Abstract
Metal-dependent nucleases that generate double-strand breaks in DNA often possess two symmetrically-equivalent subunits, arranged so that the active sites from each subunit act on opposite DNA strands. Restriction endonuclease BfiI belongs to the phospholipase D (PLD) superfamily and does not require metal ions for DNA cleavage. It exists as a dimer but has at its subunit interface a single active site that acts sequentially on both DNA strands. The active site contains two identical histidines related by 2-fold symmetry, one from each subunit. This symmetrical arrangement raises two questions: first, what is the role and the contribution to catalysis of each His residue; secondly, how does a nuclease with a single active site cut two DNA strands of opposite polarities to generate a double-strand break. In this study, the roles of active-site histidines in catalysis were dissected by analysing heterodimeric variants of BfiI lacking the histidine in one subunit. These variants revealed a novel mechanism for the scission of double-stranded DNA, one that requires a single active site to not only switch between strands but also to switch its orientation on the DNA.
INTRODUCTION
The nucleases that cleave the phosphodiester backbone of DNA to leave 5′-phosphate and 3′-hydroxyl termini participate in many varied biological processes, including DNA replication, repair, recombination, immunity, defence and apoptosis. The vast majority of nucleases use divalent metal ions as cofactors, though the number and identity of the ions can differ (1). The presence of two divalent metal ions in the active site of the 3′-5′ exonuclease domain of Escherichia coli DNA polymerase I led Steitz and co-workers to propose a reaction mechanism in which a water molecule from the coordination sphere of the first metal ion performs an in-line nucleophilic attack on the scissile phosphate to displace the 3′-leaving group, which is in turn stabilized by the second metal ion (2,3). The single step reaction proceeds with inversion of configuration at the scissile phosphate (4–6). Many of metal-dependent nucleases that generate double-strand breaks in DNA feature two identical subunits related by 2-fold symmetry. Each subunit contains a single active site: one catalyzes the cleavage of the scissile phosphodiester bond in the 3′–5′ strand and the other the equivalent bond in the 5′–3′ strand (7,8).
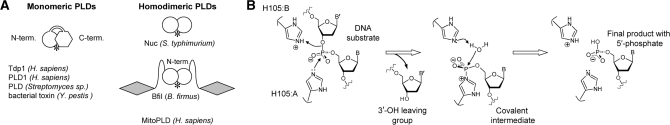
DNA nucleases of the phospholipase D (PLD) superfamily use a different mechanism for the hydrolysis of phosphodiester bonds, a two-step metal-independent scheme (9,10). The two-step scheme is conserved throughout the PLD superfamily, a large and diverse group of proteins that includes plant, mammalian and bacterial phospholipases, phospholipid synthases, bacterial toxins and poxvirus envelope proteins (11). All PLD enzymes contain two copies of a conserved sequence motif ‘HXK’; both copies contribute to the active site (11). In some instances, such as the PLD from Streptomyces species and human tyrosyl-DNA phosphodiesterase 1 (Tdp1), the enzyme is a monomeric protein with two domains, which each carry one ‘HXK’ motif that together form the active site (Figure 1A) (12,13). In contrast, the non-specific endonuclease Nuc, the restriction endonuclease BfiI and human mitochondrial phospholipase all have one ‘HXK’ motif per protein chain but form dimers with a single active site at the subunit interface that contains the ‘HXK’ motifs from both subunits (Figure 1A) (10,14,15). The histidine residues of the two ‘HXK’ motifs play key roles in catalysis by PLD enzymes (10,13,16). A His from one ‘HXK’ motif acts as the nucleophile that attacks the scissile phosphate to create a covalent phosphohistidine intermediate, while that from the second ‘HXK’ motif stabilizes the leaving group. In a monomeric PLD protein, the two active-site histidines are not equivalent and each plays a defined role in catalysis. Conversely, the active sites of the homodimeric nucleases, BfiI and Nuc, contain two identical histidines related by 2-fold symmetry, one from each subunit.
Figure 1.
Enzymes of the phospholipase D superfamily. (A). Monomeric and homodimeric PLD enzymes. The single PLD domain in the dimeric enzymes is depicted as a white circle, and the two domains in the monomeric enzymes as a circle and a hexagon. The DNA recognition domains of BfiI are marked as shaded diamonds. Both types of enzymes contain a single active site at the domain or subunit interface (marked by an asterisk). (B) The putative reaction mechanism of BfiI. During the first step of the reaction, His105 from subunit A (H105:A) attacks the scissile phosphate to generate the covalent intermediate, while His105 from subunit B (H105:B) protonates the 3′-leaving group. During the second step, a water molecule resolves the covalent intermediate releasing the first histidine (H105:A); the second histidine (H105:B) may facilitate this reaction by subtracting a proton from the water molecule.
The symmetrical arrangement of the histidines in the homodimer of restriction enzyme BfiI poses two problems. First, what are the individual roles of the two identical histidines at the active site? Does each have a distinct role, or are they interchangeable? In addition, what contribution, in quantitative terms, does each make to the catalysis of phosphodiester hydrolysis? Secondly, how does a restriction endonuclease with a single active site cut both DNA strands, despite their opposite (5′–3′ cf. 3′–5′) polarities, to generate a double-strand break.
The proposed mechanism for phosphodiester hydrolysis by dimeric PLD enzymes indicates that one of the two active-site histidines in BfiI (H105 from subunit A) should attack the scissile phosphodiester bond to form a covalent intermediate with the 5′-terminal phosphate, while the identical H105 from subunit B protonates the hydroxyl of the 3′-leaving group (Figure 1B) (9,10,13,16). In the second step, the covalent intermediate is hydrolyzed by a water molecule with the aid of H105 from subunit B, which abstracts a proton from the water, to release the cleaved DNA from the enzyme. BfiI acts sequentially on the two DNA strands, to make a double-strand break (17). It first cuts the bottom DNA strand 4-nt away from the target site (5′-ACTGGG-3′) and then switches its active site to the top strand before cleaving it 5-nt away. It was suggested that BfiI uses for the first strand cleavage H105 from one subunit as the nucleophile and the equivalent H105 from the other subunit as the proton donor/acceptor, while the role of each H105 residue is reversed in the cleavage of the complementary strand of opposite polarity (17).
In the present study, the mechanism of DNA cleavage by BfiI was elucidated by using heterodimeric variants in which the internal symmetry of its active site was disrupted by substituting one of the two H105 residues in the dimer (Figure 2). These variants were then tested against specific oligoduplexes carrying the recognition sequence for BfiI but with either a phosphodiester or a 3′-S-phosphorothiolate (with a bridging 3′-sulfur atom) linkage at the scissile bond. Analysis of the cleavage of these duplexes by WT and by the heterodimeric forms of BfiI allowed us to dissect quantitatively the contribution of the individual histidines to both the formation and the breakdown of the covalent intermediate. Surprisingly, our studies show that BfiI uses the active-site H105 from one particular subunit as the nucleophile for the cleavage of the target phosphodiester bond in both of the anti-parallel DNA strands, while the symmetry related H105 from the opposite subunit acts as the proton donor/acceptor during both strand-scission events. This represents a novel mechanism for the scission of double-stranded DNA as it requires a single active site to not only switch between strands but also to switch its orientation on the DNA.
Figure 2.
