Abstract
The Est3 subunit of yeast telomerase, which adopts a predicted OB-fold, is essential for telomere replication. To assess the possible contributions that Est3 might make to enzyme catalysis, we compared telomerase activity from wild type and est3-Δ strains of Saccharomyces castellii, which revealed that loss of the Est3 subunit results in a 2- to 3-fold decline in nucleotide addition. This effect was not primer-specific, based on assessment of a panel of primers that spanned the template of the S. castellii telomerase RNA. Furthermore, using nuclear magnetic resonance chemical shift perturbation, no chemical shift change was observed at any site in the protein upon addition of single-stranded DNA, arguing against a role for Est3 in recognition of telomeric substrates by telomerase. Addition of exogenous Est3 protein, including mutant Est3 proteins that are severely impaired for telomere replication in vivo, fully restored activity in est3-Δ telomerase reactions. Thus, Est3 performs an in vivo regulatory function in telomere replication, which is distinct from any potential contribution that Est3 might make to telomerase activity.
INTRODUCTION
In telomerase-expressing cells, telomerase is carefully controlled by both negative and positive regulation (1). These regulatory mechanisms ensure that the shortest telomeres are preferentially elongated by telomerase (2,3) and, conversely, that telomerase is repressed at over-elongated termini (4–6), thereby maintaining telomere length homeostasis. Regulation can occur at multiple levels, including controlling access to the site of catalysis (the protruding G-rich 3′ overhang present at chromosome termini) and the extent of nucleotide addition per catalytic event (7,8), as well as assembly and transport of the telomerase holoenzyme complex (9,10).
At least a subset of these regulatory steps are performed by subunits of the telomerase holoenzyme, and, thus, a full appreciation of telomerase regulation will benefit from a mechanistic understanding of biochemical properties of each telomerase subunit. However, a complete picture of the telomerase complex in various experimental systems is still emerging. In the budding yeast Saccharomyces cerevisiae, telomerase is composed of three Est (ever shorter telomere) proteins in association with the telomerase RNA, referred to as TLC1 (11–13). A secondary structure model for TLC1, constructed on the basis of both phylogenetic and in vivo analysis, reveals a central core with flexible arms protruding from this core (14,15). Est2 binds to the central core, which contains the template region of TLC1. Est1 also binds the telomerase RNA, through a bulged stem–loop structure located at the end of one of the flexible arms (16). Consistent with distinct binding sites on TLC1 for Est1 and Est2, these two subunits do not interact in the absence of TLC1 (17), arguing against a stable interaction between these two proteins. In contrast, Est3 is dependent on the Est2 subunit for complex association (11; this work), suggesting that Est2 and Est3 bind each other directly. The overall architectural features displayed by the S. cerevisiae enzyme are shared with the Schizosaccharomyces pombe telomerase RNP (18–20), although an Est3 subunit of fission yeast telomerase has not been identified. This may be a consequence of the pronounced sequence divergence exhibited by telomerase subunits among the Ascomycota phylum, which is particularly notable for Est3; so far, homologs of Est3 have only been identified in the subphylum Saccharomycotina (20; Mandell, E.K., unpublished data).
The individual functions of the components of the S. cerevisiae telomerase have been intensively studied over the past decade. Two subunits form the catalytic core of the enzyme: TLC1 provides the template that dictates the sequence of telomeric repeats synthesized by the reverse transcriptase domain of Est2 (12,13). Est1 and Est3, in contrast, are dispensable for enzyme catalysis (21,22). Instead, the primary role of the Est1 protein is to recruit telomerase to its site of action, through an interaction with the telomere-bound Cdc13 protein (8,23–25). The contribution that Est3 makes to telomere replication has remained elusive, although recent work has demonstrated that Est3 consists of a predicted oligo-saccharide/oligo-nucleotide binding (OB)-fold which is structurally similar to an OB-fold present in the mammalian TPP1 protein (26,27).
The conclusion that the Est1 and Est3 telomerase subunits are not essential for enzymatic activity has been supported by studies in multiple fungal organisms. Telomerase is capable of elongating telomeric primers in partially fractionated extracts prepared from est1-Δ strains of S. cerevisiae, Candida albicans and S. pombe, as well as est3-Δ derivatives of S. cerevisiae, Kluyveromyces lactis and C. albicans (18,19,21,22,28–30). However, telomerase activity from each of these fungal species exhibits an atypical property, when compared to the behavior of the telomerase enzyme from most other species. As was first demonstrated in Tetrahymena, a characteristic feature of telomerase is the ability to elongate a primer to the end of the RNA template, followed by translocation of the primer on the template to allow another round of elongation (31). Repeated rounds of elongation and translocation can allow synthesis of >500 nt before dissociation (32), giving rise to a periodicity in the pattern of the reaction products due to pausing that occurs at the translocation step. In contrast, telomerase extracts from S. cerevisiae, K. lactis and even the more phylogenetically distant S. pombe appear to be capable of synthesizing only up to ∼10 nt in vitro, with no evidence for translocation (19,21,30).
This does not appear to be an intrinsic property of fungal species, however, because telomerase from Saccharomyces castellii is capable of elongating a telomeric primer through multiple rounds of translocation (21). Since this provides a more sensitive assay for assessing the potential effects of regulatory factors on telomerase activity, this current study re-addresses the role of Est3 in telomerase activity. Comparison of enzyme activity prepared from EST3 and est3-Δ strains of S. castellii demonstrated that loss of the Est3 telomerase subunit resulted in a 2- to 3-fold reduction in nucleotide addition, in a primer-independent manner. Activity could be restored by adding S. castellii Est3 protein, which was affinity purified from Escherichia coli, to est3-Δ telomerase reactions. This add-back assay was used to demonstrate that Est3 proteins bearing mutations in an invariant arginine residue could fully restore enzyme activity, even though these same mutations confer a profound telomere replication defect in vivo. This demonstrates that Est3 performs a regulatory activity, which is unlinked to any contribution to enzymatic activity.
MATERIALS AND METHODS
Yeast strains
Construction of an est3-Δ derivative of S. castellii
The S. castellii strain Y258, a generous gift by Jure Piskur (33), was modified by transformation with a ura3-Δ::NATMX6 cassette, to create a Ura-derivative. In parallel, the S. castellii EST3 gene was isolated by functional complementation of an S. cerevisiae est3-Δ rad52-Δ strain, using a genomic library constructed by partial Sau3A digestion of S. castellii genomic DNA cloned in BamHI-digested YEP351 (34). A plasmid containing an ∼7-kb genomic insert which included the S. castellii EST3 gene was recovered and used to construct a est3-Δ::KANMX6 derivative with the KAN cassette replacing aa 19–50 of the Est3 ORF (pVL2617). This disruption cassette was used to construct YVL2733, which also contained pVL2722 (YCplac33 containing the S. castellii EST3 gene). Isolates of YVL2733 which had lost the covering EST3 plasmid were generated either by isolating single colonies on plates containing 5-FOA, or by passive loss by screening colonies grown on rich media (YPAD) for loss of the URA3 plasmid. In either protocol, Ura-isolates were propagated for a minimum number of generations prior to preparation of extracts for telomerase assays, to minimize possible complications when preparing extracts from senescing strains.
