Abstract
The heterogeneous nuclear ribonucleoprotein Npl3p of budding yeast is a substrate of arginine methyltransferase Hmt1p, but the role of Hmt1p in regulating Npl3p’s functions in transcription antitermination and elongation were unknown. We found that mutants lacking Hmt1p methyltransferase activity exhibit reduced recruitment of Npl3p, but elevated recruitment of a component of mRNA cleavage/termination factor CFI, to the activated GAL10-GAL7 locus. Consistent with this, hmt1 mutants displayed increased termination at the defective gal10-Δ56 terminator. Remarkably, hmt1Δ cells also exhibit diminished recruitment of elongation factor Tho2p and a reduced rate of transcription elongation in vivo. Importantly, the defects in Npl3p and Tho2p recruitment, antitermination and elongation in hmt1Δ cells all were mitigated by substitutions in Npl3p RGG repeats that functionally mimic arginine methylation by Hmt1p. Thus, Hmt1p promotes elongation and suppresses termination at cryptic terminators by methylating RGG repeats in Npl3p. As Hmt1p stimulates dissociation of Tho2p from an Npl3p-mRNP complex, it could act to recycle these elongation and antitermination factors back to sites of ongoing transcription.
INTRODUCTION
Termination of transcription by RNA polymerase II (RNAP II) is an important step of gene regulation in eukaryotes (1). Whereas premature transcription termination produces truncated and defective transcripts, expression of downstream genes is impaired if transcription reads through from upstream terminators (2–4). Transcription termination is coupled to 3′-end processing of mRNA in which cleavage and polyadenylation are performed by a multiprotein complex that contains cleavage and polyadenylation specificity factors (CPSF) and cleavage factor I (CFI) (2,3). Two models known as the `torpedo’ and ‘anti-terminator’ models have been proposed to explain the mechanism of transcription termination. According to the ‘torpedo’ model, cleavage of RNA at the poly(A) site triggers rapid degradation of the 3′ RNA by exonuclease Rat1p and subsequent release of RNAP II (4–6). In the `anti-terminator’ model, transcription through the poly(A) site evokes a conformational change in the elongation complex, involving recruitment of CPSF and CFI and attendant release of positive elongation factors and antitermination factors. The resulting decrease in the processivity of RNAP II provokes transcription termination (2). Termination on many genes was found to occur at various positions depending on the growth conditions (7), and cryptic termination sites rich in AU nucleotides are frequently found within open reading frames (ORFs) (8,9). Therefore, the selection of transcription termination sites is not determined solely by cis-acting sequence elements at the termination site and, plausibly, might also be regulated by post-translational modifications of termination and antitermination factors. However, the exact mechanism of selecting transcription termination sites is not fully understood.
Npl3p is a heterogeneous nuclear ribonucleoprotein with serine/arginine (SR) repeats. Its multiple functions include the role of an antiterminator to impede transcription termination at cryptic termination sites (8). According to the current model, Npl3p prevents transcription termination by competing with RNA cleavage factors, e.g. Hrp1p and Rna15p, for binding to nascent transcripts (8,9). Npl3p exhibits a higher affinity for RNA than do Hrp1p and Rna15p (9), and hence functions as a physical barrier impeding the binding of Hrp1p and Rna15p to nascent transcripts. On the other hand, Hrp1p and Rna15p, but not Npl3p, prefer AU-rich sequences, a characteristic of strong termination sites (9,10). Npl3p will thus be displaced by Hrp1p and Rna15p at strong termination sites, leading to the recruitment of other termination factors and cleavage of the nascent transcript. Since the competition between Npl3p and Hrp1p determines the choice of transcription termination sites (7), it would be of great interest to understand how transcription termination is regulated via post-translational modifications of Npl3p.
Transcription termination has to be well-coordinated with other steps of transcription, particularly elongation (1). As such, the elongation rate can influence termination through the action of positive elongation factors such as Spt4p, Spt6p and Paf1p, which are known to inhibit termination (1,8). Consistent with the concept that elongation and termination are co-regulated; Npl3p can interact with RNAP II and stimulate transcription elongation directly in vitro (11). However, it is not understood whether Npl3p promotes transcription elongation in vivo. In addition, the mechanisms by which Npl3p modulates transcription elongation and coordinates it with termination remain to be elucidated.
RNA-binding affinity and cellular localization of Npl3p can be modulated by post-translational modifications, including methylation (in the nucleus) (12,13) and phosphorylation (mainly in the cytoplasm) (14,15). Although RNA binding by Npl3p in vivo is apparently unaffected by arginine methylation (12), hypophosphorylation and methylation of Npl3p are suggested to be important for its export together with mRNA from the nucleus (12,15–17). Interestingly, phosphorylation of Npl3p by casein kinase II (CK2) was also suggested to contribute to Npl3p dissociation from the nascent transcript and hence to promote the recruitment of termination factor Rna15p in vitro (11). Npl3p is most heavily methylated in the nucleus (13,15), and there is evidence that its methylation is important in nuclear export of the Npl3-mRNA complex (12,13) and also in splicing (18). However, the role of Npl3p methylation in its transcription antitermination function is unclear.
Hmt1p is an arginine methyltransferase that catalyzes the methylation of arginine residues in several mRNA-binding proteins including Npl3p to facilitate their export from the nucleus (12,13). Methylation by Hmt1p also weakens Npl3p association with Tho2p, a subunit of the THO and TREX complexes, which stimulate elongation and mRNA nuclear export, respectively (13). Replacement of all 15 arginines in Npl3p’s Arg–Gly–Gly (RGG) repeats with lysines (the Npl3RKp variant) reduces Npl3p’s interaction with Tho2p and restores its nuclear export in hmt1Δ cells. This finding suggested that methylation masks the RGG repeats, and that lysine substitutions partially mimic methylation to permit Npl3p dissociation from Tho2p and its nuclear export in the absence of Hmt1p (12,13). Because Npl3RKp also restores nuclear export of Hrp1p in hmt1Δ cells, it appears that methylation of Npl3p, but not of Hrp1p itself, is crucial for Hrp1p export (12).
In this study, we hypothesized that Hmt1p might affect the selection of transcription termination sites by influencing the recruitment of Npl3p or Hrp1p to sites of transcription. We provide evidence that methylation of the RGG repeats in Npl3p by Hmt1p promotes antitermination at a weak terminator, and that it also stimulates elongation at least partly by stimulating Tho2p recruitment to sites of transcription. Our results suggest that the ability of Hmt1p to regulate the interaction between Npl3p and Tho2p on nascent mRNAs might provide an important feedback mechanism to couple the rate of transcription elongation with that of mRNA nuclear export.
MATERIALS AND METHODS
Yeast strains and growth conditions
All yeast strains and plasmids used in this work are listed in Tables 1 and 2, respectively. The BY4741 deletion derivatives were described earlier (19) and purchased from Open Biosystems. The presence of all reported deletion alleles was confirmed by PCR amplification and complementation of mutant phenotypes by plasmid-borne WT genes. Myc-tagged strains were constructed as described (20). gal10-Δ56 mutants were constructed as detailed elsewhere (21). The PGAL1-YLR454w strains were constructed as described earlier (22). All strains were verified by PCR analysis.
Table 1.
