Abstract
Using libraries of replication origins generated previously, we identified three clones that supported the autonomous replication of their respective plasmids in transformed, but not in normal cells. Assessment of their in vivo replication activity by in situ chromosomal DNA replication assays revealed that the chromosomal loci corresponding to these clones coincided with chromosomal replication origins in all cell lines, which were more active by 2–3-fold in the transformed by comparison to the normal cells. Evaluation of pre-replication complex (pre-RC) protein abundance at these origins in transformed and normal cells by chromatin immunoprecipitation assays, using anti-ORC2, -cdc6 and -cdt1 antibodies, showed that they were bound by these pre-RC proteins in all cell lines, but a 2–3-fold higher abundance was observed in the transformed by comparison to the normal cells. Electrophoretic mobility shift assays (EMSAs) performed on the most efficiently replicating clone, using nuclear extracts from the transformed and normal cells, revealed the presence of a DNA replication complex in transformed cells, which was barely detectable in normal cells. Subsequent supershift EMSAs suggested the presence of transformation-specific complexes. Mass spectrometric analysis of these complexes revealed potential new protein players involved in DNA replication that appear to correlate with cellular transformation.
INTRODUCTION
According to the replicon model, origins of DNA replication are defined by a specific DNA sequence, termed the replicator, and an initiator protein that binds to the origin (1). There is an estimated number of 104–106 replication origins per mammalian cell (2,3), clusters of which are activated at different times in S-phase and are replicated in a defined spatial and temporal order (4). Control of replication frequency and timing is exerted at the level of initiation (5). For the faithful duplication of DNA and a successful completion of one round of the cell cycle, eukaryotic cells require precise orchestration of the actions of replication proteins (6). Chromosomes replicate their DNA in units called replicons, each containing one functional origin of DNA replication (initiation start site) (2). The average size of a replicon varies from 10 to 300 kb, depending on the stage of development, growth conditions or cell transformation status (3,7,8). In the early stages of embryonic development, when fast growth occurs, the replicon size is much smaller than in somatic cells and hence the number of initiation sites may be as much as 10-fold greater (4,9). As cellular transformation and tumor progression are thought to resemble a return to the early stages of embryonic development (9), a comparative analysis of the activity of replication origins between tumor and normal cells may give us insight into the mechanisms that regulate the initiation of DNA replication in normal cells and how they may become deregulated by the transformation process.
Cellular transformation has been shown to modify the regulation of origin activation, resulting in differential origin usage (10–13) as well as a decrease in the average replicon size by approximately half (14,15). Furthermore, transformed cells exhibit a 2–10-fold increase of single-strand nuclease-sensitive regions, consistent with more origins being activated (16), while a polarity or position change of replication initiation was also observed with transformation (17). Some of the studies found the organization of DNA replication sites to be fundamentally different in normal compared to immortalized cell lines (18), but others observed no such differences (19,20).
Interestingly, the parameters governing replication kinetics are conserved between normal and transformed cells (21). A different chromatin organization and rearrangements as well as differential nuclease sensitivity between normal and transformed cells throughout the progression of S-phase have been previously reported (22,23). Furthermore, the replication timing of homologous loci was found to be more asynchronous in samples derived from transformed cells relative to their normal counterparts (24). In addition, overexpression of ORC subunits and increased ORC–chromatin association was observed in transformed compared to normal cells (25). These findings indicate a complex influence of cellular transformation on the expression and regulation of ORC subunits, extending the potential link between transformation and deregulation of pre-RC proteins as well as the subsequent pathways they affect (26–28)). Consequently, it is reasonable to suppose that more origins of DNA replication are used (activated) in transformed than normal cells, hinting at the existence of tumor-specific origins.
To date little is known about differential origin usage between normal and transformed cells. Cellular transformation resulted in a 2-fold increase of initiation sites (origins) (14,15) as well as a 2-fold increase in origin activity in transformed compared to normal cells, such as at the NOA3 origin (10), the origin at the c-myc locus (11,12), and six origins along an ∼211-kb stretch on human chromosome 19q13 (13). It was previously reported that in normal skin fibroblasts (NSFs) two prominent replication initiation sites were detected in a 5-kb region encompassing the ARSH1 locus, whereas in HeLa cells, multiple replication initiation sites were detected within the same region (29). Similarly, when analyzing initiation sites in the vicinity of the putative ribulose-5-phosphate 3-epimerase gene, NSF and normal breast epithelial cells (MCF10) had one prominent initiation site in a specific location, while HeLa cells revealed a more zonal pattern of initiation within the same region (30). There are origins, however, whose initiation activity remains unaltered in the transformed state, such as the well-characterized origin at the lamin B2 locus (31), the origin at the β-globin locus and the 343 and S14 origins (10). Altogether, these results suggest that there are at least three subsets of origins, those that are normal and remain unchanged, those with increased activity in transformed cells and those that are activated exclusively in tumor cells.
In this study, we searched for transformation-specific origins by examining the replication activity of several clones obtained from previously generated libraries enriched for human replication origins (32). Three clones were able to replicate episomally solely in transformed cells, but all three coincided with chromosomal origins of DNA replication in both transformed and normal cells, albeit with increased activity in the transformed cells. Electrophoretic mobility shift assays (EMSAs) using the most efficiently replicating clone revealed transformation-specific complexes, which were analyzed by mass spectrometry and identified potential new proteins involved in DNA replication that correlate with cellular transformation.
MATERIALS AND METHODS
Plasmid constructs
Plasmid constructs used in the transient replication assays, such as pA3/4, pLB2 and pLB2C1 were described previously (13), pCLONE 3, pCLONE 13 and pCLONE 32 were prepared by directionally cloning specifically designed polymerase chain reaction (PCR) products (Supplementary Table S1) prepared using a high fidelity Pfu DNA polymerase (Promega, San Luis Obispo, CA, USA) into the BamHI and EcoRI sites of the pBluescript vector (Stratagene, La Jolla, CA, USA), using a standard protocol (33). Supercoiled plasmid DNA of the resulting clones as well as the pM1 SEAP vector (Roche Molecular Biochemicals, Indianapolis, IN, USA) was prepared using the Qiagen Maxiprep kit (QIAGEN, Mississauga, ON, Canada) according to manufacturer’s specifications, and sequenced.
Cell culture and FACS analysis
HeLa, NSF (primary NSFs), WI38 (human lung embryo fibroblasts) and WI38 VA13 2RA (WI38 transformed with SV40 virus) were acquired from American Type Culture Collection (Manassas, VA, USA) and cultured in α-minimal essential medium supplemented with penicillin (100 μg/ml), streptomycin (100 μg/ml), 1 mM l-glutamine, tylosin (8 μg/ml) and 10% (v/v) fetal bovine serum. When the cells reached 30–50% confluence, they were harvested for the isolation of nascent DNA, while upon reaching 60–80% confluence, they were harvested for isolation of chromatin immunoprecipitated DNA, whereas upon reaching 100% confluence, they were serum-starved for 48–72 h and harvested for the isolation of genomic DNA. For flow cytometry analysis, cells were washed twice in ice-cold phosphate-buffered saline (PBS), and resuspended in Vindelov’s solution (3.4 mM Tris, 75 μM propidium iodide, 0.1% NP40, 0.01 M NaCl, 700 U/l Rnase A), overnight at 4°C, then analyzed using a Beckman flow cytometer and the CellQuest program.
Episomal DNA replication (DpnI resistance) assay
For transfections, HeLa and NSF cells were seeded in six-well plates at a density of 3 × 104 per well, and ∼16 h later were transfected with 3 μg of supercoiled plasmid DNA [2 μg of each construct (pA3/4, pLB2, pLB2C1, pCLONE 3, pCLONE 13, pCLONE 32) and 1 μg pM1 SEAP], using FuGENE 6 transfection reagent (Roche Molecular Biochemicals) as per manufacturer’s instructions. At 72 h post-transfection, low-molecular-weight DNA was isolated and half was digested with DpnI. The DpnI-digested and undigested DNA were used to transform the DH5α strain of Escherichia coli and the relative in vivo DNA replication of each transfected plasmid was determined by counting the number of colonies in a bacterial retransformation assay, as previously described (34). The levels of secreted human placental alkaline phosphatase, determined by the SEAP Reporter Gene Assay kit (Roche Molecular Biochemicals) as per manufacturer’s specifications, were used to normalize the transfection efficiency.
Isolation of genomic DNA
Genomic DNA was isolated using the GenElute Mammalian Genomic DNA Miniprep Kit (Sigma, Oakville, ON, Canada), as per instructions of the manufacturer.
Isolation of nascent DNA
Nascent DNA was prepared using the λ-exonuclease method, as previously described (11,12), with the following modifications: The λ-exonuclease digested samples were heated at 100°C for 3 min, then immediately subjected to electrophoresis on a 2% agarose gel. DNA was visualized by staining with 0.02% (w/v) methylene blue (Sigma) and the origin-containing nascent DNA, ranging between 350 and 1000 bp in size was excised from the gel, purified with the Sephaglas BandPrep Kit (GE Healthcare, Piscataway, NJ, USA), as per instructions of the manufacturer, and resuspended in TE.
Preparation of whole-cell extracts
For the preparation of whole-cell extracts (WCEs), the cells were harvested, washed twice with ice-cold PBS and resuspended in 2 × packed cell volume (pcv) hypotonic buffer [20 mM HEPES-KOH, pH 7.9, 25% glycerol, 0.42 M NaCl, 1.5 mM MgCl2, 1% Triton X-100, 20 mM EDTA, 50 mM DTT and complete protease inhibitor tablet (Roche Molecular Biochemicals, Indianapolis, IN, USA)]. Following a 1-h incubation at 4°C in constant agitation, the cells were centrifuged at 14 000g, the supernatant harvested and its protein concentration determined using the Bradford Protein Assay (Bio-Rad, Hercules, CA, USA).
Chromatin loading
Cell fractionation and preparation of the chromatin-enriched fractions were performed as previously described (35). Cells were harvested from 10-cm dishes into ice-cold PBS, centrifuged, resuspended in 1 ml of lysis buffer A [10 mM HEPES-KOH, pH 7.9, 100 mM NaCl, 300 mM sucrose, 0.1% Triton X-100 and complete protease inhibitor tablet (Roche Molecular Biochemicals)], and lysed on ice for 10 min. After centrifugation at 2000g for 3 min at 4°C, pellets were washed once more with ice-cold lysis buffer A and resuspended in lysis buffer B [10 mM HEPES-KOH, pH 7.9, 200 mM NaCl, 300 mM sucrose, 0.1% Triton X-100, 5 mM MgCl2 and complete protease inhibitor tablet (Roche Molecular Biochemicals)] containing 1000 U of DNase I (Invitrogen). Following incubation at 25°C for 30 min, the chromatin-enriched fraction was isolated in the supernatant after centrifugation at 2500g for 5 min at 4°C and its protein concentration was determined using the Bradford Protein Assay (Bio-Rad).
