and electrostatic interactions of the assembly.
The Zwit-1F octamer is best described as a pair of tetrameric “hands”, each composed of four 314 helices cupped at approximately a 90° angle to each other. The two halves of each hand are composed of symmetry-equivalent parallel dimers oriented in an antiparallel fashion. β3-Homoleucine side chains decorate the interior of each hand. Their sequestration buries a total surface area of 2385 Å2 with less than 10 Å2 of average solvent accessible surface per residue, creating a hydrophobic core highly reminiscent of globular protein structures (Figure 2B).
Packing between pairs of helices in each hand involves interactions between residues on two helical faces. Both parallel and antiparallel helical pairs exhibit packing of β3-homoleucine side chains, in accord with our design (Figure 2C). The antiparallel interaction in the center of each hand positions the salt-bridge faces of each helix to make complementary electrostatic interactions across the interface (Figure 2D). Parallel helical pairs associate the aromatic face of an internal helix with the salt-bridge face of a terminal helix, with β3-E1 and β3-O10 forming electrostatic contacts with β3-O3 and β3-D12, respectively. In addition, the β3-F side chains exhibit a degree of hydrophobic packing from the side chain carbon atoms of opposing β3-D6 and β3-O9. Despite the differences in specific interactions, both helical arrangements are tightly packed, with parallel and antiparallel interfaces burying 796 and 784 Å2 of surface area, respectively, approximately 50% of a monomer. This percentage, as well as the mass-adjusted buried surface area (480 Å2/kDa), are nearly identical to those of coiled-coil proteins (4502/kDa, 51.5%).20
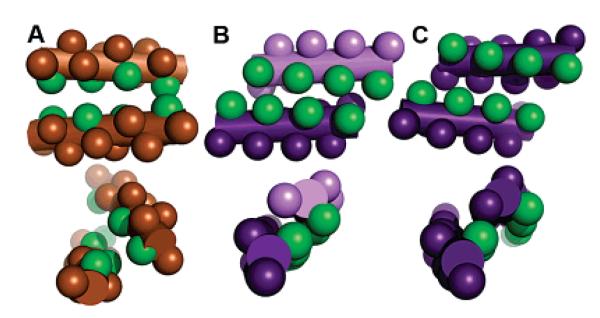
Inspiration for the design of a β3-homoleucine face to promote interhelix interaction came from the leucine zipper motif of coiled-coil proteins.20 While the 314-helical interfaces display association of leucine faces, there are significant differences in packing when compared to the knobs-into-holes pattern that allowed Crick to predict the supercoiling of leucine zippers (Figure 3).21 The periodicity of knobs and holes displayed in α-helical turns is a result of a repeat of 3.5–3.6 residues per turn. The nearly integral 3.1 residues/turn of 314 helices does not allow a similar complementary periodicity, however. Zwit-1F helices are instead offset along the helical axis, staggering the display of residues between the interacting faces. This interface allows a closer approach of the helices (~8.5 Å between helical axes) than in coiled-coil proteins (~9.5 Å) and greater main-chain contact, with direct backbone methylene contacts generating 23–33% of dimer interface. We anticipate that the participation of this additional methylene in packing interactions will be a general feature of higher order β-peptide structure.
Figure 3.
Comparison of leucine packing in (A) the leucine zipper GCN420 with the (B) antiparallel and (C) parallel interfaces of Zwit-1F. Helices are represented as solid cylinders, side chains as knobs, and interface leucines colored green.
In summary, here we describe the first high-resolution β-peptide quaternary structure. Despite significant differences between the secondary structures of discrete 314 helices and α-helices and the interactions between them, the Zwit-1F structure is remarkably protein-like. Composed of a discrete number of helices and possessing a solvent-excluded hydrophobic core, the assembly is driven only by noncovalent inter-residue interactions and is highly thermostable. Structures such as this “β-protein” promise the opportunity for more sophisticated functionality from future β-peptides.
Supplementary Material
Acknowledgment
This work was supported by the NIH and the National Foundation for Cancer Research and is based in part upon research conducted at the Cornell High Energy Synchrotron Source (CHESS), which is supported by the NSF, using the macromolecular diffraction at CHESS (MacCHESS) facility, which is supported by the NIH. This research was also supported by the Yale Center for Structural Biology, whose faculty and staff we gratefully acknowledge for use of their data collection facilities.