List of yeast strains
| Name | Parent | Relevant Genotype | Reference |
|---|---|---|---|
| BY4741 | mata his3-Δ1 leu2-Δ0 met15-Δ0 ura3-Δ0 | ||
| 3171 | BY4741 | hmt1Δ::KanMX4 | Open Biosystems |
| CMY201 | BY4741 | hmt1Δ::KanMX4 NPL3-myc13 | This work |
| CMY202 | BY4741 | hmt1Δ::KanMX4 THO2-myc13 | This work |
| CMY203 | BY4741 | hmt1Δ::KanMX4 HRP1-myc13 | This work |
| CMY127 | BY4741 | gal10-Δ56 | (23) |
| CMY223 | BY4741 | hmt1Δ::KanMX4 gal10-Δ56 | This work |
| CMY129 | BY4741 | npl3Δ::KanMX4 gal10-Δ56 | (23) |
| CMY125 | BY4741 | PGAL1-YLR454w | This work |
| CMY130 | BY4741 | hmt1Δ::KanMX4 PGAL1-YLR454w | (23) |
| W303 | matα leu2 ura3 lys2 trp1 his3 | ||
| YAM533 | W303 | hmt1Δ::HIS3 NPL3-myc::URA3 | (13) |
| YAM534 | W303 | hmt1Δ::HIS3 npl3RK- myc::URA3 | (13) |
| YAM535 | W303 | NPL3-myc::URA3 | (13) |
Table 2.
List of plasmids
| Original name | Relevant components | References | |
|---|---|---|---|
| npl3RK | pAM409 | CEN LEU2 pNop-PrA-npl3-RK1-15* | (15) |
| NPL3 | pPS2389 | CEN LEU2 pNop-PrA-NPL3* | (13) |
| hmt1-G68R | pPS1752 | CEN LEU2 hmt1-G68R | (32) |
| hmt-G68A | pPS1753 | CEN LEU2 hmt1-G68A | (32) |
| HMT1 | pPS1305 | CEN LEU2 HMT1 | (32) |
| pSCh202 | pSCh202 | 2µ URA3 PGAL-PHO5 | (25) |
| pSCh209-LAC4 | pSCh209-LAC4 | 2µ URA3 PGAL-PHO5::LAC4 | (25) |
*The plasmids express protein A (PrA) fusion protein driven by NOP1 promoter.
Biochemical methods
The ChIP experiments were conducted as described earlier (23) using the primers listed in Table 3. Northern blot analysis of total RNA was carried out as described earlier (23). Western blot analysis of whole-cell extracts was conducted using extracts prepared by trichloroacetic acid precipitation (24). Antibodies employed were monoclonal anti-Myc (Roche), anti-Rpb3p (Neoclone) and anti-Pgk1p (Invitrogen). GLAM assay was conducted as described earlier (25).
Table 3.
List of primers
| Gene (region) | Location (relative to ATG) | Sequence | References | |
|---|---|---|---|---|
| PGAL1-YLR454w | ||||
| TATA | −194/+35 | 5′-CTGGGGTAATTAATCAGCGAAGCGATG-3′ | (22) | |
| 5′-CACTTGTACAGTAGAACATTAATCGGAAAC-3′ | ||||
| 2 kb | +1986/+2199 | 5′-CATATCATCCACCCTAGGTGCTAGGTCGG-3′ | (22) | |
| 5′-GAGCTGACCAGACCTAACCATAGTAGCGTG-3′ | ||||
| 4 kb | +4069/+4268 | 5′-AGATATTACTCGTTGTTCGTGCCCAG-3′ | (22) | |
| 5′-CCCAAAACCCTAGTTAACAGAAGGATC-3′* | ||||
| 6 kb | +5904/+6074 | 5′-CGTACTGTTGAAATGGAACGAGGACGC-3′ | (22) | |
| 5′-ATCGCTTCCATACTCGTTGTATCATCAGTC-3′ | ||||
| 8 kb | +7701/+7850 | 5′-GAGGGTCACAGATCTATTACTTGCCC-3′ | (22) | |
| 5′-GTTGTGAGTTGCTTCAGTGGTGAAGTG-3′ | ||||
| Chr I | N.A. | 5′-GTTTATAGCGGGCATTATGCGTAGATCAG-3′ | (22) | |
| 5′-GTTCCTCTAGAATTTTTCCACTCGCACATTC-3′ | ||||
| GAL10-GAL7 | ||||
| A | −58/+182 | 5′-TTAAACTTCTTTGCGTCCATCC-3′ | (8) | |
| 5′-TGCTTGGTCAAGACCTCTAACC-3′ | ||||
| B | +823/+1108 | 5′-TGTCGTGAGTGGAACTTGGGTT-3′ | (8) | |
| 5′-GCATATCTTCAGCGGAAAATCTGGC-3′ | ||||
| C | +1869/+2102 | 5′-TGTCAAGGTTTTCATCCCGATTC-3′ | This work | |
| 5′-AAGAAGGATAGTAAGCTGGCAA-3′ | ||||
| D | +2323/+2604 | 5′-ATGGACGGTAGCAACAAGAATA-3′ | (8) | |
| 5′-TGGGTTAAGGAAAATGACAGAA-3′ | ||||
| E | −203/+38 | 5′-CAAAAAGCGCTCGGACAACT-3′ | (8) | |
| 5′-TGGGAATGGCTAGAAAAATCAA-3′ | ||||
| Chr V | N.A. | 5′-GGCTGTCAGAATATGGGGCCGTAGTA-3′ | (8) | |
| 5'-CACCCCGAAGCTGCTTTCACAATAC-3′ | ||||
*The sequence of this oligonucleotide was re-designed in this study.
RESULTS
Deletion of HMT1 increases termination at the defective terminator of gal10-Δ56 in vivo
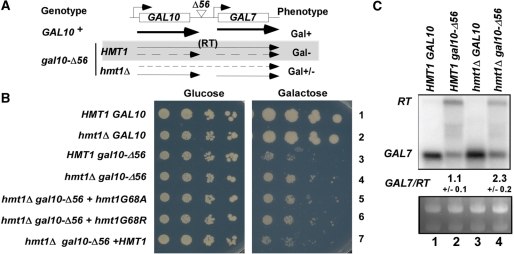
Deletion of HMT1 is synthetically lethal with deletion of NPL3 (13,26–28) or CBP80 (26). Npl3p can stimulate elongation in vitro (11) and it inhibits termination in vivo (8,9). Cbp80p has also been shown to promote antitermination in vivo (23,29). Therefore, we hypothesized that Hmt1p might play a role in transcription elongation or antitermination. To test this hypothesis, the effect of hmt1Δ on transcription of the gal10-Δ56 allele was examined. The Δ56 mutation eliminates 55 bp at the 3′ end of GAL10 and reduces the efficiency of 3′-end formation at the correct site, allowing read-through (RT) into the GAL7 gene downstream. Gal7p is not expressed from the RT bicistronic transcript, and the production of native GAL7 mRNA is impaired by transcription elongation across the GAL7 promoter—both features contributing to the Gal– phenotype of gal10-Δ56 cells (Figure 1A) (21).
Figure 1.
Hmt1p reduces termination at the defective gal10-Δ56 terminator. (A) Diagram summarizing results in (B) and (C) indicating that hmt1Δ increases termination at the gal10-Δ56 terminator, reducing the RT transcript while increasing GAL7 transcript abundance, thereby suppressing the Gal− phenotype in gal10-Δ56 cells. (B) Serially diluted strains of the indicated genotypes were spotted on synthetic complete medium lacking leucine (SC-LEU) plates containing 2% glucose (left) or 2% galactose (right) and incubated for 4 days. (C) Upper panel: northern blot analysis of GAL7 and RT transcripts. Total RNA (20 µg) from indicated strains induced with 2% galactose for 3 h were resolved and probed for GAL7 mRNA. The GAL7/RT ratios were determined by densitometry. The values indicated are the means and standard deviations of ratios calculated from three replicate experiments, which gave highly similar results. Lower panel: ethidium bromide staining of the gel.