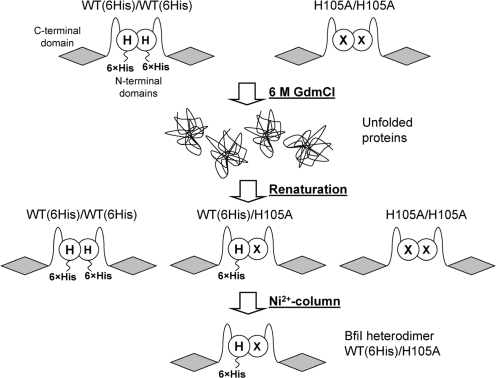
Generation of heterodimeric variants of BfiI. The dimeric forms of His-tagged WT BfiI and the H105A mutant are indicated. In both cases, the BfiI monomer is shown as two domains connected by a linker: a C-terminal DNA-binding domain (shaded diamond) and a N-terminal domain for dimerization and catalysis (unfilled circle); ‘H’ marks H105 in the WT dimer and ‘X’ marks the H105A substitution in the mutant. The His-tagged WT and the H105A homodimers are mixed and completely unfolded with 6 M GdmCl. Subsequent removal of the denaturant results in formation of three species: the two initial homodimers and the heterodimer. The heterodimer with a single His-tag is separated from the homodimeric forms of BfiI lacking the His-tag or bearing two His-tags by Ni2+-chelating chromatography.
MATERIALS AND METHODS
Protein purification
BfiI proteins without the His-tag, both the WT (wild-type) and the H105A mutant, were expressed in E. coli and purified as described in (18). BfiI proteins with an N-terminal 6 × His-tag were also expressed as before (18) and these were purified on 1 ml Ni2+ His–Trap HP and HiTrap Heparin HP columns (GE Healthcare). The fractions containing BfiI were pooled, dialyzed against Storage Buffer (200 mM KCl, 10 mM Tris–HCl (pH 8.0 at 25°C), 1 mM EDTA, 1 mM DTT and 50% v/v glycerol) and stored at −20°C. The N-terminal domains of WT BfiI and the H105A mutant (both without the His-tag) were generated by limited proteolysis in 8 mM Tris–HCl (pH 8.0 at 25°C), 200 mM NaCl and 2 mM calcium acetate (19). WT BfiI (2 mg) was digested with thermolysin (0.2 mg) for 16 h at 25°C in a final volume of 3 ml prior to adding EDTA to 10 mM, followed by heating at 40°C for 3 min. The H105A variant (1 mg) was digested with 0.01 mg of trypsin for 16 h at 25°C in a final volume of 10 ml before adding phenylmethanesulphonylfluoride to 2 mM. The digests were loaded on a HiTrap Heparin HP column pre-equilibrated in 200 mM NaCl, 20 mM Tris–HCl (pH 8.0 at 25°C), 1 mM EDTA, 7 mM 2-mercaptoethanol and 20% v/v glycerol. Bound proteins were eluted with a NaCl gradient (0.2–1.0 M). Fractions containing the N-terminal domains were pooled and dialyzed against Storage Buffer.
Generation of heterodimers
Heterodimeric forms of BfiI were constructed with subunits from two of the following: WT BfiI; the H105A mutant; WT with an N-terminal 6 × His-tag [WT(6His)]; the similarly-tagged H105A protein [H105A(6His)]; the N-terminal domain of WT BfiI [WT-N]; the N-terminal domain of H105A [H105A-N]. Two homodimeric forms of BfiI (one with the His-tag and one untagged) in Storage Buffer supplemented with 5 mM DTT were mixed together to give a 0.6-ml solution containing ∼1 mg protein, to which was added 0.6g of solid GdmCl (guanidinium monochloride). The resultant solution of protein in ∼6 M GdmCl was incubated for ∼30 min at room temperature. Successive aliquots of the denatured protein solution (10 µl) were then added to 14 ml of Renaturation Buffer (20 mM Tris–HCl (pH 8.0 at 25°C), 0.5 M KCl, 5 mM DTT, 1 mM MgCl2 and 20% v/v glycerol) at room temperature with continuous mixing. The mixture of refolded proteins was dialyzed against Renaturation Buffer with 1 mM DTT at 4°C. The various forms of BfiI were separated on a 1-ml His-Trap HP column. Protein elution was performed using an imidazole gradient (20 volumes, 15–500 mM) in 20 mM Tris–HCl (pH 8.0 at 25°C), 500 mM NaCl. The most concentrated fractions of the heterodimer were pooled and further concentrated using Ultrafree-0.5 concentrators (Millipore), dialyzed against Storage Buffer and stored at −20°C.
Concentrations of all BfiI proteins were determined from A280 measurements using extinction coefficients of 99 700 M−1cm−1 for the full-length homo/heterodimers and 80 800 M−1cm−1 for heterodimers composed of one full-length subunit and one N-terminal domain.
Oligonucleotides
The oligonucleotides used in this study are listed in Table 1. Unmodified oligonucleotides were purchased from Metabion (Martinsried, Germany). Oligonucleotides containing 3′-phosphorothiolate modifications were synthesized by similar procedures to those previously described (20). All three DNA strands containing 3′-phosphorothiolates were synthesized using 5′-dimethoxytrityl-3′-deoxy-3′-thiothymidine-phosphoramidite (0.2 M) on either an Expedite 8909 synthesizer or a MerMade 4 synthesizer. Modified oligonucleotides were validated by mass spectrometry (details are provided in Supplementary Data).
Table 1.
Oligonucleotide substrates
 |
aThe BfiI recognition sequence is underlined and sites of cleavage marked with arrows. Duplexes 14/15, 14/15s and 25/25s were radiolabeled at the 3′-termini of the bottom strand. 25s/NICK was radiolabeled at the 3′-terminus of the top DNA strand. The 13-nt segment of the bottom strand of 25s/NICK was 5′-phosphorylated.
Oligonucleotides were 3′-labeled with [α-32P]cordycepin triphosphate and terminal deoxynucleotidyl transferase (MBI Fermentas, Vilnius, Lithuania) and gel-purified. The 14/15, 14/15s and 25/25s substrates were assembled by annealing the radiolabeled bottom strand with the unlabeled top strand. The substrate 25s/NICK was assembled by annealing the radiolabeled 25 nt top strand with the 13- and 12-nt oligonucleotides of the bottom strand. To mimic the product from BfiI cleavage of the bottom strand, the 13-nt fragment was phosphorylated at its 5′-terminus using ATP and T4 polynucleotide kinase.
Reactions
DNA hydrolysis reactions were performed by manually mixing radiolabeled oligoduplexes (1–2 nM) with BfiI enzyme (50–100 nM dimer or heterodimer) in 20 mM potassium phosphate (pH 7.5), 120 mM KCl, 1 mM EDTA, 0.1 mg/ml BSA at 25°C. Samples (8 µl) were collected at timed intervals, quenched by mixing with 10 µl of 1 M NaOH and then neutralized by adding 3 µl of Neutralization Solution (1 M H3PO4, 9% SDS) and 12 µl of loading dye solution (95% v/v formamide, 25 mM EDTA, 0.01% bromphenol blue). Fast reactions were studied in a Kin-Tek RQF-3 quench-flow device: equal volumes (16 µl) of radiolabeled oligonucleotide (2–4 nM) and BfiI enzyme (100–200 nM dimer) were mixed and, after the chosen time delay, quenched with 1.0 M NaOH. The recovered samples (∼100 µl) were mixed with 8 µl of Neutralization Solution and 50 µl of loading dye. Reaction products were separated by high resolution (19:1 acrylamide/bis-acrylamide with 8 M urea in TBE buffer thermostated at 60°C). Radiolabeled DNA was detected and quantified by phosphorimager (17).
Over specified time ranges (0.25–2.5 s for WT/H105A, 0.25–1.0 s for H105A/WT-N), the covalent intermediate is the only detectable reaction product with the heterodimers of BfiI (see below: Figures 3C and 4A). Hence, no hydrolysis of the covalent intermediate occurs after the reaction is quenched with 1 M NaOH; otherwise, the reaction products would have included the final hydrolysis products. Consequently, all of the decline in the amount of the covalent intermediate observed during BfiI reactions, including a very rapid process inferred for WT BfiI (Figure 3A and B), is due to BfiI activity, and not to non-enzymatic break-down of the intermediate during sample preparation and analysis.
Figure 3.