Footnotes
Supporting Information Available: Table containing data collection and refinement statistics, experimental details for crystallography, and sedimentation equilibrium. This material is available free of charge via the Internet at http://pubs.acs.org. CCDC nnnnnn contains atomic coordinates and supplementary crystallographic data for Zwit-1F. These data can be obtained free of charge from The Cambridge Crystal-lographic Data Centre via www.ccdc.cam.ac.uk/data_request/cif.
References
- 1.Cheng RP, DeGrado WF. J. Am. Chem. Soc. 2001;123:5162–5163. doi: 10.1021/ja010438e. [DOI] [PubMed] [Google Scholar]
- 2.Arvidsson PI, Rueping M, Seebach D. Chem. Commun. 2001:649–650. [Google Scholar]
- 3.Hart SA, Bahadoor ABF, Matthews EE, Qiu XYJ, Schepartz A. J. Am. Chem. Soc. 2003;125:4022–4023. doi: 10.1021/ja029868a. [DOI] [PubMed] [Google Scholar]
- 4.Kritzer JA, Lear JD, Hodsdon ME, Schepartz A. J. Am. Chem. Soc. 2004;126:9468–9469. doi: 10.1021/ja031625a. [DOI] [PubMed] [Google Scholar]
- 5.Karlsson AJ, Pomerantz WC, Weisblum B, Gellman SH, Palecek SP. J. Am. Chem. Soc. 2006;128:12630–12631. doi: 10.1021/ja064630y. [DOI] [PubMed] [Google Scholar]
- 6.Raguse TL, Porter EA, Weisblum B, Gellman SH. J. Am. Chem. Soc. 2002;124:12774–12785. doi: 10.1021/ja0270423. [DOI] [PubMed] [Google Scholar]
- 7.Stephens OM, Kim S, Welch BD, Hodsdon ME, Kay MS, Schepartz A. J. Am. Chem. Soc. 2005;127:13126–13127. doi: 10.1021/ja053444+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Werder M, Hauser H, Abele S, Seebach D. Helv. Chim. Acta. 1999;82:1774–1783. [Google Scholar]
- 9.Cheng RP, DeGrado WF. J. Am. Chem. Soc. 2002;124:11564–11565. doi: 10.1021/ja020728a. [DOI] [PubMed] [Google Scholar]
- 10.Bruckner AM, Chakraborty P, Gellman SH, Diederichsen U. Angew. Chem., Int. Ed. 2003;42:4395–4399. doi: 10.1002/anie.200351871. [DOI] [PubMed] [Google Scholar]
- 11.Lelais G, Seebach D, Jaun B, Mathad RI, Flogel O, Rossi F, Campo M, Wortman A. Helv. Chim. Acta. 2006;89:361–403. [Google Scholar]
- 12.Raguse TL, Lai JR, LePlae PR, Gellman SH. Org. Lett. 2001;3:3963–3966. doi: 10.1021/ol016868r. [DOI] [PubMed] [Google Scholar]
- 13.Clark TD, Buehler LK, Ghadiri MR. J. Am. Chem. Soc. 1998;120:651–656. [Google Scholar]
- 14.Cheng RP, Gellman SH, DeGrado WF. Chem. Rev. 2001;101:3219–3232. doi: 10.1021/cr000045i. [DOI] [PubMed] [Google Scholar]
- 15.Seebach D, Beck AK, Bierbaum DJ. Chem. Biodivers. 2004;1:1111–1239. doi: 10.1002/cbdv.200490087. [DOI] [PubMed] [Google Scholar]
- 16.Qiu JX, Petersson EJ, Matthews EE, Schepartz A. J. Am. Chem. Soc. 2006;128:11338–11339. doi: 10.1021/ja063164+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Privalov PL, Tiktopulo EI, Venyaminov S, Griko Yu V, Makhatadze GI, Khechinashvili NN. J. Mol. Biol. 1989;205:737–750. doi: 10.1016/0022-2836(89)90318-5. [DOI] [PubMed] [Google Scholar]
- 18.Please see Supporting Information for details.
- 19.Beke T, Somlai C, Perczel A. J. Comput. Chem. 2006;27:20–38. doi: 10.1002/jcc.20299. [DOI] [PubMed] [Google Scholar]
- 20.O'Shea EK, Klemm JD, Kim PS, Alber T. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- 21.Crick FHC. Acta Crystallogr. 1953;6:689–697. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.