In agreement with previous findings, gal10-Δ56 cells could not grow on galactose medium (Figure 1B, Row 3 versus 1) (23). Interestingly, hmt1Δ gal10-Δ56 cells were able to grow on galactose medium (Figure 1B, Row 4 versus 3). The Gal+ phenotype of hmt1Δ gal10-Δ56 cells could be complemented with plasmid-borne wild-type HMT1 (Figure 1B, Row 7 versus 4), but not by hmt1-G68A or hmt1-G68R (Figure 1B, Rows 5 and 6 versus 7), whose products exhibit weakened or no arginine methyltransferase activity, respectively (13). Northern blot analysis confirmed that the Gal+ phenotype of hmt1Δ gal10-Δ56 cells could be attributed to an increased level of GAL7 mRNA level and a reduced amount of GAL10-GAL7 RT transcript; as such, the ratio of GAL7 to GAL10-GAL7 RT transcripts (GAL7/RT) was increased more than 2-fold in hmt1Δ gal10-Δ56 cells (Figure 1C, lane 4 versus 2). This suggests that elimination of Hmt1p increases termination at the defective gal10-Δ56 terminator.
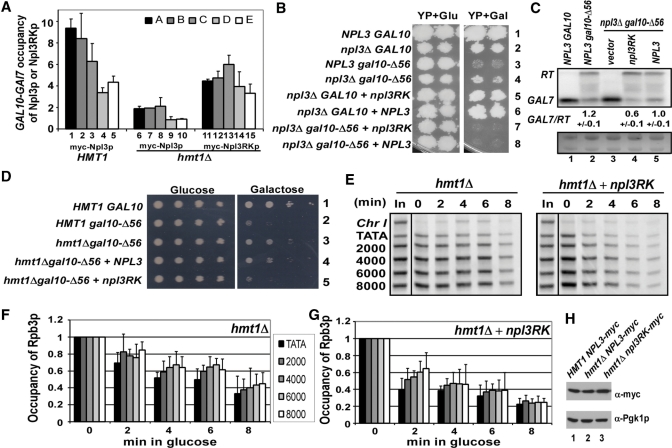
Figure 4.
Npl3RKp suppresses the antitermination and elongation rate defects caused by deletion of NPL3 or HMT1. (A) ChIP analysis of Npl3p or Npl3RKp in HMT1 or hmt1Δ strains. HMT1 NPL3-myc (YAM535, Lanes 1−5), hmt1ΔNPL3-myc (YAM533, Lanes 6−10) and hmt1Δnpl3RK-myc (YAM534, Lanes 11−15) were analyzed and the Npl3p or Npl3RKp occupancies across the GAL10-GAL7 locus were determined as described in Figure 3. Average results obtained from two independent cultures and two PCR amplifications for each culture were plotted. (B) Npl3RKp suppresses the Gal+ phenotype (antitermination defect) of npl3Δgal10-Δ56 (CMY129) cells. npl3Δgal10-Δ56 cells transformed with empty vector, NPL3 or npl3RK plasmid were spotted on YP plates containing either 2% glucose (Glu, left panel) or 2% galactose (Gal, right panels) as carbon source and incubated for 3 days in YP. Note that YP was used instead of SC-LEU medium because the extreme slow-growth phenotype conferred by npl3Δon SC-LEU+Gal medium, evident in GAL10 cells, obscures suppression of the Gal− phenotype of gal10-Δ56 by npl3Δ(data not shown). (C) Northern blot analysis of GAL7 and GAL10-GAL7 RT mRNAs conducted as in Figure 1C. The values indicated are the means and standard deviations of ratios calculated from three replicate experiments, which gave highly similar results. (D) Npl3RKp suppresses the Gal+ phenotype (antitermination defect) of hmt1Δgal10-Δ56 (CMY223) cells. HMT1 gal10-Δ56 and hmt1Δgal10-Δ56 strains with plasmid-borne NPL3 or npl3RK were spotted on SC-LEU plates containing 2% glucose (left) or 2% galactose (right) and incubated for 4 days. (E) ChIP analysis of Rpb3p occupancy across the PGAL1-YLR454w locus after imposing glucose repression. hmt1Δ cells (CMY130) with or without plasmid-borne npl3RK were induced with 2% galactose for 2 h (time = 0) and treated with 4% glucose for the indicated times. ChIP assays were done as described in Figure 2. PCR amplifications of input chromatin samples are labeled ‘In’; non-coding region on chromosome I (Chr I) was amplified as a control for non-specific immunoprecipitation. (F, G) Quantification of Rpb3p occupancies measured in (E) at the indicated positions at PGAL1-YLR454w in hmt1Δ cells (F) and hmt1Δ cells containing npl3RK (G). Average results obtained from two independent cultures and two PCR amplifications for each culture were plotted. (H) Possible effect of hmt1Δ on Myc-tagged Npl3p and Myc-tagged Np3RKp levels was examined by western blot analysis of WCEs from the relevant strains with antiMyc and antiPgk1p antibodies.
Hmt1p is required for efficient transcription elongation
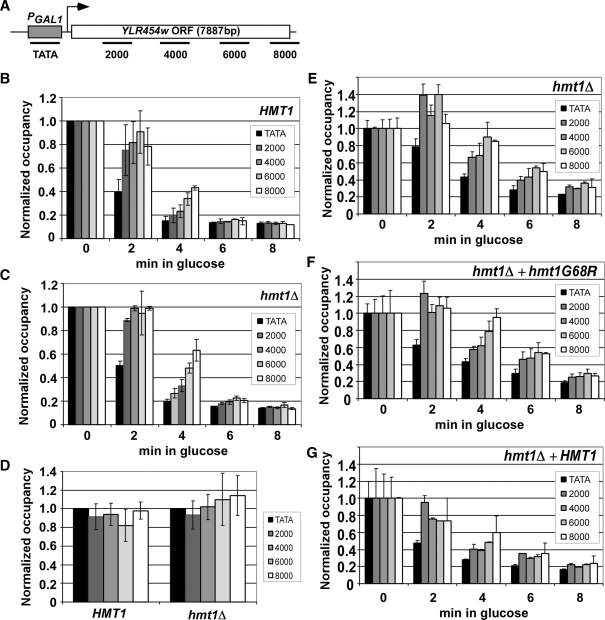
The increased termination at gal10-Δ56 in hmt1Δ cells could result from a decreased transcription elongation rate or increased termination efficiency (9). In order to dissect the role of Hmt1p in this context, the rate and processivity of transcription elongation were examined in hmt1Δ cells by using the ChIP assay to measure occupancies of RNAP II subunit Rpb3p across the PGAL1-YLR454w gene, in which the ∼8-kb YLR454w ORF is placed under the control of the GAL1 promoter (Figure 2A) (25). On glucose addition to cells growing in galactose, RNAP II recruitment to the promoter ceases and elongating RNAP II molecules finish transcribing the ORF. The kinetics of Rpb3p disappearance from the ORF during this last wave of elongation represents the elongation rate in vivo (22). Interestingly, it appeared that hmt1Δ cells exhibited a moderate decrease in the elongation rate versus wild-type HMT1 cells, which was evident at 4 min after glucose treatment (Figure 2, panel C versus B). The slower run-off of RNAP II from the YLR454w ORF in hmt1Δ cells was observed consistently in replicate experiments. To investigate whether the methyltransferase activity of Hmt1p is required for its function in transcription elongation, the ChIP analysis was repeated after introducing plasmid-borne HMT1 or mutant hmt1-G68R into hmt1Δ cells. Transformants harboring wild-type HMT1 exhibited a significantly higher elongation rate than those containing hmt1-G68R (Figure 2, Panel F versus E) or empty vector (Figure 2, panel G versus E). Thus, the arginine methyltransferase activity of Hmt1p is required for a wild-type rate of transcription elongation in vivo. The lower rate of elongation in hmt1Δ cells might contribute to the increased frequency of termination we observed at the defective terminator of gal10-Δ56.