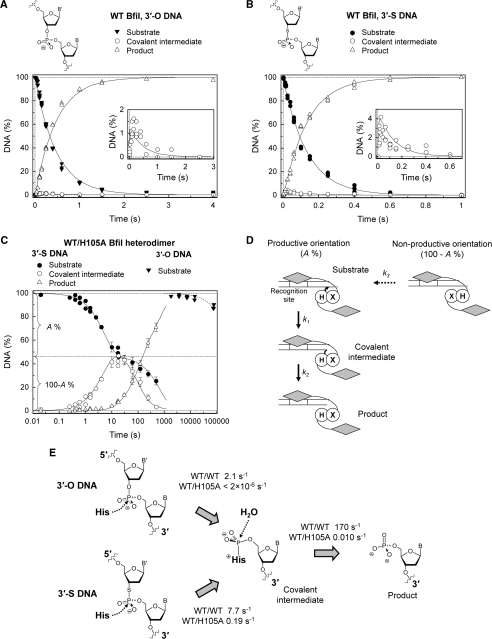
Cleavage of truncated phosphodiester and 3′-phosphorothiolate DNA substrates by WT BfiI and the WT/H105A heterodimer. The reactions contained 50–100 nM enzyme dimer and 1–2 nM radiolabeled DNA. The samples were collected and processed as described in ‘Materials and Methods’ section. (A and B) WT enzyme on the phosphodiester (3′-O) and 3′-phosphorothiolate (3′-S) substrates, 14/15 and 14/15s respectively. Cartoons above the graphs depict scissile linkages of the corresponding substrates. The time courses of intact substrate (filled symbols), covalent intermediate (open circles) and final product (open triangles) are shown: multiple data points at each time indicate values from repeat experiments. The inserts magnify the covalent intermediate data (<2 and <4% of total DNA for 3′-O and 3′-S substrates, respectively). Solid lines in both panels are the best fit of the experimental data to Equation (1) using a shared value of k2 for both substrates. The optimal fit gave k1(3′-O) = 2.1 ± 0.1 s−1, k1(3′-S) = 7.7 ± 0.1 s−1 and k2 = 170 ± 30 s−1. (C and D) The BfiI WT/H105A heterodimer on the 14/15 and 14/15s substrates. The heterodimer cleaved the 3′-S substrate 14/15s (filled circles), to form the covalent intermediate (open circles) and the final product (open triangles). The cartoon depicts the putative course of this reaction: upon mixing heterodimeric enzyme with the 14/15s substrate (the white box in the duplex marks the recognition site), only a fraction (A%) of DNA is bound by the enzyme in its active orientation capable of catalysis; the remaining fraction of the substrate (100-A%) may be cleaved only after re-binding of the enzyme in the active orientation (a slow process described by the rate constant k3). Solid lines are the optimal fit of this scheme to the 14/15s DNA cleavage data. The determined parameters are: k1 = 0.19 ± 0.02 s−1, k2 = 0.010 ± 0.001 s−1, k3 = 0.0014 ± 0.0002 s−1 and A = 54 ± 3%. Reactions of the WT/H105A heterodimer on the 3′-O substrate 14/15 are also shown (filled triangles). A single exponential fit (dashed line) gave a rate constant of 2 × 10−6 s−1 for the decline in concentration of the 14/15 substrate. (E) The impact of the single H105A substitution on BfiI activity. Compared to WT BfiI (WT/WT), the WT/H105A heterodimer lacking one active-site histidine had virtually no activity on the phosphodiester substrate, but cleaved the 3′-phosphorothiolate substrate with a 40-fold decrease in the rate of covalent intermediate formation and a 17 000-fold decrease in the rate of covalent intermediate hydrolysis.
Figure 4.
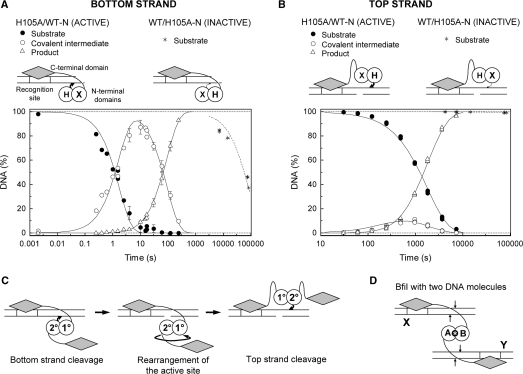
Orientation of the BfiI active site during cleavage of the bottom and the top DNA strands. Single turnover experiments were performed as in Figure 3 on oligonucleotide substrates bearing 3′-phosphorothiolate moieties at the scissile bond in either the bottom or the top strand of DNA duplex. Cartoons above the graphs depict the complexes formed by the truncated BfiI heterodimers with the corresponding duplexes. (A) Cleavage of the bottom strand of the 14/15s DNA by the BfiI heterodimers H105A/WT-N and WT/H105A-N. The heterodimer H105A/WT-N cleaved the substrate (filled circles) to the final product (open triangles) via the covalent enzyme–DNA intermediate (open circles). Solid lines are the best fit of the experimental data to Equation (1), which yielded k1 = 0.57 ± 0.07 s−1 and k2 = 0.011 ± 0.001 s−1. The preparation of WT/H105A-N caused only a residual level of DNA cleavage (asterisks). The best fit to a single exponential (dashed line) yielded a cleavage rate of 1.2 × 10−5 s−1. (B) Cleavage of the top strand of the 25s/NICK substrate. The H105A/WT-N heterodimer, that was active on the bottom strand (A), also cleaved the 25s/NICK DNA (filled circles) to the final product (open triangles) via the covalent intermediate (open circles). The fit to Equation (1) yielded k1 = 0.00056 ± 0.00001 s−1 and k2 = 0.0041 ± 0.0003 s−1. The WT/H105A-N heterodimer displayed only residual activity on this substrate (asterisks). The fit to an exponential (dashed line) yielded a cleavage rate of 1 × 10−7 s−1. (C) A model for the reactions of WT BfiI on the bottom and the top strand of a DNA duplex. The H105 residue from the same subunit of the homodimer, the 2° subunit not bound to the DNA makes the nucleophilic attacks on the target phosphodiester bonds in both bottom and top strands of the DNA. To match the anti-parallel orientation of the two strands, the N-terminal domains of BfiI must rotate by 180° between the two hydrolysis reactions. (D) Schematic representation of the synaptic complex of the BfiI endonuclease bound two copies of its recognition sequence. The two DNA duplexes are marked X and Y: boxed segments indicate recognition sites and the positions of the scissile phosphates marked by arrows. The two subunits of the dimeric protein are noted as A and B and the single active site is marked by an asterisk. All four of the target phosphodiester bonds are cleaved in this one active site in sequential reactions.
Data analysis
Non-linear regression analysis (Supplementary Data) used KYPLOT 2.0 software (21).
RESULTS
Generation of BfiI heterodimers
Each monomer of BfiI contains two domains: an N-terminal catalytic domain that belongs to the PLD superfamily and a C-terminal DNA-binding domain that specifically recognizes its target sequence, 5′-ACTGGG-3′ (19). The functionally active form of BfiI is a homodimer made through the association of two catalytic domains (Figure 1A). The homodimer has two DNA-binding surfaces located at the C-terminal domains but only one active site, at the dimer interface between the N-terminal domains (17,18). Consequently, a mutation in the gene for BfiI that substitutes an active-site residue results in a homodimeric mutant with two substitutions at the active site (Figure 2). Site-directed mutagenesis therefore cannot be applied directly to probe the roles of individual active-site residues in this enzyme. To solve this problem, a method was developed to obtain heterodimers of BfiI composed of WT and mutant subunits (Figure 2).
