Abstract
We report novel inhibitors of Gli1-mediated transcription as potential anticancer agents. Focused chemical libraries were designed and assessed for inhibition of functional cell-based Gli1-mediated transcription and selective toxicity toward cancer cells. The SAR was revealed and the selectivity of the lead compounds’ inhibition of Gli1-mediated transcription over that of Gli2 was determined. Compound 63 (NMDA298-1), which inhibited Gli1-mediated transcription in C3H10T1/2 cells with an IC50 of 6.9 µM, showed 3-fold selectivity for inhibiting transcription mediated by Gli1 over that by Gli2. Cell-viability assays were performed to evaluate the chemical library in a normal cell line and a panel of cancer cell lines with or without upregulated expression of the Gli1 gene. These compounds decreased the viability of several cancer cell lines but were less active in that of noncancerous BJ-hTERT cells.
Keywords: anticancer, Sonic Hedgehog, Gli, benzophenone
Introduction
The Sonic hedgehog (Shh) -Gli signaling pathway regulates the patterning and cellular growth of embryos. Inappropriate activation of this pathway is implicated in diverse cancers.1–6 Shh signaling is initiated by the binding of Shh to the transmembrane protein Patched (Ptch) and the subsequent release of Smoothened (Smo) from the Ptch-Smo complex. This event facilitates nuclear translocation of the Gli family of zinc finger transcription factors, which direct the transcriptional output of the Shh signal.7–10 Three Gli transcription factors, Gli1, Gli2, and Gli3, are reported in vertebrates. Gli1 appears to be a transcriptional activator, and Gli2 and Gli3 can act as both activators and repressors of transcription.11, 12 Shh expression is not the only activator of Gli1-mediated transcription; other regulatory mechanisms downstream of Smo also upregulate Shh signaling.2, 13–16 Elevated expression of Gli1 and its central role in the formation of several cancers has been well documented,2, 12, 17, 18 especially in childhood sarcoma.19 Gli1 and Gli2 are required for the tumorigenicity of human glioma stem cells, but Gli3 has very little or no reported role in tumorogenesis.12, 20 Gli1-knockout mice show no obvious phenotypic defects. In contrast, Gli2- and Gli3-knockout mice show skeletal and neural defects and embryonic or perinatal lethality.1, 17, 21, 22 In juvenile mice, inhibition of the Shh-Gli pathway upstream by a Smo inhibitor causes severe bone defects.23 These observations suggest that selective inhibitors of Gli1-mediated transcription, a down-stream event of the Shh-pathway, would be useful anticancer drugs24 tolerated well by pediatric patients who need hedgehog activity for somatic development and growth.
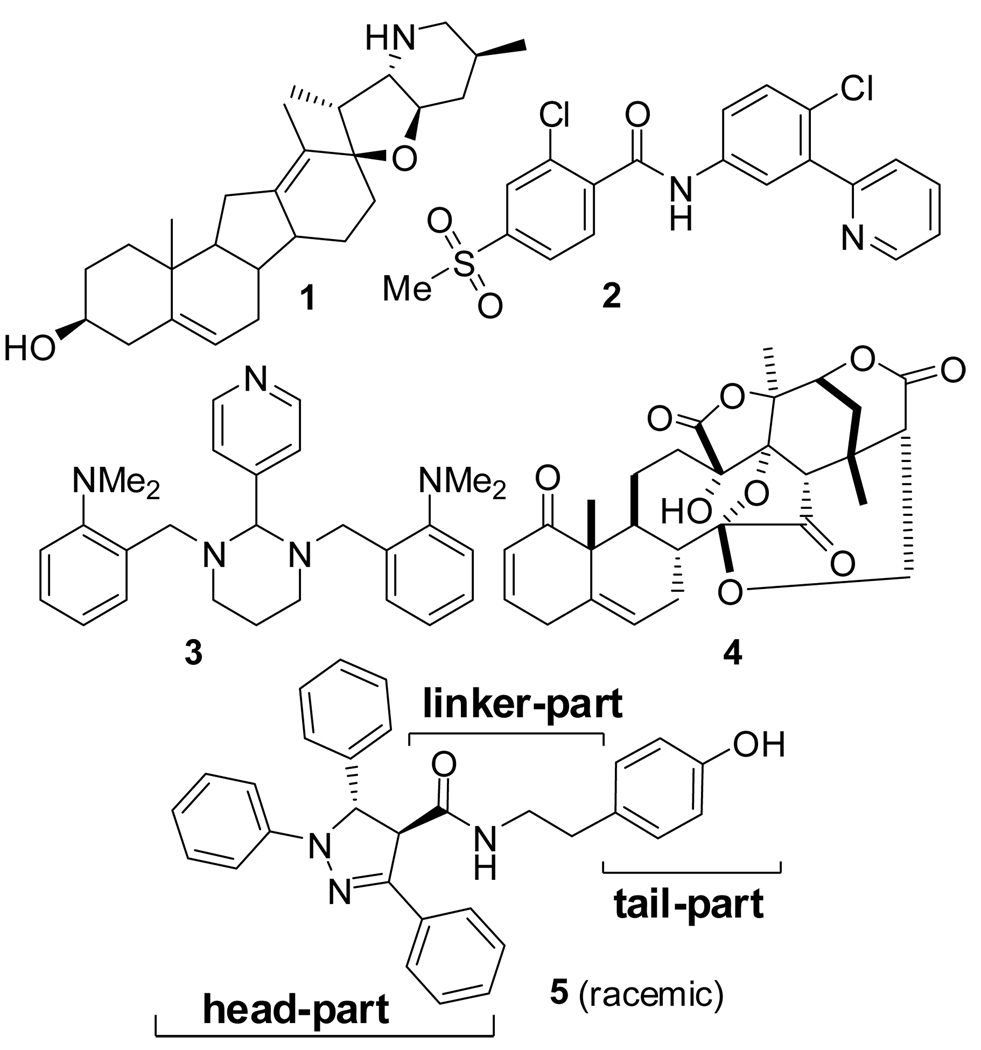
Smo receptor antagonists such as compounds 1 (cyclopamine) and 2 (GDC-0449) (Figure 1), which inhibit Shh signaling at the level of Smo, are in clinical trials.24 Small-molecule inhibitors of Glimediated transcription, compounds 3 (Gant61),16 4 (Physalin),25 and 5 (FN1–8),26 have also been reported (Figure 1). We designed novel inhibitors of Gli1-mediated transcription based on 5 by replacing the compound’s pyrazoline moiety while retaining a tyramine amide moiety that is an essential pharmacophorefor 5. 24, 26
Figure 1.
Inhibitors of the Shh-Gli signaling pathway.24 Compound 5 was chosen as a seed compound for the structure-activity relationship (SAR) investigation. Each partial structure (i.e., the head-, linker-, and tail-parts) discussed in this paper is shown in 5.
Chemistry
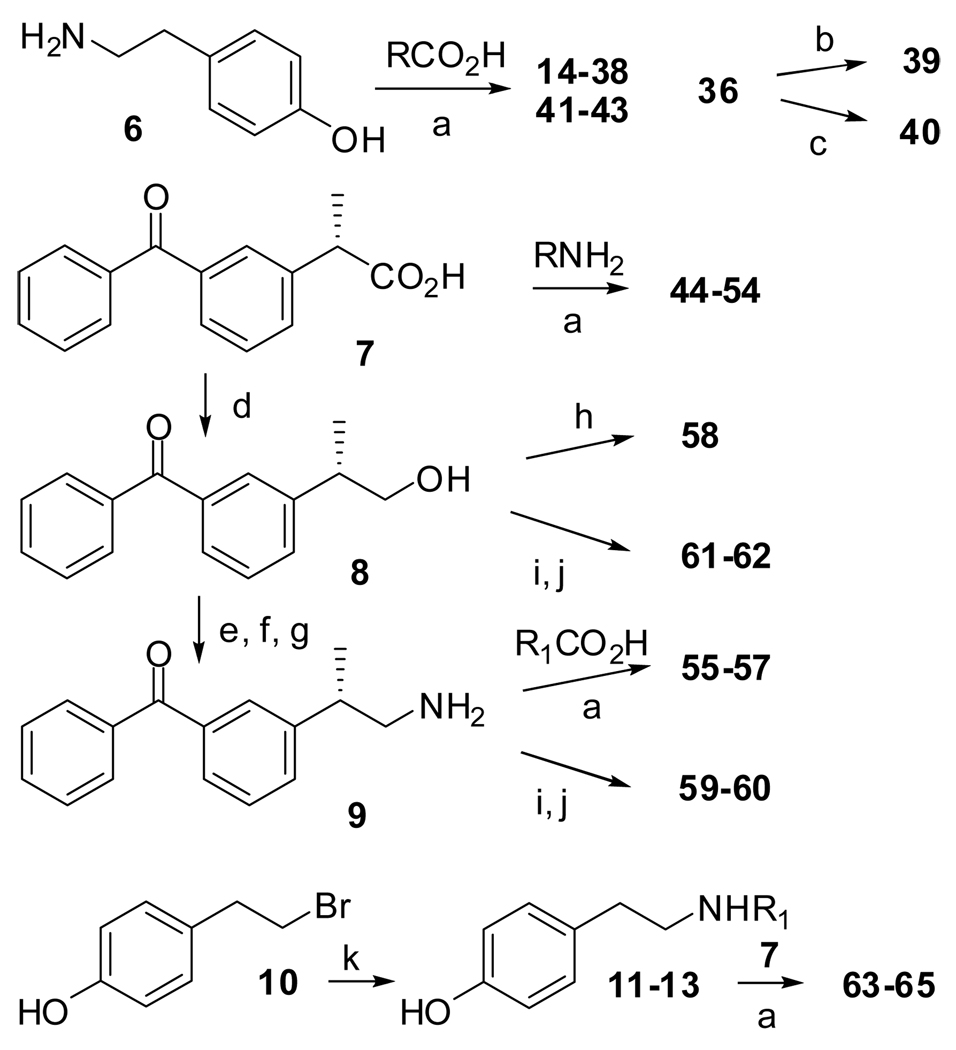
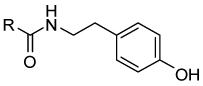
The compounds 14–38 and 41–54 were synthesized by coupling diverse carboxylic acids with appropriate tyramine derivatives (Scheme 1). Compounds 39 and 40 were prepared from 36. Carboxylic acid 7 was reduced to give alcohol 8, which on mesylation followed by azide formation and reduction gave the amine 9. The reverse amides 55–57 were synthesized from 9 by reacting with appropriate carboxylic acids. A compound with an ether linker (58) was prepared by alkylating 8. Analogues with a urea linker (59–60) were synthesized from 9 by first preparing the p-nitrophenylcarbamate, which was reacted with corresponding amines. Carbamates (61–62) were similarly synthesized from alcohol 8. N-substituted amides 63–65 were synthesized by reacting 10 with appropriate primary amines and then coupling the resulting secondary amines with 7.
Scheme 1a.
a Reagent and Conditions: (a) HBTU, DIPEA, DMF, rt; (b) NaBH4, MeOH, 2 h, rt; (c) Pd/H2, MeOH, 18 h, rt. (d) BH3-THF, THF, −20 °C – rt; (e) MsCl, Et3N, CH2Cl2, 1 h, 0 °C; (f) NaN3, DMF, 2 h, 80 °C; (g) PPh3, NH4OH, pyridine, rt; (h) R1-Br, NaH, DMF, rt; (i) 4-nitrophenyl chloroformate, Et3N, THF, 0 °C – rt; (j) R1NH2, Et3N, THF, 0 °C – rt. (k) R1NH2, DIPEA, acetonitrile, 16 h, 60 °C. Structures of compounds 11–13 are shown in the Experimental Section, and those of 14–65 are shown in Table 1 and Table 2 and in Figure 2 and Figure 4.