In vivo cross-linking and chromatin fragmentation
Cells cultured in complete media were washed with pre-warmed PBS and treated with 1% formaldehyde for 10 min to cross-link proteins and DNA in vivo (36); they were then washed and scraped into ice-cold PBS and resuspended in lysis buffer (50 mM HEPES-KOH pH 7.5, 140 mM NaCl, 1% Triton X-100, 2 mM EDTA) supplemented with a complete protease inhibitor tablet (Roche Molecular Biochemicals). Following passage through a 21-G needle three times, the nuclei were harvested, resuspended in one packed nuclear volume of lysis buffer, and sonicated until DNA fragments of <1 kb were obtained. Chromatin size was monitored by electrophoresis. For cell counting, one untreated plate was scraped into PBS and resuspended well. The cells were then counted with a hematocytometer, and this number was used to derive the total number of treated cells. In addition, the concentrations of the extracts were determined using the Bradford protein assay (BioRad, Hercules, CA, USA) in order to normalize samples across cell lines.
Immunoprecipitation and DNA isolation
Immunoprecipitation (IP) was carried out as previously described (36), with the following modifications: Briefly, sheared chromatin lysates (500 μg) were pre-cleared by incubation with 50 μl of protein A/G agarose (Roche Molecular Biochemicals) to reduce background caused by non-specific adsorption to the beads, incubated for 6 h with either 20 µg of anti-ORC2 (Santa Cruz, sc-32734), anti-cdc6 (Santa Cruz, sc-8341), anti-cdt1 (Santa Cruz, sc-28262), anti-EEF1D (gift from Dr. William C. Merrick)/(Santa Cruz, sc-68483), anti-Fe65 (Santa Cruz, sc-33155), anti-FKBP-25 (Santa Cruz, sc-81089), anti-REF-1 (Santa Cruz, sc-5572) or normal rabbit serum (NRS) at 4°C with constant rotation. Protein A/G agarose (50 µl) was added and incubated overnight at 4°C. The pelleted beads were washed successively twice with 1 ml of lysis buffer for 15 min each at 4°C, followed by 1 ml of WB1 (50 mM Tris–HCl pH 7.5, 500 mM NaCl, 0.1% NP40, 0.05% sodium deoxycholate, complete protease inhibitor tablet), 1 ml of WB2 (50 mM Tris–HCl pH 7.5, 0.1% NP40, 0.05% sodium deoxycholate, complete protease inhibitor tablet) and 1 ml of sterile TE. The beads were resuspended in 200 µl TE/1% SDS, incubated at room temperature (rt) for 15 min and centrifuged at 3000 r.p.m. for 1 min at rt. Half of the supernatant was then incubated overnight at 65°C to reverse the cross-links, followed by 100 µg of proteinase K at 55°C for 2 h. The DNA was purified using QIAquick PCR purification kit (Qiagen, Valencia, CA, USA) and eluted in 100 µl TE. The remaining half of the supernatant was boiled for 10 min in SDS-PAGE loading buffer and subjected to electrophoresis on a 5% stacking/8% separating SDS-PAGE gel for western blot analysis.
Western blot analysis
Western blot analysis was carried out according to standard protocols (33). Briefly, the indicated amounts of WCEs, chromatin-enriched fractions, cross-linked immunoprecipitated samples and the EMSA samples obtained for the verification of the proteins identified in the complex by mass spectrometry were resuspended in SDS loading buffer (50 mM Tris–HCl, pH 6.8, 100 mM DTT, 2% SDS, 0.1% bromophenol blue, 10% glycerol), boiled for 10 min and loaded onto a 5% stacking/8% separating SDS-PAGE gel. Following electrophoresis and transfer onto a PVDF membrane, the membrane was immunoblotted with the indicated primary and corresponding HRP-conjugated secondary antibodies. The following antibodies were used: a 1/500 dilution of anti-ORC2 (Santa Cruz, sc-32734), anti-cdc6 (Santa Cruz, sc-8341), anti-cdt1 (Santa Cruz, sc-28262), anti-EEF1D (gift from Dr. William C. Merrick)/(Santa Cruz, sc-68483), anti-Fe65 (Santa Cruz, sc-33155), anti-FKBP-25 (Santa Cruz, sc-81089), anti-ORC1 (Santa Cruz, sc-23887) and a 1/1000 dilution of anti-REF-1 (Santa Cruz, sc-5572) and anti-actin (Sigma, A2066). Proteins were visualized using the enhanced chemiluminescence kit according to the manufacturer's; instructions (Amersham Biosciences, Arlington Heights, IL, USA).
Real-time PCR quantification analyses
PCRs were carried out in a total volume of 20 μl with 5 μl of genomic, nascent or immunoprecipitated DNA, using the LightCycler (Roche Diagnostics), as previously described (13). The sequences and amplification conditions for all primer sets are shown in Supplementary Table S2. Genomic DNA (1, 2, 3 and 4 ng) from NSF cells was used to generate the standard curves needed for quantification of the PCR products. A negative control without template DNA was included with each set of reactions. PCR products were also resolved on 2% agarose gels, visualized with ethidium bromide, and photographed with an Eagle Eye apparatus (Speed Light/BT Sciencetech-LT1000). No extraneous bands were generated with any of the primer sets.
Nuclear extract preparation
Cells cultured in complete media were harvested, washed once with complete medium and twice with PBS. The pelleted cells were resuspended in buffer 1 (100 mM Tris–HCl pH 7.4, 100 mM NaCl, 30 mM MgCl2, 0.5% NP40, 0.5 mM DTT, complete protease inhibitor tablet), incubated for 5 min at 4°C and pelleted by centrifugation for 5 min at 1500 r.p.m. to separate them from the cytoplasmic fraction. The remaining nuclei were washed with buffer 1, then resuspended in buffer 2 (20 mM HEPES pH 7.4, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol, 0.5 mM DTT, complete protease inhibitor tablet) and incubated at 4°C for 40 min and finally spun for 10 min at 4°C (14 000 r.p.m.). The concentrations of the nuclear extracts (NEs) were determined using the Bradford protein assay (BioRad).
EMSA
The genomic region corresponding to the clone 3 origin sequence (probe 3P) (AL031123.14 nucleotides 67323–67467) and non-origin sequence (probe 3C) (AL031123.14 nucleotides 68610–68786) were amplified by PCR and 32 P-end-labeled by the enzymatic activity of T4 polynucleotide kinase. NEs (10 μg) prepared from each cell line were incubated with 0.4 fmol of 32P-labeled probes 3P or 3C, in the presence of 1μg non-specific inhibitor poly(dI–dC) (Amersham Biosciences), and the binding reaction was performed in binding buffer (10 mM Tris–HCl, pH 7.5, 80 mM NaCl, 1 mM EDTA, 10 mM β-mercaptoethanol, 0.1% Triton X-100, 4% glycerol) for 1 h on ice. Subsequently, the mixtures were subjected to electrophoresis on a native 5% PAGE at 140 V in 0.5×TBE and the gels were then dried and subjected to autoradiography. For the competitive EMSA, increasing molar excess amounts of cold specific probe 3P or non-specific probe 3C were included in the reactions. For the supershift EMSA, 4 μg of anti-ORC1 (Santa Cruz, sc-23887/sc-28741), anti-ORC2 (Santa Cruz, sc-13238), anti-ORC3 (Santa Cruz, sc-23888), anti-ORC4 (Santa Cruz, sc-20634), anti-ORC5 (Santa Cruz, sc-20635), anti-cdc6 antibodies (Santa Cruz, sc-8341/sc-9964) or NRS were added to the EMSA reaction for two additional hours after the binding reaction.
In-solution digest
Gel pieces were destained (100 mM ammonium bicarbonate pH8, acetonitrile) then dehydrated with acetonitrile. Samples were reduced (in the dark) with 50 µl DTT for 30 min followed by alkylation with 50 µl iodoacetamide (20 min). Protein digestion was done for 4.5 h with 6 ng/µl of trypsin Gold (Promega) in 100 mM ammonium bicarbonate. Peptides were extracted with 30 µl formic acid (FA) solution [1% FA: 2% acetonitrile (Acn)] for 30 min followed by two 1% FA: 50% Acn solutions for 30 min.
Chromatography
Sample injection and HPLC separation was performed using an Agilent 1100 series system. Twenty microliters of digest solution was loaded onto a Zorbax 300SB-C18 5 × 0.3 mm trapping column then washed for 5 min at 15 ul/min with 3% Acn:0.1% FA. Nano-HPLC peptide separation was done using a New Objective (Woburn, MA, USA) Biobasic C18 10 × 0.075 mm picofrit analytical column. The gradient was 10% Acn:0.1% FA to 95% Acn:0.1% FA in 30 min at 200 nl/min.
Mass spectrometry
The instrument used was a Micro-Qtof from Waters (Milford, MA, USA). Data-dependent analysis was done on the most intense ions from each full scan mass spectrometry (MS) with dynamic exclusion for 60 s. Simple MS scan range was from 350 to 1600 m/z for 1 s. MS/MS data was acquired for up to three 1-s scans from 50 to 2000 m/z.
Database searches
Peak picking for MS/MS peaklist generation was done with Mascot distiller 2.1 from Matrixscience (Boston, MA, USA). Protein identification was done with Mascot 2.1 by searching against sequences from the July 2007 NCBInr database, Mascot results were processed using a bio-informatics program (CellMapBase) used to cluster and group peptides and proteins based on peptides identified at the 95% confidence level by Mascot so as to generate a non-redundant minimal list of identified proteins.
RESULTS
Location of clones 3, 13 and 32 within the human genome
Clones 3, 13 and 32 used in this study were derived from libraries enriched for human origins of DNA replication previously generated in our laboratory (32). These sequences were subjected to a BLAST search to reference them against GenBank nucleotide databases that were obtained from the National Center for Biotechnology Information (NCBI) in order to determine the location of the clones within the human genome. Clone 3 is a 690-bp sequence located on chromosome 6, clone 13 is a 1727-bp sequence located on chromosomes 1 and 4 (the two sequences located within these chromosomes are 99% homologous), and clone 32 is a 247-bp sequence located on chromosome 2 (Table 1).
Table 1.