The dimerization interface at the N-terminal domains of BfiI is disrupted by 4 M GdmCl, but the protein is known to reassemble into a functionally active dimer upon removal of the denaturant (19). To generate a variant of BfiI with a substitution at one of the two His105 residues at the active site, an equimolar mixture of WT(6His) (the native protein with a His-tag) and the H105A mutant were unfolded in 6 M GdmCl and, after renaturation, the protein solution was applied to a Ni2+-NTA column and eluted with an imidazole gradient. Three different forms of BfiI were obtained: the two initial homodimers and the heterodimer with a single His-tag (Figure 2). The heterodimer with a single tag forms a weaker complex with the Ni2+-NTA resin than the homodimer with two His-tags and eluted at a lower imidazole concentration (Supplementary Figure S1A) while the untagged homodimer did not bind to the Ni2+ column at all. To check whether the His-tag or protein refolding had any influence on BfiI function, the above procedure was used to generate a control heterodimer, WT(6His)/WT, which had a native active site and a single His-tag. The WT(6His)/WT heterodimer showed the same specific activity and DNA binding as the native WT homodimer (data not shown). In addition, the WT/H105A heterodimer was constructed in two possible configurations with the His-tag on the WT or the mutant subunit, H105A(6His)/WT and WT(6His)/H105A, respectively: no significant differences were observed between these alternatives.
WT BfiI on phosphodiester and 3′-phosphorothiolate substrates
The homodimeric WT BfiI enzyme and the heterodimer containing a single His residue at its active site were tested against truncated oligoduplex substrates that lacked the site of top-strand cleavage 5-nt downstream of the recognition sequence but which possessed the site of bottom-stand cleavage 4-nt away: the latter site featured either a conventional phosphodiester bond (the 14/15 duplex: Table 1) or a 3′-phosphorothiolate linkage (14/15s). Our first set of experiments investigated the cleavage of these two duplexes by WT BfiI (Figure 3A and B). The reactions were initiated by mixing equal volumes of 3′-radiolabeled DNA and enzyme in a quench-flow device. The reactions were stopped after the requisite time delays by mixing with 1 M NaOH and the samples then neutralized before analysis by denaturing PAGE. To ensure the binding of only one DNA molecule to the BfiI dimer, the reactions were performed with enzyme (50–100 nM BfiI dimer) in large excess over the substrate (1–2 nM). Reaction rates at both enzyme concentrations tested (50 and 100 nM) were identical (data not shown), suggesting that all of the substrate was bound to enzyme and the observed reaction rates are not limited by the association step but must instead reflect just the conversion of enzyme-bound substrate to product.
WT BfiI rapidly converted both the unmodified and the 3′-phosphorothiolate substrates to the hydrolysis product (Figure 3A and B, respectively). Nonetheless, a small fraction of the 3′-radiolabeled substrates remained in the wells of the polyacrylamide gels used for sample analysis. The amount of this reaction product increased at the beginning of the reaction but declined upon longer incubation (Figure 3A and B). Moreover, this novel product was resistant to heating at 90°C. The slowly migrating product is shown below (Supplementary Figure S2) to be the predicted covalent BfiI–DNA intermediate which was not detected in previous studies, most likely due to its low yield and to the sub-optimal procedure used previously for sample neutralization (9).
According to the reaction mechanism in Figure 1B, the time course for DNA hydrolysis by BfiI can be described by Equation (1):
| 1 |
where k1 is the rate constant for the conversion of the substrate S into the covalent intermediate Ci and k2 the rate constant for the hydrolysis of the covalent intermediate into the final product P. The phosphorothiolate substrate differs from the standard oligoduplex only in a single sulfur atom, and this sulfur forms part of the dinucleotide released upon the formation of the covalent intermediate. Consequently, the covalent intermediates formed with both the 14/15 and 14/15s substrates are identical, so the hydrolysis rates (k2) of both complexes should be the same. Therefore, the kinetics of both phosphodiester and 3′-phosphorothiolate cleavage by WT BfiI (Figure 3A and B) were fitted to Equation (1) using the same value for k2, as described in Supplementary Data. The fit yielded comparable values of k1 for both the 14/15 and the 14/15s substrates (2.1 and 7.7 s−1, respectively), implying that the better leaving-group ability of the 3′-phosphorothiolate in the 14/15s substrate has only a minor effect on the enzyme with a native active site. The value of 170 s−1 for k2 indicates that the covalent intermediate formed with either substrate is hydrolyzed at an extremely rapid rate.
WT/H105A BfiI heterodimer on phosphodiester and 3′-phosphorothiolate substrates
The WT/H105A heterodimer was also tested against both the phosphodiester (14/15) and phosphorothiolate (14/15s) substrates (Figure 3C). The heterodimer cleaved only a small fraction of the phosphodiester substrate even after prolonged reactions: the rate of utilization of the 14/15 duplex was 106-fold slower with the WT/H105A heterodimer than with the WT homodimer. This underscores the necessity of both active-site histidines for phosphodiester hydrolysis by BfiI. In contrast, the WT/H105A heterodimer readily cleaved the substrate with the 3′-phosphorothiolate substitution, albeit in an unusual biphasic reaction (Figure 3C). Approximately half of the DNA was converted rapidly into the covalent intermediate, which was then hydrolyzed at a relatively slow rate into the final product, while the decline in the concentration of the remaining substrate occurred at an even slower rate.
The WT/H105A heterodimer contains a single active-site histidine but two equivalent DNA-binding domains. A model that can account for the biphasic reaction (Figure 3C) is that the BfiI heterodimer binds to its recognition site in either of two alternative and equally probable orientations (Figure 3D). In the productive orientation, the subunit carrying the single H105 residue is juxtaposed against the scissile phosphodiester bond in the bottom strand, which allows for the formation of the covalent phosphohistidine intermediate and results in the fast cleavage of ∼50% of DNA. In the alternative non-productive orientation, the subunit with the H105A mutation is placed against the target bond and so cannot form the covalent intermediate. In the latter case, the heterodimer has to dissociate and then re-bind in the productive orientation before cleavage can only occur. This rearrangement is slow and therefore impedes cleavage of the remaining 50% of the substrate. However, the heterodimer configurations assigned to the productive and to the non-productive orientations in Figure 3D are arbitrary. There exist two possible arrangements of the heterodimer: the C-terminal domain bound to the recognition site can come from either the subunit carrying H105 or it can come from the subunit with A105, but which of these is the productive and which the non-productive configuration cannot be determined from the data in Figure 3C.
The kinetics of the reactions of the WT/H105A heterodimer on the 14/15s duplex (Figure 3C) were fitted to the rate equations (Supplementary Data) for the reaction scheme in Figure 3D. This scheme, with two binding orientations of the heterodimer on the DNA, accounted quantitatively for the experimental data. As predicted by the model, the heterodimer initially binds ∼50% of DNA in the productive orientation (parameter A = 54%) and rapidly converts the 14/15s substrate into the covalent intermediate followed by the slow release of the final hydrolysis product, with rate constants of 0.19 and 0.010 s−1 for the formation and decay of the intermediate (k1 and k2 respectively). The dissociation of the non-productive heterodimer-DNA complex and formation of the productive complex is the slowest step in this scheme (k3 = 0.0014 s−1). The rate constants k1 and k2 are intrinsic to the active orientation of the WT/H105A heterodimer and therefore can be directly compared to corresponding parameters of WT BfiI (Figure 3E).
Analysis of BfiI–DNA covalent intermediate
The high yield of the presumed covalent intermediate formed by the BfiI heterodimers on the 14/15s substrate enabled the biochemical characterization of this novel species. First, samples from the reactions of the WT(6His)/H105A and the H105A(6His)/WT heterodimers on radiolabeled 14/15s were analyzed by SDS–PAGE (Supplementary Figure S2A). Phosphorimager records of the gels revealed, in addition to the DNA itself, a radiolabeled species with the mobility expected for the covalent intermediate: the mass of the BfiI monomer with or without the 6 × His-tag (40.0 and 42.5 kDa, respectively) bound to a 14-nt DNA fragment (∼4.2 kDa).