Figure 2.
hmt1Δ reduces the rate of transcription elongation in vivo. (A) Diagram of the PGAL1-YLR454w gene with positions of PCR primers indicated (in bp) from nucleotide A of the start codon. (B, C) Quantification of Rpb3p occupancy measured by ChIP with antiRpb3p antibodies at the five positions across PGAL1-YLR454w indicated in (A) for HMT1 (CMY125) (B) or hmt1Δ (CMY130) (C). Cells grown in YP containing galactose were treated with 4% glucose for the indicated times. DNA extracted from immunoprecipitates and input chromatin samples was amplified by PCR in the presence of [33P]dATP for the different regions of PGAL1-YLR454w, together with a fragment from a non-transcribed region of Chr I to control for non-specific immunoprecipitation. PCR products were quantified with by phosphorimaging and ratios of experimental-to-control signals in the immunoprecipitates were normalized for the corresponding ratios for input samples to yield the occupancy values. Occupancies were normalized to those derived from cells grown in 2% galactose for 2 h (time = 0). (D) hmt1Δ does not detectably affect RNAP II processivity in vivo. HMT1 and hmt1Δ cells were grown in galactose medium (time = 0) and quantification of Rpb3p occupancy was performed as above. The value at each position was normalized to that of the WT strain. (E−G) Methyltransferase activity of Hmt1p stimulates the rate of transcription elongation in vivo. hmt1Δ cells containing empty vector (E) or plasmids harboring hmt1-G68R (F) or HMT1 (G) were induced with SC-LEU 2% galactose for 2 h (time = 0) and treated with 4% glucose for the indicated times. ChIP analysis of Rpb3p across the PGAL1-YLR454w gene was conducted as described earlier. Average results obtained from two independent cultures and two PCR amplifications for each culture were plotted in the histograms.
Processivity of RNAP II during elongation can be examined by comparing the Rpb3p occupancy at the 5′ and 3′ ends of YLR454w in cells grown in galactose (22). Because no appreciable difference was found in Rpb3p occupancy at PGAL1-YLR454w between wild-type and hmt1Δ cells grown in galactose (Figure 2D), we conclude that elimination of Hmt1p does not produce a detectable decrease in the processivity of elongating RNAP II.
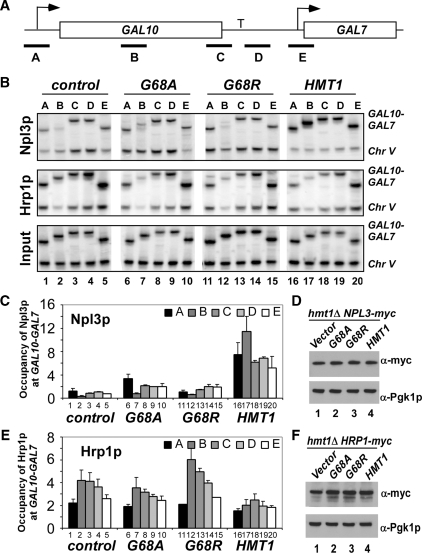
Recruitment of Npl3p and Hrp1p is regulated by the arginine methyltransferase activity of Hmt1p
Apart from stimulating transcription elongation, Hmt1p might reduce recognition of defective terminators by promoting recruitment of Npl3p, or impeding that of Hrp1p, to the transcribed ORF. Hmt1p methylates Npl3p (13) and Hrp1p (12), and deletion of HMT1 provokes accumulation of both proteins in the nucleus (30), although the accumulation of Hrp1p is dependent on Npl3p (15). In addition, a genome-wide binding study revealed a slight change in Npl3p genomic occupancy and a more dramatic alteration in Hrp1p recruitment in hmt1Δ cells (31). To determine whether Npl3p and Hrp1p recruitment to a native termination site might be regulated by Hmt1p, ChIP assays were performed to examine the effect of eliminating Hmt1p on occupancy of Npl3p and Hrp1p at GAL10-GAL7. Consistent with previous findings (8), in hmt1Δ cells harboring wild-type HMT1, Npl3p was found in all regions of GAL10 (Figure 3B, Lanes 16–20; also see Figure 3C, Lanes 16–20 for quantification). Remarkably, the hmt1Δ cells containing empty vector showed greatly reduced Npl3p recruitment (Figure 3B and C, Lanes 1–5 versus 16–20) particularly in the promoter (region A) and middle region (region B) of the GAL10 ORF (Figure 3B and C, Lanes 1 and 2 versus 16 and 17). Moreover, Npl3p recruitment could not be restored by either hmt1-G68A or hmt1-G68R (Figure 3B and C, Lanes 6 and 7, and 11 and 12). In agreement with previous findings (13, 31), Western blot analysis revealed no differences in myc-Npl3p levels among these different strains (Figure 3D). These results suggested that the arginine methyltransferase activity of Hmt1p is required for strong Npl3p recruitment to the GAL10-GAL7 locus.
Figure 3.
Loss of HMT1 affects Npl3p and Hrp1p occupancies at activated GAL10-GAL7. (A) Diagram of the GAL10-GAL7 locus, with PCR primer pairs indicated with black bars and terminator labeled as ‘T’. (B) ChIP analysis of Npl3p (upper panel) and Hrp1p (middle panel) occupancies across the GAL10-GAL7 locus. ChIP analysis of Myc-Npl3p or Myc-Hrp1p occupancies was conducted using antimyc antibodies as described in Figure 2, except using primers to amplify different regions of GAL10-GAL7 in panel A and to amplify non-transcribed region of Chr V was conducted to control for non-specific immunoprecipitation. Upper panel, transformants of hmt1Δ NPL3-myc strain (CMY201) containing empty vector (Lanes 1–5), or plasmids harboring hmt1-G68A (Lanes 6–10), hmt1-G68R (Lanes 11–15) or HMT1 (Lanes 16–20) were grown in SC-LEU galactose medium. Middle panel, transformants of hmt1Δ HRP1-myc strain (CMY203) harboring the same plasmids described for the upper panel were analyzed as described earlier. Lower panel: Analysis of input chromatin samples for the CMY201 transformants. (C) Quantification of Npl3p occupancy measured at the indicated positions. The ratios of experimental-to-control signals in the immunoprecipitate samples (B, upper panel) were normalized for the corresponding ratios for input samples (B, lower panel) to yield the occupancy values. Average results obtained from two independent cultures and two PCR amplifications for each culture were plotted. (D) Possible effect of mutation or deletion of HMT1 on expression of Myc-tagged Npl3p was analyzed by western blot analysis of WCEs of the relevant strains with antiMyc and antiPgk1p antibodies (to provide a loading control). (E) Quantification of Hrp1p occupancy measured at the indicated positions. The ratios of experimental to-control signals in the immunoprecipitate samples (B, middle panel) were normalized for the corresponding ratios for input samples (B, lower panel) to yield the occupancy values. Average results obtained from two independent cultures and two PCR amplifications for each culture were plotted. (F) Possible effect of mutation or deletion of HMT1 on expression of Myc-tagged Hrp1p was analyzed by western blot analysis of WCEs of the relevant strains with antiMyc and antiPgk1p antibodies (to provide a loading control).