Results and Discussion
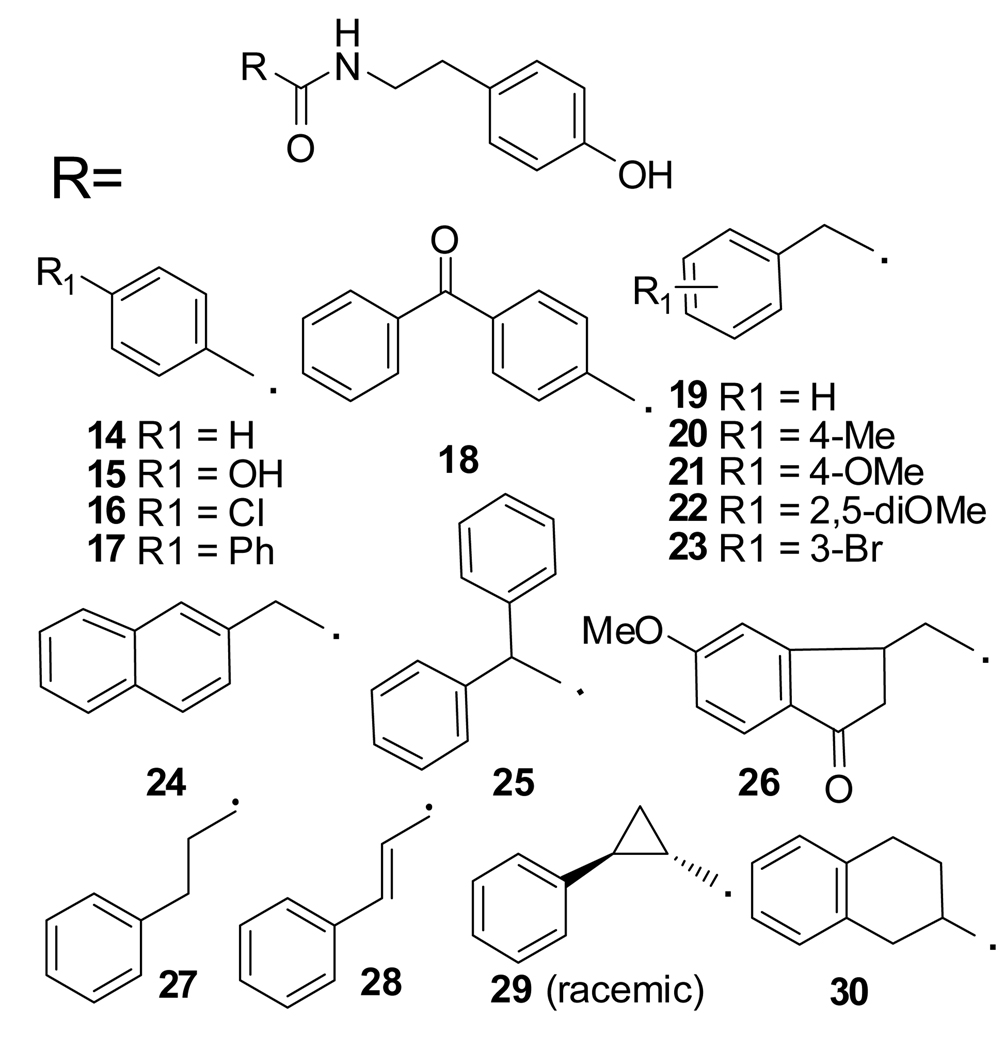
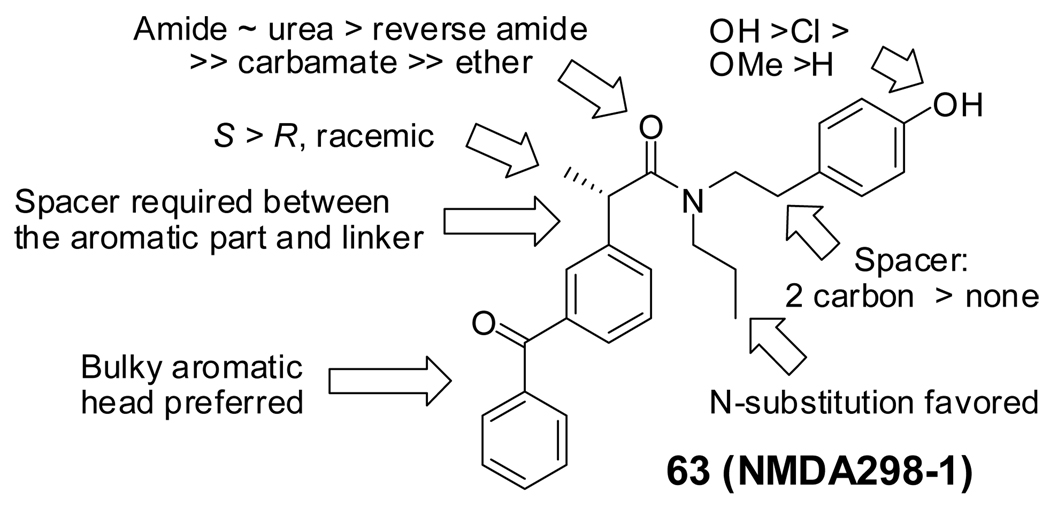
We started our SAR investigation by replacing the head-part of 5 (Figure 1). To assay compounds for selective inhibition of Gli1-mediated transcription, we used C3H10T1/2 mouse embryo fibroblasts with exogenously transfected vectors encoding human Gli1 and a Gli-luciferase reporter vector27. Because the Gli-reporter activities in these cells are activated solely by the exogenous Gli1, compounds that downregulate reporter activity in these cells are believed to target Gli1-mediated transcription but not upstream components such as Smo. Consistently, cyclopamine (1), an inhibitor of Smo, is inactive in this assay. Compounds with a small aromatic group as the head-part (14–17, 19–23) (Figure 2) also showed no inhibition of Gli1-mediated transcription (data not shown). We thus increased the size of the aromatic group (17, 18, 24–26) or the distance between the aromatic group and the amide linker (27–30). The compounds with bulkier aromatic groups and a methylene spacer between the aromatic group and amide (24–26) showed slight inhibition of Gli1-mediated transcription (data not shown), a finding that suggested the importance of the methylene spacer. Therefore, we next prepared compounds 31–36 with the bulkier aromatic group separated from the amide linker by a methylene spacer (Table 1).
Figure 2.
Inactive compounds in the Gli1-mediated transcription assay.
Table 1.
Compounds with different R groups at the head-part of 5
 | |||
|---|---|---|---|
| R | R | ||
| 5 | 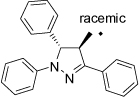 |
37 | 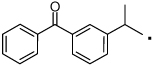 |
| 31 | 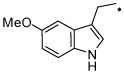 |
38 | 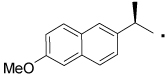 |
| 32 | 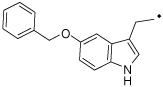 |
39 | 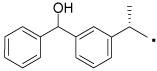 |
| 33 | 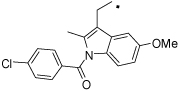 |
40 | 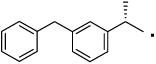 |
| 34 | 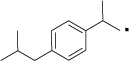 |
41 | 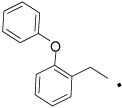 |
| 35 | 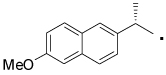 |
42 | |
| 36 | 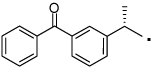 |
43 | 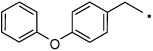 |
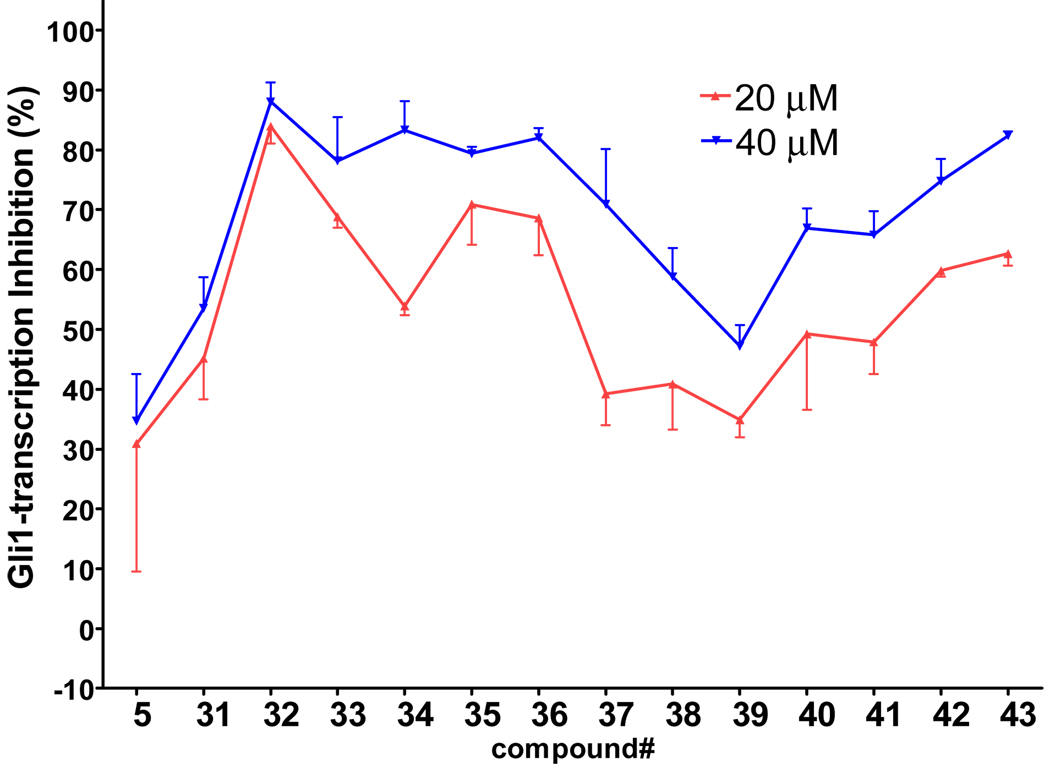
Compounds with a bulky group (32 and 33) showed better activity than did an analogue with a less bulky group (31) (Figure 3). Compound 34, which has a methyl substitution on the methylene spacer, also showed modest inhibition despite a comparatively smaller aromatic head-part. Compounds that included the methyl group on the methylene spacer and increased bulk of the aromatic group (35 and 36) showed higher activity than that of 34 at 20 µM and equivalent activity at 40 µM. To evaluate the effect of the chirality on the methylene spacer, we compared the activity of 35 with its (R)-enantiomer 38 and that of 36 with the racemic mixture 37. Compound 38 showed a lower activity than did 35, and 37 showed a lower activity than did 36, suggesting a preference of the (S)-enantiomer. The hydroxyl and methylene analogues (39 and 40) were less potent than 36. Replacement of the benzoyl moiety of 36 with a phenoxy group afforded equipotent compounds 42 and 43, but shifting the phenoxy group to the ortho position (41) decreased activity (Figure 3).
Figure 3.
Activity of the head-part library compounds. Percent inhibition of Gli-reporter activity in Gli1-transfected C3H10T1/2 cells 24 h after the addition of 20 µM (red plot) or 40 µM (blue plot) of the test compound (5, 31–43). DMSO control = 0%. Error bars represent the SEs of triplicated data.
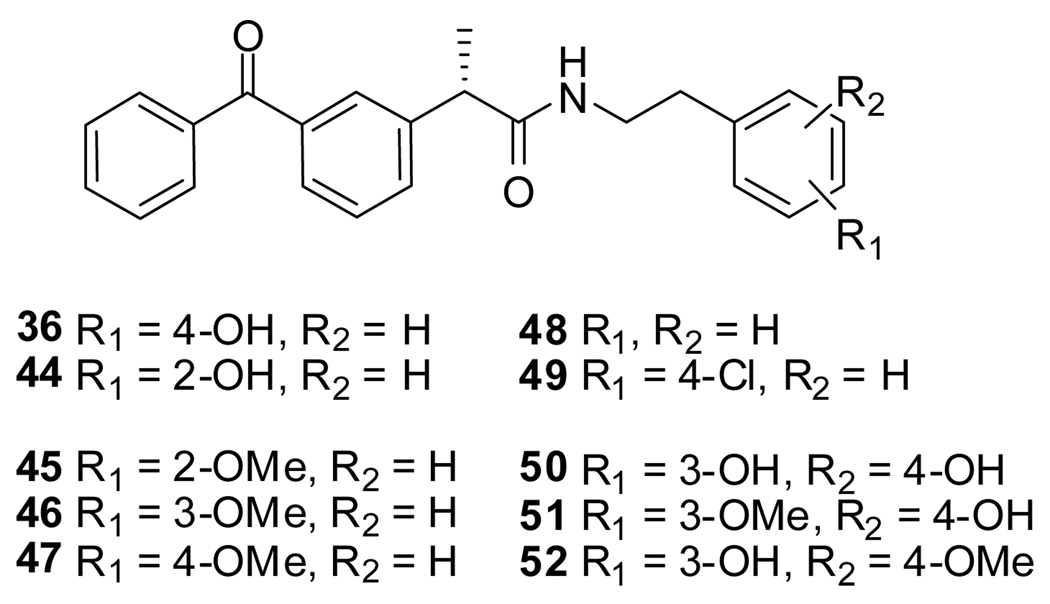
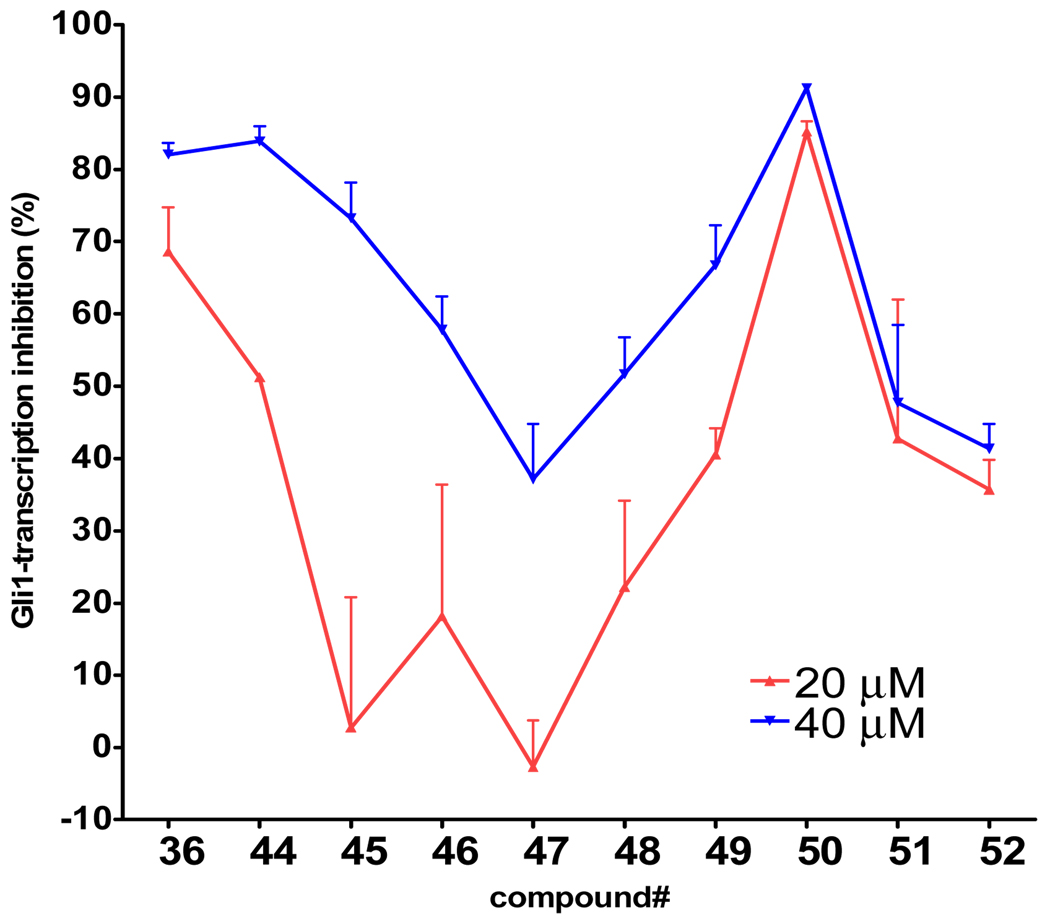
Next, we focused on 36 to investigate the SAR of the tail-part, because this compound has high activity and minimal toxicity as compared to 32 towards the C3H10T1/2 cells in the reporter assay (data not shown). Compound 7, in which the whole tail-part was removed, had no activity. Inhibition of Gli1-mediated transcription was slightly decreased at 20 µM when the hydroxyl group was moved to ortho position (44). Replacement of the hydroxyl group with a methoxy group (45–47) decreased activity. The unsubstituted derivative 48 also showed significantly lower activity than 36, and the 4-chloro analogue 49 showed slightly lower activity than 36. The catechol analog 50 afforded a higher activity than the phenol analog 36, but methylation of the catechol (51 and 52) reduced the activity by about half. All other substitutions on the benzene ring that were tested, including dichloro, amino, and trifluoromethyl group or saturation of the benzene ring to a cyclohexyl ring, decreased the activity substentially (data not shown). Overall, the tail-part showed little tolerance for change from phenol (36) or catechol (50) to any another substituent. (Figure 4 and Figure 5)
Figure 4.