Reactions were also carried out on the unlabeled 14/15s substrate using both variants of the WT/H105A heterodimer, with the His-tag on either the WT or the mutant subunit. Samples from the reactions were then analyzed by western bloting using a polyclonal anti-BfiI antibody, to visualize simultaneously the unreacted BfiI proteins and the postulated covalent intermediate (Supplementary Figure S2B). The anti-BfiI antibody revealed that both the WT(6His)/H105A and the H105A(6His)/WT heterodimers gave additional bands with reduced mobilities upon the addition of the DNA, presumably due to a protein–DNA adduct. Formation of the presumed adduct concomitantly decreased the amount of unreacted WT BfiI subunits but did not affect the amount of mutant H105A subunits (Supplementary Figure S2B).
As expected for a phosphohistidine intermediate (22–24), the low-mobility radiolabeled species formed by the heterodimers of BfiI are labile in acid but are stable at alkaline pH (Supplementary Figure S2C); in acid, they were converted into the final hydrolysis product. Furthermore, proteinase K digestion converted the low-mobility radiolabeled band into a novel high-mobility species, presumably the DNA fragment covalently attached to a proteolytic fragment of BfiI (Supplementary Figure S2C). Taken together, the above data indicate that the low-mobility species is indeed the covalent BfiI–DNA intermediate.
Orientation of the active site of BfiI for cutting bottom and top strands
The reactions of the WT/H105A heterodimer of BfiI on 14/15s, a DNA substrate with a target phosphodiester bond only in the bottom strand (Figure 3C), suggests that the heterodimer must bind to its recognition site in a defined orientation in order to cleave the bottom strand (Figure 3D). However, it is unclear whether in the productive binding orientation the single H105 in this heterodimer, that must act as a nucleophile in the reaction, comes from the subunit directly bound via its C-terminal domain to the recognition site [the primary, or 1°, subunit: (25)] or from the subunit whose C-terminal domain is not bound to the recognition site (the secondary, or 2°, subunit). To address this question, two truncated forms of the WT/H105A heterodimer were generated in which either the WT or the H105A mutant subunit lacked the C-terminal DNA-binding domain, the H105A/WT-N or the WT/H105A-N variants respectively: both still posses the N-terminal catalytic domain carrying residue 105.
Contrary to the full length WT/H105A heterodimer that can bind in two alternative configurations, productive or non-productive (Figure 3D), only the full-length subunit of a truncated heterodimer can act as the 1° subunit, thus fixing its configuration on the DNA to one particular orientation. Consequently, if the His105 residue that mounts the in-line attack on the scissile phosphate in the bottom strand comes from the 1° subunit bound to the recognition site, then the WT/H105A-N variant must form the productive complex, as the WT subunit is the only subunit that can bind to the recognition site: its H105A-N subunit lacks the DNA-binding domain and so can only act as the 2° monomer. A further prediction from the scheme in which the 1° subunit makes the attack is that the H105A/WT-N heterodimer forms exclusively the non-productive complex as only its H105A subunit can bind to the site. In the productive complex, the 3′-phosphorothiolate DNA should be cleaved in a monophasic reaction, while no cleavage should occur in the non-productive complex. On the other hand, if the key histidine comes from the 2° subunit not bound to the recognition site, then H105A/WT-N will be active on the bottom strand but not WT/H105A-N. Furthermore, if the two H105 residues from the two subunits in the BfiI dimer exchange functions between cutting the two DNA strands of opposite polarity, as suggested previously (17), then the truncated heterodimer that can attack the bottom strand will be inactive towards the top strand and, conversely, the truncated species that is inactive on the bottom strand should be active towards the top strand.
Truncated heterodimers were constructed as shown in Figure 2. To produce the H105A/WT-N heterodimer, the denaturation/refolding procedure was applied to a mixture of His-tagged H105A homodimer and the N-terminal domain of WT BfiI lacking a His-tag: the latter was obtained by limited proteolysis [(19) and ‘Materials and methods’ section). The heterodimer with a single His-tag was separated from the mixture by Ni2+-chelating chromatography (Supplementary Figure S1B). The WT/H105A-N heterodimer was generated from a mixture of WT BfiI with a His-tag and the N-terminal domain of the H105A mutant. Both variants of truncated heterodimer retained the ability to bind cognate DNA in gel shift assays (data not shown).
The truncated heterodimers were tested against the 14/15s duplex, a substrate that lacks a scissile bond in the top strand of the DNA but which carries a 3′-phosphorothiolate at the cleavage site in the bottom strand (Table 1). One of the truncated heterodimers, H105A/WT-N, cleaved the 14/15s duplex 5000-fold more rapidly than the other, WT/H105A-N (Figure 4A). Furthermore, the H105A/WT-N dimer formed a significant amount of covalent intermediate, and consumed all substrate in a single phase, in a reaction that could be fitted to Equation (1) (Figure 4A). Analysis of the covalent intermediate formed between the 14/15s substrate and the H105A/WT-N heterodimer revealed that the DNA fragment was attached to the ∼26 kDa N-terminal domain WT of BfiI (Supplementary Figure S2A and D).
In the heterodimer H105A/WT-N, only the H105A subunit can act as the 1° subunit on the recognition site as only this chain possesses a DNA recognition domain, but this subunit carries an Ala in place of H105 so cannot form the phosphohistidine intermediate. Therefore, the H105 residue that performs the nucleophilic attack on the scissile phosphate in the bottom DNA strand must come from the WT-N subunit, but this cannot bind to the recognition site as it lacks the DNA recognition domain, so can only function as the 2° subunit. Accordingly, during the reaction of the WT homodimer on the bottom strand, the H105 residue of the 2° subunit makes the nucleophilic attack while the H105 residue from the DNA-bound 1° subunit most likely protonates the DNA leaving group and activates the water molecule in the second step of the reaction (Figure 1B).
The very slow cleavage of 14/15s DNA by the WT/H105A-N heterodimer (Figure 4A) suggests that it binds the substrate in the non-productive orientation, with the A105 residue in the 2° subunit positioned for the in-line attack on the scissile phosphate. One might have expected this arrangement to result in zero rather than severely reduced activity. The observed residual activity of the WT/H105A-N heterodimer may, however, be due to a minor contamination in the preparation of the heterodimer with WT homodimer. This scheme is consistent with the observation that the WT/H105A-N preparation also displayed detectable activity on DNA substrates lacking the 3′-phosphorothiolate substitution (data not shown).
The WT BfiI homodimer uses its single active site at the dimer interface to cut both DNA strands in a fixed order: it has to cut the bottom strand before it can cut the top strand (17). Tests to determine which of the two H105 residues attacks the scissile phosphate during cleavage of the top DNA strand were carried out with a full-length oligoduplex, 25s/NICK (Table 1). The top strand of this substrate was made susceptible to heterodimer cleavage by introducing a 3′-phosphorothiolate at the scissile position 5-nt downstream of the recognition site and a nick at the bottom DNA strand, in order to mimic the natural intermediate during BfiI-cleavage of double-stranded DNA (17). When the H105A/WT-N and the WT/H105A-N heterodimers were tested against the 25s/NICK duplex, the H105A/WT-N variant formed first the covalent intermediate which was then converted at a relatively rapid rate to the final reaction product, while the WT/H105A-N heterodimer showed only trace activity (Figure 4B). The H105A/WT-N heterodimer is thus active on both the bottom and the top strands while the alternative truncated heterodimer, WT/H105-N, is essentially inactive on both strands.
In the H105A/WT-N enzyme, the single copy of H105 is located in the shortened WT subunit that cannot bind the recognition sequence as it lacks the DNA-binding domain. Hence, the H105 residue in the 2° subunit must mount the nucleophilic attack on the scissile phosphate not only in the bottom strand but also the top. In contrast, in the WT/H105A-N dimer, the single H105 is located in the full-length WT subunit bound to the DNA, and this residue seemingly cannot be utilized to attack either the bottom or the top strands.