Location of clones 3, 13 and 32 within the human genome
| Clone name | Genbank Accession No.a | Nucleotide location (bp) on ( + ) or (–) strand (5′ → 3′) b | Length (bp) |
|---|---|---|---|
| 3 | AL031123.14 | 67215-67904 | 690 |
| 13 | AC092670.2/ AC092782.2 | 62462-64188/ 90186-88460 | 1727 |
| 32 | AC023672.9 | 19334-19580 | 247 |
aNucleotide blast revealed that for clone 13, initially found within AC092670.2 was also found at 99% homology within AC092782.2
bNucleotide position 5′→ 3′ of each clone sequence (ascending numbers indicate presence on the + strand and descending numbers indicate presence on the − strand) (strand labeling is arbitrary)
Autonomous replication activity of plasmids containing clones 3, 13 and 32
The ability of clones 3, 13 and 32 to confer autonomous replication activity when cloned into a plasmid was analyzed by the DpnI-resistance assay, which is an indicator of semiconservative DNA replication, as previously described (34).
HeLa (transformed) and NSF (normal) cells were transfected with one of the following constructs: pA3/4, containing a version of the 36-bp mammalian consensus [positive control; (37)]; pLB2, an origin containing sequence within the lamin B2 locus [positive control; (13)]; pLB2C1, a non-origin containing sequence within the lamin B2 locus [negative control for the lamin B2 locus; (13,38)]; pCLONE 3, containing clone 3; pCLONE 13, containing clone 13; pCLONE 32, containing clone 32; pBluescript vector without an insert (negative control); also pM1 SEAP vector was co-transfected with each construct to normalize for the transfection efficiency; finally, a mock transfection without a construct was done as an additional negative control. At 72 h post-transfection, plasmid DNA was isolated, digested with DpnI and the DpnI-digested DNA was used to transform E. coli. After 18 h, the number of colonies produced was counted, corrected for the amount of DNA recovered, and related to the most efficiently replicating plasmid in both cell lines (pA3/4), which was taken as 100% (Figure 1). Interestingly, all three clones conferred autonomous replication activity to their respective plasmids solely in HeLa and not in NSF cells, suggesting that the chromosomal sequences contained in these clones may represent transformation-specific origins. In HeLa cells, the plasmids containing clone 3 replicated with a 3-fold higher efficiency by comparison to clones 13 and 32, which replicated with equal efficiency. In contrast, pLB2 was able to replicate autonomously at nearly the same efficiency in both HeLa and NSF cells, unlike pLB2C1, which was unable to replicate autonomously in either cell line. Similarly, no bacterial colonies were obtained with DpnI-digested DNA recovered when either the backbone vector (pBluescript) had been transfected, as before (34), or from the mock transfections. A relatively small number of colonies (ranging from ∼7 to ∼34) was produced, on average, by even the most efficiently replicating plasmid (pA3/4 produced 33.5 colonies), as expected of mammalian replication origins, which are activated only once per cell cycle. This ‘low’ signal may also be the result of weaker replication activity, resulting from the insert lacking sequences that would normally be present on the chromosomal DNA, whose presence may be necessary to increase replication efficiency (39).
Figure 1.
Episomal replication activity of clones 3, 13 and 32 in HeLa and NSF cells. Histogram plot of the number of bacterial colonies produced after transformation of E. coli with DpnI-digested Hirt extracts of HeLa and NSF cells transfected for 72 h with one of the following constructs: pA3/4, pLB2, pLB2C1, pCLONE 3, pCLONE 13 and pCLONE 32. The average number of DpnI-resistant colonies per plate was corrected for the amount of DNA recovered and normalized to the number of colonies obtained with pA3/4 (most efficiently replicating clone), which was taken as 100%. The number at the bottom of each bar denotes the average number of colonies obtained from three experiments. The error bars represent the average of three experiments performed in triplicate and 1 SD.
Copy number of clone 3, 13 and 32 chromosomal sites
To assess the copy number per haploid genome of all regions examined in this study, equal amounts of genomic DNA from each cell line was amplified by real-time PCR. Figure 2 (top sections of panels A–C), shows representative ethidium bromide-stained 2% agarose gels of the expected amplification products of 362 bp, 270 bp and 425 bp, obtained with the 3 M, 13 M and 32 M primer sets (Supplementary Table S2) of clones 3, 13 and 32, respectively. The histograms depicted in Figure 2 (bottom sections of panels A–C), were produced after normalization of the results by making NSF equal to one copy per haploid genome (3 M and 32 M) or two copies per haploid genome (13 M), as sequence verification of the chromosomal locus of clone 13 by BLAST analysis revealed its presence twice in the genome (on chromosomes 1 and 4 at a level of 99% homology). The results show that in all cell lines 3 M and 32 M were present at one copy per haploid genome, while 13 M was present at two copies per haploid genome. Similar results were obtained for all primer sets amplifying these regions in every cell line examined in this study (data not shown), indicating that all the 3 and 32 primer sets target a single chromosomal locus per haploid genome and all 13 primer sets target two chromosomal loci per haploid genome.
Figure 2.
Copy number per haploid genome of clones 3, 13 and 32 at their respective chromosomal loci in normal and transformed cell lines (A–C) and nascent DNA preparation (D, E). (Top section of A–C) PCR amplification products (362 bp, 425 bp, 270 bp) produced by primer sets 3 M, 13 M and 32 M, respectively, resolved on a 2% agarose gel and stained by ethidium bromide. Template DNA was as follows: lanes 1–3, genomic DNA (1, 2, 3 ng, respectively) from NSF cells [used to build the standard curve for quantification of DNA by real-time PCR; positive control (+ve)]; lanes 4–7, 10 ng of genomic DNA from: HeLa, NSF, WI38, WI38(SV40) cells; lane 8, water [no template DNA added; negative control (−ve)]. (Bottom section of A–C) Histogram plot of copy number/haploid genome of clones 3, 13 and 32 in all cell lines used in the study. The results were normalized by making NSF equal to one copy/haploid genome for primer sets 3 and 32 (or equal to two copies/haploid genome for primer set 13). The error bars represent the average of at least two experiments performed in triplicate and 1 SD. (NB: these are representative graphs, as the copy number of all the primer sets used in this study was measured; Supplementary Table S2.) (D) Nascent DNA preparation. Lane 1: GeneRuler DNA Ladder Mix; lane 2: ∼600 ng total cellular (DNA + RNA) sheared with a fine needle (26G3/8); lane 3: ∼1200 ng sheared and denatured total cellular (DNA + RNA); the single band was the internal control, linear pCR-XL-TOPO (∼50 ng); lane 4: sheared total cellular (DNA + RNA) and internal linear control digested by λ-exonuclease; lane 5: the nascent DNA sample after further treatment with RNase A; lane 6: HindIII-digested λ phage DNA marker. The nucleic acids were separated by electrophoresis on a 1% agarose native gel. (E) Assessment of quality of nascent DNA, exemplified by the lamin B2 origin, in HeLa, NSF, WI38 and WI38(SV40) cells. Histogram plot of the nascent DNA abundance (ng), measured by real-time PCR, at the lamin B2 peak region (dark gray) and a non-origin-containing (control) region, located ∼4 kb downstream (light gray). The error bars represent the average of at least two experiments performed in triplicate and 1 SD.
Nascent DNA abundance
The above results allowed quantitative comparisons between the nascent DNA abundance among the transformed and normal cell lines, thus permitting measurements of replication origin activity.
Nascent DNA was prepared by the λ-exonuclease method. Briefly, λ-exonuclease digests the 5′ phosphorylated parental and broken DNA strands, but the nascent DNA strands bearing 5′ RNA primers are resistant to digestion by this enzyme (11). Representative results of all nascent DNA preparations carried out for each cell line are shown in Figure 2D. Total cellular DNA (together with total RNA) was isolated and sheared with a fine needle. The average size of the sheared total cellular DNA was ∼30 kb, seen as a band on a 1% agarose gel, whereas the smear of lower molecular weight represented RNA (Figure 2D, lane 2). Phosphorylation and λ-exonuclease digestion were monitored by an internal control of dephosphorylated linear plasmid (Figure 2D, lane 3); the plasmid DNA was completely phosphorylated and digested by λ-exonuclease (Figure 2D, lane 3 versus lane 4), and residual RNA (Figure 2D, lane 4) was removed by RNase A digestion (Figure 2D, lane 5). To eliminate any Okazaki fragments that might contaminate the nascent DNA preparations, the samples were subjected to electrophoresis on agarose gel, stained with methylene blue, and only nascent DNA ranging between 350 and 1000 bp was excised from the gel and purified.
To assess the quality of nascent DNA samples, the distribution of nascent DNA from an origin region associated with the lamin B2 locus was quantified. This origin has been shown to be constitutively active in a number of different proliferating human cells (11–13,31,38). Nascent DNA was measured at two reference points, one located at the center of the origin, (LB2 primer set = peak activity) and the other located ∼4 kb away from it (LB2C1 primer set = background activity). The ratio of highest to lowest abundance in the region to be studied is indicative of the quality of the nascent DNA preparation and signal-to-noise estimates (11,12,31). Moreover, a ratio of signals from the initiation site to distant non-initiation sites of ≥10, is indicative of good quality nascent DNA (40). The ratio of highest abundance at the lamin B2 peak region to lowest abundance at the lamin B2 control region ranged from 10.2 to 13.7, indicating good-quality nascent DNA preparations for all cell lines (Figure 2E), and thus suitable for mapping replication origins in vivo. These samples were used to measure the nascent DNA abundance across the chromosomal loci that coincided with the sequences of clones 3, 13 and 32, in each of the transformed and normal cell lines.
Origin activities at the chromosomal loci of clones 3, 13 and 32 in transformed and normal cell lines
Origin activity in vivo was measured in two transformed [HeLa and WI38(SV40)] and two normal (NSF and WI38) human cell lines, by quantification of nascent DNA abundance across the chromosomal loci of clones 3 (Figure 3A), 13 (Figure 3B) and 32 (Figure 3C). Each of the 15 primer sets used amplified a fragment of ∼200–400 bp in size (Supplementary Table S2). Chromosomal nascent DNA abundance at each locus corresponding to clones 3, 13 and 32 was measured using five primer sets: M, which amplifies a region containing part of the autonomously replicating sequence (Table 1); L, which amplifies a region located ∼500–1000 bp 5′ (upstream) of the M region; R, which amplifies a region located ∼500–1000 bp 3′ (downstream) of the M region; FL, which amplifies a region located ∼5–7 kb 5′ (upstream) of the M region; and FR, which amplifies a region located ∼5–7 kb 3′ (downstream) of the M region. The FL and FR primers, designed to amplify regions not containing replication origins (negative controls), were used to measure the lowest abundance of nascent DNA at the chromosomal loci of their respective clones.
Figure 3.