SAR library of modified tail-parts of 36.
Figure 5.
Activity of the tail-part library compounds. Percent inhibition of Gli-reporter activity in Gli1-transfected C3H10T1/2 cells 24 h after addition of 20 µM (red plot) or 40 µM (blue plot) of the test compound (36, 44–52). DMSO control = 0%. Error bars represent the SEs of triplicated data.
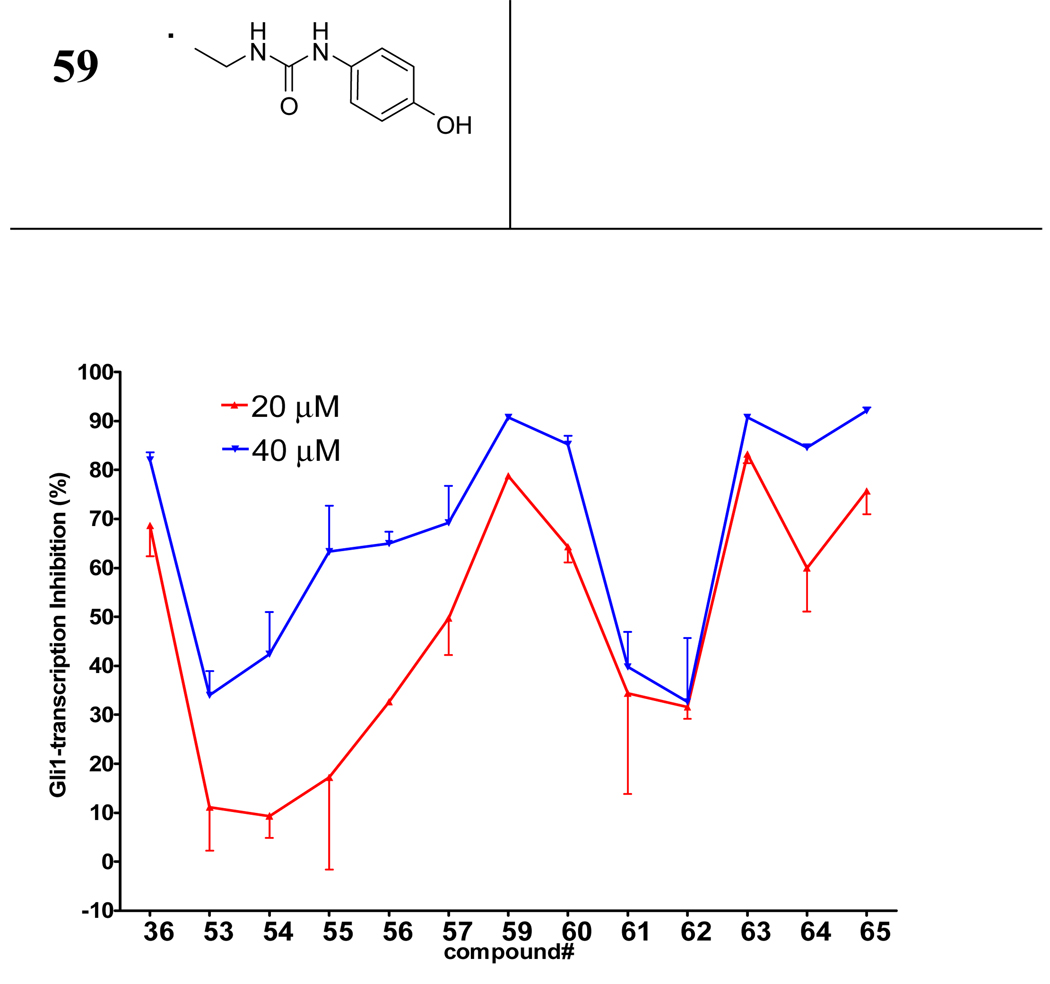
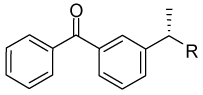
Finally, we studied the linker-part by shortening or replacing the amide linker with a substituted amide, reverse amide, ether, urea, or carbamate. (Table 2 and Figure 6) Decrease in the length of the linker-part of 36 decreased activity (53 and 54). Moving the amide carbonyl of 53 to the reverse position afforded better activity in 55; however, extension of the linker length of 55 afforded only a modest increase in the potency of 56. Despite decrease in the activity of 47 by methylation of the phenol in the tail-part of 36, methylation of the reverse amide 55 retained the activity (in 57). However, the corresponding ether analogue 58 showed almost no activity (data not shown) suggesting the importance of the amide structure in 36 analogues. The potency of compounds with a urea group in the linker-part (59 and 60) was comparable to that of 36, but they later appeared to be toxic to non-cancerous BJ-hTERT cells (Figure 10E). Exchanging the urea with a carbamate substantially decreased the activity (in 61 and 62). Substitution of the amide nitrogen in 63–65 showed activity comparable to or greater than that of 36.
Table 2.
Analogues for studying SARs at the linker-part of 36.
 | |||
|---|---|---|---|
| R | R | ||
| 53 |  |
60 |  |
| 54 | 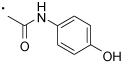 |
61 | 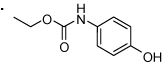 |
| 55 | 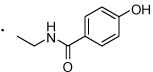 |
62 | 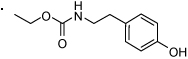 |
| 56 | 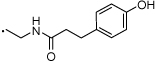 |
63 | 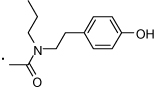 |
| 57 | 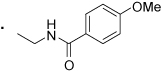 |
64 | 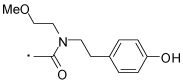 |
| 58 | 65 | 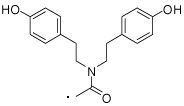 |
|
| 59 | 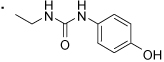 |
||
Figure 6.
Activity of the linker-part library compounds. Percent inhibition of Gli-reporter activity in Gli1-transfected C3H10T1/2 cells 24 h after addition of 20 µM (red plot) or 40 µM (blue plot) of the test compound (36, 53–57, 59–65). DMSO control = 0%. Error bars represent the SEs of triplicated data.
Figure 10.
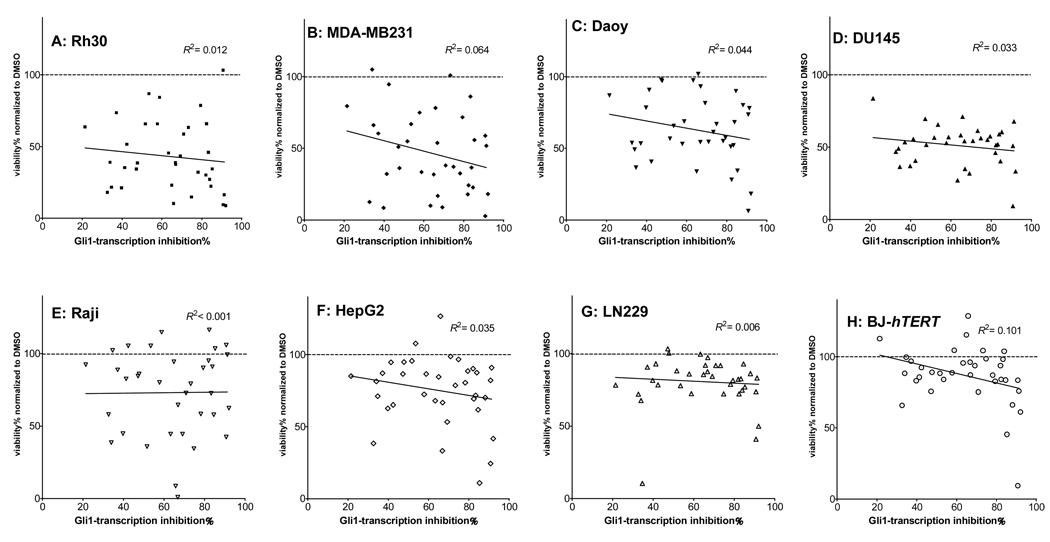
Scatter plots of the inhibition% of Gli1-mediated transcription in the C3H10/T1/2 cells 24 h after treatment with 40 µM of each compound vs. the cell viability% of each cell line 72 h after treatment in an AlmarBlue assay. Each plot represents each single compound shown in Figure 3, Figure 5, and Figure 6. Both data sets were normalized to those of DMSO as 0% transcription inhibition and 100% cell viability. (A) Rh30, (B) MDA-MB231, (C) Daoy, (D) DU145, (E) Raji.. (F) HepG2, (G) LN229, and (H) BJ-hTERT. Linear regressions were calculated by Prism software (GraphPad). Data from individual compounds are shown in Supplement Figure 2.
In summary, the SAR was clearly observed (Figure 7). The scatter plots showed that the library generally followed Lipinski’s Rule of Five, and that the activity in Gli1-mediated transcription inhibition does not relate to the calculated values of logP or total polar surface area for each compound (Supplemental Figure 1). These findings suggest that the inhibition of Gli1-mediated transcription is not due to hydrophobic aggregation by the compound. To exclude the possibility that these compounds are inhibitors of transcription/translation of the exogenously transfected Gli1 in the assayed C3H10T1/2 cells, we determined the Gli1 protein levels by Western blot analysis. The Gli1-transfected C3H10T1/2 cells have confirmed to overexpress the exogenous Gli1 protein and it is not reduced by 36 or 63 (Supplemental Figure 5). This finding suggests that they do not affect the protein expression from the exogenous Gli1 vector or the transgenic promoter. We have also tested the possibility that these compounds are inhibitors of assay reporter.28 Instead of adding compounds to the cells, they were added to the C3H10T1/2 lysate in which luciferase is highly expressed by the exogenous Gli1/Gli-Luc reporter. The luciferase activity in these samples was not changed by 36 or 63 (Supplemental Figure 6), confirming that they are not inhibitors of luciferase reporter.
Figure 7.
Summary of the structure-activity relationships.
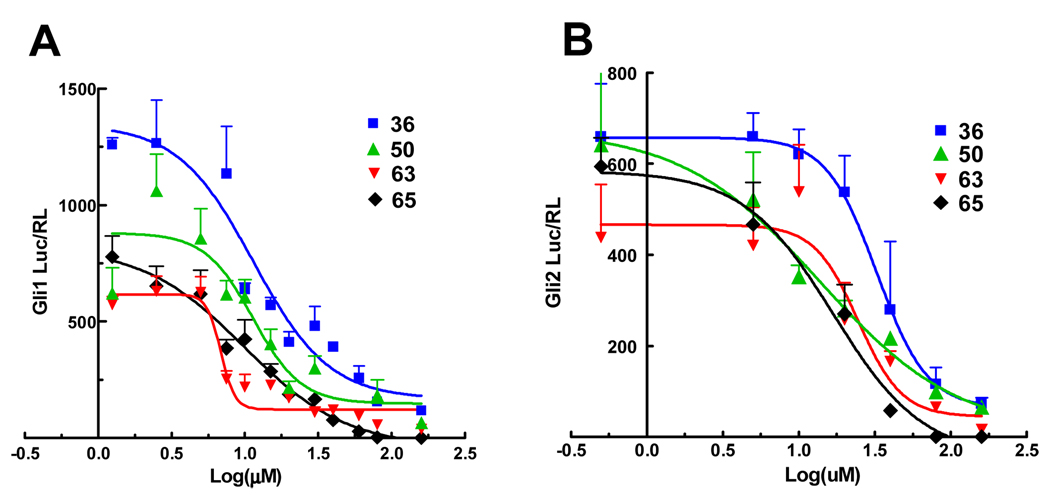
Selected compounds were evaluated for their selectivity in inhibiting the Gli1-mediated transcription over Gli2-mediated transcription by detemining their IC50 values (Figure 8 and Table 3). The compounds showed 3 to 4 times weaker inhibition of Gli2-mediated transcription than that of Gli1 (Table 3). Compound 65 was least selective for Gli1. We also tested the stability of compounds 36, 50, 63 and 65 in the C3H10T1/2 cell cultures. Catechol derivative 50 showed significant degradation, while phenol derivatives 36, 63, and 65 were stable for 24 hours. (Supplemental Figure 4)
Figure 8.
Dose-response curve of Gli-reporter activity in Gli1-transfected (A) or ΔNGli229-transfected (B) C3H10T1/2 cells 24 h after addition of the test compound 36 (blue), 50 (green), 63 (red), 65 (black). Error bars represent the SEs of triplicated data. Response curve fittings were calculated by Prism software (GraphPad). IC50 in these curves is summarized in Table 3.
Table 3.