For co-immunoprecipitation experiments, two isogenic protease-deficient strains (YVL3187 and YVL3188) were constructed with (myc)13::TRP1 tags at the C-terminus of Est1 and Est2, respectively, and with tlc1-Δ148-440 integrated in place of the wild-type TLC1 gene. Both strains contained pVL2076, which expressed Est3-(Flag)3 and either pVL4061 (URA3 CEN tlc1-47) or pVL4062 (URA3 CEN tlc1-59), respectively. The strains used for Figure 6C were constructed similarly, with genomic myc-tagged versions of Est1 or Est2, and containing pVL2076 and pVL4126 (URA3 CEN tlc1-Δ535-707).
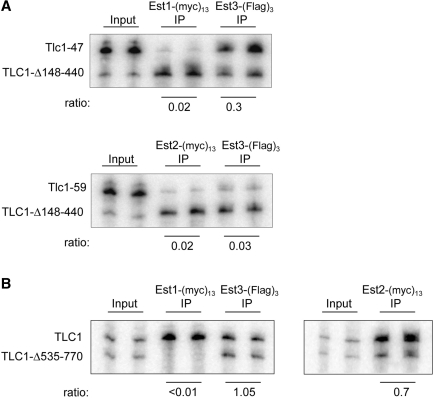
Figure 6.
Association of Est3 with the S. cerevisiae telomerase complex is Est2-dependent. (A) Anti-myc and anti-Flag immunoprecipitations of Est1-(myc)13 and Est3-(Flag)3 (YVL3187, upper panel) or Est2-(myc)13 and Est3-(Flag)3 (YVL3187, lower panel) were examined for association with the indicated TLC1 RNAs by northern analysis, as described previously (39). The efficiency of co-immunoprecipitation of all three Est proteins with the wild-type TLC1 RNA was between 12 and 18%. (B) Anti-myc and anti-Flag immunoprecipitations of Est1-(myc)13 and Est3-(Flag)3, expressed in the same strain, were monitored for association with TLC1 versus TLC1-Δ535-770, in parallel with Est2-(myc)13 association with the same two RNAs.
Telomerase assays
Extracts were minimally fractionated, using a previously published protocol (21). Briefly, cell pellets were lysed under liquid nitrogen by mechanical disruption, with a mortar and pestle, extracts were clarified by two successive 15-min spins at 20 000 g, loaded onto a DEAE column which was washed 2 × with 0.5 M Na acetate and eluted with 0.7 M Na acetate; eluates were desalted and concentrated on a Microcon-30 column. The levels of TLC1 RNA in eluates prepared from wild type and est3-Δ strains was monitored by northern analysis in more than a half dozen experiments, and always found to be equivalent (Figure 4A, Supplementary Figure S3). Freshly prepared eluates were assayed for telomerase activity in 40 µl reactions incubated at 30°C for 30 min, in 50 mM Tris pH 8.0, 100 mM K glutamate, 1 mM spermidine, 2.5 mM MgCl2, 1 mM DDT, 1 µM oligo, 0.25 µM [α-32P]-dGTP (3000 Ci/mol), 5 µM dGTP, 50 µM dCTP and 50 µM dTTP. Oligomers, which were polyacrylamide gel electrophoresis (PAGE)-purified, were obtained from Integrated DNA Technologies; the sequences of the 16-mer oliogomers used in this study are primer 1, (GTCTGGGT)2; primer 2, (GTGTCTGG)2; primer 3,(GGTGTCTG)2; primer 4, (GGGTGTCT)2; primer 5, (TGTCTGGG)2; primer 6, (TGGGTGTC)2; and primer 7, (CTGGGTGT)2. For chain termination assays, dTTP was replaced with 50 µM ddTTP. Completed reactions were treated with 0.5 mg/ml proteinase K + 0.5% SDS at 55°C for 30 min, 500 cpm of a [γ-32P]-labeled oligomer was added as a loading control, and the mixture was extracted with phenol/chloroform; reactions were run on 10% acrylamide plus 7M urea sequencing gels.
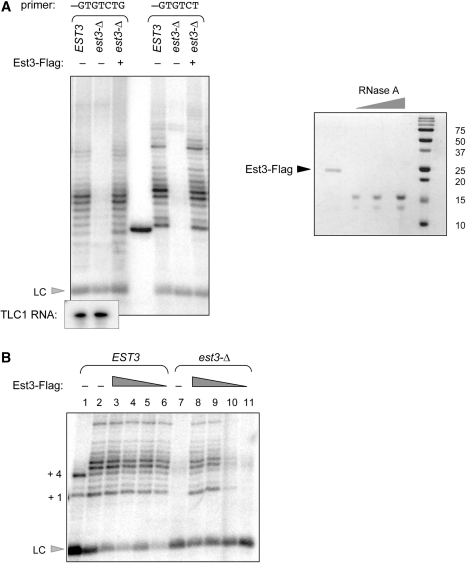
Figure 4.