Nascent DNA abundance of clones 3, 13 and 32 at their respective chromosomal loci in normal and transformed cell lines. Histogram plots of the quantification by real-time PCR of nascent DNA abundance (ng) at the chromosomal loci of clones 3, 13 and 32, respectively, of two transformed [HeLa and WI38(SV40)] (gray bars) and two normal (NSF and WI38) (white bars) cell lines. Each of the 15 primer sets amplifies a region of ∼200–400 bp in size: M, a region containing the autonomously replicating sequence (predicted origin sequences) of clone 3 (A), 13 (B) and 32 (C); L, a region located ∼500–1000 bp 5′ (upstream) of the M region; R, a region located ∼500–1000 bp 3′ (downstream) of the M region; FL (negative control), a region located ∼5–7 kb 5′ (upstream) of the M region; FR (negative control), a region located ∼5–7 kb 3′ (downstream) of the M region. For the location of all primers used in the study, see Supplementary Table S2. The error bars represent the average of at least two experiments performed in triplicate and 1 SD.
The nascent strand abundance across the chromosomal loci of clones 3, 13 and 32 was determined in the same preparation of short nascent DNA and normalized to that of an internal reference, the lamin B2 locus, to control for the possibility of a greater recovery of nascent DNA from the transformed cells compared to the normal ones. Specifically, amplification of nascent DNA with primer set LB2C1 (background activity compared to lamin B2 peak region, primer set LB2) gave baseline values that were used to normalize results from all the nascent DNA preparations of all cell lines, permitting comparison of data between different preparations and different cell lines.
Histogram plots of the nascent DNA abundance measured across the chromosomal loci of clones 3 (Figure 3A), 13 (Figure 3B) and 32 (Figure 3C) in all the cell lines used show a peak of origin activity at the M region of each clone, decreasing to ∼71–53% less origin activity at the L & R regions immediately flanking the M region and reaching background levels ∼10% or less origin activity at the FL & FR negative regions located at least 5 kb away from any of the M regions. These data indicate that the M region of each clone is located at a site of initiation (origin) of DNA replication.
The results also indicated a 2–3-fold higher origin activity in the transformed cell lines compared to the normal ones, suggesting a transformation-related activation of these origins. The same result was obtained with the isogenic pair of WI38 (normal) and WI38(SV40) (transformed) cells, ruling out the possibility that the observed increased frequency of initiation in the transformed cell lines might be due to cell type. In all the cell lines examined, the highest abundance of nascent DNA was located at the position of primer set 3 M, then 13 M and finally 32 M, suggesting that clone 3 was the most efficient replication origin, followed by the clone 13 origin and finally the clone 32 origin. Moreover, the lowest abundance was located at the position of the FL & FR primer sets for all three clones (Figure 3A–C), which is most distant from any M region, indicating that no origins were located at these regions. Furthermore, for all three clones the ratio of the highest abundance at primer set M to lowest abundance at primer set FL/FR for all the cell lines ranged from 10.4 to 21.0 (clone 3), 11.0 to 18.1 (clone 13) and 10.4 to 21.4 (clone 32), indicating that all the nascent DNA preparations were of good quality. Finally, all clones exhibited ratios of highest abundance (peak activity) to lowest abundance (background activity) of values >10, confirming that these are true start sites of DNA replication (40).
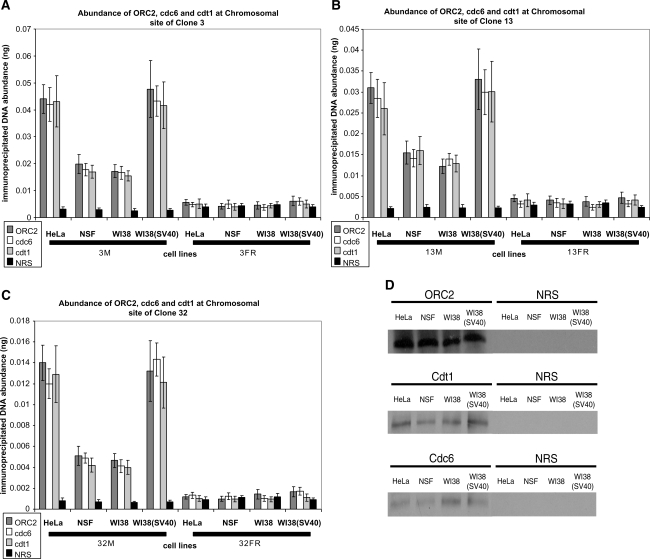
In vivo association of ORC2, cdc6 and cdt1 at the chromosomal loci of clones 3, 13 and 32 in transformed and normal cell lines
The in vivo association of ORC2, cdc6, and cdt1 was measured in the transformed [HeLa and WI38(SV40)] and normal (NSF and WI38) cell lines, by quantification of chromatin immunoprecipitated DNA corresponding to the origin regions (3 M, 13 M and 32 M) of the three clones (3, 13 and 32, respectively) compared to non-origin containing (control) regions (3FR, 13FR and 32FR) (Figure 4A–C). The cell cycle distribution of a population of asynchronously growing cells from all cell lines was found to be the same by FACS analysis (Supplementary Figure S1), permitting the subsequent comparisons between the cell types. Equal amounts of cross-linked chromatin extracts (500 µg) were immunoprecipitated with anti-ORC2, anti-cdc6 and anti-cdt1 antibodies and an equivalent amount of NRS as a negative control to give baseline values that were used to normalize results from all ChIP preparations of all cell lines, permitting comparison of data between different preparations and different cell lines. Immunobloting of the immunoprecipitated lysates verified that all antibodies did immunoprecipitate their respective proteins in all cell lines, while no proteins were immunoprecipitated by the NRS, indicating the specificity of the IP reactions (Figure 4D).
Figure 4.
In vivo association of ORC2, cdc6 and cdt1 to the chromosomal loci of clones 3, 13 and 32 in normal and transformed cell lines. Histogram plots of the quantification by real-time PCR of immunoprecipitated DNA abundance (ng) at the chromosomal loci of clones 3 (A), 13 (B) and 32 (C) respectively, in two transformed cell lines [HeLa and WI38(SV40)] and two normal cell lines (NSF and WI38). Chromatin IP was performed with antibodies directed against ORC2 (dark gray bars), cdc6 (white bars) and cdt1 (light gray bars); normal rabbit serum (NRS) (black bars) was used as a negative control. The primer sets of clones 3 (A), 13 (B) and 32 (C) are explained in the legend of Figure 3 and Supplementary Table S2. The error bars represent the average of at least two experiments performed in triplicate and 1 SD. (D) Western blot analysis using anti-ORC2, anti-cdc6 and anti-cdt1 antibodies to verify the IP of ORC2, cdc6 and cdt1 proteins in all cell lines used. IP with NRS was used as a negative control.
Histogram plots of the immunoprecipitated DNA abundance of ORC2, cdc6 and cdt1 measured at the chromosomal loci of clones 3 (Figure 4A), 13 (Figure 4B) and 32 (Figure 4C) in all the cell lines used show an increased in vivo association of ORC2, cdc6 and cdt1 at each of the origin sequences (M region) of the three clones compared to non-origin sequences (FR region) located at least 5 kb away from any of the origins. The association of ORC2, cdc6 and cdt1 to the clone 3 origin varied from 5.9- to 17.2-fold enrichment over NRS and was 5.5- to 12.2-fold greater than the non-origin containing sequence (Figure 4A), while their association to the clone 13 origin varied from 5.3- to 14.8-fold enrichment over NRS and was 4.9- to 12.4-fold greater than the non-origin containing sequence (Figure 4B), and their association to the clone 32 origin varied from 5.8- to 20.7-fold enrichment over NRS and was 6.1- to 14.5-fold greater than the non-origin containing sequence (Figure 4C). The low abundance of DNA detected in the NRS immunoprecipitates indicated that the ChIP conditions were stringent enough to prevent substantial non-specific association of DNA with the immunoglobulins or agarose beads.
The results also indicated a 2- to 3-fold higher in vivo association of ORC2, cdc6 and cdt1 in the transformed cell lines compared to the normal ones, suggesting a transformation-related increased association of these initiator proteins with the origins located at the chromosomal loci of clones 3, 13 and 32. Use of the isogenic pair of WI38 and WI38 (SV40) showed that the origins were bound by at least twice as much ORC2, cdc6 and cdt1 in the transformed compared to the normal cell lines, again ruling out the possibility that the observed increased binding of these pre-RC proteins in transformed cells might be due to cell type. In all the cell lines examined, the highest binding of ORC2, cdc6 and cdt1 was obtained at the clone 3 origin, followed by the clone 13 origin and lastly the clone 32 origin, in agreement with their replication profile (Figure 3A–C). Background levels of ORC2, cdc6 and cdt1 were observed at the position of the FR primer sets, which is most distant from any clone origin, confirming that no origins were located at these regions (Figure 3A–C).
MS analysis of replication complexes formed on the origin sequence of clone 3
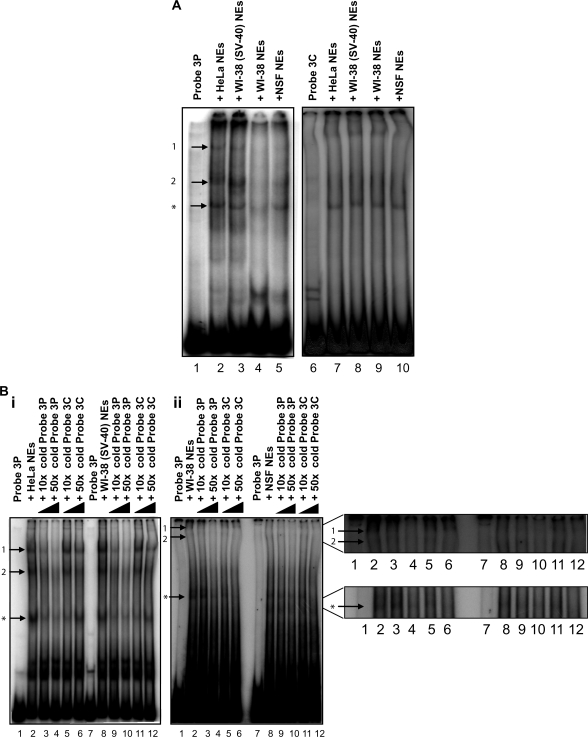
In view of clone 3 being the most active replication origin and its higher association with the pre-RC proteins (ORC2, cdc6 and cdt1), it was further analyzed by EMSA to find out whether additional proteins bound to this region. Equal amounts of nuclear extract from the transformed and normal cell lines were incubated with radioactive probe 3P (a subset of region 3M) or radioactive probe 3C (a non-origin containing sequence that has a similar AT content as probe 3P). EMSA analyses (Figure 5A) revealed the formation of protein–DNA complexes on both probes in all cell lines (Figure 5A, lanes 2–5 and 7–10, complex indicated by arrow asterisk), but additional protein–DNA complexes could be detected on probe 3P (Figure 5A, lanes 2–5, complexes indicated by arrows 1 and 2) and not on probe 3C (Figure 5A, lanes 7–10), which were prominent in the transformed (Figure 5A, lanes 2 and 3), but barely detectable in the normal (Figure 5A, lanes 4 and 5) cells. Competitive EMSA analyses (Figure 5B) showed that the DNA–protein complexes formed solely on probe 3P (complexes 1 an 2 indicated by arrows) were sequence specific, as 10- to 50-fold excess of cold probe 3P (origin sequence), but not of cold probe 3C (non-origin sequence), could efficiently compete the formation of these complexes (Figure 5B i and ii, lanes 2–6 and 8–12, respectively). In contrast, the lower complex asterisk was found to be sequence non-specific since its formation was similarly competed by the specific (3P) and non-specific (3C) probes (Figure 5B i and ii, lanes 2–6 and 8–12, respectively). This is in agreement with its formation when 3C was used as a probe in EMSA Figure 5A, lanes 7–10.