The IC50 (µM) and 95% confidence intervals (in parenthesis) of selected compounds for inhibiting transcriptional activation by exogenous human Gli1 or ΔNGli229 in C3H10T1/2 cells 24 h after addition of the test compound (Figure 8).
| Gli1 | Gli2 | |
|---|---|---|
| 36 | 11.4 (7.5 – 17.5) | 35.2 (17.8 – 59.9) |
| 50 | 11.4 (7.9 – 16.3) | 29.4 (2.9 – 66.6) |
| 63 | 6.9 (6.1 – 7.8) | 23.9 (12.3 – 49.0) |
| 65 | 9.4 (6.40 – 15.4) | 20.5 (9.2 – 30.9) |
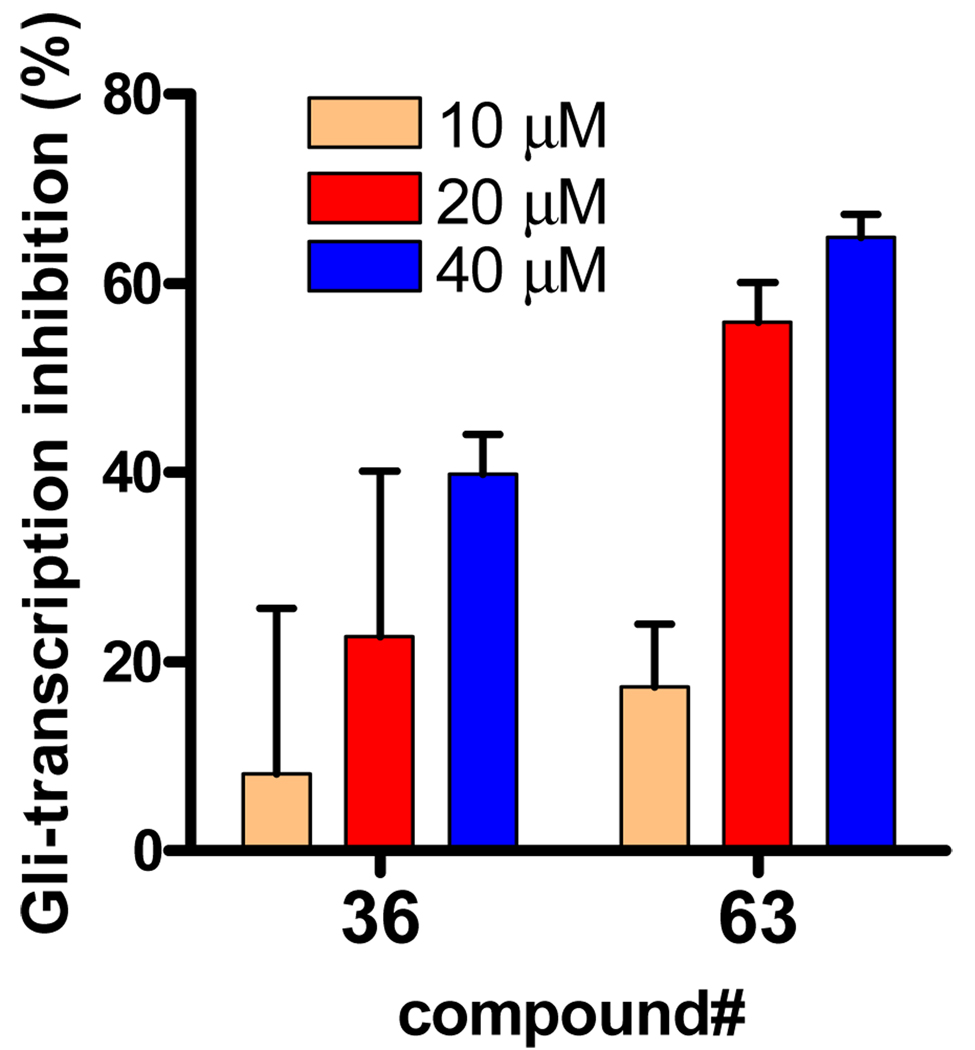
To prove that inhibitors of transcription in C3H10T1/2 cells with exogenously overexpressed Gli1 are also active in cells that endogenously overexpressed Gli1, we assayed compounds 36 and 63 for inhibition of Gli-reporter activity in Rh30, a rhabdomyosarcoma cell line that overexpresses Gli1 endogenously,3 without contransfection of exogenous Gli1. This assay confirmed that 36 and 63 inhibit transcription by the endogenous Gli1 in this cell line (Figure 9); thus, these compounds are expected to suppress the growth of cancer cells in which growth control is Gli1-dependent, such as meduloblastoma progenitor cells.17
Figure 9.
Percent inhibition of Gli-reporter activity in Rh30 cells 24 h after the addition of 10 µM (peach), 20 µM (red), or 40 µM (blue) of the test compound 36 or 63. DMSO control = 0%. Error bars represent the SEs of triplicated data.
To determine whether inhibitors of Gli1-mediated transcription inhibit the growth of cancer cells, we tested them on the viability of several cancer cell lines. Cell viability was reduced in Rh30 (rhabdomyosarcoma, Figure 10A), MDA-MB231 (breast cancer, Figure 10B), Daoy (medulloblastoma, Figure 10C), DU145 (prostate cancer, Figure 10D), and Raji (Burkitt’s Lymphoma, Figure 10E), but not in HepG2 (hepatocellular carcinoma, Figure 10F), LN229 (glioma, Figure 10G), and hTERT-immortalized BJ (noncancerous human skin fibroblasts30, Figure 10H). This finding suggests that inhibitors of Gli1-mediated transcription could suppress the growth of cancer cells. However, it is difficult to observe simple relationship between reduction of cell viability and inhibition of Gli1-mediated transcription, likely due to a non-specific effect of some of the compounds.
It is interesting to see if the growth inhibition effect correlates to the level of Gli1 protein in these cell lines. However, in our hand, the Western blot analysis reliably detects exogenous Gli1 protein expression in the C3H10T1/2 cells (Supplementary Figure 5) but not endogenous Gli1 protein in these cell lines for the viability study, with the only exception being Rh30 (data not shown). Expression of Gli1 mRNA has been previously shown in Rh303, MDA-MB23131, Daoy32, 33, and DU14534 cell lines. However, it is well-known that Gli1 protein is down-regulated post-translationally32, 35 even when Gli1 mRNA is expressed36, thus the level of Gli1 protein does not linearly correlate to that of Gli1 mRNA. A statistical approach with a simple two-category analysis (e.g., Gli1 mRNA -overexpression or non-overexpression) suggests that the Gli1-mediated transcription inhibitors decrease the viability of Rh30, MDA-MB231, Daoy, and DU145 cell lines by 0.19% more than that of the other four cell lines, per 1% increase in the inhibition (see Supporting Information for detail).
Conclusion
In this study, we report the discovery of novel inhibitors of Gli1-mediated transcription that are potential anticancer agents. The amide analogue 36 and substituted amide 63 inhibited Gli1-mediated transcription without showing toxicity against a normal cell line, downregulated endogenous Gli-mediated transcription in Rh30 cells, and demonstrated inhibition selectivity of Gli1-mediated transcription that approximately 3 times greater than that of Gli2-mediated transcription. Cell viability is reduced by analogues of those compounds in cancer cells. These compounds might be the basis to design potential lead compounds that function as anticancer agents by selectively inhibiting Gli1-mediated transcription.
Experimental Section
General Procedure
Compounds 14–38, 41–54, and 55–56 were synthesized as follows: A mixture appropriate carboxylic acid (1 eq), HBTU (2.5 eq), and DIPEA (3 eq) in DMF (0.3 mL) was allowed to stand for 30 min at rt. Appropriate amine (3.0 eq) was then added to the mixture and stirred for 24 h at rt. Water was added to the reaction mixture and extracted with ethyl acetate followed by successive washings with water and brine. The organic layer was dried over anhydrous sodium sulphate. The solvent was removed in vacuo to give the residue, which was chromatographed over silica gel (Biotage SP4, 12+S Column, eluting with hexane ethyl acetate gradient 10–80%) to give the desired compound.
The purity of all the compounds was determined by HPLC on a Waters Alliance HT LC-MS system (Waters 2795 Separation Module linked to a Waters 2996 Photodiode Array Detector) using a Waters XBridge C18, 3.5 µm (4.6 × 50 mm) column by running a 0 to 95% gradient for Water (+ 0.05% TFA)/MeOH (Supplemental Figure 3). All the compounds showed ≥ 95% purity, unless otherwise mentioned.
N-(4-Hydroxyphenethyl)benzamide (14)
1H NMR (400 MHz, CDCl3 + CD3OD) δ 7.72 (m, 2H), 7.49 (m, 1H), 7.41 (t, 2H, J = 7.4 Hz), 7.08 (d, 2H, J = 8.5 Hz), 6.79 (d, 2H, J = 8.5 Hz), 3.61 (t, 2H, J = 7.2 Hz), 2.84 (t, 2H, J = 7.2 Hz). HRMS (ESI (M+H)+ m/z) calcd for C15H16NO2 242.1181, found 242.1175.
4-Hydroxy-N-(4-hydroxyphenethyl)benzamide (15)
1H NMR (400 MHz, CDCl3 + CD3OD) δ 7.57 (d, 2H, J = 8.7 Hz), 7.07 (d, 2H, J = 8.4 Hz), 6.80 (m, 4H), 3.59 (t, 2H, J = 7.0 Hz), 2.82 (t, 2H, J = 7.0 Hz). HRMS (ESI (M+H)+ m/z) calcd for C15H16NO3 258.1130, found 258.1125. HPLC purity 93%.
4-Chloro-N-(4-hydroxyphenethyl)benzamide (16)
1H NMR (400 MHz, CDCl3 + CD3OD) δ 7.70 (d, 2H, J = 8.3 Hz), 7.40 (d, 2H, J = 8.3 Hz), 7.07 (d, 2H, J = 8.2 Hz), 6.78 (d, 2H, J = 8.1 Hz), 3.58 (t, 2H, J = 7.3 Hz), 2.83 (t, 2H, J = 7.3 Hz). HRMS (ESI (M+H)+ m/z) calcd for C15H15NO2Cl 276.0791, found 276.0781.
N-(4-Hydroxyphenethyl)biphenyl-4-carboxamide (17)
1H NMR (400 MHz, CDCl3 + CD3OD) δ 7.83 (d, 2H, J = 8.2 Hz), 7.64 (dd, 4H, J = 7.7, 16.9 Hz), 7.47 (t, 2H, J = 7.5 Hz), 7.39 (t, 1H, J = 7.4 Hz), 7.11 (d, 2H, J = 7.8 hz), 6.79 (d, 2H, J = 7.6 Hz), 3.62 (t, 2H, J = 6.9 hz), 2.87 (t, 2H, J = 7.2 Hz). HRMS (ESI (M+H)+ m/z) calcd for C21H20NO2 318.1494, found 318.1491.
4-Benzoyl-N-(4-hydroxyphenethyl)benzamide (18)
1H NMR (400 MHz, CDCl3 + CD3OD) δ 7.83 (m, 6H), 7.65 (t, 1H, J = 7.4 Hz), 7.52 (t, 2H, J = 7.6 Hz), 7.10 (d, 2H, J = 8.5Hz), 6.83 - 6.76 (m, 2H), 3.63 (t, 2H, J = 7.2Hz), 2.87 (t, 2H, J = 7.2Hz). HRMS (ESI (M+H)+ m/z) calcd for C22H20NO3 346.1443, found 346.1436.
N-(4-Hydroxyphenethyl)-2-phenylacetamide (19)
1H NMR (400 MHz, CDCl3) δ 7.31 (m, 3H), 7.17 (d, 2H, J = 7.2 Hz), 6.86 (d, 2H, J = 8.4 Hz), 6.71 (d, 2H, J = 8.3 Hz), 5.86 (s, 1H), 3.50 (s, 2H), 3.39 (t, 2H, J = 6.7 Hz), 2.63 (t, 2H, J = 6.8 Hz). HRMS (ESI (M+H)+ m/z) calcd for C16H18NO2 256.1338, found 256.1335.
N-(4-Hydroxyphenethyl)-2-p-tolylacetamide (20)
1H NMR (400 MHz, CDCl3) δ 7.12 (d, 2H, J = 7.7 Hz), 7.03 (d, 2H, J = 8.0 Hz), 6.86 (d, 2H, J = 8.5 Hz), 6.72 (m, 2H), 5.46 (s, 1H), 3.50 (s, 2H), 3.42 (q, 2H, J = 6.8 Hz), 2.63 (t, 2H, J = 6.9 Hz), 2.34 (s, 3H). HRMS (ESI (M+H)+ m/z) calcd for C17H20NO2 270.1494, found 270.1483.
N-(4-Hydroxyphenethyl)-2-(4-methoxyphenyl)acetamide (21)
1H NMR (400 MHz, CDCl3) δ 7.05 (d, 2H, J = 6.8 Hz), 6.85 (t, 4H, J = 7.2 Hz), 6.72 (d, 2H, J = 6.5 Hz), 5.45 (s, 1H), 3.80 (s, 3H), 3.47 (s, 2H), 3.42 (dd, 2H, J = 6.4, 13.0 Hz), 2.63 (t, 2H, J = 6.7 Hz). HRMS (ESI (M+H)+ m/z) calcd for C17H20NO3 286.1443, found 286.1436.
2-(2,5-Dimethoxyphenyl)-N-(4-hydroxyphenethyl)acetamide (22)
1H NMR (400 MHz, CDCl3) δ 6.87 (d, 2H, J = 7.4 Hz), 6.78 (d, 3H, J = 14.2 Hz), 6.70 (d, 2H, J = 7.4 Hz), 5.84 (s, 1H), 3.77 (s, 3H), 3.68 (s, 7H), 3.38 (m, 2H), 2.61 (t, 2H, J = 6.8 Hz). HRMS (ESI (M+H)+ m/z) calcd for C18H22NO4 316.1549, found 316.1545.
2-(3-Bromophenyl)-N-(4-hydroxyphenethyl)acetamide (23)
1H NMR (400 MHz, CDCl3) δ 7.42 (d, 1H, J = 7.9 Hz), 7.35 (d, 1H, J = 1.7 Hz), 7.19 (t, 1H, J = 7.8 Hz), 7.11 (d, 1H, J = 7.7 Hz), 6.90 (d, 2H, J = 8.5 Hz), 6.77 - 6.70 (m, 2H), 5.51 (s, 1H), 5.35 (s, 1H), 3.48 (s, 2H), 3.45 (dd, 2H, J = 6.7, 12.8 Hz), 2.67 (t, 2H, J = 6.7 Hz).HRMS (ESI (M+H)+ m/z) calcd for C16H16BrNO2Na 356.0262, found 356.0255.
N-(4-Hydroxyphenethyl)-2-(naphthalen-2-yl)acetamide (24)
1H NMR (400 MHz, CDCl3) δ 7.82 (dt, 3H, J = 7.2, 16.9 Hz), 7.61 (s, 1H), 7.50 (m, 2H), 7.26 (m, 1H), 6.77 (d, 2H, J = 8.4 Hz), 6.54 (dd, 2H, J = 2.4, 8.9 Hz), 5.41 (s, 1H), 3.69 (s, 2H), 3.41 (q, 2H, J = 6.6 Hz), 2.60 (t, 2H, J = 6.8 Hz). HRMS (ESI (M+H)+ m/z) calcd for C20H20NO2 306.1494, found 306.1486.
N-(4-Hydroxyphenethyl)-2,2-diphenylacetamide (25)
1H NMR (400 MHz, CDCl3) δ 7.26 (m, 6H), 7.15 (d, 4H, J = 6.7 Hz), 6.85 (d, 2H, J = 8.3 Hz), 6.68 (d, 2H, J = 8.3 Hz), 5.82 (s, 1H), 4.87 (s, 1H), 3.48 (t, 2H, J = 6.8 Hz), 2.66 (t, 3H, J = 6.8 Hz). HRMS (ESI (M+H)+ m/z) calcd for C22H22NO2 332.1651, found 332.1642.