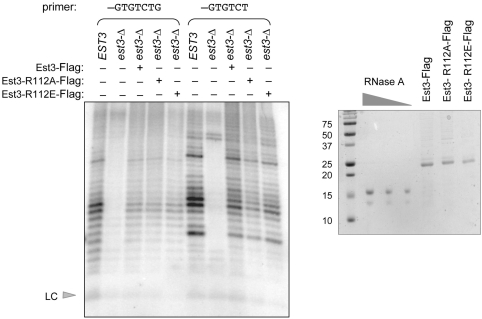
Exogenously added S. castellii Est3 protein can restore telomerase activity. (A) Telomerase activity from wild-type and est3-Δ extracts was examined with primer 3 (first three lanes) and primer 4 (last three lanes); affinity-purified S. castellii Est3-Flag protein was added to the telomerase reactions at ∼500 nM, as estimated by the RNaseA standards (0.5, 0.25 and 0.125 µg) shown in the Coomassie-stained gel on the right. The middle lane contains a [γ-32P]-labeled 15-mer oligomer used as an additional size marker control. Northern analysis indicates the levels of the S. castellii TLC1 RNA in the wild-type and est3-Δ eluates used for this experiment. (B) Ten-fold serial dilutions of affinity-purified S. castellii Est3-Flag protein were added to 40 µl telomerase reactions with wild-type and est3-Δ extracts, assessed with primer 4; LC = loading control ([γ-32P]-labeled 10-mer oligomer); lane 1, telomerase activity in the presence of ddTTP. The concentration of Est3-Flag protein added to lanes 4–6 and lanes 8–11 was 100 nM, 10 nM, 1 nM and 0.1 nM. Since the stoichiometry of the telomerase complex in these reactions was not determined, we do not know whether the recombinant Est3 protein was added in molar excess (nor do we know what fraction of the affinity purified Est3-Flag protein is properly folded and/or biologically active). However, the reduction in telomerase activity between lanes 9 and 10, which correspond to 10 and 1 nM protein, respectively, provides an empirical estimate of the amount of Est3 protein required to restore activity to an est3-Δ extract. This can be used to demonstrate that when 10-fold excess of this empirically determined amount of Est3-Flag protein is added to a wild-type telomerase extract (lane 3), there is no enhancement of activity, when compared to the wild-type telomerase reaction shown in lane 2.
For add-back assays, a frame-shift corrected version of the S. castellii Est3 protein was cloned in pRSET, with a C-terminal Flag epitope (pVL3057). Protein was expressed and affinity purified in BL21(DE3) E. coli, as described previously (35). In brief, cultures were grown to OD600 ∼0.6 and induced with 0.5 mM isopropyl-β-thio galactopyranoside (IPTG) for 3.5 h at 26°C. Pelleted cells were resuspended in lysis buffer, lysed by sonication and cleared by two successive spins at 20 000 g for 10 min. Clarified extracts were incubated with anti-Flag M2 beads (Sigma) for 2–3 h at 4°C, and bound protein was eluted from washed beads with 0.5 µg/ml Flag peptide (Sigma). Protein concentration was assessed on Coomassie-stained 15% sodium dodecyl sulfate (SDS)-PAGE gels by comparison with an RNase A serial dilution series.
1H-15N minimal chemical shift perturbation analysis of Est3
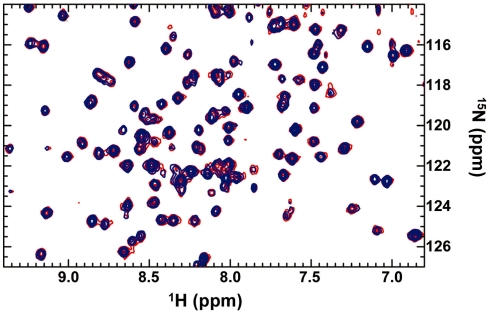
15N-labeled S. cerevisiae Est3 bearing an N-terminal (His)10 tag was expressed in E. coli BL21(DE3) cells in minimal growth medium containing 1.5 g/l of (15NH4)2SO4. Following lysis, the His-tagged protein was isolated with Ni–NTA agarose beads (Qiagen) and size-exclusion chromatography to yield final a sample concentration of ∼200 µM. 1H-15N TROSY-HSQC spectra were collected on Varian Inova 800-MHz nuclear magnetic resonance (NMR) spectrometer at the W.M. Keck High Field NMR facility at the University of Colorado, Boulder, using standard Varian pulse sequences with minor modifications. Overnight spectra were obtained for identical samples of 15N-labeled S. cerevisiae Est3 in the presence and absence of 40-fold molar excess of a 17-mer telomeric oligomer (5′-GTGTGGGTGTGGTGTGG-3′) obtained from Integrated DNA Technologies and used without further purification. Spectra were collected at 25°C equipped with a salt-tolerant HCN cryoprobe. NMR data were processed in NMRPipe and spectra overlaid using CcpNmr analysis software.
RESULTS
Loss of Est3 function in S. castellii results in a telomere replication defect
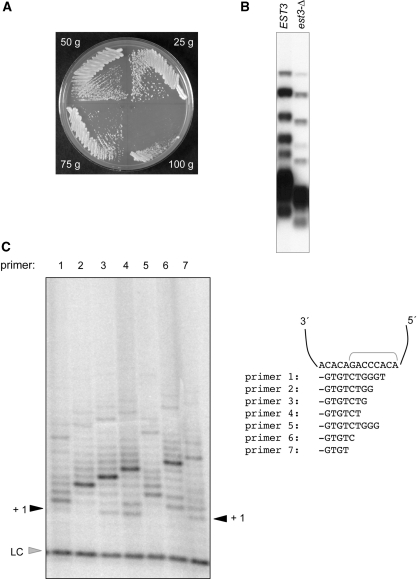
In order to generate an est3-Δ strain of S. castellii, the EST3 gene was cloned by functional complementation of an S. cerevisiae est3-Δ strain (described in ‘Materials and Methods’ section), and used to generate an est3-Δ::kanMX6 disruption cassette. A haploid S. castellii strain was transformed with this cassette and isolates were recovered that had replaced the wild-type EST3 gene with est3-Δ::kanMX6. The resulting S. castellii est3-Δ strain exhibited a senescence phenotype that was strikingly similar to that of an S. cerevisiae strain, which also lacks EST3: a growth defect was apparent by 50–75 generations, with senescence becoming pronounced by 100 generations (Figure 1A). As expected, telomere length was shorter in the est3-Δ strain and showed a progressive decline with continued propagation (Figure 1B and Supplementary Figure S1). These observations, combined with the ability of the S. castellii EST3 gene to partially complement the telomere replication defect of an S. cerevisiae est3-Δ strain (34), indicate that the S. castellii and S. cerevisiae Est3 proteins perform functionally equivalent roles in telomere replication in these two species. The S. castellii est3-Δ strain was maintained with a covering EST3 plasmid; isolates that had newly lost the EST3 plasmid were used for the biochemical experiments presented below.
Figure 1.
An est3-Δ strain of S. castellii is defective for telomere replication. (A) Successive streak-outs of an S. castellii est3-Δ, following loss of a covering EST3 plasmid. (B) Southern blot analysis of genomic DNA from EST3 and est3-Δ strains of S. castellii digested with HindIII and probed with a radiolabeled (TCTGGGTG)10 oligomer. (C) Telomerase activity from a wild-type S. castellii strain, with a panel of 16-mer single-stranded telomeric oligomers; the position of the 3′ terminus of each oligo when annealed with the template region of the S. castellii telomerase RNA is indicated. LC = loading control (a [γ-32P]-labeled 12-mer oligomer).