Figure 5.
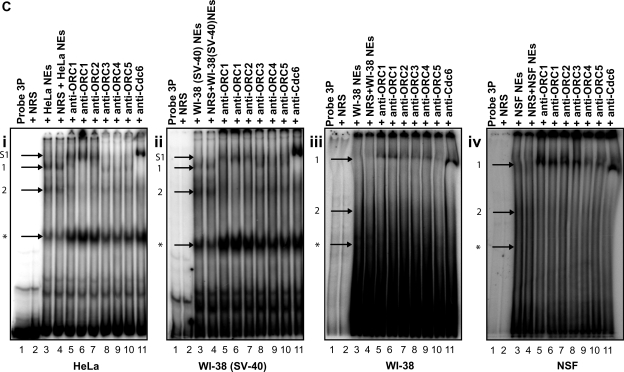
Formation of DNA replication complexes on the clone 3 origin containing sequence. (A) EMSA showing the formation of protein–DNA complexes on probe 3P (ori) and probe 3C (control); 0.4 fmol of radiolabeled probes 3P (lanes 1–5) or 3C (lanes 6–10) was incubated with 10 μg of NEs prepared from HeLa (lanes 2 and 7), WI38(SV40) (lanes 3 and 8), WI38 (lanes 4 and 9), NSF (lanes 5 and 10) cells and then subjected to non-denaturing polyacrylamide electrophoresis. Complexes 1 and 2 (indicated by arrows) represent protein–DNA complexes that only form in the presence of probe 3P and not probe 3C, while complex asterisk (indicated by an arrow) represents a non-specific protein–DNA complex formed in the presence of both probes 3P and 3C. (B) Competition EMSAs showing the sequence specificity of the protein-DNA complexes 1 and 2 but not complex asterisk formed onto probe 3P (lanes 1 and 7) in (i) HeLa (lanes 2–6) and WI38(SV40) (lanes 8–12) NEs and in (ii) WI38 (lanes 2–6) and NSF (lanes 8–12) NEs. Inset shows a magnification of the area of interest in Figure 5Bii. (C) Supershift EMSAs showing that the protein-DNA complex 1 on probe 3P (ori) contains pre-RC proteins in (i) HeLa and (ii) WI38(SV40) NEs (compare complex 1 to its supershifted position S1), but not in (iii) WI38 and (iv) NSF NEs. Lane 1: Probe 3P alone; lane 2: Probe 3P incubated with NRS; lane 3: Probe 3P incubated with NEs; lane 4: Probe 3P incubated with NEs + NRS; lane 5: Probe 3P incubated with NEs + anti-ORC1; lane 6: Probe 3P incubated with NEs + anti-ORC1; lane 7: Probe 3P incubated with NEs + anti-ORC2; lane 8: Probe 3P incubated with NEs + anti-ORC3; lane 9: Probe 3P incubated with NEs + anti-ORC4; lane 10: Probe 3P incubated with NEs + anti-ORC5; lane 11: Probe 3P incubated with NEs + anti-cdc6. Probe incubated with NRS and probe incubated with NEs and NRS were used as controls to confirm the specific binding of the antibodies.
Supershift EMSA analyses (Figure 5C) were performed using antibodies against various members of the pre-RC (ORC1-5 and cdc6), after the incubation of probe 3P with transformed (Figure 5C i and ii) and normal (Figure 5C iii and iv) NEs. The results (Figure 5C i and ii) showed that the addition of anti-ORC1, anti-ORC2 (lanes 5–7) and anti-cdc6 (lane 11) antibodies resulted in a supershift of the complexes formed [Figure 5C i and ii, compare complex 1 (lanes 3 and 4) to complex S1 (lanes 5–7 and 11), indicated by arrows] indicating the presence of these proteins in complex 1 within the transformed NEs. In contrast, addition of the same antibodies did not change the mobility of the complex ‘1’ formed within the normal NEs (Figure 5C iii and iv, compare lanes 3 and 4 to lanes 5–7 and 11) indicating that these proteins were not present in the complex, but only resulted in its stabilization due to the effect that the BSA contained in the antibodies has on the stability of some protein–DNA interactions. Addition of NRS to probe 3P alone or to probe 3P and NEs (Figure 5C i–iv, lanes 2 and 4, negative controls) as well as of anti-ORC 3,4,5 antibodies (Figure 5C i–iv, lanes 8–10), did not affect the mobility of the complexes confirming the antibody specificity of the supershift.
The inability of the anti-ORC 3,4,5 antibodies to supershift the complex may reflect the unavailability of the antibody-targeted protein epitope due to the conformation of the proteins. To determine the composition of the transformation-specific complex, multiple bands corresponding to the most prominent supershift (Figure 5C i and ii, lane 11) (T) obtained with transformed NEs and the exact same portion of the gel (Figure 5C iii and iv, lane 11) (N) using normal NEs as well as the exact same portion of the gel in a non-supershift reaction using transformed NEs (negative control) (C) were excised from the gel and analyzed by mass spectrometry to verify the constituents of the complex. The following proteins were identified uniquely in the DNA replication complex obtained from lane 11 of Figure 5C i and ii of the transformed cells: Ku86, DNA-PKcs, Fe65, REF-1, U5 snRNP-specific 200 kD protein, FKBP-25 and EEF1D (Table 2 for more information), while the composition of the transformation-specific complex was also verified by western blot (Figure 6A and B) showing the presence of the above-mentioned proteins in the T but not the N or C samples, indicating their involvement in DNA replication during cellular transformation.
Table 2.
Proteins identified by mass spectrometric analysis of the transformation-specific DNA replication complexes obtained by means of the supershift EMSA
| Locus | Symbol | Official full name | Mass (kDa) | Function | Processes |
|---|---|---|---|---|---|
| P13010 | XRCC5 | X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining; Ku autoantigen, 80 kDa) (Ku86) | 82.7 | ATP-binding / ATP-dependent DNA helicase activity/DNA binding/helicase activity/hydrolase activity/nucleotide binding/protein C-terminus binding | double-strand break repair via non-homologous end joining/ initiation of viral infection/ provirus integration / regulation of DNA repair / DNA replication |
| O00213 | APBB1 | amyloid beta (A4) precursor protein-binding, family B, member 1(Fe65) | 77.0 | beta-amyloid binding / transcription factor binding | axonogenesis / cell cycle arrest / negative regulation of S-phase of mitotic cell cycle/negative regulation of cell growth/negative regulation of thymidylate synthase biosynthetic process/ regulation of transcription / signal transduction |
| P78527 | PRKDC | protein kinase, DNA-activated, catalytic polypeptide (DNA-PKcs) | 469.1 | ATP binding / DNA binding / DNA-dependent protein kinase activity / protein binding / transferase activity | DNA recombination / DNA repair /double-strand break repair via non-homologous end joining/ peptidyl-serine phosphorylation/ protein modification process |
| Q5TZP7 | APEX1 | APEX nuclease (multifunctional DNA repair enzyme) 1 (APEX-1) (REF-1) | 35.6 | 3′-5′ exonuclease activity/DNA binding/DNA-(apurinic or apyrimidinic site) lyase activity/ endodeoxyribonuclease activity / lyase activity/magnesium ion binding/oxidoreductase activity/ phosphodiesterase I activity/ protein binding/ribonuclease H activity/transcription coactivator activity/ transcription corepressor activity/ uracil DNA N-glycosylase activity | base-excision repair / regulation of DNA binding/regulation of cell redox homeostasis/ transcription from RNA polymerase II promoter |
| O75643 | ASCC3L1 | activating signal cointegrator 1 complex subunit 3-like 1 (U5-200KD)(U5 snRNP-specific 200-kDa protein) | 244.5 | ATP binding / ATP-dependent helicase activity / hydrolase activity / nucleic acid binding/ nucleoside-triphosphatase activity/nucleotide binding / protein binding | RNA splicing/cis assembly of pre-catalytic spliceosome/nuclear mRNA splicing, via spliceosome |
| Q53GD8 | FKBP3 | FK506 binding protein 3 (FKBP-25) | 25.2 | FK506 binding/isomerase activity/peptidyl-prolyl cis-trans isomerase activity/receptor activity | Protein folding |
| Q4VBZ6 | EEF1D | eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) (EF-1D) (EEF1D) | 28.6 | protein binding/signal transducer activity/translation elongation factor activity | positive regulation of I-kappaB kinase/NF-kappaB cascade/translational elongation |
Figure 6.
Verification of the proteins in the complex identified by EMSA and mass spectrometry. Western blot analysis performed on (A) T (electroeluted excised cdc6 supershift bands from transformed NEs) and N (electroeluted excised cdc6 supershift bands from normal NEs) samples as well as (B) T (electroeluted excised cdc6 supershift bands from transformed NEs) and C (electroeluted excised control bands) samples after SDS-PAGE and transfer onto a PVDF membrane to confirm the composition of the transformation-specific complex with the following antibodies: anti-cdc6, anti-EEF1D, anti-Fe65, anti-FKBP-25, anti-ORC1, anti-ORC2, and anti-REF-1. The protein complexes have been analyzed independently a total of 4 times and the experiments were performed from different cell batches giving the same result each time.
Endogenous comparative analysis of the newly identified proteins in transformed and normal cells
To assess the expression levels of these proteins, increasing amounts of WCEs from transformed [HeLa and WI38(SV40)] and normal (NSF and WI38) cells were immunoblotted for Fe65, REF-1, FKBP-25 and EEF1D (Figure 7A). All these proteins had a higher level of expression in transformed compared to normal cells, possibly accounting for the higher prevalence of this complex in transformed compared to normal cells.