N-(4-Hydroxyphenethyl)-2-(6-methoxy-3-oxo-2,3-dihydro-1H-inden-1-yl)acetamide (26)
1H NMR (400 MHz, CDCl3) δ 7.66 (d, 1H, J = 8.5 Hz), 7.01 (d, 2H, J = 8.3 Hz), 6.93 (dd, 2H, J = 3.8, 12.3 Hz), 6.77 (d, 2H, J = 8.4 Hz), 5.32 (s, 1H), 3.88 (s, 3H), 3.80 (dd, 1H, J = 7.6, 14.7 Hz), 3.54 (m, 1H), 3.41 (m, 2H), 2.87 (dd, 1H, J = 7.3, 18.7 Hz), 2.73 (m, 2H), 2.63 (dd, 1H, J = 4.8, 14.4 Hz), 2.27 (dd, 1H, J = 9.8, 13.9 Hz). HRMS (ESI (M+H)+ m/z) calcd for C20H22NO4 340.1549, found 340.1533.
N-(4-Hydroxyphenethyl)-3-phenylpropanamide (27)
1H NMR (400 MHz, CDCl3) δ 7.28 (m, 2H), 7.19 (dd, 3H, J = 6.5, 13.3 Hz), 6.94 (d, 2H, J = 8.3 Hz), 6.75 (d, 2H, J = 8.4 hz), 5.32 (m, 1H), 3.43 (dd, 2H, J = 6.6, 12.9 Hz), 2.94 (t, 2H, J = 7.6 Hz), 2.65 (t, 2H, J = 6.8 Hz), 2.42 (t, 2H, J = 7.6 Hz). HRMS (ESI (M+H)+ m/z) calcd for C17H20NO2 270.1494, found 270.1504.
N-(4-Hydroxyphenethyl)cinnamamide (28)
1H NMR (400 MHz, CDCl3) δ 7.62 (d, 1H, J = 15.6 Hz), 7.48 (s, 2H), 7.36 (m, 3H), 7.09 (d, 2H, J = 8.5 Hz), 6.80 (d, 2H, J = 8.5 Hz), 6.31 (d, 1H, J = 15.6 Hz), 5.58 (m, 1H), 3.62 (d, 2H, J = 6.1 Hz), 2.82 (t, 2H, J = 6.9 Hz). HRMS (ESI (M+H)+ m/z) calcd for C17H18NO2 268.1338, found 268.1327. HPLC purity 92%.
(1SR, 2SR)-N-(4-Hydroxyphenethyl)-2-phenylcyclopropanecarboxamide (29)
1H NMR (400 MHz, CDCl3) δ 7.26 (t, 2H, J = 7.6 Hz), 7.18 (t, 1H, J = 7.3 Hz), 7.05 (m, 4H), 6.78 (d, 2H, J = 8.3 Hz), 5.70 (s, 1H), 3.51 (m, 2H), 2.75 (t, 2H, J = 7.0 Hz), 2.49 (m, 1H), 1.62 (m, 1H), 1.52 (m, 1H), 1.23 (m, 2H). HRMS (ESI (M+H)+ m/z) calcd for C18H20NO2 282.1494, found 282.1491.
N-(4-Hydroxyphenethyl)-1,2,3,4-tetrahydronaphthalene-2-carboxamide (30)
1H NMR (400 MHz, CDCl3) δ 7.06 (m, 6H), 6.79 (d, 2H, J = 8.5 Hz), 5.64 (t, 1H, J = 5.1 Hz), 3.52 (dd, 2H, J = 6.9, 14.3 Hz), 2.85 (m, 6H), 2.46 (m, 1H), 2.03 (d, 1H, J = 10.2 Hz), 1.86 (m, 1H). HRMS (ESI (M+H)+ m/z) calcd for C19H22NO2 296.1651, found 296.1641.
N-(4-Hydroxyphenethyl)-2-(5-methoxy-1H-indol-3-yl)acetamide (31)
1H NMR (400 MHz, CDCl3) δ 8.88 (s, 1H), 7.27 (d, 1H, J = 8.7 Hz), 6.89 (m, 3H), 6.62 (dd, 4H, J = 8.5, 22.2 Hz), 5.88 (s, 1H), 3.64 (s, 2H), 3.35 (t, 2H, J = 5.7 Hz), 2.53 (t, 2H, J = 6.7 Hz). HRMS (ESI (M+H)+ m/z) calcd for C19H21N2O3 325.1552, found 325.1545.
2-(5-(Benzyloxy)-1H-indol-3-yl)-N-(4-hydroxyphenethyl)acetamide (32)
1H NMR (400 MHz, CDCl3) δ 7.48 (d, 1H, J = 7.5 Hz), 7.38 (t, 1H, J = 7.5 Hz), 7.31 (dd, 1H, J = 4.5, 8.0 Hz), 6.99 (dd, 1H, J = 9.1, 11.5 Hz), 6.64 (d, 1H, J = 8.4 Hz), 6.55 (d, 1H, J = 8.3 Hz), 5.08 (s, 1H), 3.64 (s, 0H), 3.33 (m, 1H), 2.52 (t, 1H, J = 6.6 Hz). HRMS (ESI (M+H)+ m/z) calcd for C25H25N2O3 401.1865, found 401.1855.
2-(1-(4-Chlorobenzoyl)-5-methoxy-2-methyl-1H-indol-3-yl)-N-(4-hydroxyphenethyl) acetamide (33)
1H NMR (400 MHz, CDCl3 + CD3OD) δ 7.64 (d, 2H, J = 8.3 Hz), 7.52 (d, 2H, J = 8.4 Hz), 6.91 (dd, 2H, J = 5.7, 8.1 Hz), 6.80 (d, 2H, J = 8.4 Hz), 6.72 (dd, 1H, J = 2.5, 9.0 Hz), 6.62 (d, 2H, J = 8.4 Hz), 6.29 (s, 1H), 3.84 (s, 3H), 3.58 (s, 2H), 3.40 (dd, 2H, J = 6.6, 12.7 Hz), 2.62 (t, 2H, J = 6.8 Hz), 2.28 (s, 3H). HRMS (ESI (M+H)+ m/z) calcd for C27H26N2O4Cl 477.1581, found 477.1571.
N-(4-Hydroxyphenethyl)-2-(4-isobutylphenyl)propanamide (34)
1H NMR (400 MHz, CDCl3) δ 7.10 (m, 4H), 6.83 (d, 2H, J = 8.5 Hz), 6.72 (d, 2H, J = 8.6 Hz), 5.44 (t, 1H, J = 5.4 Hz), 3.43 (ddq, 3H, J = 7.0, 13.3, 26.0 Hz), 2.61 (m, 2H), 2.46 (d, 2H, J = 7.2 Hz), 1.85 (d p, 1H, J = 6.8, 13.6 Hz), 1.49 (d, 3H, J = 7.2 Hz), 0.91 (d, 6H, J = 6.6 Hz). HRMS (ESI (M+H)+ m/z) calcd for C21H28NO2 326.2120, found 326.2109.
(S)-N-(4-Hydroxyphenethyl)-2-(6-methoxynaphthalen-2-yl)propanamide (35)
1H NMR (400 MHz, CDCl3) δ 7.68 (t, 2H, J = 7.8 Hz), 7.56 (s, 1H), 7.27 (d, 1H, J = 8.4 Hz), 7.14 (m, 2H), 6.74 (d, 2H, J = 8.3 Hz), 6.59 (d, 2H, J = 8.4 Hz), 5.47 (s, 1H), 3.91 (s, 3H), 3.64 (dd, 1H, J = 6.3, 13.4 Hz), 3.38 (ddd, 2H, J = 6.4, 16.3, 19.2 Hz), 2.57 (t, 2H, J = 6.5 Hz), 1.57 (d, 3H, J = 7.1 Hz). HRMS (ESI (M+H)+ m/z) calcd for C22H24NO3 350.1756, found 350.1754.
(S)-2-(3-Benzoylphenyl)-N-(4-hydroxyphenethyl)propanamide (36)
1H NMR (400 MHz, CDCl3) δ 7.77 (m, 2H), 7.66 (m, 2H), 7.59 (m, 1H), 7.46 (m, 4H), 6.81 (d, 2H, J = 8.5 Hz), 6.68 (m, 2H), 5.54 (t, 1H, J = 5.6 Hz), 3.56 (q, 1H, J = 7.1 Hz), 3.41 (m, 2H), 2.62 (t, 2H, J = 6.8 Hz), 1.51 (d, 3H, J = 7.2 Hz). HRMS (ESI (M+H)+ m/z) calcd for C24H24NO3 374.1756, found 374.1749.
(±)-2-(3-Benzoylphenyl)-N-(4-hydroxyphenethyl)propanamide (37)
1H NMR (400 MHz, CDCl3) δ 7.77 (m, 2H), 7.66 (m, 2H), 7.60 (m, 1H), 7.47 (m, 4H), 6.82 (m, 2H), 6.68 (m, 2H), 5.48 (t, 1H, J = 6.0 Hz), 3.55 (q, 1H, J = 7.1 Hz), 3.46 (m, 1H), 3.36 (dq, 1H, J = 6.8, 13.4 Hz), 2.63 (t, 2H, J = 6.8 Hz), 1.51 (d, 3H, J = 7.2 Hz). HRMS (ESI (M+H)+ m/z) calcd for C24H24NO3 374.1756, found 374.1750.
(R)-N-(4-Hydroxyphenethyl)-2-(6-methoxynaphthalen-2-yl)propanamide (38)
1H NMR (400 MHz, CDCl3) δ 7.69 (dd, 2H, J = 5.9, 8.6 Hz), 7.57 (s, 1H), 7.28 (m, 1H), 7.16 (m, 2H), 6.74 (d, 2H, J = 8.5 Hz), 6.56 (d, 2H, J = 8.5 Hz), 5.41 (s, 1H), 3.92 (s, 3H), 3.64 (q, 1H, J = 7.2 Hz), 3.45 (dq, 1H, J = 6.6, 13.1 Hz), 3.34 (td, 1H, J = 6.7, 13.0 Hz), 2.58 (t, 2H, J = 6.8 Hz), 1.58 (d, 3H, J = 7.2 Hz). HRMS (ESI (M+H)+ m/z) calcd for C22H24NO3 350.1756, found 350.1743.
(2S)-2-(3-(Hydroxy(phenyl)methyl)phenyl)-N-(4-hydroxyphenethyl)propanamide (39)
A solution of 36 (0.016 g, 0.043 mmol) in MeOH (2 mL) was treated with NaBH4 (0.002 g, 0.047 mmol). The resulting mixture was stirred for 2 h at rt. The reaction mixture was quenched with 4 mL of water and extracted with EtOAc (10 mL). The extract was washed with water (3 × 5 mL) and brine (1 × 5 mL). The organic layer was dried over Na2SO4, filtered, and concentrated in vacuo. The residue was purified by flash-column chromatography over silica gel (Biotage SP4, 12+S Column, eluting with hexane ethyl acetate gradient 10%–80 %) to give 39 (0.013 g, 81%). 1H NMR (400 MHz, CDCl3) δ 7.27 (m, 8H), 7.10 (dd, 1H, J = 3.8, 7.1 Hz), 6.76 (dd, 2H, J = 4.3, 8.4 Hz), 6.66 (d, 2H, J = 8.4 Hz), 5.74 (s, 1H), 5.56 (m, 1H), 3.42 (m, 2H), 3.27 (dt, 1H, J = 6.5, 13.1 Hz), 2.56 (m, 2H), 1.44 (dd, 3H, J = 2.7, 7.2 Hz). HRMS (ESI (M+H)+ m/z) calcd for C24H26NO3 376.1913, found 376.1912.
(S)-2-(3-Benzylphenyl)-N-(4-hydroxyphenethyl)propanamide (40)
1H NMR (400 MHz, CDCl3) δ 7.21 (m, 6H), 7.06 (dd, 3H, J = 7.8, 16.8 Hz), 6.79 (d, 2H, J = 8.4 Hz), 6.70 (d, 2H, J = 8.3 Hz), 5.41 (t, 1H, J = 5.5 Hz), 3.94 (s, 2H), 3.44 (m, 2H), 3.32 (td, 1H, J = 6.8, 13.0 Hz), 2.57 (m, 2H), 1.47 (d, 3H, J = 7.2 Hz). HRMS (ESI (M+H)+ m/z) calcd for C24H26NO2 360.1964, found 360.1951.
N-(4-Hydroxyphenethyl)-2-(2-phenoxyphenyl)acetamide (41)
1H NMR (400 MHz, CDCl3) δ 7.27 (m, 4H), 7.08 (m, 2H), 6.82 (m, 5H), 6.69 (d, 2H, J = 8.5 Hz), 5.77 (s, 1H), 3.57 (s, 2H), 3.39 (q, 2H, J = 6.7 Hz), 2.61 (t, 2H, J = 6.8 Hz). HRMS (ESI (M+H)+ m/z) calcd for C22H22NO3 348.1600, found 348.1599.
N-(4-Hydroxyphenethyl)-2-(3-phenoxyphenyl)acetamide (42)
1H NMR (400 MHz, CDCl3) δ 7.31 (ddd, 4H, J = 4.9, 8.2, 15.8 Hz), 7.12 (t, 1H, J = 7.4 Hz), 7.00 (m, 2H), 6.90 (m, 4H), 6.83 (s, 1H), 6.71 (d, 2H, J = 8.5 Hz), 5.41 (bs, 1H), 3.49 (s, 2H), 3.43 (dd, 2H, J = 6.6, 12.8 Hz), 2.65 (t, 2H, J = 6.8 Hz). HRMS (ESI (M+H)+ m/z) calcd for C22H22NO3 348.1600, found 348.1592.
N-(4-Hydroxyphenethyl)-2-(4-phenoxyphenyl)acetamide (43)
A solution of 36 (0.040 g, 0.107 mmol) and 10% Pd/C (0.002 g) in methanol (3 mL) was stirred under H2 for 18 h at rt and then filtered through a short pad of celite. The solvent was removed in vacuo, and the residue was flash chromatographed over silica gel (Biotage SP4, 12+S Column, eluting with hexane ethyl acetate gradient 10%–80 %) to give 43 (0.035 g, 0.097 mmol, 91% yield). 1H NMR (400 MHz, CDCl3) δ 7.35 (t, 2H, J = 7.9 Hz), 7.11 (m, 3H), 7.01 (m, 2H), 6.94 (dd, 2H, J = 2.9, 8.5 Hz), 6.88 (dd, 2H, J = 2.6, 8.4 Hz), 6.73 (dd, 2H, J = 2.9, 8.4 Hz), 5.47 (s, 1H), 3.49 (s, 2H), 3.44 (dd, 2H, J = 6.6, 12.8 Hz), 2.66 (t, 2H, J = 6.6 Hz). HRMS (ESI (M+H)+ m/z) calcd for C22H22NO3 348.1600, found 348.1600.