S. castellii telomerase activity does not exhibit a primer-specific defect in the absence of Est3
To monitor S. castellii telomerase activity, extracts were prepared according to the protocol described by Cohn and Blackburn (21) and monitored for telomerase activity in the presence of radiolabeled dGTP and unlabeled dCTP and dTTP. Single-stranded 16-mer telomeric oligomers could be elongated, in an RNase-sensitive manner, by the addition of >30 nt, with products exhibiting an 8-nt periodicity. This periodicity corresponds to translocation of the elongating primer on the template RNA; the identity of this pause position was confirmed by the pattern of products generated in the presence of the chain-terminating dideoxynucleotide analog ddTTP (ref. 21; Figure 1C; Supplementary Figure S2).
The behavior of telomerase activity from partially fractionated extracts prepared from S. castellii EST3 and est3-Δ strains was assessed with a panel of seven 16-mer telomeric primers (Figure 2A and Supplementary Figure S3). Somewhat surprisingly, enzyme activity from the est3-Δ strain appeared to be substantially diminished, relative to wild-type activity. This was an unexpected result, as telomerase activity could be readily detected in extracts prepared from an S. cerevisiae est3-Δ strain (22; Lee,J.S. and Lundblad,V., unpublished data). The reduction in activity was reproducible, based on more than a dozen experiments that compared independently prepared extracts from wild type and est3-Δ: in every case, elongation of telomeric primers was reduced in the absence of the Est3 telomerase subunit. One possible explanation for the apparent reduction in activity might be the presence of an inhibitor in the est3-Δ extracts that masked enzyme activity; however, addition of an est3-Δ extract to a wild-type extract did not inhibit wild-type telomerase activity (data not shown), arguing against this possibility. Activity was not restored in the est3-Δ extracts when longer primers (32 nt) were used, nor was activity responsive to several variations in reaction conditions (data not shown). The reduction in activity also did not appear to reflect a primer-specific defect, as no notable differences could be observed in the activity of telomerase isolated from an est3-Δ extract with seven primers that spanned the template of the S. castellii telomerase RNA (Figure 2A and Supplementary Figure S3).
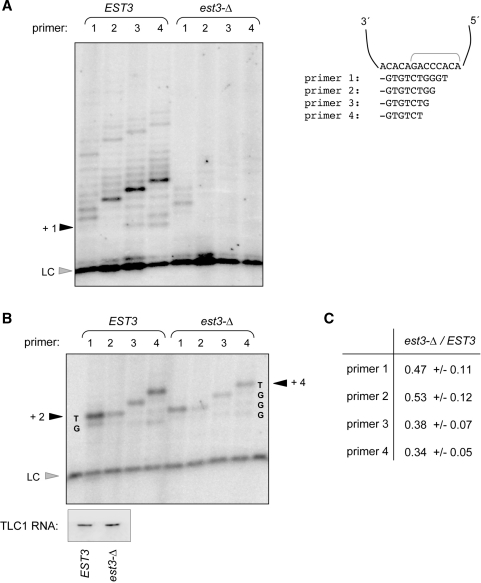
Figure 2.
Assessing S. castellii telomerase activity from wild-type and est3-Δ strains. (A) Telomerase assay, with primers 1–4, comparing enzyme activity from partially fractionated wild-type and est3-Δ extracts, assayed as described in Figure 1C and ‘Materials and Methods’ section. (B) Telomerase assay with the same set of primers, in the presence of the chain-terminating dideoxynucleotide analog ddTTP. Northern analysis indicates the levels of the S. castellii TLC1 RNA in the wild-type and est3-Δ eluates used for this experiment. The relative level of activity with these four primers was reproducible; for example, signal with primer 1 was always ∼2-fold more intense than that of primer 2, with telomerase isolated from both wild-type and est3-Δ strains. (C) The ratio of signal recovered from est3-Δ extracts, relative to EST3, for primers 1 through 4. Signal was calculated by monitoring the + 2 band for primers 1 and 2, and the +3 and +4 bands for primers 3 and 4, respectively, and normalizing to the loading control. Results are calculated from three independent experiments, using freshly prepared extracts from EST3 and est3-Δ strains for each repeat.
As an alternative means of assessing the effects of the Est3 protein on telomerase, enzyme activity was monitored in a chain termination assay, in the presence of either ddTTP or ddGTP. This assay has the advantage that reaction products are concentrated in essentially a single band, providing an easier direct comparison between mutant and wild-type extracts. Telomerase activity was assayed under exactly the same conditions as in Figure 2A, except that dTTP or dGTP was replaced by the equivalent dideoxynucleotide analog. Figure 2B shows the results with four primers, where elongation was terminated by the addition of ddTTP. This more sensitive assay clearly demonstrated that the telomerase preparations from the est3-Δ extracts were catalytically active, consistent with prior results in S. cerevisiae (22). However, S. castellii telomerase activity displayed a modest decrease in activity in the absence of the Est3 protein: activity was reduced 2–3-fold in est3-Δ extracts with each of the four oligos, relative to wild type (Figure 2C). This behavior was not specific to termination by ddTTP; when elongation by primers 5 and 6 in the presence of ddGTP was examined, a similar ∼2-fold decline in activity in the absence of Est3 was observed (data not shown). Thus, the six primers examined in this chain termination assay behaved the same in response to loss of the Est3 protein. This argues against a role for S. castellii Est3 in mediating primer specificity in telomerase activity, as was previously observed for telomerase activity isolated from est3-Δ derivatives of C. albicans (29).
The Est3 protein does not bind telomeric single-strand DNA
Although telomerase lacking the Est3 subunit did not exhibit a primer-specific defect, loss of the Est3 protein might confer a general reduction in the ability of telomerase to interact with telomeric primers. This possibility was raised by a prior study which reported that an S. cerevisiae GST-Est3 fusion protein, when expressed in E. coli and affinity purified, exhibited an apparent (albeit weak) ability to bind telomeric DNA substrates, as assessed by gel shifts (36). We re-examined this, using NMR chemical shift perturbation, which should reveal chemical shift changes at proximal amide sites, as well as any sites that undergo a conformational change, upon binding of DNA. This has several advantages over a gel-shift assay, particularly when probing for weak binding. NMR is very sensitive to even a small degree of binding, since it is a true equilibrium technique and not impacted by fast off rates characteristic of weak binding interactions. In addition, this approach provides the ability to probe for binding at very high concentrations of protein and single-stranded DNA. Finally, because the spectrum of the Est3 protein is monitored for a change upon addition of single-stranded DNA, this unequivocally establishes that we are probing the activity of the correct protein. In contrast, when assessing a very weak DNA-binding activity by gel shifts, it is difficult to rule out that a positive result is not due to a contaminating protein in the extract.