As Npl3p is thought to function as an antitermination factor by competing with cleavage factors such as Hrp1p for binding to cryptic termination sites (8), we also examined the effect of eliminating Hmt1p on Hrp1p occupancy at GAL10-GAL7. Interestingly, deletion of HMT1 led to increased Hrp1p recruitment at GAL10, particularly to the middle region B (Figure 3B and 3E, Lane 2 versus 17). Again, the change in Hrp1p occupancy was dependent on the arginine methyltransferase activity of Hmt1p, as the Hrp1p occupancy in the middle to 3′ region of GAL10 in hmt1-G68A and hmt1-G68R cells was higher than in HMT1 cells (Figure 3B and E, Lanes7–8 and 12–13 versus 17–18). As expected from previous findings (31), we observed no differences in Hrp1p levels associated with the different HMT1 alleles (Figure 3F). Thus, although both Npl3p and Hrp1p are concentrated in the nucleus of hmt1Δ cells (32), Npl3p recruitment to the highly transcribed GAL10-GAL7 locus was reduced, whereas Hrp1p recruitment was increased in hmt1Δ cells. Plausibly, Hmt1p could suppress Hrp1p recruitment indirectly by stimulating Npl3p occupancy at GAL10. The dramatically decreased ratio of Npl3p to Hrp1p occupancy found in hmt1Δ cells (Figure 3C and E) might contribute to the increased recognition of the defective gal10-Δ56 terminator we observed in hmt1Δ gal10-Δ56 cells (Figure 1B and C).
Hmt1p regulates Npl3p recruitment, anti-termination and transcription elongation by methylating RGG repeats in Npl3p
Replacing all 15 RGG repeats in Npl3p with KGG repeats partially mimics the effect of methylation by Hmt1p and facilitates Npl3p dissociation from Tho2p and nuclear export in hmt1Δ cells (12,13). We wished to determine whether the KGG substitutions in Npl3RKp would also reduce the requirement for Hmt1p in efficient Npl3p recruitment, as this would imply that Hmt1p stimulates Npl3p recruitment directly by methylating RGG repeats. In agreement with results in Figure 3, deletion of HMT1 impaired Npl3p recruitment to GAL10-GAL7 (Figure 4A, Lanes 6–10 versus 1–5). Interestingly, however, recruitment of Npl3RKp was considerably less dependent on Hmt1p than is wild-type Npl3p (Figure 4A, Lanes 11–15 versus 6–10). Comparing Npl3RKp recruitment in hmt1Δ cells to that of wild-type Npl3p in HMT1 cells reveals lower occupancies for Npl3RKp at the 5′ and middle regions of GAL10 (regions A and B, respectively) (Figure 4A, Lanes 11–12 versus 1–2), but similar occupancies in the 3′ region of GAL10 and promoter region of GAL7 (regions D and E, respectively) (Figure 4A, Lanes 14–15 versus 4–5). In agreement with previous findings (13, 31), western analysis indicated that there are no differences in expression levels of myc-Npl3p or myc-npl3RKp among the three strains (Figure 4H). These findings suggested that Npl3RKp might function as an antitermination factor in hmt1Δ cells as effectively as wild-type Npl3p does in HMT1 cells.
To address this possibility, we first asked whether the KGG substitutions would impair the antitermination function of Npl3p. As described in our previous study (23), npl3Δ suppressed the Gal− phenotype conferred by gal10-Δ56, allowing modest growth on YP+Gal medium (Figure 4B, right panel, Row 4 versus 3) despite the fact that npl3Δ confers a strong slow growth phenotype on this medium even in the presence of wild-type GAL10 (Figure 4B, right panel, Row 2 versus 1). Importantly, the npl3RK mutant was indistinguishable from wild-type NPL3 in reversing the ability of npl3Δ gal10-Δ56 cells to grow on YP+Gal (Figure 4B, right panel, Rows 7–8 versus 4), as well as in complementing the slow growth phenotype of npl3ΔGAL10 cells in this medium (Figure 4B, right panel Rows 5 and 6 versus 2). In line with the Gal− phenotypes, northern blot analysis showed that both npl3RK and NPL3 conferred a strong reduction in GAL7 mRNA and elevated levels of GAL10-GAL7 RT transcripts in npl3Δ gal10-Δ56 cells (Figure 4C, Lanes 4–5 versus 3). In fact, npl3RK provoked a lower GAL7/RT ratio than did wild-type NPL3 (Figure 4C, Lane 4 versus 5), indicating that Npl3RKp actually has a higher level of antitermination activity than wild-type Npl3p.
Having found that Npl3RKp has antitermination function and is recruited independently of Hmt1p, we tested whether Npl3RKp could suppress the reduced antitermination phenotype in hmt1Δ cells. In agreement with our findings in Figure 1B, hmt1Δ partially suppressed the Gal− phenotype of gal10Δ-56 cells, which we attributed to increased recognition of the defective GAL10 terminator and greater production of GAL7 mRNA (Figure 1C). Interestingly, over-expression of Npl3RKp but not wild-type Npl3p, reversed this effect of hmt1Δ and restored the Gal− phenotype in hmt1Δgal10Δ-56 cells (Figure 4D, Rows 5 versus. 3, and Rows 4 versus 3). This finding suggests that the antitermination function of Npl3RKp is independent of Hmt1p.
We also asked whether Npl3RKp could rescue the transcription elongation defect in hmt1Δ cells. Using the same ChIP assay employed in Figure 2 to measure the transcription elongation rate at PGAL1-YLR454w, we found that overexpressing Npl3RKp significantly reduced the elongation defect in hmt1Δ cells (Figure 4E–G), while this was not observed in cells over-expressing wild-type Npl3p (data not shown).
Methylation of Npl3p by Hmt1p facilitates co-transcriptional recruitment of THO/TREX
There is evidence that Hmt1p methylation of Npl3p reduces its interaction with transcription elongation factor Tho2p (13,31), and that the KGG substitutions in Npl3p partially bypass the requirement for Hmt1p in reducing Npl3p-Tho2p association (13). We hypothesized that the elongation defect in hmt1Δ cells involves sequestration of Npl3p and Tho2p in nuclear mRNPs that are not competent for export with attendant impairment of Tho2p recruitment by elongating RNAP II. Consistent with this idea, ChIP analysis showed that cells lacking Hmt1p or containing Hmt1p variants lacking arginine methyltransferase activity displayed reduced Tho2p occupancy in the middle region of the GAL10 ORF compared with that seen in cells containing wild-type Hmt1p (Figure 5A, Lanes 1–3 versus 4). Importantly, over-expression of Npl3RKp, but not Npl3p, restored Tho2p recruitment in hmt1Δ cells to the same level observed in HMT1 cells with native levels of Npl3p (Figure 5A, Lanes 5–6 versus 1 and 4). This effect was not due to differences in RNAP II occupancy at GAL10 in these strains (see comment) or Tho2p expression level (data not shown) (13). These findings support the idea that the transcription elongation defect in hmt1Δ cells involves binding of non-methylated Npl3p to the THO or TREX complex, thereby reducing the recruitment of one or both of these elongation factor complexes to elongating RNAP II.