Figure 7.
Comparative analysis of the expression, chromatin-bound levels as well as in vivo association of Fe65, REF-1, FKBP-25 and EEF1D to the chromosomal locus of clone 3 in normal and transformed cell lines. (A) Expression levels and (B) Chromatin bound levels of Fe65, REF-1, FKBP-25 and EEF1D. Twenty and forty micrograms of whole-cell extracts or chromatin-enriched fractions were subjected to SDS-PAGE and transferred onto a PVDF membrane and probed with anti-Fe65, anti-REF-1, anti-FKBP-25 and anti-EEF1D antibodies. Anti-actin was used as a loading control. (C) Histogram plot of the quantification by real-time PCR of immunoprecipitated DNA abundance (ng) at the chromosomal locus of clone 3 in two transformed [HeLa and WI38(SV40)] and two normal (NSF and WI38) cell lines. Chromatin IP of the clone 3 chromosomal region was performed with antibodies directed against Fe65 (dark gray bars), REF-1 (white bars), FKBP-25 (light gray bars) and EEF1D (spotted grey bars); normal rabbit serum (NRS) (black bars) was used as a negative control. The clone 3 primer sets are described in the legend of Figure 3 and Supplementary Table S2. The error bars represent the average of at least two experiments performed in triplicate and one standard deviation. (D) Western blot analysis using anti-Fe65, anti-REF-1, anti-FKBP-25 and anti-EEF1D antibodies to verify the IP of Fe65, REF-1, FKBP-25 and EEF1D proteins in all cell lines used. IP with NRS was used as a negative control.
To assess the global chromatin association of these proteins, increasing amounts of chromatin-enriched extracts from transformed [HeLa and WI38(SV40)] and normal (NSF and WI38) cells were immunoblotted for Fe65, REF-1, FKBP-25 and EEF1D (Figure 7B). The results showed that Fe65 had similar chromatin bound levels in HeLa, WI38 and NSF cells, but increased in WI38(SV40) cells; REF-1 and FKBP-25 had a higher chromatin bound levels in transformed [HeLa and WI38(SV40)] compared to normal cells (NSF and WI38); and EEF1D had similar chromatin bound levels in WI38 and WI38(SV40) cells, less so in NSF cells and the most in HeLa cells.
The local origin-specific in vivo association of Fe65, REF-1, FKBP-25, and EEF1D was measured in the transformed [HeLa and WI38(SV40)] and normal (NSF and WI38) cell lines, by quantification of chromatin immunoprecipitated DNA corresponding to the clone 3 origin region (3 M) compared to non-origin containing (control) region (3FR) (Figure 7C). Equal amounts of cross-linked chromatin extracts (500 µg) were immunoprecipitated with anti-Fe65, anti-REF-1, anti-FKBP-25 and anti-EEF1D antibodies and an equivalent amount of NRS as a negative control. Immunoblots of the immunoprecipitated lysates verified that all antibodies did immunoprecipitate their respective proteins in all cell lines (Figure 7D). No proteins were immunoprecipitated by the NRS, indicating the specificity of the IP reactions.
Histogram plots of the immunoprecipitated DNA abundance of Fe65, REF-1, FKBP-25 and EEF1D measured at the chromosomal locus of clone 3 (Figure 7C) in all the cell lines used show an increased in vivo association of Fe65, REF-1, FKBP-25 and EEF1D at the clone 3 origin sequence (3 M) compared to the non-origin sequence (3FR) located at ∼6 kb away from the origin. The binding of Fe65, REF-1, FKBP-25 and EEF1D to the clone 3 origin varied from 6.6- to 19.5-fold enrichment over NRS for the transformed cells and from 2.4- to 3.1-fold enrichment over NRS for the normal cells, while it was 4.3–10.3-fold greater than the non-origin containing sequence for the transformed cells and was 2.0–2.2-fold greater than the non-origin containing sequence for the normal cells (Figure 7C). The low abundance of DNA detected in the NRS immunoprecipitates indicated that the ChIP conditions were stringent enough to prevent substantial non-specific association of DNA with the immunoglobulins or agarose beads.
The results also indicated a 3–6-fold higher in vivo association of Fe65, REF-1, FKBP-25 and EEF1D in the transformed cell lines compared to the normal ones, suggesting a transformation-related increased association of these proteins with the clone 3 origin. Use of the isogenic pair of WI38 and WI38(SV40) ruled out the possibility that the observed increased binding of these proteins in transformed cell lines might be due to cell type.
DISCUSSION
Clones 3, 13 and 32 coincide with chromosomal replication origins in both transformed and normal cells, but support the autonomous replication of plasmids solely in transformed cells
In this study, we took advantage of origin libraries that we had generated previously, which contain clones with autonomous replication activity that have been sequenced and characterized (32). We focused on three of these clones (3, 13 and 32) that had been found to replicate autonomously only in transformed cells and examined both their ectopic an in situ replication activity in transformed and normal cells, by transient episomal replication assays (Figure 1) and nascent strand abundance analysis (Figure 3), respectively. Interestingly, all three clones were able to support autonomous replication of their respective plasmids after transfection into HeLa (transformed) cells but not NSF (normal) cells, suggesting the presence of a more favorable environment (factors) present in transformed cells that is absent in normal ones, which allows for transformation-specific ectopic replication. Trans-acting factors are apparently responsible for the transformation-specific episomal replication of the clones, as the cis-acting sequences transfected into both the transformed and normal cells were identical. The presence of such factor(s) possibly alters the DNA topology, facilitating episomal replication, as also suggested in a recent study of DNA synthesis of extrachromosomal DNA (41).
Origin activities in normal versus transformed cells
The above results suggested a transformation-specific replication and the existence of transformation-specific origins. We thus examined the ability of the three clones to act as chromosomal replication origins by in situ replication assays in two transformed and two normal cell lines, using real-time PCR to quantify the abundance of nascent DNA (Figure 3). All three clones at their respective chromosomal loci corresponded to replication origins in both the transformed and normal cell lines tested, but these origins were 2–3-fold more active in the transformed cells compared to the normal ones at the chromosomal loci of all three clones.
The replication origin associated with clone 3 exhibited the highest level of nascent DNA abundance followed by clone 13 then clone 32. One possible explanation is that the origin at clone 3 may be activated in more cells per population by comparison to the origins corresponding to clones 13 and 32. Alternatively, there may be other initiation sites present in the vicinity of the origins associated with clones 13 and 32, causing origin interference (42), which may result in a broader and flatter peak by comparison to the origin associated with clone 3, leading to their observed lower level of nascent DNA abundance. Regarding the origin associated with clone 13, which is present at two copies per haploid genome at a level of 99% homology, it was not possible to design primer sets to distinguish between the two chromosomal locations due to the high homology and thus it could not be determined whether the origin activity stemmed from chromosome 1 or chromosome 4 or both.
These results indicated that the deregulated increase in replication initiation may be an early event in the stepwise progression to cancer occurring at the transformation stage, causing replication stress, which may lead to DNA double-strand breaks and activation of the DNA damage checkpoint, increased genomic instability, and tumor progression (43–45).
The differential origin activities observed between the transformed and normal cells might be due to some origins being fired more than once, but the repeated initiations be subsequently aborted to prevent gene amplification. It has been speculated that the early events of genomic instability in a cancer cell might entail unregulated origin firing, providing substrates for genetic recombination and further amplification (46). Alternatively, the differential origin activities might be due to the clone origins not always being used in normal cells, but used in a higher percentage of transformed cells. It is conceivable that in normal cells, at least during some S phases, the DNA at the clone origins might be replicated by upstream or downstream origins flanking this region.
In vivo association of ORC2, cdc6 and cdt1 at the clone origins in normal versus transformed cells
Comparative analysis of the in vivo association of ORC2, cdc6 and cdt1 with the chromosomal replication origins corresponding to clones 3, 13, and 32 between two transformed cell lines and two normal cell lines (Figure 4A–C) showed that they were bound by these pre-RC proteins in all cell lines. As expected from bona fide replication origins, the abundance of each pre-RC protein at any given clone origin was enriched by at least 5.3-fold over NRS and by at least 4.9-fold over any non-origin containing (negative control) region located at least 5 kb away from any origin. The highest level of association of ORC2, cdc6 and cdt1 was at the clone 3 origin, the next highest at the clone 13 origin and the lowest at the clone 32 origin, in agreement with their replication profiles. Together, the nascent DNA and ChIP data indicate that the chromosomal clone origins behave as such and are bound by members of the pre-RC, as observed in other well-characterized origins, such as those at the lamin B2 locus (47), c-myc locus (48) and β-globin locus (36). Furthermore, the results revealed a 2–3-fold higher abundance of these pre-RC proteins bound at all the clone chromosomal origins of the transformed cells by comparison to the normal ones, once more correlating with the origin mapping results.
The differential association of ORC2, cdc6 and cdt1 observed between the transformed and normal cells is in agreement with the finding that ORC2 is overexpressed and bound at a higher frequency to chromatin in transformed compared to normal cells (25) and that cdc6 as well as cdt1 overexpression promotes malignant behavior (49). The impact that these findings have on pre-RC proteins playing a role during oncogenesis is reviewed in (27), and have led to the suggestion that there is a pre-RC checkpoint lacking in cancer cells (26). The elevated binding of the pre-RC proteins to the clone origins in transformed compared to normal cells may be an event resulting from the deregulation of the pre-RC checkpoint by two likely scenarios: origins firing more than once per cell cycle, due to an improper licensing of these origins, leading to re-replication, causing genetic instability and then cancer (28), or, alternatively, the differential origin activities might be due to the clone origins being activated at a lower frequency per cell cycle in normal cells, but at a higher frequency in transformed cells.
Transformation-specific DNA replication complexes reveal possible new protein players involved in DNA replication and cellular transformation
Comparative EMSA (Figure 5A) analysis of the ability of NEs from transformed and normal cells to form complexes on the origin sequence of clone 3 revealed the formation of DNA–protein complexes 1 and 2 that were not formed on the non-origin sequence. These complexes were easily detectable in transformed NEs but barely detectable in normal NEs. This was consistently observed even after prolonged autoradiograph exposure times (up to 14 days), suggesting that these complexes exist in very low abundance and/or are highly unstable in normal NEs (Figure 5A, B ii, C iii and iv). Competitive EMSAs (Figure 5B i and ii) confirmed that these complexes were sequence-specific and supershift EMSAs when using antibodies against known replication initiation proteins (ORC1, ORC2 and cdc6) (Figure 5C i and ii) confirmed that complex 1 was indeed a DNA replication complex formed when using transformed NEs but not when using normal NEs. (Figure 5 C iii and iv).