The 15N-HSQC spectra of 15N-labeled S. cerevisiae Est3 (200 µM) were obtained in the presence and absence of a 17-mer telomeric oligomer. Superposition of these spectra revealed no chemical shift changes at any of the 160 resolved crosspeaks in the protein upon addition of single-stranded DNA, even at 8 mM (Figure 3). We conservatively estimate that we would be able to detect as little as 10% bound species, placing a minimal limit on the binding affinity of Est3 for single-stranded DNA in the high millimolar range. These observations indicate that any interaction between Est3 and telomeric single-stranded DNA is weaker than ∼10 mM, arguing against a role for the Est3 protein in binding telomeric substrates.
Figure 3.
The S. cerevisiae Est3 protein does not bind single-stranded telomeric DNA. Superposition of 1H-15N HSQC-TROSY spectra of 15N-labeled S. cerevisiae Est3 (200 µM) in the presence (blue) and absence (red) of 40-fold excess of a 17-mer single-strand telomeric oligomer.
Telomerase activity in est3-Δ extracts can be restored by exogenously added Est3 protein
The reduced activity observed in the S. castellii est3-Δ extracts might be due to altered assembly of the telomerase enzyme in the absence of Est3; for example, the Est3 protein could be required to mediate binding of Est2 to the TLC1 RNA or otherwise contribute to RNP assembly. If so, this predicts that addition of purified Est3 protein to extracts prepared from an est3-Δ strain would not restore telomerase activity. To test this, the S. castellii Est3 protein, bearing a C-terminal Flag epitope, was expressed in E. coli and purified by immunoprecipitation. When this affinity-purified Est3-Flag protein preparation was added to extracts prepared from the S. castellii est3-Δ strain, telomerase activity was restored to wild type levels (Figure 4A). This did not appear to be due to stimulatory effects of the exogenously added Est3 protein, as no enhancement of enzyme activity was observed when the Est3 protein was added to extracts prepared from a wild-type extract, even at >10-fold excess of the amounts required to restore activity to an est3-Δ extract (Figure 4B). This result indicates that the DEAE eluate prepared from both wild-type and est3-Δ extracts contains an intact catalytic core (which is also consistent with the fact that the level of the TLC1 RNA subunit was equivalent in telomerase preparations from EST3 and est3-Δ strains; Figure 4A and Supplementary Figure S3).
This assay was also dependent on whether the exogenously added Est3 protein was capable of associating with the telomerase RNP, as revealed by the behavior of two mutant versions of the S. castellii Est3 protein (Est3-D168A and Est3-D168R) in this assay. These two mutations were chosen based on prior work examining the effect of missense mutations in the S. cerevisiae Est3 protein on telomerase association, using co-immunoprecipitation with the TLC1 RNA (26). In these experiments, the S. cerevisiae Est3-D164A protein (D164 is the equivalent residue to D168 in S. castellii) retained wild-type levels of association, whereas the Est3-D164R protein had a greatly reduced association with the S. cerevisiae telomerase RNP (20; Supplementary Figure S4A). The ability to form a complex with the telomerase RNP was mirrored in the S. castellii add-back assay: the Est3-D168A protein behaved like wild-type Est3, with regard to restoration of telomerase activity, whereas the Est3-D168R was unable to restore activity (Supplementary Figure S4B).
The above results argue that this add-back assay provides a sensitive means of probing whether mutant Est3 proteins that still retain association with telomerase would exhibit a reduction in telomerase activity, similar to that displayed by the est3-Δ null strain. We therefore examined the behavior of a fully defective Est3 protein, bearing a separation-of-function mutation in a highly conserved arginine residue (R110 or R112, in the S. cerevisiae or S. castellii proteins, respectively). Previous work demonstrated that S. cerevisiae est3-R110E and est3-R110A strains exhibit a pronounced in vivo telomere replication defect, even though the Est3-R110E and Est3-R110A mutant proteins, which are expressed at levels comparable to that of wild type, remain bound to the telomerase complex (26). The severity of the defect of the est3-R110E and est3-R110A mutant strains—which is equivalent to that of the null phenotype—indicate that mutations in this residue impair a key biochemical property of the Est3 protein; consistent with this, the S. castellii Est3-R112A was also defective in vivo (Supplementary Figure S5). Therefore, we examined S. castellii telomerase activity following addition of affinity purified S. castellii Est3-R112E-Flag and Est3-R112A-Flag mutant proteins to an est3-Δ extract. As shown in Figure 5 and Supplementary Figure S5, these two mutant proteins were indistinguishable from wild type in their ability to restore telomerase activity. Thus, we conclude that the surface of Est3 that is defined by this arginine residue (and presumably adjacent residues) performs a function which is essential for Est3 function but which is independent of telomerase activity.
Figure 5.
The invariant R112 (R110 in S. cerevisiae) of Est3 is not required for telomerase activity. Telomerase activity from wild type and est3-Δ extracts were examined with primers 3 and 4 (lanes 1–5 and lanes 6–10, respectively), with ∼500 nM of wild type S. castellii Est3, Est3-R112A and Est3-R112E added back to the est3-Δ telomerase reactions, as assessed by the Coomassie-stained gel shown on the right. Supplementary Figure S5, which shows a serial dilution series of these three proteins added back to est3-Δ telomerase reactions, provides further support that these three proteins exhibit equal activity in restoring enzyme activity.
The Est3 protein associates with telomerase in an Est2-dependent manner
The previously shown results raise the possibility that the apparent reduction in telomerase activity in the S. castellii est3-Δ strain might not be the result of the loss of a specific biochemical property of the Est3 protein. An alternative possibility is that the effects are due to non-specific effects on the catalytic core of the enzyme, as a result of (partial) destabilization of the Est2 catalytic subunit when the Est3 protein is not present. Such a model implies a direct interaction between the Est2 and Est3 proteins. However, the prior published literature is contradictory on this point. Experiments that examined the pattern of co-immunoprecipitation of S. cerevisiae Est3 with TLC1, in the presence or absence of the Est1 and Est2 subunits, led to the conclusion that association of Est3 with the telomerase complex was Est2-dependent and Est1-independent (11). However, this observation was contradicted by a subsequent report, which concluded that association of the S. cerevisiae Est3 subunit with the telomerase complex was dependent on Est1 (37).