(S)-2-(3-Benzoylphenyl)-N-(2-hydroxyphenethyl)propanamide (44)
1H NMR (400 MHz, CDCl3) δ 7.88 (m, 2H), 7.73 (t, 1H, J = 1.6 Hz), 7.65 (m, 1H), 7.55 (m, 4H), 7.42 (t, 1H, J = 7.6 Hz), 7.05 (m, 1H), 7.00 (dd, 1H, J = 1.6, 7.5 Hz), 6.85 (dd, 1H, J = 1.0, 8.0 Hz), 6.77 (td, 1H, J = 1.1, 7.4 Hz), 6.25 (s, 1H), 3.65 (q, 1H, J = 7.2 Hz), 3.39 (m, 2H), 2.76 (m, 2H), 1.54 (d, 3H, J = 7.3 Hz). HRMS (ESI (M+H)+ m/z) calcd for C24H24NO3 374.1756, found 374.1746.
(S)-2-(3-Benzoylphenyl)-N-(2-methoxyphenethyl)propanamide (45)
1H NMR (400 MHz, CDCl3) δ 7.77 (m, 2H), 7.67 (m, 2H), 7.60 (t, 1H, J = 7.4 Hz), 7.45 (m, 4H), 7.15 (td, 1H, J = 1.6, 7.9 Hz), 6.94 (m, 1H), 6.80 (t, 2H, J = 7.3 Hz), 5.60 (s, 1H), 3.75 (s, 3H), 3.53 (q, 1H, J = 7.1 Hz), 3.45 (m, 2H), 2.77 (m, 2H), 1.50 (d, 3H, J = 7.2 Hz). HRMS (ESI (M+H)+ m/z) calcd for C25H26NO3 388.1913, found 388.1903.
(S)-2-(3-Benzoylphenyl)-N-(3-methoxyphenethyl)propanamide (46)
1H NMR (400 MHz, CDCl3) δ 7.77 (m, 2H), 7.67 (m, 2H), 7.60 (m, 1H), 7.46 (m, 4H), 7.12 (t, 1H, J = 7.7 Hz), 6.70 (m, 1H), 6.60 (d, 2H, J = 7.3 Hz), 5.44 (s, 1H), 3.73 (s, 3H), 3.49 (m, 3H), 2.70 (t, 2H, J = 7.1 Hz), 1.52 (d, 3H, J = 7.2 Hz). HRMS (ESI (M+H)+ m/z) calcd for C25H26NO3 388.1913, found 388.1903.
(S)-2-(3-Benzoylphenyl)-N-(4-methoxyphenethyl)propanamide (47)
1H NMR (400 MHz, CDCl3) δ 7.78 (d, 2H, J = 7.2 Hz), 7.68 (d, 2H, J = 8.6 Hz), 7.60 (t, 1H, J = 7.4 Hz), 7.47 (m, 4H), 6.92 (d, 2H, J = 8.5 Hz), 6.75 (d, 2H, J = 8.5 Hz), 5.39 (s, 1H), 3.75 (s, 3H), 3.55 (q, 1H, J = 7.1 Hz), 3.48 (dq, 1H, J = 6.5, 13.1 Hz), 3.38 (dt, 1H, J = 6.6, 13.3 Hz), 2.66 (td, 2H, J = 2.0, 6.8 Hz), 1.52 (d, 3H, J = 7.2 Hz). HRMS (ESI (M+H)+ m/z) calcd for C25H26NO3 388.1913, found 388.1901.
(S)-2-(3-Benzoylphenyl)-N-phenethylpropanamide (48)
1H NMR (400 MHz, CDCl3) δ 7.77 (dd, 2H, J = 1.1, 8.2 Hz), 7.67 (dd, 2H, J = 1.2, 7.2 Hz), 7.60 (t, 1H, J = 7.4 Hz), 7.47 (m, 4H), 7.19 (m, 3H), 7.02 (d, 2H, J = 6.8 Hz), 5.39 (s, 1H), 3.48 (m, 3H), 2.73 (td, 2H, J = 2.0, 6.8 Hz), 1.52 (d, 3H, J = 7.2 Hz). HRMS (ESI (M+H)+ m/z) calcd for C245H24NO2 358.1807, found 358.1794.
(S)-2-(3-Benzoylphenyl)-N-(4-chlorophenethyl)propanamide (49)
1H NMR (400 MHz, CDCl3) δ 7.78 (d, 2H, J = 6.9 Hz), 7.64 (m, 3H), 7.46 (dt, 4H, J = 5.9, 23.4 Hz), 7.17 (d, 2H, J = 8.1 Hz), 6.94 (d, 2H, J = 7.9 Hz), 5.44 (s, 1H), 3.45 (m, 3H), 2.70 (t, 2H, J = 6.0 Hz), 1.52 (d, 3H, J = 6.8 Hz). HRMS (ESI (M+H)+ m/z) calcd for C24H23NO2Cl 392.1417, found 392.1414.
(S)-2-(3-Benzoylphenyl)-N-(3,4-dihydroxyphenethyl)propanamide (50)
1H NMR (400 MHz, CDCl3) δ 7.81 (d, 2H, J = 7.8 Hz), 7.64 (dd, 3H, J = 9.6, 15.6 Hz), 7.46 (dt, 4H, J = 7.7, 27.0 Hz), 6.73 (d, 1H, J = 7.6 Hz), 6.43 (d, 2H, J = 7.6 Hz), 5.51 (s, 1H), 3.54 (m, 2H), 3.28 (dt, 1H, J = 6.6, 13.3 Hz), 2.53 (m, 2H), 1.52 (d, 3H, J = 7.2 Hz). HRMS (ESI (M+H)+ m/z) calcd for C24H24NO4 390.1705, found 390.1703.
(S)-2-(3-Benzoylphenyl)-N-(4-hydroxy-3-methoxyphenethyl)propanamide (51)
1H NMR (400 MHz, CDCl3) δ 7.77 (m, 2H), 7.69 (s, 1H), 7.66 (d, 1H, J = 7.5 Hz), 7.60 (t, 1H, J = 7.4 Hz), 7.48 (t, 3H, J = 7.6 Hz), 7.42 (t, 1H, J = 7.6 Hz), 6.75 (d, 1H, J = 8.0 Hz), 6.59 (d, 1H, J = 1.8 Hz), 6.49 (dd, 1H, J = 1.8, 8.0 Hz), 5.46 (s, 1H), 3.80 (s, 3H), 3.55 (q, 1H, J = 7.1 Hz), 3.44 (ddt, 2H, J = 6.4, 13.3, 19.4 Hz), 2.66 (t, 2H, J = 7.0 Hz), 1.52 (d, 3H, J = 7.2 Hz). HRMS (ESI (M+H)+ m/z) calcd for C25H26NO4 404.1862, found 404.1851.
(S)-2-(3-Benzoylphenyl)-N-(3-hydroxy-4-methoxyphenethyl)propanamide (52)
1H NMR (400 MHz, CDCl3) δ 7.77 (m, 2H), 7.67 (d, 2H, J = 7.6 Hz), 7.60 (t, 1H, J = 7.4 Hz), 7.47 (m, 4H), 6.67 (d, 1H, J = 8.2 Hz), 6.61 (d, 1H, J = 2.0 Hz), 6.45 (dd, 1H, J = 2.0, 8.2 Hz), 5.41 (s, 1H), 3.82 (s, 3H), 3.55 (q, 1H, J = 7.1 Hz), 3.41 (ddt, 2H, J = 6.7, 13.2, 34.1 Hz), 2.61 (td, 2H, J = 3.4, 6.7 Hz), 1.52 (d, 3H, J = 7.2 Hz). HRMS (ESI (M+H)+ m/z) calcd for C25H26NO4 404.1862, found 404.1854.
(S)-2-(3-Benzoylphenyl)-N-((4-hydroxyphenyl)methyl)propanamide (53)
1H NMR (400 MHz, CDCl3) δ 7.75 (m, 3H), 7.65 (d, 1H, J = 6.5 Hz), 7.59 (t, 2H, J = 8.4 Hz), 7.45 (m, 3H), 7.00 (d, 2H, J = 6.7 Hz), 6.71 (d, 2H, J = 6.6 Hz), 5.78 (s, 1H), 4.29 (m, 2H), 3.64 (q, 1H, J = 7.0 Hz), 1.56 (d, 3H, J = 7.1 Hz). HRMS (ESI (M+H)+ m/z) calcd for C23H22NO3 360.1600, found 360.1593.
(S)-2-(3-Benzoylphenyl)-N-(4-hydroxyphenyl)propanamide (54)
1H NMR (400 MHz, CDCl3) δ 7.79 (m, 3H), 7.62 (m, 3H), 7.46 (td, 3H, J = 3.2, 7.7 Hz), 7.32 (s, 1H), 7.19 (d, 2H, J = 8.7 Hz), 6.70 (d, 2H, J = 8.7 Hz), 3.76 (q, 1H, J = 7.0 Hz), 1.59 (d, 3H, J = 7.1 Hz). HRMS (ESI (M+H)+ m/z) calcd for C22H20NO3 346.1443, found 346.1429.
(S)-(3-(1-Hydroxypropan-2-yl)phenyl)(phenyl)methanone (8)
A stirred solution of 7 (0.500 g, 1.966 mmol) in THF (5 mL) at −20 °C was treated dropwise with borane-tetrahydrofuran complex (2.36 mL, 2.36 mmol), and the cooling bath was allowed to equilibrate overnight to rt. The solution was cooled to 5 °C and treated with 1 mL of methanol, diluted with 5 mL of water and concentrated in vacuo to remove the organic solvents. The residue was diluted with water (10 mL) and dichloromethane (10 mL) and washed with NaHCO3. The organic phase was dried over sodium sulfate and concentrated in vacuo. The residue was chromatographed over silica (Biotage SP4, 25+M Column, eluting with hexane ethyl acetate gradient 10%-100 %) to give 8 (0.470 g, 1.956 mmol, 99% yield). 1H NMR (400 MHz, CDCl3) δ 7.80 (dd, 2H, J = 1.2, 8.2 Hz), 7.70 (s, 1H), 7.65 - 7.55 (m, 2H), 7.47 (dd, 3H, J = 4.7, 10.3 Hz), 7.42 (t, 1H, J = 7.6 Hz), 3.80 - 3.68 (m, 2H), 3.02 (sext., 1H, J = 6.9 Hz), 1.87 (s, 1H), 1.30 (d, 3H, J = 7.0 Hz).
(S)-(3-(1-Aminopropan-2-yl)phenyl)(phenyl)methanone (9)
To a solution of 8 (0.400 g, 1.665 mmol) in dry dichloromethane (15 mL) at 0 °C under nitrogen atmosphere, triethylamine (0.7 mL, 5 mmol) was added followed by methanesulfonyl chloride (0.19 mL, 2.5 mmol) dropwise. The reaction mixture was stirred at 0 °C for 1 h and was worked-up by adding water, followed by extraction with dichloromethane. The combined organic extracts were washed with water and brine and dried. The solvent was evaporated to give the mesylate as a gum, which was taken-up for the next step without any purification.
To a solution of the above mesylate (0.450 g, 1.413 mmol) in dry DMF (10 mL) under nitrogen atmosphere, sodium azide (0.092 g, 1.413 mmol) was added, and the resulting mixture was stirred at 70 °C for 2 h. The reaction mixture was allowed to cool to rt and was worked-up by addition of water (20 mL) followed by extraction with ethyl acetate (3 × 20 mL). The combined organic extracts were washed with water (5 × 20 mL) and brine (1 × 20 mL) and dried over sodium sulphate. The residue obtained upon evaporation of solvent was chromatographed over silica gel to get the desired azide (0.350 g, 1.319 mmol, 93% yield).
Azide from the previous step (0.350 g, 1.319 mmol) was dissolved in THF (10 mL), and triphenylphosphine (0.519 g, 1.979 mmol) was added to the solution. After being stirred for 48 h at rt, the reaction mixture was concentrated in vacuo. The residue was dissolved in 2N sodium hydroxide (15 mL), and the product was extracted with ethyl acetate (3 × 20 mL) and washed with water (2 × 20 mL) followed by brine (1 × 20 mL). The orgaic layer was dried over sodium sulfate, evaporated to dryness, and purified by flash chromatography (Biotage SP4, 25+M silica gel column, eluting with hexane ethyl acetate gradient 10%–100 %) to give 52 (0.288 g, 1.203 mmol, 91% yield). 1H NMR (400 MHz, MeOD) δ 7.77 (dd, 2H, J = 3.2, 5.2 Hz), 7.69 - 7.59 (m, 3H), 7.56 - 7.45 (m, 4H), 2.94 - 2.79 (m, 3H), 1.30 (d, 3H, J = 6.6 Hz).
Compounds 55–57 were synthesized from 9 and the appropriate carboxylic acid following the general procedure mentioned above.
(S)-N-(2-(3-Benzoylphenyl)propyl)-4-hydroxybenzamide (55)
1H NMR (400 MHz, CDCl3) δ 7.78 (d, 2H, J = 7.2 Hz), 7.71 (s, 1H), 7.65 (d, 1H, J = 7.3 Hz), 7.63 - 7.51 (m, 3H), 7.51 - 7.41 (m, 4H), 6.81 (d, 2H, J = 8.5 Hz), 3.87 - 3.72 (m, 1H), 3.53 - 3.40 (m, 1H), 3.23 - 3.10 (m, 1H), 1.37 (d, 3H, J = 7.0 Hz). HRMS (ESI (M+H)+ m/z) calcd for C23H22NO3 360.1600, found 360.1589.
(S)-N-(2-(3-Benzoylphenyl)propyl)-3-(4-hydroxyphenyl)propanamide (56)
1H NMR (400 MHz, CDCl3) δ 7.85 - 7.76 (m, 2H), 7.61 (dd, 2H, J = 2.9, 6.7 Hz), 7.55 (s, 1H), 7.49 (t, 2H, J = 7.6 Hz), 7.40 (t, 1H, J = 7.6 Hz), 7.32 (d, 1H, J = 7.7 Hz), 6.96 (d, 2H, J = 8.3 Hz), 6.69 (d, 2H, J = 8.5 Hz), 5.18 (s, 1H), 3.58 (dt, 1H, J = 6.3, 12.8 Hz), 3.21 (dd, 1H, J = 11.2, 16.2 Hz), 2.89 (d, 1H, J = 8.3 Hz), 2.81 (t, 2H, J = 7.2 Hz), 2.35 (dd, 2H, J = 5.9, 9.1 Hz), 1.22 (d, 3H, J = 7.0 Hz). HRMS (ESI (M+H)+ m/z) calcd for C25H26NO3 388.1913, found 388.1906.