Figure 5.
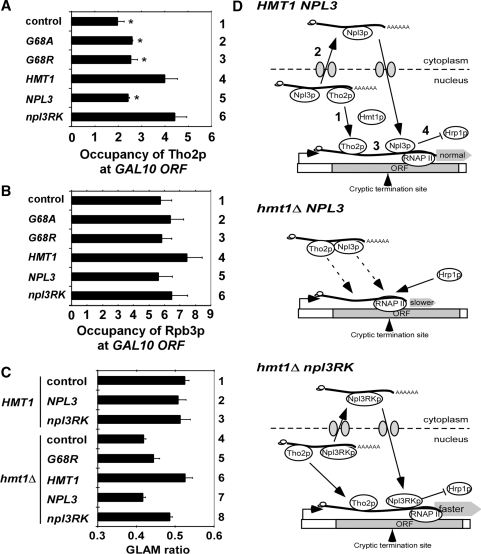
Tho2p recruitment and gene length-dependent transcription defects in hmt1Δ cells are suppressed by npl3RK, and a hypothetical model for the role of Hmt1p in recycling Tho2p and Npl3p from mRNPs to sites of transcription. ChIP analysis of Tho2p occupancy at the 5′ region of GAL10 gene (region B in Fig. 3A) in hmt1Δ THO2-myc13 (CMY202) cells. Cells transformed with empty vector, or plasmids containing hmt1-G68A, hmt1-G68R, HMT1, NPL3 or npl3RK were grown in SC-LEU with 2% galactose and ChIP analysis was conducted using antiMyc antibodies (to measure Tho2p occupancy) or antibodies against Rpb3p (to measure RNAP II occupancy). Non-coding region on chromosome V (Chr V) was amplified as control for non-specific immunoprecipitation. The occupancies were quantified and plotted for Tho2p (A) and Rpb3p (B). Average results obtained from two independent cultures and two PCR amplifications for each culture were plotted. The occupancies of Tho2p at the GAL10 ORF are significantly lower in control (hmt1Δ), G68A and G68R versus HMT1 cells, and in cells containing extra copies of NPL3 versus extra copies of npl3RK, as judged by the Student’s t-test, with P < 0.01(*). (C) HMT1 (BY4741) and hmt1Δ (3171) cells transformed with reporter plasmid pSCH209-LAC4 plus plasmids harboring hmt1-G68A, NPL3, npl3RK or empty vector were grown in SC-LEU-URA with 2% galactose. HMT1 (BY4741) and hmt1Δ (3171) cells harboring pSCH202 were analyzed as controls. Acid phosphatase activity was measured in cell lysates and the GLAM ratio was calculated as the ratio of activities in transformants harboring pSCH209-LAC4 versus those containing pSCH202. Results are the averages from three independent experiments and error bars indicate SD (D) Model depicting how Hmt1p regulates transcription elongation and termination by methylating Npl3p. Upper panel: In WT cells, methylation of Npl3p by Hmt1p allow dissociation the TREX(Tho2p)−Npl3p−mRNP to facilitate (1) TREX(Tho2p) recruitment to sites of transcription and (2) nuclear export of the Npl3p−mRNP. Subsequent nuclear import and methylation of Npl3p by Hmt1p enables its recruitment to sites of transcription (3). Npl3p and TREX both stimulate elongation and, by impeding recruitment of Hrp1p (4), Npl3p prevents transcription termination at cryptic terminators. Middle panel, in hmt1Δ NPL3 cells, the failure to methylate Npl3p causes Tho2p (TREX) and Npl3p to be sequestered in mRNPs that are not exported efficiently; the ensuing impaired recruitment of TREX(Tho2p) and Npl3p to sites of transcription reduces the rate of elongation, and the resulting increased Hrp1p occupancy enables transcription termination at cryptic terminators. Lower panel, in hmt1Δ npl3RK cells, TREX(Tho2p)-Npl3RKp-mRNP can dissociate independently of Npl3p methylation to recycle TREX(Tho2p) and Npl3RKp back to sites of transcription where they stimulate elongation and block termination at cryptic terminators, as occurs in WT cells.
Finally, we employed an assay for gene-length dependent accumulation of mRNA (GLAM), which reveals defects in transcription elongation, to provide further support that Hmt1p stimulates elongation by methylating Npl3p. The GLAM assay compares the expression of reporters with the same promoter (PGAL) and PHO5 coding sequences for acid phosphatase, but harboring 3′ untranslated regions that differ by 3 kb. Mutations in elongation factors produce a greater reduction in the steady-state amount of the long versus short transcript, and their encoded Pho5p enzyme activities (decreased GLAM ratio) (25). In agreement with our findings from the kinetic ChIP assay in Figures 2, and 4, we found that hmt1Δ cells exhibit a significantly lower GLAM ratio than do HMT1 cells (Figure 5C, Lanes 1 versus 4), and the reduced GLAM ratio of hmt1Δcells was complemented with plasmid-borne HMT1 but not by hmt1G68R (Figure 5C, Lanes 5–6 versus 4). Importantly, the reduced GLAM ratio of hmt1Δ cells was suppressed by over-expressing npl3RK, but not by over-expressing wild-type NPL3 (Figure 5C, Lanes 7–8 versus 4).
DISCUSSION
Achieving efficient transcription termination at the correct termination sites and blocking termination at cryptic terminators involves a complex interplay between elongation factors, anti-termination factors and termination factors, all recruited by elongating RNAP II (8,10). In this study, we have explored the role of arginine methylation by Hmt1p in regulating the transition from transcription elongation to termination. We obtained evidence that Hmt1p suppresses recognition of the mutationally weakened terminator at gal10-Δ56 (Figure 1) and set out to elucidate the mechanism of its antitermination activity. The increased termination at gal10-Δ56 in hmt1Δ cells could arise from less efficient transcription elongation, diminished recruitment of an antitermination factor or increased recruitment of a termination factor. We found evidence for all three mechanisms. Results from two different in vivo assays revealed that mutants lacking Hmt1p or impaired for its arginine methyltransferase activity exhibit transcription elongation defects (Figure 5C), involving a reduced rate of elongation (Figure 2). This is noteworthy because, apart from mutation of RNAP II itself, inactivation of various elongation factors was not found to reduce the elongation rate in vivo (22).
The hmt1 mutants also display decreased recruitment to GAL10 of the antitermination factor Npl3p, known to block recognition of the defective gal10-Δ56 terminator (8), and they further exhibit elevated occupancies of cleavage/termination factor Hrp1p (Figure 3). A previous genome-wide analysis did not reveal an important role for Hmt1p arginine methyltransferase activity in modulating co-transcriptional recruitment of Npl3p or Hrp1p, although a non-enzymatic function of Hmt1p was implicated in down-regulating Hrp1p recruitment at many genes (31). Perhaps the marked effects of Hmt1p catalytic function on Npl3p and Hrp1p recruitment observed here are restricted to highly induced genes like GAL10.