MS analysis of the most prominent supershifted complex that was produced by the anti-cdc6 antibody revealed seven proteins that were present uniquely in the transformation-specific DNA replication complex of interest (Table 2): (i) Ku86, shown to be involved in DNA replication (50) and references therein; (ii) DNA–PKcs, which is the catalytic subunit of DNA–PK that forms a complex with Ku70/Ku86 and has been involved in DNA replication (51) and cellular transformation, as it has been found to modulate the stability of c-Myc oncoprotein (52); (iii) Fe65, which has a role in the response of cells to DNA damage (53) as well as interacts with Tip60 (a histone acetyltransferase) (54); (iv) REF-1, which is a multi-functional DNA repair enzyme (55) and is elevated in human tumors (56); (v) U5 snRNP-specific 200-kDa protein, which was found in a complex with Ku86 and DNA–PKcs as a transcriptional activator (57) and may be involved in chromatin remodeling through its interaction with N-CoR deacetylase (58); (vi) FKBP-25, which may play a regulatory role in cell growth through its interaction with casein kinase II complex (59) and also involved in chromatin remodeling through its association with histone deacetylases (HDAC1 and HDAC2) and the HDAC-binding transcriptional regulator YY1 (60); and (vii) eukaryotic translation elongation factor 1 delta (EEF1D), which has been implicated in cellular transformation, as a higher level of EEF1D expression is associated with a more invasive phenotype for breast cancer cells (61) and poorer prognosis for esophageal carcinoma (62). In addition, a block of the translation of EEF1D resulted in a significant reversal of its oncogenic potential (63). These proteins may represent new players involved in DNA replication, as they seem to be involved in overlapping processes such as response to DNA damage and chromatin modification, which may be indirectly implicated in the process of cellular transformation. With the exception of Ku86 and DNA–PKcs, these proteins have not been previously considered in the context of DNA replication and their potential involvement in this process in relation to cellular transformation requires further investigation. Thus far, in support of this notion we know that several of these proteins have a higher level of expression and chromatin association as well as exhibit a ∼3–6-fold higher abundance in vivo at the clone 3 origin in transformed compared to normal cells.
OVERALL SUMMARY AND CONCLUSION
Previous studies have suggested that there are at least two types of changes in the activation of replication origins during cell transformation and malignancy: an increase in origin activity at some loci (10–13) and the activation of origins that are silent in normal cells (14,15). The origins examined in this study fall under the category of those whose activity is increased in tumor/transformed cells by comparison to normal ones. Thus, it appears that there are at least three subsets of origins: those that are normal and remain unchanged, those with increased origin activity in transformed/immortalized or malignant cells, and those that are activated uniquely in tumor cells.
In summary, we have identified three clones that confer autonomous replication of their respective plasmids in transformed cells but not normal cells. These clones act as chromosomal origins of DNA replication and are bound by members of the pre-RC in both transformed and normal cells, but they exhibit both differential origin activity and pre-RC binding between transformed and normal cells. Use of the most efficiently replicating clone as bait to analyze protein–DNA complexes revealed a transformation-specific DNA replication complex from which we identified seven proteins that are overexpressed as well as bound to chromatin at a higher level in transformed compared to normal cells, in addition to being associated ∼3–6-fold higher in vivo with the clone 3 origin in transformed compared to normal cells, indicating that these proteins may be involved in the pathways leading to deregulated DNA replication and cellular transformation. The present data indicate that replication origins (both active and cryptic) are used more frequently in transformed cells than in normal ones. Although this study did not identify any transformation-specific origins, their existence cannot be ruled out. Experiments addressing these possibilities as well as the role of the proteins present in the transformation-specific complex are ongoing.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This research was supported by grants from the Canadian Prostate Cancer Research Initiative and NCIC—Canadian Cancer Society. D.D.P is a past recipient of a Canadian Institutes of Health Research (CIHR) Cancer Consortium Training Grant Award, a McGill Faculty of Medicine internal studentship award, and present recipient of a Fonds de la Recherche en Santé du Québec (FRSQ) studentship award.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
This work is dedicated to the memory of Dr. Gerald B. Price, whose expert intellectual and technical advice and most of all friendship is greatly missed. We also thank the proteomics platform of the McGill University and Genome Quebec Innovation Centre for their aid with the mass spectroscopy experiments.
REFERENCES
- 1.Jacob F, Brenner S, Cuzin F. On the regulation of DNA replication in bacteria. Cold Spring Harbor Symp. Quant. Biol. 1963;28:329–348. [Google Scholar]
- 2.Edenberg HJ, Huberman JA. Eukaryotic chromosome replication. Annu. Rev. Genet. 1975;9:245–284. doi: 10.1146/annurev.ge.09.120175.001333. [DOI] [PubMed] [Google Scholar]
- 3.Hand R. Eucaryotic DNA: organization of the genome for replication. Cell. 1978;15:317–325. doi: 10.1016/0092-8674(78)90001-6. [DOI] [PubMed] [Google Scholar]
- 4.Zannis-Hadjopoulos M, Price GB. Eukaryotic DNA replication. J. Cell. Biochem. 1999;Supplements 32/33:1–14. doi: 10.1002/(sici)1097-4644(1999)75:32+<1::aid-jcb2>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 5.Machida YJ, Hamlin JL, Dutta A. Right place, right time, and only once: replication initiation in metazoans. Cell. 2005;123:13–24. doi: 10.1016/j.cell.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 6.Teer JK, Dutta A. Regulation of S phase. Results Probl. Cell Differ. 2006;42:31–63. doi: 10.1007/b137221. [DOI] [PubMed] [Google Scholar]
- 7.Martin R. The transformation of cell growth and transmogrification of DNA synthesis by simian virus 40. Adv. Cancer Res. 1981;34:1–68. doi: 10.1016/s0065-230x(08)60238-9. [DOI] [PubMed] [Google Scholar]
- 8.Anglana M, Apiou F, Bensimon A, Debatisse M. Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell. 2003;114:385–394. doi: 10.1016/s0092-8674(03)00569-5. [DOI] [PubMed] [Google Scholar]
- 9.Zannis-Hadjopoulos M, Price G. Regulatory parameters of DNA replication. Crit. Rev. Eukary. Gene Exp. 1998;8:81–106. doi: 10.1615/critreveukargeneexpr.v8.i1.40. [DOI] [PubMed] [Google Scholar]
- 10.Tao L, Nielsen T, Friedlander P, Zannis-Hadjopoulos M, Price G. Differential DNA replication origin activities in human normal skin fibroblast and HeLa cell lines. J. Mol. Biol. 1997;273:509–518. doi: 10.1006/jmbi.1997.1352. [DOI] [PubMed] [Google Scholar]
- 11.Tao L, Dong Z, Leffak M, Zannis-Hadjopoulos M, Price G. Major DNA replication initiation sites in the c-myc locus in human cells. J. Cell. Biochem. 2000;78:442–457. doi: 10.1002/1097-4644(20000901)78:3<442::aid-jcb9>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 12.Tao L, Dong Z, Zannis-Hadjopoulos M, Price GB. Immortalization of human WI38 cells is associated with differential activation of the c-myc origins. J. Cell. Biochem. 2001;82:522–534. doi: 10.1002/jcb.1173. [DOI] [PubMed] [Google Scholar]
- 13.Di Paola D, Price GB, Zannis-Hadjopoulos M. Differentially active origins of DNA replication in tumor versus normal cells. Cancer Res. 2006;66:5094–5103. doi: 10.1158/0008-5472.CAN-05-3951. [DOI] [PubMed] [Google Scholar]
- 14.Martin RG, Oppenheim A. Initiation points for DNA replication in nontransformed and simian virus 40-transformed Chinese hamster lung cells. Cell. 1977;11:859–869. doi: 10.1016/0092-8674(77)90297-5. [DOI] [PubMed] [Google Scholar]
- 15.Oppenheim A, Martin RG. Initiation points for DNA replication in nontransformed and simian virus 40-transformed BALB/c 3T3 cells. J. Virol. 1978;25:450–452. doi: 10.1128/jvi.25.1.450-452.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collins J, Glock M, Chu A. Nuclease S1 sensitive sites in potential deoxyribonucleic acid of cold-and temperature-sensitive mammalian cells. Biochemistry. 1982;21:3414–3419. doi: 10.1021/bi00257a026. [DOI] [PubMed] [Google Scholar]
- 17.Itoh-Lindstrom Y, Leffak M. Alteration of in vitro DNA synthesis in the alpha globin locus of embryo fibroblasts due to in vivo activity of Rous Sarcoma Virus. Nucleic Acids Res. 1994;22:498–508. [PMC free article] [PubMed] [Google Scholar]
- 18.Kennedy BK, Barbie DA, Classon M, Dyson N, Harlow E. Nuclear organization of DNA replication in primary mammalian cells. Genes Dev. 2000;14:2855–2868. doi: 10.1101/gad.842600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dimitrova DS, Berezney R. The spatio-temporal organization of DNA replication sites is identical in primary, immortalized and transformed mammalian cells. J. Cell Sci. 2002;115:4037–4051. doi: 10.1242/jcs.00087. [DOI] [PubMed] [Google Scholar]
- 20.Panning MM, Gilbert DM. Spatio-temporal organization of DNA replication in murine embryonic stem, primary, and immortalized cells. J. Cell. Biochem. 2005;95:74–82. doi: 10.1002/jcb.20395. [DOI] [PubMed] [Google Scholar]
- 21.Conti C, Sacca B, Herrick J, Lalou C, Pommier Y, Bensimon A. Replication fork velocities at adjacent replication origins are coordinately modified during DNA replication in human cells. Mol. Biol. Cell. 2007;18:3059–3067. doi: 10.1091/mbc.E06-08-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Campos Vidal B, Russo J, Mello ML. DNA content and chromatin texture of benzo[a]pyrene-transformed human breast epithelial cells as assessed by image analysis. Exp. Cell Res. 1998;244:77–82. doi: 10.1006/excr.1998.4171. [DOI] [PubMed] [Google Scholar]