(S)-N-(2-(3-Benzoylphenyl)propyl)-4-methoxybenzamide (57)
1H NMR (400 MHz, CDCl3) δ 7.82 - 7.74 (m, 2H), 7.71 (s, 1H), 7.68 - 7.54 (m, 4H), 7.52 - 7.38 (m, 4H), 6.89 - 6.84 (m, 2H), 6.04 (s, 1H), 3.87 - 3.75 (m, 4H), 3.47 (ddd, 1H, J = 5.3, 8.5, 13.6 Hz), 3.18 (dd, 1H, J = 6.8, 15.0 Hz), 1.37 (d, 3H, J = 7.0 Hz). HRMS (ESI (M+H)+ m/z) calcd for C24H24NO3 374.1756, found 374.1746. HPLC purity 93%.
(S)-(3-(1-(4-Methoxybenzyloxy)propan-2-yl)phenyl)(phenyl)methanone (58)
1H NMR (400 MHz, CDCl3) δ 7.79 (dd, 2H, J = 3.2, 5.2 Hz), 7.68 (s, 1H), 7.65 - 7.55 (m, 2H), 7.46 (t, 3H, J = 7.5 Hz), 7.40 (t, 1H, J = 7.5 Hz), 7.17 (d, 2H, J = 8.6 Hz), 6.84 (d, 2H, J = 8.6 Hz), 4.43 (s, 2H), 3.79 (s, 3H), 3.54 (m, 2H), 3.12 (dd, 1H, J = 6.9, 13.8 hz), 1.31 (d, 3H, J = 7.0 Hz). HRMS (ESI (M+H)+ m/z) calcd for C24H25O3 361.1804, found 361.1799. HPLC purity 91%.
(S)-4-Nitrophenyl 2-(3-benzoylphenyl)propylcarbamate (66)
To a solution of 4-nitrophenyl chloroformate (0.111 g, 0.552 mmol) and triethylamine (0.064 mL, 0.460 mmol) in anhydrous THF (5 mL), a solution of 9 in THF (5 mL) was added dropwise at 0 °C to 5 °C under nitrogen atmosphere. The reaction mixture was then allowed to warm up to rt and was stirred for 2 h at rt. The resulting mixture was evaporated to dryness, and residue chromatographed over silicagel (Biotage SP4, 25+M silica gel column, eluting with dichlomethane ethanol gradient 0%–7%) to give 66 (0.135 g, 0.334 mmol, 72.6% yield). 1H NMR (400 MHz, CDCl3) δ 8.26 - 8.17 (m, 2H), 7.85 - 7.78 (m, 2H), 7.72 (s, 1H), 7.66 (dt, 1H, J = 1.7, 6.8 Hz), 7.61 (t, 1H, J = 7.4 Hz), 7.48 (q, 4H, J = 7.5 Hz), 7.29 - 7.20 (m, 2H), 5.16 (s, 1H), 3.58 (dt, 1H, J = 6.5, 13.3 Hz), 3.42 (ddd, 1H, J = 5.6, 8.4, 13.8 Hz), 3.13 (dd, 1H, J = 6.9, 14.8 Hz), 1.37 (d, 3H, J = 7.0 Hz).
(S)-2-(3-Benzoylphenyl)propyl 4-nitrophenyl carbonate (67)
was made following a procedure similar to that for making 66 starting from 8 in 79% yield. 1H NMR (400 MHz, CDCl3) δ 8.28 - 8.21 (m, 2H), 7.81 (d, 2H, J = 7.3 Hz), 7.75 (s, 1H), 7.68 (d, 1H, J = 7.4 Hz), 7.63 - 7.57 (m, 1H), 7.52 - 7.47 (m, 4H), 7.35 - 7.28 (m, 2H), 4.41 (qd, 2H, J = 7.0, 10.6 Hz), 3.32 (dd, 1H, J = 7.0, 14.0 Hz), 1.42 (d, 3H, J = 7.0 Hz).
(S)-1-(2-(3-Benzoylphenyl)propyl)-3-(4-hydroxyphenyl)urea (59)
To a solution of 66 (0.020 g, 0.049 mmol) and 4-aminophenol (6.48 mg, 0.059 mmol) in anhydrous THF (2 mL), triethylamine (8.27 µl, 0.059 mmol) was added. The resulting mixture was stirred at rt for 15 h and then evaporated to dryness and residue chromatographed over silica gel (Biotage SP4, 25+M silica gel column, eluting with dichlomethane ethanol gradient 0%–7%) to 59 (0.011 g, 0.029 mmol, 59.4% yield). 1H NMR (400 MHz, CDCl3 + CD3OD) δ 7.79 (d, 2H, J = 8.2 Hz), 7.61 (dd, 3H, J = 7.1, 14.5 Hz), 7.55 - 7.40 (m, 4H), 7.00 (d, 2H, J = 8.8 Hz), 6.77 (d, 2H, J = 8.8 Hz), 3.48 (dd, 1H, J = 6.4, 13.4 Hz), 3.26 (dd, 1H, J = 8.5, 13.5 Hz), 3.04 (dd, 1H, J = 7.0, 14.4 Hz), 1.31 (d, 3H, J = 7.0 Hz). HRMS (ESI (M+H)+ m/z) calcd for C23H23N2O3 375.1709, found 375.1706. HPLC purity 87%.
(S)-1-(2-(3-Benzoylphenyl)propyl)-3-(4-hydroxyphenethyl)urea (60)
was synthesized by following the procedure for synthesis of 59 using 6. 1H NMR (400 MHz, CDCl3 + CD3OD) 1H NMR (400 MHz, CDCl3 + CD3OD) δ 7.82 - 7.75 (m, 2H), 7.64 - 7.54 (m, 3H), 7.47 (t, 2H, J = 7.7 Hz), 7.43 - 7.35 (m, 2H), 6.91 (d, 2H, J = 8.4 Hz), 6.71 (d, 2H, J = 8.4 Hz), 3.42 (dd, 1H, J = 6.2, 13.5 Hz), 3.29 (t, 2H, J = 6.7 Hz), 3.18 (dd, 1H, J = 8.3, 13.4 Hz), 2.95 (dd, 1H, J = 6.9, 14.5 Hz), 2.62 (t, 2H, J = 6.7 Hz), 1.25 (d, 3H, J = 7.0 Hz). HRMS (ESI (M+H)+ m/z) calcd for C25H27N2O3 403.2022, found 403.2015. HPLC purity 91%.
Compounds 61–62 were synthesized from 67 following the procedure for synthesis of 59 using the appropriate amine.
(S)-2-(3-Benzoylphenyl)propyl 4-hydroxyphenylcarbamate (61)
1H NMR (400 MHz, CDCl3 + CD3OD) δ 7.80 (d, 2H, J = 7.2 Hz), 7.72 (s, 1H), 7.65 (d, 1H, J = 7.4 Hz), 7.59 (t, 1H, J = 7.4 Hz), 7.54 - 7.38 (m, 4H), 7.19 (m, 2H), 6.75 (d, 2H, J = 8.3 Hz), 6.43 (bs, 1H), 4.33 - 4.21 (m, 2H), 3.22 (dd, 1H, J = 7.0, 14.0 Hz), 1.35 (d, 3H, J = 7.1 Hz). HRMS (ESI (M+H)+ m/z) calcd for C23H22NO4 376.1549, found 376.1554.
(S)-2-(3-Benzoylphenyl)propyl 4-hydroxyphenethylcarbamate (62)
1H NMR (400 MHz, CDCl3 + CD3OD) δ 7.80 (d, 2H, J = 7.4 Hz), 7.71 - 7.55 (m, 3H), 7.52 - 7.37 (m, 4H), 6.98 (d, 2H, J = 8.0 Hz), 6.74 (d, 2H, J = 8.3 Hz), 4.65 (s, 1H), 4.17 (d, 2H, J = 7.0 Hz), 3.35 (dd, 2H, J = 6.4, 12.7 Hz), 3.20 - 3.08 (m, 1H), 2.69 (t, 2H, J = 6.7 Hz), 1.30 (d, 3H, J = 6.9 Hz). HRMS (ESI (M+H)+ m/z) calcd for C25H26NO4 404.1862, found 404.1854.
4-(2-(1-Propylamino)ethyl)phenol (11)
To a solution of 4-hydroxyphenethyl bromide (0.200 g, 0.995 mmol) in acetonitrile (5 mL), propylamine (0.817 mL, 9.95 mmol) was added, followed by DIPEA (0.520 mL, 2.98 mmol). The resulting solution was stirred at 60 °C for 6 h. The reaction mixture was allowed to cool to rt and was filtered. The solvent was removed in vacuo, and residue was flash chomatographed over silica gel (Biotage SP4, 25+S silica gel column, eluting with hexan and ethylacetate gradient 10%-100%) to give 11 (0.150 g, 0.837 mmol, 84% yield). 1H NMR (400 MHz, MeOD) δ 7.13 (d, 2H, J = 8.6 Hz), 6.83 - 6.74 (m, 2H), 3.23 - 3.16 (m, 2H), 3.04 - 2.96 (m, 2H), 2.93 (dd, 2H, J = 6.9, 9.3 Hz), 1.74 (dq, 2H, J = 7.5, 15.1 Hz), 1.04 (t, 3H, J = 7.5 Hz).
4-(2-(2-Methoxyethylamino)ethyl)phenol (12)
was synthesized in a manner similar to 11 starting from 4-hydroxyphenethyl bromide and 2-methoxyethylamine. 1H NMR (400 MHz, CDCl3 + CD3OD) δ 7.00 (d, 2H, J = 8.4 Hz), 6.69 (d, 2H, J = 8.5 Hz), 5.43 (s, 1H), 3.50 (dd, 2H, J = 5.8, 10.9 Hz), 3.30 (s, 3H), 2.89 (t, 2H, J = 7.0 Hz), 2.83 (t, 2H, J = 5.1 Hz), 2.75 (t, 2H, J = 7.1 Hz).
4,4'-(2,2'-Azanediylbis(ethane-2,1-diyl))diphenol (13)
was synthesized in a manner similar to 11 starting from 4-hydroxyphenethyl bromide and tyramine. 1H NMR (400 MHz, CDCl3 + CD3OD) δ 6.94 (dd, 2H, J = 4.6, 6.6 Hz), 6.77 - 6.68 (m, 2H), 2.83 (t, 2H, J = 6.9 Hz), 2.70 (t, 2H, J = 7.0 Hz).
(S)-2-(3-Benzoylphenyl)-N-(4-hydroxyphenethyl)-N-propylpropanamide (63)
was synthesized from 7 and 11 following the general procedure mentioned above. 1H NMR (400 MHz, CDCl3) (a mixture of rotamers) δ 7.84 - 7.73 (m, 2H), 7.72 - 7.51 (m, 4H), 7.51 - 7.37 (m, 3H), 7.01 - 6.89 (m, 2H), 6.78 (d, 1H, J = 8.2 Hz), 6.70 (d, 1H, J = 8.1 Hz), 6.00 and 5.61 (2 s, 1H, -OH), 3.97 - 3.69 (m, 1H), 3.45 - 2.85 (m, 4H), 2.81 - 2.52 (m, 2H), 1.58 - 1.24 (m, 5H with two overlaping doublets of rotamers), 0.91 - 0.77 (m, 3H). HRMS (ESI (M+H)+ m/z) calcd for C27H30NO3 416.2226, found 416.2222.
(S)-2-(3-Benzoylphenyl)-N-(4-hydroxyphenethyl)-N-(2-methoxyethyl)propanamide (64)
was synthesized from 7 and 12 following the general procedure mentioned above. 1H NMR (400 MHz, CDCl3) (a mixture of rotamers) δ 7.83 - 7.74 (m, 2H), 7.72 - 7.51 (m, 4H), 7.44 (m, 3H), 6.94 (dd, 2H, J = 2.1, 8.5 Hz), 6.77 (d, 1H, J = 8.5 Hz), 6.67 (d, 1H, J = 8.5 Hz), 5.93 and 5.63 (2s, 1H), 4.17 - 3.01 (m, 9H, with overlaping singlet at 3.28), 2.80 - 2.53 (m, 2H), 1.47 and 1.30 (2d, J = 6.9, 3H). HRMS (ESI (M+H)+ m/z) calcd for C27H30NO4 432.2175, found 432.2173.
(S)-2-(3-Benzoylphenyl)-N,N-bis(4-hydroxyphenethyl)propanamide (65)
was synthesized from 7 and 13 following the general procedure mentioned above. 1H NMR (400 MHz, CDCl3) δ 7.77 (d, 2H, J = 7.1 Hz), 7.65 - 7.55 (m, 3H), 7.49 - 7.39 (m, 4H), 6.93 (dd, J = 8.4, 14.7, 4H), 6.76 (d, 2H, J = 8.5 Hz), 6.67 (d, 2H, J = 8.5 Hz), 3.85 - 3.74 (m, 1H), 3.57 (q, 1H, J = 6.7 Hz), 3.45 - 3.35 (m, 1H), 3.28 - 3.19 (m, 1H), 3.17 - 3.07 (m, 1H), 2.82 - 2.63 (m, 3H), 2.58 - 2.49 (m, 1H), 1.29 (d, J = 6.9, 4H). HRMS (ESI (M+H)+ m/z) calcd for C32H32NO4 494.2331, found 494.2311.
Gli-mediated transcription reporter assay in C3H10T1/2 cells with exogenous Gli1 or Gli2
C3H10/T1/2 cells were plated at 2.5×105 cells/dish in two 60-mm culture dishes in 5 mL of basal medium Eagle (BME) (Invitrogen Corporation, Carlsbad, CA) containing 2 mM L-glutamine and 10% fetal bovine serum (FBS, Hyclone Laboratories, Logan, UT). The cells were maintained in 5% CO2 at 37 °C in a humidified incubator. After 24 h, the cells were transfected at approximately 75% confluence by replacing the media with 5 mL BME (10% FBS) containing the DNA mixture of GliLuc-BS (250 ng/mL), hGli1 (250 ng/mL), and pRL-TK (12.5 ng/mL) (Promega, Madison, WI) and the transfection reagent Fugene6 (1:3 w/v of DNA) (F. Hoffmann-La Roche, Basel, Switzerland) and incubated overnight. A simultaneous control was transfected with empty vector, pcDNA3, GliLuc, pRL-TK, and Fugene6 in similar ratios as used for hGli1. Eighteen hours after transfection, the cells were trypsinized and reconstituted in 10 mL BME (10% FBS) and plated in a white 96-well cell culture plate at 100 µL/well. Six hours after plating, the media in the wells was replaced with 100 µL BME (10% FBS) containing the compound or the DMSO control. After exposing the cells to compound for 24 h at 37 °C in 5% CO2, the media was aspirated, and the cells were lysed with 20 µL/well of Passive Lysis Buffer (1×) by placing the plate on a shaking platform for 15 min at 600 rpm. The luciferase activity was determined using Dual-Luciferase Reporter Assay System (Promega) according to manufacturer’s instructions. The activity was determined by dividing the luminescence of the luciferase by that of Renilla.