Substituting all of the RGG repeats in Npl3p with KGG (in the Npl3RKp variant) was shown previously to mimic the effects of Hmt1p on nuclear export of Npl3p and its interaction with Tho2p (12,13). Interestingly, we found that the KGG substitutions also partially rescue Npl3p recruitment to GAL10 (Figure 4A) and, consistently, restore the antitermination function of Npl3p at gal10-Δ56 in hmt1Δ cells (Figure 4D). Remarkably, overexpressing Npl3RKp also rescues efficient recruitment of Tho2p (Figure 5A) and mitigates the transcription elongation defects in hmt1Δ cells (Figure 4E–G and Figure 5C). The fact that substituting the RGG repeats in Npl3p suppresses the effects of hmt1Δ on antitermination, elongation and recruitment of Npl3p implies that Hmt1p influences all of these events by methylating Npl3p rather than another substrate. The ability of the KGG substitutions to rescue Npl3p recruitment and transcription elongation fits with previous findings that Npl3p directly stimulates elongation by purified RNAP II (11). However, the reduced recruitment of elongation factor Tho2p might also contribute to the elongation defect in hmt1Δ cells, as Tho2p recruitment was likewise rescued by overexpressing Npl3RKp (Figure 5A). This last result is consistent with previous findings that Npl3p, but not Npl3RKp, is bound tightly to Tho2p in an mRNP complex in hmt1Δ cells. It was proposed that Hmt1p methylation of Npl3p is required for its dissociation from Tho2p and other TREX components to facilitate nuclear export of the Npl3p-mRNP complex (13,31).
Based on our findings and the previous results, we propose that methylation of the RGG repeats in Npl3p by Hmt1p facilitates mRNA export and leads to dissociation of TREX (Tho2p) from Npl3p-containing mRNPs recycling TREX (Tho2p) back to sites of transcription where it can stimulate elongation. Following mRNA export, the Npl3p re-enters the nucleus, and is methylated by Hmt1p (Figures 3C and 4A). Methylated Npl3p is recruited co-transcriptionally to enhance elongation and also block recruitment of CFI at cryptic terminators (Figure 5D, top panel). In hmt1Δ cells, the TREX–Npl3p–mRNA complex persists in the nucleus (13,31) and co-transcriptional recruitment of both Npl3p and THO/TREX complex are diminished, thereby reducing the elongation rate and increasing utilization of cryptic terminators (Figure 5D, middle panel). The KGG substitutions in Npl3p partially rescue mRNA export and thereby dissociate TREX from the Npl3p–mRNA complexes independently of Npl3p methylation by Hmt1p, thus suppressing the elongation and antitermination defects of hmt1Δ cells (Figure 5D, bottom). This model is consistent with the finding that the defect in mRNA nuclear export in hmt1Δ cells is rescued by overexpressing Npl3RKp (13). It is also supported by the fact that the transient Tho2p–Npl3p–mRNA complex has been detected by coimmunoprecipitation only in hmt1Δ cells, which are defective for mRNA nuclear export (13). Methylation of Npl3p might enhance its recruitment to sites of elongation by another more active mechanism besides dissociation of the putative TREX–Npl3p–mRNA complex postulated in our model.
The proposed role of Hmt1p in regulating interaction of Tho2p and Npl3p on nascent mRNAs may provide a feedback mechanism for coordinating the rate of transcription elongation with nuclear export. If methylation of Npl3p is required to enable nuclear export of the Npl3p–mRNP and recycle TREX (Tho2p), as suggested in Figure 5D, then the availability of TREX (Tho2p) for new rounds of transcription elongation will be dependent on the completion of mRNA export of existing nascent transcripts.
In addition to methylation, Npl3p can be phosphorylated by Sky1p (14,15,33) and CK2 (11). Phosphorylation of Npl3p (on Ser-411) by Sky1p reduces Npl3p RNA binding affinity and promotes Npl3p dissociation from mRNA (14). Hence, it is thought that phosphorylation of Npl3p by Sky1p in the cytoplasm is important for recycling Npl3p back to the nucleus (14,15,33) and releasing the transcripts for translation (34). However, there is evidence that Npl3p methylation by Hmt1p antagonizes Sky1p-mediated phosphorylation of Npl3p in vitro (15). Therefore, an unknown Npl3p demethylase might have to operate in the cytoplasm to permit Sky1p phosphorylation of Npl3p. Interestingly, Npl3p is also phosphorylated on Ser-411 by CK2 in the nucleus (11). This phosphorylation is suggested to promote Npl3p dissociation from the nascent transcript, and hence promote the recruitment of termination factors (11). Our results indicate that the Npl3p recruited to sites of transcription is methylated by Hmt1p (Figures. 3C and 4C). Since CK2 phosphorylates the same site as Sky1p, it is possible that Npl3p methylation would also antagonize CK2-mediated phosphorylation at Ser-411 and prevent the proposed function in dissociation of Npl3p at termination sites. It is unclear whether methylation of Npl3p would antagonize its phosphorylation by CK2 in vivo and the attendant proposed effects of CK2 on termination. It is clearly of great interest to fully elucidate the sequence and precise timing of post-translational modifications of Npl3p (including methylation, demethylation, phosphorylation and dephosphorylation) during the transcription cycle, splicing and mRNA export.
The human homologue of yeast Hmt1p, Prmt1p, was suggested to affect transcription elongation via methylation of Spt5p (35) and histone H4 Arg-3 (36,37). Presumably Npl3RKp does not affect the methylation of other molecules in hmt1Δ cells, and thus Npl3RKp likely rescues the transcription elongation defect conferred by hmt1Δ through mechanisms distinct from those described for Prmt1p, e.g. by promoting TREX (Tho2p) recruitment (Figure 5D). Hmt1p might also stimulate elongation via methylation of Npl3p by enhancing recruitment of Npl3p to elongating RNAP II, as Npl3p can stimulate elongation in a purified system lacking TREX (11,30).
In summary, we have provided the first direct evidence that the arginine methyltransferase activity of Hmt1p enhances transcription elongation and antitermination in vivo and that both functions involve its methylation of RGG repeats in Npl3p, enhancing recruitment of Npl3p and THO/TREX while impeding recruitment of CFI to sites of transcription elongation.
FUNDING
The University Research Committee of the University of Hong Kong; Hong Kong Research Grants Council (project HKU 7486/06M); Intramural Research Program of the NIH.
Conflict of interest statement. None declared.
ACKNOWLEDGMENTS
We thank Dr Sebastián Chávez, Anne E. McBride, Kevin Struhl and Fred Winston for reagents, and members of Jin laboratory for critical reading of manuscript.