It is possible that BfiI might cleave the bottom strand in a full-length 25-bp duplex, that had scissile bonds in both strands, by a different mechanism compared to the abbreviated 14/15 duplexes that have scissile sites only in the bottom strand. To examine this possibility, the activity of heterodimers H105A/WT-N and WT/H105A-N were tested against a full length duplex, 25/25s, with a 3′-phosphorothiolate linkage at the target bond in the bottom strand (Table 1). As with the truncated 14/15s substrate, only the H105A/WT-N heterodimer cleaved the bottom strand of the 25/25s duplex, to form a detectable amount of the covalent intermediate (Supplementary Figure S3). Hence, BfiI employs the same orientation of its active site during bottom strand cleavage in both abbreviated and full length DNA substrates.
DISCUSSION
Monomeric PLD-superfamily enzymes like human Tdp1 and PLD from Streptomyces species (12,13) are bi-lobed monomers (Figure 1A) which contain in the active site two His residues from duplicated ‘HXK’ sequence motifs located distantly in the protein chain. The active-site histidines of these enzymes are not equivalent and perform pre-defined roles in catalysis. One particular His mounts the nucleophilic attack on the scissile phosphate to make a covalent intermediate (Figure 1B). Not surprisingly, its replacement by site-directed mutagenesis renders the enzyme completely inactive (13,16,26). The other His plays a supporting role—it protonates the leaving group during the formation the covalent intermediate and subsequently facilitates the hydrolysis of the phosphohistidine linkage. Mutations of the latter histidine residue often compromise catalytic activity and result in the accumulation of the covalent intermediate (27).
The structurally characterized PLD-family nucleases—Nuc and the restriction endonuclease BfiI (10,14)—are homodimers which contain a single active site structurally similar to that of human Tdp1 and PLD from Streptomyces species. However, in contrast to monomeric PLD enzymes, the active site of BfiI is fully symmetric, as it contains two His residues related by the 2-fold symmetry axis of the dimer, each donated by one enzyme subunit (Figure 1A). This precludes the assignment of the individual roles of each His residue in catalysis. To solve this problem, we disrupted the 2-fold symmetry intrinsic to BfiI by constructing heterodimeric forms of the enzyme (Figure 2).
The roles of active-site histidines in catalysis
WT BfiI forms the covalent intermediate on truncated phosphodiester and 3′-phosphorothiolate substrates with comparable rates (2.1 and 7.7 s−1, respectively: Figure 3E), despite the substantially more acidic leaving group of the 3′-phosphorothiolate substrate [the pKa values of the 3′-SH and 3′-OH groups are ∼11 and ∼16, respectively (28)], suggesting that protonation of the 3′-leaving group is not a rate-determining factor for WT BfiI. The ability of BfiI to cleave the 3′-phosphorothiolate linkage more rapidly than the all-oxygen substrate sharply contrasts with most metal-dependent nucleases (5,29,30). These enzymes, unlike the metal independent BfiI, are inhibited by the 3′-S substitution due to its impaired interaction of Mg2+ ions with sulfur; however, some of these enzymes are rescued by the more thiophilic Mn2+ ion (5,29).
The replacement of one of the active-site histidines with alanine, in the WT/H105A heterodimer, resulted in a dramatic 106-fold decrease in the rate of cleavage of the oxyester bond in the 14/15 oligoduplex (Figure 3E). The residual activity could have been due to a small amount of WT BfiI present in the sample of heterodimer but different preparations of the heterodimer, including the alternative variants WT(6His)/H105A and H105A(6His)/WT, all gave the same low level of activity. Therefore, the observed activity is most likely intrinsic to the WT/H105A heterodimer. Thus, the second His105 residue in the active site of BfiI accelerates the formation of the covalent intermediate on the phosphodiester substrate by a factor of at least 106.
Strikingly, the heterodimer cleaved the 3′-phosphorothiolate linkage in the 14/15s duplex 105-fold more rapidly than the phosphodiester group in the original 14/15 substrate (Figure 3E). The resultant rate of covalent intermediate formation by the heterodimer (0.19 s−1) is only 40-fold lower than that for the WT enzyme on the same substrate (7.7 s−1, Figure 3E). Furthermore, the rate enhancement over the all-oxygen substrate by a factor of 105 coincides with the pKa difference between the 3′-OH and 3′-SH leaving groups [∼5 units (28)]. These observations argue that one of the two H105 residues in the WT homodimer of BfiI protonates the 3′-leaving group during the first reaction step (Figure 1B): upon its removal, in the WT/H105A heterodimer, the stability of the conjugate base of the 3′-leaving group becomes a major factor governing the reaction rate. The difference in rate between the heterodimeric and WT enzymes on the 3′-phosphorothiolate substrate implies that even the relatively acidic 3′-thio leaving group must be protonated to achieve the maximum rate of covalent intermediate formation.
The second step in the reaction, the hydrolysis of the covalent enzyme–DNA intermediate (Figure 1B), is an extremely rapid process for WT BfiI (k2 = 170 s−1, Figure 3E). However, the rate is reduced by a factor of 17 000 for the WT/H105A heterodimer (k2 = 0.010 s−1). The dramatic effect of the H105A substitution confirms the direct involvement of the second active-site histidine in the hydrolysis of the covalent phosphohistidine intermediate; presumably, the second histidine activates the water molecule that hydrolyzes the intermediate by removing a proton (Figure 1B).
The single H105A substitution also reverses the ratio of the reaction rates for the formation (k1) and decay (k2) of the covalent intermediate. For WT BfiI on the 14/15s substrate, the ratio of k1/k2 is 0.05, i.e. the covalent intermediate is formed 20 times more slowly than it is hydrolyzed. For the WT(6His)/H105A and H105A/WT-N heterodimers, the ratio of k1/k2 is ≥20, which leads to the accumulation of the covalent intermediate during the reaction (Figures 3C and 4A). Biochemical analysis of the low-mobility species postulated to be the covalent intermediate (Supplementary Figure S2) indicated that this was the expected phosphohistidine adduct. The covalent intermediate is formed only by the wild type but not the H105A subunit of the heterodimer (Supplementary Figure S2B and D). Moreover, as expected for a phosphohistidine compound (22,24), the BfiI–DNA adduct is stable at alkaline pH but decomposes in acid (Supplementary Figure S2C).
A novel mechanism for double-stranded DNA cleavage
Nucleases that cut double-stranded DNA often contain two identical subunits related by rotational symmetry, so that the active site from one subunit cleaves the 5′–3′ strand while that from the oppositely-oriented subunit attacks the anti-parallel 3′-5′ strand (7). However, this strategy cannot be generalized for all nucleases that act on double-stranded DNA. A number of enzymes including the homing endonuclease I-TevI (31), the RecBCD complex of E. coli (32) and the BfiI restriction enzyme (17) all utilize single active site to cut both DNA strands, despite their opposite polarities.
The intron-encoded endonuclease I-TevI is a monomer and contains a single active site but it cuts both DNA strands at the recipient site for intron homing, leaving in both cases products with 3′-hydroxyl and 5′-phosphate termini (33). It is thought that it first cleaves its target phosphodiester bond in the bottom strand and then distorts the DNA to guide into the active site the scissile phosphate from the top strand (31). However, it is not yet clear how its single active site can accommodate and cut phosphodiester bonds from both the 3′–5′ and the 5′–3′ strands of the DNA, as in both cases it has to displace the leaving group on the 3′ side of the phosphorous at the scissile bond: i.e., in opposite directions on the 3′–5′ compared to the 5′–3′ strand. Moreover, given the crystal structure of the catalytic domain of I-TevI, alternative reaction schemes, including transient dimerization, cannot be excluded (34). The mode of action of another monomeric endonuclease, FokI, involves transient dimerization (35,36), to give a protein assembly at the recognition site with two catalytic domains juxtaposed in anti-parallel alignment (37,38), which each cut one strand of the DNA. In this case, the 1° monomer bound directly to the recognition site cleaves the bottom strand while the 2° monomer recruited to the site by protein–protein interactions cuts the top strand (25), but the symmetry within the dimer of catalytic domains (37) enables one to cut the 3′–5′ strand and the other the 5′–3′ strand.