- 23.Mello ML, Russo P, Russo J, Vidal BC. 17-beta-estradiol affects nuclear image properties in MCF-10F human breast epithelial cells with tumorigenesis. Oncol. Rep. 2007;18:1475–1481. [PubMed] [Google Scholar]
- 24.Amiel A, Litmanovitch T, Lishner M, Mor A, Gaber E, Tangi I, Fejgin M, Avivi L. Temporal differences in replication timing of homologous loci in malignant cells derived from CML and lymphoma patients. Genes Chromosomes Cancer. 1998;22:225–231. doi: 10.1002/(sici)1098-2264(199807)22:3<225::aid-gcc8>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 25.McNairn AJ, Gilbert DM. Overexpression of ORC subunits and increased ORC-chromatin association in transformed mammalian cells. J. Cell. Biochem. 2005;96:879–887. doi: 10.1002/jcb.20609. [DOI] [PubMed] [Google Scholar]
- 26.Lau E, Jiang W. Is there a pre-RC checkpoint that cancer cells lack? Cell Cycle. 2006;5:1602–1606. doi: 10.4161/cc.5.15.3124. [DOI] [PubMed] [Google Scholar]
- 27.Lau E, Tsuji T, Guo L, Lu SH, Jiang W. The role of pre-replicative complex (pre-RC) components in oncogenesis. FASEB J. 2007;21:3786–3794. doi: 10.1096/fj.07-8900rev. [DOI] [PubMed] [Google Scholar]
- 28.Dutta A. Chaotic license for genetic instability and cancer. Nat Genet. 2007;39:10–11. doi: 10.1038/ng0107-10. [DOI] [PubMed] [Google Scholar]
- 29.Hu L, Xu X, Valenzuela MS. Identification of novel initiation sites for human DNA replication around ARSH1, a previously characterized yeast replicator. Biochem. Biophys. Res. Commun. 2004;313:1058–1064. doi: 10.1016/j.bbrc.2003.12.040. [DOI] [PubMed] [Google Scholar]
- 30.Hu L, Xu X, Valenzuela MS. Initiation sites for human DNA replication at a putative ribulose-5-phosphate 3-epimerase gene. Biochem. Biophys. Res. Commun. 2004;320:648–655. doi: 10.1016/j.bbrc.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 31.Kumar S, Giacca M, Norio G, Biamonti G, Riva S, Falaschi A. Utilization of the same DNA replication origin by human cells of different derivation. Nucleic Acids Res. 1996;24:3289–3294. doi: 10.1093/nar/24.17.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nielsen T, Bell D, Lamoureux C, Zannis-Hadjopoulos M, Price GB. A reproducible method for identification of human genomic DNA autonomously replicating sequences. Mol. Gen. Genet. 1994;242:280–288. doi: 10.1007/BF00280417. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Matheos D, Novac O, Price GB, Zannis-Hadjopoulos M. Analysis of the DNA replication competence of the xrs-5 mutant cells defective in Ku86. J. Cell Sci. 2003;116:111–124. doi: 10.1242/jcs.00156. [DOI] [PubMed] [Google Scholar]
- 35.Tatsumi Y, Tsurimoto T, Shirahige K, Yoshikawa H, Obuse C. Association of human origin recognition complex 1 with chromatin DNA and nuclease-resistant nuclear structures. J. Biol. Chem. 2000;275:5904–5910. doi: 10.1074/jbc.275.8.5904. [DOI] [PubMed] [Google Scholar]
- 36.Sibani S, Price GB, Zannis-Hadjopoulos M. Decreased origin usage and initiation of DNA replication in haploinsufficient HCT116 Ku80+/– cells. J. Cell Sci. 2005;118:3247–3261. doi: 10.1242/jcs.02427. [DOI] [PubMed] [Google Scholar]
- 37.Price GB, Allarakhia M, Cossons N, Nielsen T, Diaz-Perez M, Friedlander P, Tao L, Zannis-Hadjopoulos M. Identification of a cis-element that determines autonomous DNA replication in eukaryotic cells. J. Biol. Chem. 2003;278:19649–19659. doi: 10.1074/jbc.M207002200. [DOI] [PubMed] [Google Scholar]
- 38.Giacca M, Zentilin L, Norio P, Diviacco S, Dimitrova D, Contreas G, Biamonti G, Perini G, Weighardt F, Riva S, et al. Fine mapping of a replication origin of human DNA. Proc. Natl Acad. Sci. USA. 1994;91:7119–7123. doi: 10.1073/pnas.91.15.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DePamphilis M. How transcription factor regulate origins of DNA replication in eukaryotic cells. Trends Cell. Biol. 1993;3:161–167. doi: 10.1016/0962-8924(93)90137-p. [DOI] [PubMed] [Google Scholar]
- 40.DePamphilis ML. The search for origins of DNA replication. Methods. 1997;13:211–219. doi: 10.1006/meth.1997.0521. [DOI] [PubMed] [Google Scholar]
- 41.Wang J, Lindner SE, Leight ER, Sugden B. Essential elements of a licensed, mammalian plasmid origin of DNA synthesis. Mol. Cell. Biol. 2006;26:1124–1134. doi: 10.1128/MCB.26.3.1124-1134.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lebofsky R, Heilig R, Sonnleitner M, Weissenbach J, Bensimon A. DNA replication origin interference increases the spacing between initiation events in human cells. Mol. Biol. Cell. 2006;17:5337–5345. doi: 10.1091/mbc.E06-04-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 44.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio R.A., Jr, Kastrinakis NG, Levy B, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 45.Bartek J, Bartkova J, Lukas DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene. 2007;26:7773–7779. doi: 10.1038/sj.onc.1210881. [DOI] [PubMed] [Google Scholar]
- 46.Bandura JL, Calvi BR. Duplication of the genome in normal and cancer cell cycles. Cancer Biol. Ther. 2002;1:8–13. doi: 10.4161/cbt.1.1.31. [DOI] [PubMed] [Google Scholar]
- 47.Abdurashidova G, Danailov MB, Ochem A, Triolo G, Djeliova V, Radulescu S, Vindigni A, Riva S, Falaschi A. Localization of proteins bound to a replication origin of human DNA along the cell cycle. EMBO J. 2003;22:4294–4303. doi: 10.1093/emboj/cdg404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ghosh M, Kemp M, Liu G, Ritzi M, Schepers A, Leffak M. Differential binding of replication proteins across the human c-myc replicator. Mol. Cell. Biol. 2006;26:5270–5283. doi: 10.1128/MCB.02137-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liontos M, Koutsami M, Sideridou M, Evangelou K, Kletsas D, Levy B, Kotsinas A, Nahum O, Zoumpourlis V, Kouloukoussa M, et al. Deregulated overexpression of hCdt1 and hCdc6 promotes malignant behavior. Cancer Res. 2007;67:10899–10909. doi: 10.1158/0008-5472.CAN-07-2837. [DOI] [PubMed] [Google Scholar]
- 50.Rampakakis E, Di Paola D, Zannis-Hadjopoulos M. Ku is involved in cell growth, DNA replication and G1-S transition. J. Cell Sci. 2008;121:590–600. doi: 10.1242/jcs.021352. [DOI] [PubMed] [Google Scholar]
- 51.Guan J, DiBiase S, Iliakis G. The catalytic subunit DNA-dependent protein kinase (DNA-PKcs) facilitates recovery from radiation-induced inhibition of DNA replication. Nucleic Acids Res. 2000;28:1183–1192. doi: 10.1093/nar/28.5.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.An J, Yang DY, Xu QZ, Zhang SM, Huo YY, Shang ZF, Wang Y, Wu DC, Zhou PK. DNA-dependent protein kinase catalytic subunit modulates the stability of c-Myc oncoprotein. Mol. Cancer. 2008;7:32. doi: 10.1186/1476-4598-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Minopoli G, Stante M, Napolitano F, Telese F, Aloia L, De Felice M, Di Lauro R, Pacelli R, Brunetti A, Zambrano N, et al. Essential roles for Fe65, Alzheimer amyloid precursor-binding protein, in the cellular response to DNA damage. J. Biol. Chem. 2007;282:831–835. doi: 10.1074/jbc.C600276200. [DOI] [PubMed] [Google Scholar]
- 54.Yang Z, Cool BH, Martin GM, Hu Q. A dominant role for FE65 (APBB1) in nuclear signaling. J. Biol. Chem. 2006;281:4207–4214. doi: 10.1074/jbc.M508445200. [DOI] [PubMed] [Google Scholar]
- 55.Ono Y, Furuta T, Ohmoto T, Akiyama K, Seki S. Stable expression in rat glioma cells of sense and antisense nucleic acids to a human multifunctional DNA repair enzyme, APEX nuclease. Mutat. Res. 1994;315:55–63. doi: 10.1016/0921-8777(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 56.Bobola MS, Blank A, Berger MS, Stevens BA, Silber JR. Apurinic/apyrimidinic endonuclease activity is elevated in human adult gliomas. Clin. Cancer Res. 2001;7:3510–3518. [PubMed] [Google Scholar]
- 57.Chen Y, Schnetz MP, Irarrazabal CE, Shen RF, Williams CK, Burg MB, Ferraris JD. Proteomic identification of proteins associated with the osmoregulatory transcription factor TonEBP/OREBP: functional effects of Hsp90 and PARP-1. Am. J. Physiol. Renal Physiol. 2007;292:F981–F992. doi: 10.1152/ajprenal.00493.2005. [DOI] [PubMed] [Google Scholar]
- 58.Dellaire G, Makarov EM, Cowger JJ, Longman D, Sutherland HG, Luhrmann R, Torchia J, Bickmore WA. Mammalian PRP4 kinase copurifies and interacts with components of both the U5 snRNP and the N-CoR deacetylase complexes. Mol. Cell. Biol. 2002;22:5141–5156. doi: 10.1128/MCB.22.14.5141-5156.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jin YJ, Burakoff SJ. The 25-kDa FK506-binding protein is localized in the nucleus and associates with casein kinase II and nucleolin. Proc. Natl Acad. Sci. USA. 1993;90:7769–7773. doi: 10.1073/pnas.90.16.7769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang WM, Yao YL, Seto E. The FK506-binding protein 25 functionally associates with histone deacetylases and with transcription factor YY1. EMBO J. 2001;20:4814–4825. doi: 10.1093/emboj/20.17.4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang S, Du J, Wang Z, Yuan W, Qiao Y, Zhang M, Zhang J, Gao S, Yin J, Sun B, et al. BMP-6 promotes E-cadherin expression through repressing deltaEF1 in breast cancer cells. BMC Cancer. 2007;7:211. doi: 10.1186/1471-2407-7-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ogawa K, Utsunomiya T, Mimori K, Tanaka Y, Tanaka F, Inoue H, Murayama S, Mori M. Clinical significance of elongation factor-1 delta mRNA expression in oesophageal carcinoma. Br. J. Cancer. 2004;91:282–286. doi: 10.1038/sj.bjc.6601941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lei YX, Chen K, Wu ZL. Blocking the translation elongation factor-1 delta with its antisense mRNA results in a significant reversal of its oncogenic potential. Teratog. Carcinog. Mutagen. 2002;22:377–383. doi: 10.1002/tcm.10034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.