REFERENCES
- 1.Richard P, Manley JL. Transcription termination by nuclear RNA polymerases. Genes Dev. 2009;23:1247–1269. doi: 10.1101/gad.1792809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buratowski S. Connections between mRNA 3′ end processing and transcription termination. Curr. Opin. Cell Biol. 2005;17:257–261. doi: 10.1016/j.ceb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 4.Tollervey D. Molecular biology: termination by torpedo. Nature. 2004;432:456–457. doi: 10.1038/432456a. [DOI] [PubMed] [Google Scholar]
- 5.Lykke-Andersen S, Jensen TH. Overlapping pathways dictate termination of RNA polymerase II transcription. Biochimie. 2007;89:1177–1182. doi: 10.1016/j.biochi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Koch F, Jourquin F, Ferrier P, Andrau JC. Genome-wide RNA polymerase II: not genes only! Trends Biochem. Sci. 2008;33:265–273. doi: 10.1016/j.tibs.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 7.Sparks KA, Dieckmann CL. Regulation of poly(A) site choice of several yeast mRNAs. Nucleic Acids Res. 1998;26:4676–4687. doi: 10.1093/nar/26.20.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bucheli ME, Buratowski S. Npl3 is an antagonist of mRNA 3′ end formation by RNA polymerase II. EMBO J. 2005;24:2150–2160. doi: 10.1038/sj.emboj.7600687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bucheli ME, He X, Kaplan CD, Moore CL, Buratowski S. Polyadenylation site choice in yeast is affected by competition between Npl3 and polyadenylation factor CFI. RNA. 2007;13:1756–1764. doi: 10.1261/rna.607207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim Guisbert K, Duncan K, Li H, Guthrie C. Functional specificity of shuttling hnRNPs revealed by genome-wide analysis of their RNA binding profiles. RNA. 2005;11:383–393. doi: 10.1261/rna.7234205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dermody JL, Dreyfuss JM, Villen J, Ogundipe B, Gygi SP, Park PJ, Ponticelli AS, Moore CL, Buratowski S, Bucheli ME. Unphosphorylated SR-like protein Npl3 stimulates RNA polymerase II elongation. PLoS ONE. 2008;3:e3273. doi: 10.1371/journal.pone.0003273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu C, Henry MF. Nuclear export of hnRNP Hrp1p and nuclear export of hnRNP Npl3p are linked and influenced by the methylation state of Npl3p. Mol. Cell Biol. 2004;24:10742–10756. doi: 10.1128/MCB.24.24.10742-10756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McBride AE, Cook JT, Stemmler EA, Rutledge KL, McGrath KA, Rubens JA. Arginine methylation of yeast mRNA-binding protein Npl3 directly affects its function, nuclear export, and intranuclear protein interactions. J. Biol. Chem. 2005;280:30888–30898. doi: 10.1074/jbc.M505831200. [DOI] [PubMed] [Google Scholar]
- 14.Gilbert W, Siebel CW, Guthrie C. Phosphorylation by Sky1p promotes Npl3p shuttling and mRNA dissociation. RNA. 2001;7:302–313. doi: 10.1017/s1355838201002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yun CY, Fu XD. Conserved SR protein kinase functions in nuclear import and its action is counteracted by arginine methylation in Saccharomyces cerevisiae. J. Cell. Biol. 2000;150:707–718. doi: 10.1083/jcb.150.4.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Y, Yario TA, Steitz JA. A molecular link between SR protein dephosphorylation and mRNA export. Proc. Natl Acad. Sci. USA. 2004;101:9666–9670. doi: 10.1073/pnas.0403533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, Guo W, Tartakoff PY, Tartakoff AM. A Crm1p-independent nuclear export path for the mRNA-associated protein, Npl3p/Mtr13p. Proc. Natl Acad. Sci. USA. 1999;96:6739–6744. doi: 10.1073/pnas.96.12.6739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kress TL, Krogan NJ, Guthrie C. A single SR-like protein, Npl3, promotes pre-mRNA splicing in budding yeast. Mol. Cell. 2008;32:727–734. doi: 10.1016/j.molcel.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 20.Longtine MS, McKenzie A., 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 21.Kaplan CD, Holland MJ, Winston F. Interaction between transcription elongation factors and mRNA 3′-end formation at the Saccharomyces cerevisiae GAL10-GAL7 locus. J. Biol. Chem. 2005;280:913–922. doi: 10.1074/jbc.M411108200. [DOI] [PubMed] [Google Scholar]
- 22.Mason PB, Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol. Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 23.Wong CM, Qiu H, Hu C, Dong J, Hinnebusch AG. Yeast cap binding complex impedes recruitment of cleavage factor IA to weak termination sites. Mol. Cell. Biol. 2007;27:6520–6531. doi: 10.1128/MCB.00733-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reid GA, Schatz G. Import of proteins into mitochondria. Extramitochondrial pools and post-translational import of mitochondrial protein precursors in vivo. J. Biol. Chem. 1982;257:13062–13067. [PubMed] [Google Scholar]
- 25.Morillo-Huesca M, Vanti M, Chavez S. A simple in vivo assay for measuring the efficiency of gene length-dependent processes in yeast mRNA biogenesis. FEBS J. 2006;273:756–769. doi: 10.1111/j.1742-4658.2005.05108.x. [DOI] [PubMed] [Google Scholar]
- 26.Weiss VH, McBride AE, Soriano MA, Filman DJ, Silver PA, Hogle JM. The structure and oligomerization of the yeast arginine methyltransferase, Hmt1. Nat. Struct. Biol. 2000;7:1165–1171. doi: 10.1038/82028. [DOI] [PubMed] [Google Scholar]
- 27.Henry MF, Silver PA. A novel methyltransferase (Hmt1p) modifies poly(A)+-RNA-binding proteins. Mol. Cell. Biol. 1996;16:3668–3678. doi: 10.1128/mcb.16.7.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen EC, Stage-Zimmermann T, Chui P, Silver PA. The yeast mRNA-binding protein Npl3p interacts with the cap-binding complex. J. Biol. Chem. 2000;275:23718–23724. doi: 10.1074/jbc.M002312200. [DOI] [PubMed] [Google Scholar]
- 29.Das B, Butler JS, Sherman F. Degradation of normal mRNA in the nucleus of Saccharomyces cerevisiae. Mol. Cell. Biol. 2003;23:5502–5515. doi: 10.1128/MCB.23.16.5502-5515.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jobert L, Argentini M, Tora L. PRMT1 mediated methylation of TAF15 is required for its positive gene regulatory function. Exp. Cell Res. 2009;315:1273–1286. doi: 10.1016/j.yexcr.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 31.Yu MC, Bachand F, McBride AE, Komili S, Casolari JM, Silver PA. Arginine methyltransferase affects interactions and recruitment of mRNA processing and export factors. Genes Dev. 2004;18:2024–2035. doi: 10.1101/gad.1223204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McBride AE, Weiss VH, Kim HK, Hogle JM, Silver PA. Analysis of the yeast arginine methyltransferase Hmt1p/Rmt1p and its in vivo function. Cofactor binding and substrate interactions. J. Biol. Chem. 2000;275:3128–3136. doi: 10.1074/jbc.275.5.3128. [DOI] [PubMed] [Google Scholar]
- 33.Siebel CW, Feng L, Guthrie C, Fu XD. Conservation in budding yeast of a kinase specific for SR splicing factors. Proc. Natl Acad. Sci. USA. 1999;96:5440–5445. doi: 10.1073/pnas.96.10.5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Windgassen M, Sturm D, Cajigas IJ, Gonzalez CI, Seedorf M, Bastians H, Krebber H. Yeast shuttling SR proteins Npl3p, Gbp2p, and Hrb1p are part of the translating mRNPs, and Npl3p can function as a translational repressor. Mol. Cell. Biol. 2004;24:10479–10491. doi: 10.1128/MCB.24.23.10479-10491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwak YT, Guo J, Prajapati S, Park KJ, Surabhi RM, Miller B, Gehrig P, Gaynor RB. Methylation of SPT5 regulates its interaction with RNA polymerase II and transcriptional elongation properties. Mol. Cell. 2003;11:1055–1066. doi: 10.1016/s1097-2765(03)00101-1. [DOI] [PubMed] [Google Scholar]
- 36.Strahl BD, Briggs SD, Brame CJ, Caldwell JA, Koh SS, Ma H, Cook RG, Shabanowitz J, Hunt DF, Stallcup MR, et al. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr. Biol. 2001;11:996–1000. doi: 10.1016/s0960-9822(01)00294-9. [DOI] [PubMed] [Google Scholar]
- 37.Huang S, Litt M, Felsenfeld G. Methylation of histone H4 by arginine methyltransferase PRMT1 is essential in vivo for many subsequent histone modifications. Genes Dev. 2005;19:1885–1893. doi: 10.1101/gad.1333905. [DOI] [PMC free article] [PubMed] [Google Scholar]