The E. coli RecBCD enzyme acts in the repair of double-stranded DNA breaks as a trimeric protein with multiple catalytic activities that include two helicase functions, both 3′→5′ and 5′→3′ in the B and D subunits respectively and a single endonuclease function, located in B from where it degrades both strands (32). To account for how the nuclease cleaves both of the newly unwound strands despite their opposite polarities, it was suggested that the nascent 3′-terminus generated by RecB progresses directly into the nuclease centre, which is also in B, while the nascent 5′-terminus generated by RecD forms a loop before entering the nuclease centre with the same 3′–5′ orientation as the 3′-strand (39). However, this model has yet to be confirmed experimentally, though it can readily be reconciled to the crystal structure of RecBCD (40).
The BfiI restriction enzyme employs yet another strategy. It had been shown previously that it uses a single active site to cut both DNA strands downstream of its recognition site in sequential steps, in a fixed order; first the bottom and only then the top strand (17). The BfiI endonuclease contains two symmetrically-positioned His residues at the active site, so it was proposed that BfiI cuts one strand by using the histidine from one subunit as the nucleophile and that from the other subunit as the proton donor/acceptor, while these roles are reversed for cutting the complementary strand of opposite polarity (17).
In this article, this hypothesis was tested experimentally by using truncated heterodimers of BfiI that lack the DNA-binding domain from one subunit: from either the subunit carrying the inactivating H105A mutation, WT/H105A-N; or from the WT subunit, H105A/WT-N. Contrary to the full length heterodimers bearing both DNA recognition domains (Figure 3D), each truncated heterodimer has to bind DNA in a specified orientation: either the productive orientation in which the His105 residue from the WT subunit is positioned for the in-line attack on the scissile phosphate; or the non-productive orientation where the residue in position for the in-line attack is the alanine from the H105A subunit (Figure 4A and B). Hence, if the WT/H105-N heterodimer shows catalytic activity, the covalent intermediate is formed by the histidine from the full-length enzyme subunit bound to the target site on DNA, the 1° subunit. Alternatively, if the H105A/WT-N variant displays activity, the histidine nucleophile comes from the truncated subunit that is not bound to the recognition sequence, the 2° subunit. Thus, by analyzing the activities of the truncated heterodimers, we were able to identify directly which histidine residue forms the covalent intermediate during the cleavage of the bottom (3′–5′) and the top (5′–3′) DNA strands.
Contrary to the suggestion that H105 from one particular subunit of BfiI attacks the scissile bond in the bottom (3′–5′) DNA strand while the symmetry-related H105 from the opposite subunit takes this role for cutting the top (5′–3′) strand, it was found here the His from the 2° subunit not bound to the recognition site attacks sequentially the target phosphodiester bonds in both 3′–5′ and 5′–3′ strands. The equivalent histidine from the DNA-bound 1° subunit presumably acts as the proton donor/acceptor for the reactions on both strands. To match the anti-parallel polarity of the two DNA strands, the catalytic center of BfiI therefore must rotate by 180° between the two hydrolysis reactions (Figure 4C). Thus, we demonstrate here a novel mechanism for the scission of double-stranded DNA as it requires a single active site to not only switch between strands but also to switch its orientation on the DNA.
The above reactions all contained BfiI in excess over the DNA, to favor binding of a single DNA molecule to each enzyme dimer. However, BfiI is optimally active when bound to two copies of its recognition sequence (18). To cut four phosphodiester bonds across two target sites, the single active site in the BfiI dimer must relocate between the scissile phosphates in the two sites, cleaving one phosphodiester bond at a time. In the synaptic complex of the BfiI dimer with two recognition sites (Figure 4D), one subunit (B) is attached via its DNA-binding domain to recognition site X while the other subunit (A) is attached to site Y. This leaves subunit A as the 2° subunit with respect to recognition site X so H105 from A presumably forms the covalent intermediate during the sequential cutting of both strands at site X, while H105 from subunit B fulfils the proton donor/acceptor roles in both strand-scission events. Conversely, subunit B is the 2° subunit for site Y so, for cutting this second site, the H105 residues from the B and the A subunits should fulfill the same roles as those played by, respectively, the A and the B subunits when cutting site X. Hence, the two His residues may switch roles while cleaving two specific sites bound to an enzyme dimer (Figure 4D), though it will always be the histidine from one particular subunit that attacks both bottom and top strands at each DNA site (Figure 4C).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Wellcome Trust Collaborative Research Initiative Grant 074498/Z/04 (to V.S. and S.E.H.); Lithuania Science and Studies Foundation Grant T-17/08 (to G.S.); EPSRC research grant EP/F011938 (to R.C. and J.W.G.); Marie Curie Research Training Network ‘DNA Enzymes’. Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank D. Golovenko and E. Manakova for samples of WT BfiI and H105A mutant without His-tags.
REFERENCES
- 1.Horton NC. In: Protein-Nucleic Acid Interactions: Structural Biology. Rice PA, Correll CC, editors. Cambridge, UK: RSC Publishing; 2008. [Google Scholar]
- 2.Beese LS, Steitz TA. Structural basis for the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991;10:25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Derbyshire V, Grindley ND, Joyce CM. The 3′-5′ exonuclease of DNA polymerase I of Escherichia coli: contribution of each amino acid at the active site to the reaction. EMBO J. 1991;10:17–24. doi: 10.1002/j.1460-2075.1991.tb07916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Connolly BA, Eckstein F, Pingoud A. The stereochemical course of the restriction endonuclease EcoRI-catalyzed reaction. J. Biol. Chem. 1984;259:10760–10763. [PubMed] [Google Scholar]
- 5.Elliott SL, Brazier J, Cosstick R, Connolly BA. Mechanism of the I DNA T:G-mismatch endonuclease (Vsr protein) probed with thiophosphate-containing oligodeoxynucleotides. J. Mol. Biol. 2005;353:692–703. doi: 10.1016/j.jmb.2005.08.048. [DOI] [PubMed] [Google Scholar]
- 6.Mizuuchi K, Nobbs TJ, Halford SE, Adzuma K, Qin J. A new method for determining the stereochemistry of DNA cleavage reactions: application to the SfiI and HpaII restriction endonucleases and to the MuA transposase. Biochemistry. 1999;38:4640–4648. doi: 10.1021/bi990054p. [DOI] [PubMed] [Google Scholar]
- 7.Pingoud A, Fuxreiter M, Pingoud V, Wende W. Type II restriction endonucleases: structure and mechanism. Cell Mol. Life Sci. 2005;62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galburt EA, Stoddard BL. Catalytic mechanisms of restriction and homing endonucleases. Biochemistry. 2002;41:13851–13860. doi: 10.1021/bi020467h. [DOI] [PubMed] [Google Scholar]
- 9.Sasnauskas G, Connolly BA, Halford SE, Siksnys V. Site-specific DNA transesterification catalyzed by a restriction enzyme. Proc. Natl Acad. Sci. USA. 2007;104:2115–2120. doi: 10.1073/pnas.0608689104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stuckey JA, Dixon JE. Crystal structure of a phospholipase D family member. Nat. Struct. Biol. 1999;6:278–284. doi: 10.1038/6716. [DOI] [PubMed] [Google Scholar]