Therefore, we reexamined this issue, using several approaches. First, we examined the pattern of association of the three Est proteins in S. cerevisiae cells expressing both the wild-type TLC1 RNA and either of two mutant RNAs (tlc1-47 and tlc1-59). A number of studies have established that Est1 and Est2 independently associate with TLC1, through separate structures on the RNA. Est1 interacts with TLC1 through a bulged stem–loop, and removal of this 5-nt bulge (referred to as tlc1-47 here) eliminates the ability of Est1 (but not Est2) to bind to the telomerase RNA (16). Est2 interacts with a separate region of the RNA (38), and association of Est2 (but not Est1) is eliminated by a 65-nt deletion (tlc1-59) in this region (39). If the Est3 protein binds the telomerase RNP through an interaction with Est2, as originally proposed, then Est3 should exhibit the same pattern of association with TLC1-47 and TLC1-59 as the Est2 protein. To test this, two S. cerevisiae strains were constructed (YVL3187 and YVL3188), which co-expressed a version of the wild-type TLC1 RNA (TLC1-Δ148-440) as well as either of the two mutant RNAs. The deletion of ∼350 non-essential nucleotides allows the TLC1-Δ148-440 RNA to be readily distinguished from the full-length mutant RNAs; as shown previously, Est1 and Est2 retain full association with the TLC1-Δ148-440 RNA (38). These two strains also contained tagged versions of the three Est proteins: YVL3187, which expressed the mutant TLC1-47 RNA, contained Est1-(myc)13 and Est3-(Flag)3, whereas YVL3188, which expressed the mutant TLC1-59, contained Est2-(myc)13 and Est3-(Flag)3. Parallel anti-myc and anti-FLAG immunoprecipitations were performed with both strains, and the immunoprecipitates were examined by northern analysis for the pattern of association with the two sets of TLC1 RNAs present in each strain. As expected, the Est1 and Est2 proteins exhibited a roughly 50-fold reduction in their ability to associate with TLC1-47 or TLC1-59, respectively, relative to TLC1-Δ148-440 (39). Notably, Est3 exhibited the same behavior as the Est2 protein, as would be predicted if these two proteins directly interact: co-precipitation of the Est3 protein with TLC1-59 was dramatically reduced, whereas Est3 retained association with the TLC1-47 RNA (Figure 6A).
We also repeated an experiment from the study by the Friedman laboratory (37), which examined the relative association of the Est1 and Est3 proteins with a different mutant RNA, TLC1-Δ535-770, which is impaired for binding the Est1 protein, but not the Est2 protein (38). Our experimental design employed the same strategy as that employed above: two S. cerevisiae strains were constructed which co-expressed tagged versions of Est1 and Est3, or Est2 and Est3, as well as the full-length TLC1 and TLC1-Δ535-770 RNAs. Immunoprecipitation, followed by northern analysis, demonstrated that Est3 retained wild-type levels of association with TLC1-Δ535-770, whereas, within the exact same strain, Est1 had lost association with TLC1-Δ535-770 (Figure 6B). As expected, the Est2 protein could be co-immunoprecipitated with both the wild type TLC1 and TLC1-Δ535-770 RNAs. These observations with TLC1-Δ535-770 are consistent with our original report (11), as well as the experiments with the mutant TLC1-47 and TLC1-59 RNAs shown in Figure 6A, but inconsistent with prior results by the Friedman laboratory (37), which reported that the Est3 protein had lost association with the TLC1-Δ535-770 RNA. We suspect that the experimental design employed here, where the pattern of Est1 and Est3 association with mutant and wild-type RNAs was examined in the same strain, which provides an internal control to ensure that the predicted results for Est1 and Est2 were as expected, may explain the differing conclusions.
DISCUSSION
This study, together with our previous investigations on the role of the Est3 telomerase subunit in telomere function, combines the strengths of two related model organisms, S. cerevisiae and S. castellii. Est3 is a rapidly diverging protein at the primary amino acid sequence (26), such that the percent identity between the S. cerevisiae and S. castellii proteins is only 46%. Nevertheless, the S. castellii EST3 gene partially compensates for the loss of Est3 function in S. cerevisiae (34), supporting the idea that observations in both organisms can contribute to models about the role of Est3 in telomere replication. Therefore, we exploited the ability of the S. castellii telomerase to elongate an oligo through several rounds of translocation, in contrast to the non-processive activity of the S. cerevisiae telomerase enzyme (21). Our initial observations, using the standard telomerase elongation assay, indicated that the S. castellii Est3 protein might make a substantial contribution to enzyme activity. However, when telomerase activity was monitored in the presence of a dideoxynucleotide analog, the consequence of the loss of the Est3 protein was much more modest. We suggest that the difference in the results obtained in these two assay conditions can be explained by the cumulative effects of a modest reduction at each nucleotide addition step, which can translate to an apparent dramatic reduction in telomerase activity. This raises a cautionary note when interpreting potential differences in telomerase activity between two different genotypes, when relying solely on assay conditions that monitor multiple rounds of elongation.
This study raises the issue of why an S. castellii telomerase complex lacking the Est3 subunit exhibits a decrease in nucleotide addition. One possibility is that this effect is the result of non-specific destabilization of the complex due to the absence of a telomerase subunit, as mentioned already. Alternatively, the Est3 protein may contribute a specific biochemical property to enzyme catalysis, for example as a processivity factor. The S. castellii telomerase assay provides a sensitive assay for potentially differentiating between these possible models. Since the ability to add telomeric repeats onto a primer through multiple rounds of elongation is fully restored by the exogenous addition of Est3 protein affinity purified from E. coli, this provides a biochemical means of assessing the properties of separation-of-function mutations in Est3. Such an approach alleviates the potential concern that a defect in this assay is simply due to destabilization in the absence of the Est3 subunit, rather than loss of a specific property such as processivity. To pursue this, we turned to a cluster of residues on the predicted surface of the S. cerevisiae Est3 protein which exhibit an in vivo telomere replication defect when mutated (26). In particular, strains expressing missense mutations in the invariant R110 residue (R112 in S. castellii) display an Est-phenotype that is indistinguishable from that of an est3-Δ null strain, even though the mutant Est3-R110A and Est3-R110E proteins remain associated with the telomerase RNP at wild-type levels. Furthermore, the properties of strains that overexpress either of these two mutant proteins are consistent with the proposal that mutations at amino acid R110 confer a defect in a specific biochemical property of the Est3 protein, rather than a defect due to an unfolded protein (26). Notably, however, addition of the S. castellii Est3-R112A or Est3-R112E proteins back to a telomerase extract prepared from an est3-Δ strain fully restored telomerase activity. Thus, we conclude that Est3 performs an essential function in telomere replication, as defined by R110 (or R112 in S. castellii), which is distinct from any potential contribution that Est3 might make to enzyme activity.