To enhance the luciferase signal in the assay readout of Gli2 transcription, we used the ΔNhGli2, an hGli2 construct from which the amino-terminal region encoding a transcription repressor domain was deleted29. The assay for ΔNhGli2 was carried out following the protocol for the hGli1-transactivation assay; however, hGli1 was replaced with ΔNhGli2.
Gli-mediated transcription reporter assay in Rh30 cells
Rh30 cells were plated at 3 ×105 cells/dish in two 60-mm culture dishes in 5 mL RPMI-1640 (ATCC, Manassas, VA) containing 10% FBS (Hyclone Laboratories) and were maintained in 5% CO2 at 37 °C in a humidified incubator. After 24 h, the cells were transfected at approximately 70% confluence by replacing the media with 5 mL RPMI-1640 (10% FBS) containing the DNA mixture of GliLuc-BS (500 ng/mL) and pRL-CMV (2.5 ng/mL) (Promega), and Fugene6 (1:3 w/v of DNA) (F. Hoffmann-La Roche, Basel, Switzerland) and incubated overnight. Six hours after transfection, the cells were trypsinized and reconstituted in 10 mL RPMI-1640 (10% FBS) and plated in a white 96-well cell culture plate at 100 µL/well. Eighteen hours after plating, the media was replaced with 100 µL RPMI-1640 (10% FBS) containing the compound or the DMSO control. After cells were exposed to compound for 24 h at 37 °C in 5% CO2, the media was aspirated, and the cells were lysed with 20 µL/well of Passive Lysis Buffer (1×) by placing the plate on a shaking platform for 15 min at 600 rpm. The luciferase activity was determined using Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instuctions. The activity was determined by dividing the luminescence of luciferase by that of Renilla.
Cell viability assay
The cells were plated in the appropriate medium in a 384-well plate 24 h before the addition of the compounds. The 10-mM stock solution of each compound in DMSO was diluted in EMEM media to prepare 160-µM solution (4×), and 10 µL of that solution was added in triplicate to get a final concentration of 40 µM/well. The cells were allowed to incubate at 37 °C for 68 h. Alamar Blue (4 µL) (Biotium, Inc., Hayward, CA) was added to each well, and the plates were incubated for 4 h at 37 °C. The fluorescence was measured with excitation wavelength at 510 nm and emission wavelength at 590 nm using an EnVision Multilabel Plate Reader (PerkinElmer, Waltham, MA, USA). The cell viability was determined by comparing the fluorescence of experimental wells with that of the DMSO control wells.
Supplementary Material
Acknowledgments
We thank Dr. Hiroshi Sasaki (RIKEN, Kobe, Japan) for the gift of human Gli1 and Gli-Luc plasmids, Dr. Erich Roessler (NIH, Bethesda, MD) for the ΔNhGli2 plasmid (through Addgene, Cambridge, MA), Dr. Anand Mayasundari for excellent technical support, and Dr. Angela McArthur for editorial advice. This work was supported by the American Lebanese Syrian Associated Charities (ALSAC) and NIH Cancer Center Support Grant P30CA021765-30.
Abbreviations
- Gli1
glioma-associated oncogene homolog 1
- Shh
Sonic Hedgehog
- Ptch
Patched
- Smo
Smoothened
- HBTU
2-(1H-benzo[d][1,2,3]triazol-1-yl)-1,1,3,3-tetramethyl-uronium hexafluorophosphate
- DIPEA
N,N-Diisopropylethylamine
- rt
room temperature
Footnotes
Supporting Information Available: The scatter plots of activity vs the calculated logP or tPSA values, the cell viability data for each compound on each cell line, HPLC traces for the compounds, data of stability test, counter-assays for non-specific transcription inhibition, and detail of statistic analysis, are available online, free of charge at http://pubs.acs.org.
References
- 1.Ruiz i Altaba A, Sanchez P, Dahmane N. Gli and hedgehog in cancer: tumours, embryos and stem cells. Nat Rev Cancer. 2002;2:361–372. doi: 10.1038/nrc796. [DOI] [PubMed] [Google Scholar]
- 2.Bian YH, Huang SH, Yang L, Ma XL, Xie JW, Zhang HW. Sonic hedgehog-Gli1 pathway in colorectal adenocarcinomas. World J Gastroenterol. 2007;13:1659–1665. doi: 10.3748/wjg.v13.i11.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerber AN, Wilson CW, Li YJ, Chuang PT. The hedgehog regulated oncogenes Gli1 and Gli2 block myoblast differentiation by inhibiting MyoD-mediated transcriptional activation. Oncogene. 2007;26:1122–1136. doi: 10.1038/sj.onc.1209891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pasca di Magliano M, Hebrok M. Hedgehog signalling in cancer formation and maintenance. Nat Rev Cancer. 2003;3:903–911. doi: 10.1038/nrc1229. [DOI] [PubMed] [Google Scholar]
- 5.Sanchez P, Hernandez AM, Stecca B, Kahler AJ, DeGueme AM, Barrett A, Beyna M, Datta MW, Datta S, Ruiz i Altaba A. Inhibition of prostate cancer proliferation by interference with SONIC HEDGEHOG-GLI1 signaling. Proc Natl Acad Sci U S A. 2004;101:12561–12566. doi: 10.1073/pnas.0404956101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Remsberg JR, Lou H, Tarasov SG, Dean M, Tarasova NI. Structural analogues of smoothened intracellular loops as potent inhibitors of Hedgehog pathway and cancer cell growth. Journal of Medicinal Chemistry. 2007;50:4534–4538. doi: 10.1021/jm0705657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 8.Lum L, Beachy PA. The Hedgehog response network: sensors, switches, and routers. Science. 2004;304:1755–1759. doi: 10.1126/science.1098020. [DOI] [PubMed] [Google Scholar]
- 9.Briscoe J, Therond P. Hedgehog signaling: from the Drosophila cuticle to anti-cancer drugs. Dev Cell. 2005;8:143–151. doi: 10.1016/j.devcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 10.Hooper JE, Scott MP. Communicating with Hedgehogs. Nat Rev Mol Cell Biol. 2005;6:306–317. doi: 10.1038/nrm1622. [DOI] [PubMed] [Google Scholar]
- 11.Stecca B, Ruiz i Altaba A. The therapeutic potential of modulators of the Hedgehog-Gli signaling pathway. J Biol. 2002;1:9. doi: 10.1186/1475-4924-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruiz i Altaba A, Mas C, Stecca B. The Gli code: an information nexus regulating cell fate, stemness and cancer. Trends Cell Biol. 2007;17:438–447. doi: 10.1016/j.tcb.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Marcotullio L, Ferretti E, De Smaele E, Argenti B, Mincione C, Zazzeroni F, Gallo R, Masuelli L, Napolitano M, Maroder M, Modesti A, Giangaspero F, Screpanti I, Alesse E, Gulino A. REN(KCTD11) is a suppressor of Hedgehog signaling and is deleted in human medulloblastoma. Proc Natl Acad Sci U S A. 2004;101:10833–10838. doi: 10.1073/pnas.0400690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sasai K, Romer JT, Lee Y, Finkelstein D, Fuller C, McKinnon PJ, Curran T. Shh pathway activity is down-regulated in cultured medulloblastoma cells: implications for preclinical studies. Cancer Res. 2006;66:4215–4222. doi: 10.1158/0008-5472.CAN-05-4505. [DOI] [PubMed] [Google Scholar]
- 15.Dellovade T, Romer JT, Curran T, Rubin LL. The hedgehog pathway and neurological disorders. Annu Rev Neurosci. 2006;29:539–563. doi: 10.1146/annurev.neuro.29.051605.112858. [DOI] [PubMed] [Google Scholar]
- 16.Lauth M, Bergstrom A, Shimokawa T, Toftgard R. Inhibition of GLI-mediated transcription and tumor cell growth by small-molecule antagonists. Proc Natl Acad Sci U S A. 2007;104:8455–8460. doi: 10.1073/pnas.0609699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimura H, Stephen D, Joyner A, Curran T. Gli1 is important for medulloblastoma formation in Ptc1+/− mice. Oncogene. 2005;24:4026–4036. doi: 10.1038/sj.onc.1208567. [DOI] [PubMed] [Google Scholar]
- 18.Goyette P, Allan D, Peschard P, Chen CF, Wang W, Lohnes D. Regulation of gli activity by all-trans retinoic acid in mouse keratinocytes. Cancer Res. 2000;60:5386–5389. [PubMed] [Google Scholar]
- 19.Roberts WM, Douglass EC, Peiper SC, Houghton PJ, Look AT. Amplification of the gli gene in childhood sarcomas. Cancer Res. 1989;49:5407–5413. [PubMed] [Google Scholar]
- 20.Clement V, Sanchez P, de Tribolet N, Radovanovic I, Ruiz i Altaba A. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr Biol. 2007;17:165–172. doi: 10.1016/j.cub.2006.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park HL, Bai C, Platt KA, Matise MP, Beeghly A, Hui CC, Nakashima M, Joyner AL. Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development. 2000;127:1593–1605. doi: 10.1242/dev.127.8.1593. [DOI] [PubMed] [Google Scholar]
- 22.Mo R, Freer AM, Zinyk DL, Crackower MA, Michaud J, Heng HH, Chik KW, Shi XM, Tsui LC, Cheng SH, Joyner AL, Hui C. Specific and redundant functions of Gli2 and Gli3 zinc finger genes in skeletal patterning and development. Development. 1997;124:113–123. doi: 10.1242/dev.124.1.113. [DOI] [PubMed] [Google Scholar]
- 23.Kimura H, Ng JM, Curran T. Transient inhibition of the Hedgehog pathway in young mice causes permanent defects in bone structure. Cancer Cell. 2008;13:249–260. doi: 10.1016/j.ccr.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 24.Mahindroo N, Punchihewa C, Fujii N. Hedgehog-Gli Signaling Pathway Inhibitors as Anticancer Agents. J Med Chem. 2009 doi: 10.1021/jm801420y. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hosoya T, Arai MA, Koyano T, Kowithayakorn T, Ishibashi M. Naturally occurring small-molecule inhibitors of hedgehog/GLI-mediated transcription. Chembiochem. 2008;9:1082–1092. doi: 10.1002/cbic.200700511. [DOI] [PubMed] [Google Scholar]
- 26.He B, Fujii N, You L, Xu Z, Jablons DM. Dihydropyrazolecarboxamides and their preparation, pharmaceutical compositions and use in the targeting GLI proteins in human cancer by small molecules. 2007 2006-US47231 WO2007067814, 20061208. [Google Scholar]
- 27.Sasaki H, Hui C, Nakafuku M, Kondoh H. A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development. 1997;124:1313–1322. doi: 10.1242/dev.124.7.1313. [DOI] [PubMed] [Google Scholar]
- 28.Auld DS, Thorne N, Nguyen DT, Inglese J. A specific mechanism for nonspecific activation in reporter-gene assays. ACS Chem Biol. 2008;3:463–470. doi: 10.1021/cb8000793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roessler E, Ermilov AN, Grange DK, Wang A, Grachtchouk M, Dlugosz AA, Muenke M. A previously unidentified amino-terminal domain regulates transcriptional activity of wild-type and disease-associated human GLI2. Hum Mol Genet. 2005;14:2181–2188. doi: 10.1093/hmg/ddi222. [DOI] [PubMed] [Google Scholar]
- 30.Jiang XR, Jimenez G, Chang E, Frolkis M, Kusler B, Sage M, Beeche M, Bodnar AG, Wahl GM, Tlsty TD, Chiu CP. Telomerase expression in human somatic cells does not induce changes associated with a transformed phenotype. Nat Genet. 1999;21:111–114. doi: 10.1038/5056. [DOI] [PubMed] [Google Scholar]
- 31.Kubo M, Nakamura M, Tasaki A, Yamanaka N, Nakashima H, Nomura M, Kuroki S, Katano M. Hedgehog signaling pathway is a new therapeutic target for patients with breast cancer. Cancer Res. 2004;64:6071–6074. doi: 10.1158/0008-5472.CAN-04-0416. [DOI] [PubMed] [Google Scholar]
- 32.Di Marcotullio L, Ferretti E, Greco A, De Smaele E, Po A, Sico MA, Alimandi M, Giannini G, Maroder M, Screpanti I, Gulino A. Numb is a suppressor of Hedgehog signalling and targets Gli1 for Itch-dependent ubiquitination. Nat Cell Biol. 2006;8:1415–1423. doi: 10.1038/ncb1510. [DOI] [PubMed] [Google Scholar]
- 33.Bar EE, Chaudhry A, Farah MH, Eberhart CG. Hedgehog signaling promotes medulloblastoma survival via Bc/II. Am J Pathol. 2007;170:347–355. doi: 10.2353/ajpath.2007.060066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanchez P, Hernandez AM, Stecca B, Kahler AJ, DeGueme AM, Barrett A, Beyna M, Datta MW, Datta S, Ruiz i Altaba A. Inhibition of prostate cancer proliferation by interference with SONIC HEDGEHOG-GLI1 signaling. Proc. Natl. Acad. Sci. U S A. 2004;101:12561–12566. doi: 10.1073/pnas.0404956101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huntzicker EG, Estay IS, Zhen H, Lokteva LA, Jackson PK, Oro AE. Dual degradation signals control Gli protein stability and tumor formation. Genes Dev. 2006;20:276–281. doi: 10.1101/gad.1380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sasai K, Romer JT, Lee Y, Finkelstein D, Fuller C, McKinnon PJ, Curran T. Shh pathway activity is down-regulated in cultured medulloblastoma cells: implications for preclinical studies. Cancer Res. 2006;66:4215–4222. doi: 10.1158/0008-5472.CAN-05-4505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.