- 11.Ponting CP, Kerr ID. A novel family of phospholipase D homologues that includes phospholipid synthases and putative endonucleases: identification of duplicated repeats and potential active site residues. Protein Sci. 1996;5:914–922. doi: 10.1002/pro.5560050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies DR, Interthal H, Champoux JJ, Hol WG. The crystal structure of human tyrosyl-DNA phosphodiesterase, Tdp1. Structure. 2002;10:237–248. doi: 10.1016/s0969-2126(02)00707-4. [DOI] [PubMed] [Google Scholar]
- 13.Leiros I, McSweeney S, Hough E. The reaction mechanism of phospholipase D from Streptomyces sp. strain PMF. Snapshots along the reaction pathway reveal a pentacoordinate reaction intermediate and an unexpected final product. J. Mol. Biol. 2004;339:805–820. doi: 10.1016/j.jmb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 14.Grazulis S, Manakova E, Roessle M, Bochtler M, Tamulaitiene G, Huber R, Siksnys V. Structure of the metal-independent restriction enzyme BfiI reveals fusion of a specific DNA-binding domain with a nonspecific nuclease. Proc. Natl Acad. Sci. USA. 2005;102:15797–15802. doi: 10.1073/pnas.0507949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi SY, Huang P, Jenkins GM, Chan DC, Schiller J, Frohman MA. A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat. Cell Biol. 2006;8:1255–1262. doi: 10.1038/ncb1487. [DOI] [PubMed] [Google Scholar]
- 16.Davies DR, Interthal H, Champoux JJ, Hol WG. Insights into substrate binding and catalytic mechanism of human tyrosyl-DNA phosphodiesterase (Tdp1) from vanadate and tungstate-inhibited structures. J. Mol. Biol. 2002;324:917–932. doi: 10.1016/s0022-2836(02)01154-3. [DOI] [PubMed] [Google Scholar]
- 17.Sasnauskas G, Halford SE, Siksnys V. How the BfiI restriction enzyme uses one active site to cut two DNA strands. Proc. Natl Acad. Sci. USA. 2003;100:6410–6415. doi: 10.1073/pnas.1131003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lagunavicius A, Sasnauskas G, Halford SE, Siksnys V. The metal-independent type IIs restriction enzyme BfiI is a dimer that binds two DNA sites but has only one catalytic centre. J. Mol. Biol. 2003;326:1051–1064. doi: 10.1016/s0022-2836(03)00020-2. [DOI] [PubMed] [Google Scholar]
- 19.Zaremba M, Urbanke C, Halford SE, Siksnys V. Generation of the BfiI restriction endonuclease from the fusion of a DNA recognition domain to a non-specific nuclease from the phospholipase D superfamily. J. Mol. Biol. 2004;336:81–92. doi: 10.1016/j.jmb.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 20.Gaynor JW, Bentley J, Cosstick R. Synthesis of the 3′-thio-nucleosides and subsequent automated synthesis of oligodeoxynucleotides containing a 3′-S-phosphorothiolate linkage. Nat. Protoc. 2007;2:3122–3135. doi: 10.1038/nprot.2007.451. [DOI] [PubMed] [Google Scholar]
- 21.Yoshioka K. KyPlot – a user-oriented tool for statistical data analysis and visualization. CompStat. 2002;17:425–437. [Google Scholar]
- 22.Gottlin EB, Rudolph AE, Zhao Y, Matthews HR, Dixon JE. Catalytic mechanism of the phospholipase D superfamily proceeds via a covalent phosphohistidine intermediate. Proc. Natl Acad. Sci. USA. 1998;95:9202–9207. doi: 10.1073/pnas.95.16.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hultquist DE, Moyer RW, Boyer PD. The preparation and characterization of 1-phosphohistidine and 3-phosphohistidine. Biochemistry. 1966;5:322–331. doi: 10.1021/bi00865a041. [DOI] [PubMed] [Google Scholar]
- 24.Rudolph AE, Stuckey JA, Zhao Y, Matthews HR, Patton WA, Moss J, Dixon JE. Expression, characterization, and mutagenesis of the Yersinia pestis murine toxin, a phospholipase D superfamily member. J. Biol. Chem. 1999;274:11824–11831. doi: 10.1074/jbc.274.17.11824. [DOI] [PubMed] [Google Scholar]
- 25.Sanders KL, Catto LE, Bellamy SR, Halford SE. Targeting individual subunits of the FokI restriction endonuclease to specific DNA strands. Nucleic Acids Res. 2009;37:2105–2115. doi: 10.1093/nar/gkp046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raymond AC, Rideout MC, Staker B, Hjerrild K, Burgin A.B., Jr Analysis of human tyrosyl-DNA phosphodiesterase I catalytic residues. J. Mol. Biol. 2004;338:895–906. doi: 10.1016/j.jmb.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 27.Interthal H, Chen HJ, Kehl-Fie TE, Zotzmann J, Leppard JB, Champoux JJ. SCAN1 mutant Tdp1 accumulates the enzyme – DNA intermediate and causes camptothecin hypersensitivity. EMBO J. 2005;24:2224–2233. doi: 10.1038/sj.emboj.7600694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith MB, March J. March's; Advanced Organic Chemistry. 5th edn. New York: John Wiley and Sons; 2001. [Google Scholar]
- 29.Curley JF, Joyce CM, Piccirilli JA. Functional evidence that the 3‘-5’exonuclease domain of Escherichia coli DNA polymerase I employs a divalent metal ion in leaving group stabilization. J. Am. Chem. Soc. 1997;119:12691–12692. [Google Scholar]
- 30.Vyle JS, Connolly BA, Kemp D, Cosstick R. Sequence- and strand-specific cleavage in oligodeoxyribonucleotides and DNA containing 3′-thiothymidine. Biochemistry. 1992;31:3012–3018. doi: 10.1021/bi00126a024. [DOI] [PubMed] [Google Scholar]
- 31.Mueller JE, Smith D, Bryk M, Belfort M. Intron-encoded endonuclease I-TevI binds as a monomer to effect sequential cleavage via conformational changes in the td homing site. EMBO J. 1995;14:5724–5735. doi: 10.1002/j.1460-2075.1995.tb00259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dillingham MS, Kowalczykowski SC. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 2008;72:642–671. doi: 10.1128/MMBR.00020-08. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chu FK, Maley F, Wang AM, Pedersen-Lane J, Maley G. Purification and substrate specificity of a T4 phage intron-encoded endonuclease. Nucleic Acids Res. 1991;19:6863–6869. doi: 10.1093/nar/19.24.6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Roey P, Meehan L, Kowalski JC, Belfort M, Derbyshire V. Catalytic domain structure and hypothesis for function of GIY-YIG intron endonuclease I-TevI. Nat. Struct. Biol. 2002;9:806–811. doi: 10.1038/nsb853. [DOI] [PubMed] [Google Scholar]
- 35.Bitinaite J, Wah DA, Aggarwal AK, Schildkraut I. FokI dimerization is required for DNA cleavage. Proc. Natl Acad. Sci. USA. 1998;95:10570–10575. doi: 10.1073/pnas.95.18.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Catto LE, Ganguly S, Milsom SE, Welsh AJ, Halford SE. Protein assembly and DNA looping by the FokI restriction endonuclease. Nucleic Acids Res. 2006;34:1711–1720. doi: 10.1093/nar/gkl076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wah DA, Bitinaite J, Schildkraut I, Aggarwal AK. Structure of FokI has implications for DNA cleavage. Proc. Natl Acad. Sci. USA. 1998;95:10564–10569. doi: 10.1073/pnas.95.18.10564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vanamee ES, Santagata S, Aggarwal AK. FokI requires two specific DNA sites for cleavage. J. Mol. Biol. 2001;309:69–78. doi: 10.1006/jmbi.2001.4635. [DOI] [PubMed] [Google Scholar]
- 39.Wang J, Chen R, Julin DA. A single nuclease active site of the Escherichia coli RecBCD enzyme catalyzes single-stranded DNA degradation in both directions. J. Biol. Chem. 2000;275:507–513. doi: 10.1074/jbc.275.1.507. [DOI] [PubMed] [Google Scholar]
- 40.Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004;432:187–193. doi: 10.1038/nature02988. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.