The observations reported in this study differ in one key aspect from prior work investigating the role of Est3 in C. albicans telomerase activity. In contrast to the lack of primer-specific defects in the absence of Est3 reported here, the Est3-deficient C. albicans telomerase exhibits substantial primer-specific differences (29), even with primers that align at comparable positions on the templates of the C. albicans and S. castellii telomerase RNAs (Supplementary Figure S6). This disparity may be a reflection of the fact that the C. albicans telomerase assays monitored addition onto primers that were only 12 nt in length. Previous work has shown that the Tetrahymena enzyme, which is normally highly processive in vitro, is non-processive with 10–12-nt primers (40). Thus, if the C. albicans enzyme active site is partially destabilized by the loss of the Est3 telomerase subunit, such an effect may be further exaggerated with very short telomeric primers. The unusually long (28 nt) length of the C. albicans RNA template (41) may also impose special requirements that differentiate the behavior of the C. albicans telomerase from telomerases of other species.
This study also addresses several contradictory observations, in S. cerevisiae and C. albicans, with regard to how Est3 associates with the telomerase complex (11,29,37). A common approach that has been used in S. cerevisiae has been to examine the pattern of association of tagged versions of Est1, Est2 and Est3 with TLC1 or with each other, comparing wild type with strains deleted for one of the EST genes (11,17,37). However, past work in our lab has indicated that such experiments can be problematic in telomerase-defective strains that are undergoing senescence, as false negatives can result from immunoprecipitations performed with cultures that are severely senescent (42), presumably due to a high degree of cell inviability. Thus, such experiments require strain manipulations to minimize senescence, as well as ensuring that the immunoprecipitations are performed well above detection limits. To alleviate this potential problem, we instead employed a set of strains that expressed mutant versions of TLC1 that were impaired for association with either Est1 or Est2, in the presence of a functional copy of TLC1, thereby avoiding any problems with senescence. Since Est1 and Est2 independently associate with TLC1, via separate structures on the RNA, this provides an internal control for assessing whether Est3 is dependent on either Est1 or Est2 for its association. The results of this experiment clearly establish that Est3 relies on Est2 in order to assemble with the telomerase complex, suggesting that Est2 and Est3 directly interact. If so, this provides a potential explanation for the reduction in telomerase activity observed when the Est3 telomerase subunit is missing.
However, the results presented in this study do not rule out the possibility that Est3 contributes a specific biochemical activity, such as processivity. Speculation about this possibility is driven by prior observations indicating that the Est3 protein consists of a predicted OB-fold, which exhibits potential structural similarity to the OB-fold found in the mammalian TPP1 protein (26,27). TPP1 is one subunit of a telomere end-binding complex, called POT1/TPP1 (43), and in vitro studies have shown that this heterodimeric complex acts as a processivity factor for human telomerase (44). However, there are some notable differences between Est3 and TPP1. Defects in these two proteins result in substantially different in vivo consequences (45–47), consistent with the fact that Est3 is a subunit of telomerase, whereas TPP1 is a subunit of an end protection complex. Furthermore, although the very small Est3 telomerase protein consists solely of the predicted OB-fold, TPP1 is a larger protein possessing additional domains. These distinctions argue that, despite the predicted structural similarities in their OB-folds, Est3 and TPP1 are not functional orthologs of each other and instead may reflect the repeated usage of OB-folds in telomere-associated proteins.
Although the behavior of the mutant Est3 proteins that were analyzed in this study does not support a role for Est3 in processivity, it is possible that additional function(s) of Est3 are yet to be identified, particularly since the entire surface of Est3 has not yet been completely surveyed by mutagenesis (26). If so, the S. castellii telomerase assay presented here provides a sensitive assay for assessing whether mutant Est3 proteins defective for a postulated second function will influence the ability of telomerase to elongate telomeric primers in vitro.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health AG11728 (to V.L.); National Science Foundation 0617956 (to D.S.W.); and National Institutes of Health T32 GM08759 (to T.R.). Funding for open access charge: National Institutes of Health AG11728.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Jure Piskur for providing us with the initial S. castellii strain used in these studies, and we thank Geoffrey Armstrong for assistance with the NMR data collection.
REFERENCES
- 1.Smogorzewska A, de Lange T. Regulation of telomerase by telomeric proteins. Ann. Rev. Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- 2.Marcand S, Brevet V, Gilson E. Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J. 1999;18:3509–3519. doi: 10.1093/emboj/18.12.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teixeira MT, Arneric M, Sperisen P, Lingner J. Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell. 2004;117:323–335. doi: 10.1016/s0092-8674(04)00334-4. [DOI] [PubMed] [Google Scholar]
- 4.Marcand S, Gilson E, Shore D. A protein-counting mechanism for telomere length regulation in yeast. Science. 1997;275:986–990. doi: 10.1126/science.275.5302.986. [DOI] [PubMed] [Google Scholar]
- 5.Smogorzewska A, van Steensel B, Bianchi A, Oelmann S, Schaefer MR, Schnapp G, de Lange T. Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol. 2000;20:1659–1668. doi: 10.1128/mcb.20.5.1659-1668.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- 7.Chang M, Arneric M, Lingner J. Telomerase repeat addition processivity is increased at critically short telomeres in a Tel1-dependent manner in Saccharomyces cerevisiae. Genes Dev. 2007;21:2485–2494. doi: 10.1101/gad.1588807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nugent CI, Hughes TR, Lue NF, Lundblad V. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 9.Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 10.Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell. 2008;132:945–957. doi: 10.1016/j.cell.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes TR, Evans SK, Weilbaecher RG, Lundblad V. The Est3 protein is a subunit of yeast telomerase. Curr. Biol. 2000;10:809–812. doi: 10.1016/s0960-9822(00)00562-5. [DOI] [PubMed] [Google Scholar]
- 12.Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 13.Singer MS, Gottschling DE. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 14.Zappulla DC, Cech TR. Yeast telomerase RNA: a flexible scaffold for protein subunits. Proc. Natl Acad. Sci. USA. 2004;101:10024–10029. doi: 10.1073/pnas.0403641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dandjinou AT, Levesque N, Larose S, Lucier JF, Abou Elela S, Wellinger RJ. A phylogenetically based secondary structure for the yeast telomerase RNA. Curr. Biol. 2004;14:1148–1158. doi: 10.1016/j.cub.2004.05.054. [DOI] [PubMed] [Google Scholar]
- 16.Seto AG, Livengood AJ, Tzfati Y, Blackburn EH, Cech TR. A bulged stem tethers Est1p to telomerase RNA in budding yeast. Genes Dev. 2002;16:2800–2812. doi: 10.1101/gad.1029302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans SK, Lundblad V. The Est1 subunit of Saccharomyces cerevisiae telomerase makes multiple contributions to telomere length maintenance. Genetics. 2002;162:1101–1115. doi: 10.1093/genetics/162.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beernink HT, Miller K, Deshpande A, Bucher P, Cooper JP. Telomere maintenance in fission yeast requires an Est1 ortholog. Curr. Biol. 2003;13:575–580. doi: 10.1016/s0960-9822(03)00169-6. [DOI] [PubMed] [Google Scholar]
- 19.Leonardi J, Box JA, Bunch JT, Baumann P. TER1, the RNA subunit of fission yeast telomerase. Nat. Struct. Mol. Biol. 2008;15:26–33. doi: 10.1038/nsmb1343. [DOI] [PubMed] [Google Scholar]
- 20.Webb CJ, Zakian VA. Identification and characterization of the Schizosaccharomyces pombe TER1 telomerase RNA. Nat. Struct. Mol. Biol. 2008;15:34–42. doi: 10.1038/nsmb1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohn M, Blackburn EH. Telomerase in yeast. Science. 1995;269:396–400. doi: 10.1126/science.7618104. [DOI] [PubMed] [Google Scholar]
- 22.Lingner J, Cech TR, Hughes TR, Lundblad V. Three Ever Shorter Telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc. Natl Acad. Sci. USA. 1997;94:11190–11195. doi: 10.1073/pnas.94.21.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pennock E, Buckley K, Lundblad V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104:387–396. doi: 10.1016/s0092-8674(01)00226-4. [DOI] [PubMed] [Google Scholar]
- 24.Bianchi A, Negrini S, Shore D. Delivery of yeast telomerase to a DNA break depends on the recruitment functions of Cdc13 and Est1. Mol. Cell. 2004;16:139–146. doi: 10.1016/j.molcel.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 25.Chan A, Boule JB, Zakian VA. Two pathways recruit telomerase to Saccharomyces cerevisiae telomeres. PLoS Genet. 2008;4:e1000236. doi: 10.1371/journal.pgen.1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee J, Mandell EK, Tucey TM, Morris DK, Lundblad V. The Est3 protein associates with yeast telomerase through an OB-fold domain. Nat. Struct. Mol. Biol. 2008;15:990–997. doi: 10.1038/nsmb.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu EY, Wang F, Lei M, Lue NF. A proposed OB-fold with a protein-interaction surface in Candida albicans telomerase protein Est3. Nat. Struct. Mol. Biol. 2008;15:985–989. doi: 10.1038/nsmb.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh SM, Lue NF. Ever shorter telomere 1 (EST1)-dependent reverse transcription by Candida telomerase in vitro: evidence in support of an activating function. Proc. Natl Acad. Sci. USA. 2003;100:5718–5723. doi: 10.1073/pnas.1036868100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsu M, Yu EY, Singh SM, Lue NF. Mutual dependence of Candida albicans Est1p and Est3p in telomerase assembly and activation. Eukaryot. Cell. 2007;6:1330–1338. doi: 10.1128/EC.00069-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kabaha MM, Zhitomirsky B, Schwartz I, Tzfati Y. The 5' arm of Kluyveromyces lactis telomerase RNA is critical for telomerase function. Mol. Cell. Biol. 2008;28:1875–1882. doi: 10.1128/MCB.01683-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 32.Greider CW. Telomerase is processive. Mol. Cell. Biol. 1991;11:4572–4580. doi: 10.1128/mcb.11.9.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petersen RF, Langkjaer RB, Hvidtfeldt J, Gartner J, Palmen W, Ussery DW, Piskur J. Inheritance and organisation of the mitochondrial genome differ between two Saccharomyces yeasts. J. Mol. Biol. 2002;318:627–636. doi: 10.1016/S0022-2836(02)00037-2. [DOI] [PubMed] [Google Scholar]
- 34.Chappell AS. Houston, Texas: Baylor College of Medicine; 2004. Molecular and phylogenetic analysis of the Saccharomyces cerevisiae EST3 and TLC1 telomerase subunits. Thesis Dissertation. [Google Scholar]
- 35.Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. RPA-like proteins mediate yeast telomere function. Nat. Struct. Mol. Biol. 2007;14:208–214. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- 36.Sharanov YS, Zvereva MI, Dontsova OA. Saccharomyces cerevisiae telomerase subunit Est3p binds DNA and RNA and stimulates unwinding of RNA/DNA heteroduplexes. FEBS Lett. 2006;580:4683–4690. doi: 10.1016/j.febslet.2006.07.048. [DOI] [PubMed] [Google Scholar]
- 37.Osterhage JL, Talley JM, Friedman KL. Proteasome-dependent degradation of Est1p regulates the cell cycle-restricted assembly of telomerase in Saccharomyces cerevisiae. Nat. Struct. Mol. Biol. 2006;13:720–728. doi: 10.1038/nsmb1125. [DOI] [PubMed] [Google Scholar]
- 38.Livengood AJ, Zaug AJ, Cech TR. Essential regions of Saccharomyces cerevisiae telomerase RNA: separate elements for Est1p and Est2p interaction. Mol. Cell. Biol. 2002;22:2366–2374. doi: 10.1128/MCB.22.7.2366-2374.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chappell AS, Lundblad V. Structural elements required for association of the Saccharomyces cerevisiae telomerase RNA with the Est2 reverse transcriptase. Mol. Cell. Biol. 2004;24:7720–7736. doi: 10.1128/MCB.24.17.7720-7736.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee MS, Blackburn EH. Sequence-specific DNA primer effects on telomerase polymerization activity. Mol. Cell. Biol. 1993;13:6586–6599. doi: 10.1128/mcb.13.10.6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsu M, McEachern MJ, Dandjinou AT, Tzfati Y, Orr E, Blackburn EH, Lue NF. Telomerase core components protect Candida telomeres from aberrant overhang accumulation. Proc. Natl Acad. Sci. USA. 2007;104:11682–11687. doi: 10.1073/pnas.0700327104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Evans SK. Houston, Texas: Baylor College of Medicine; 2000. Analysis of EST1 and CDC13 as regulators of telomerase access in Saccharomyces cerevisiae. Thesis Dissertation. [Google Scholar]
- 43.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 44.Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 45.Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hockemeyer D, Palm W, Else T, Daniels JP, Takai KK, Ye JZ, Keegan CE, de Lange T, Hammer GD. Telomere protection by mammalian Pot1 requires interaction with Tpp1. Nat. Struct. Mol. Biol. 2007;14:754–761. doi: 10.1038/nsmb1270. [DOI] [PubMed] [Google Scholar]
- 47.Miyoshi T, Kanoh J, Saito M, Ishikawa F. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science. 2008;320:1341–1344. doi: 10.1126/science.1154819. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.